Abstract
Members of the Thermococcales are anaerobic Archaea belonging to the kingdom Euryarchaea that are studied in many laboratories as model organisms for hyperthermophiles. We describe here a molecular analysis of 86 new Thermococcales isolates collected from six different chimneys of a single hydrothermal field located in the 13°N 104°W segment of the East Pacific ridge at a depth of 2,330 m. These isolates were sorted by randomly amplified polymorphic DNA (RAPD) fingerprinting into nine groups, and nine unique RAPD profiles were obtained. One RAPD group corresponds to new isolates of Thermococcus hydrothermalis, whereas all other groups and isolates with unique profiles are different from the 22 reference strains included in this study. Analysis of 16S rRNA gene sequences of representatives of each RAPD group and unique profiles showed that one group corresponds to Pyrococcus strains, whereas all the other isolates are Thermococcus strains. We estimated that our collection may contain at least 11 new species. These putative species, isolated from a single area of hydrothermal deep-sea vents, are dispersed in the 16S rRNA tree among the reference strains previously isolated from diverse hot environments (terrestrial, shallow water, hydrothermal vents) located around the world, suggesting that there is a high degree of dispersal of Thermococcales. About one-half of our isolates contain extrachromosomal elements that could be used to search for novel replication proteins and to develop genetic tools for hyperthermophiles.
All hyperthermophiles that thrive at temperatures above 95°C belong to the domain Archaea (19, 45). These extremophiles have been extensively studied during the last two decades for both academic and biotechnological reasons (16, 34, 40). Many type species that belong to new genera, orders, and families in the kingdoms Crenarchaea and Euryarchaea have been described. Some of these organisms, members of the order Thermococcales (genera Thermococcus, Pyrococcus, and Palaeococcus) (15, 17, 47), have been widely used as model organisms for biochemical or physiological studies (1). Members of the Thermococcales are anaerobic heterotrophs that grow at temperatures between 70 and 105°C, depending on the species, and their optimal growth temperatures range from 80 to 100°C. The genomes of three Pyrococcus species (Pyrococcus horikoshii, Pyrococcus abyssi, and Pyrococcus furiosus) have been completely sequenced (9, 20, 30), which allowed the first comparative studies of the genomes of several closely related species in the archaeal domain (22, 48). Two plasmids from Pyrococcus species have been characterized (13, 42). Plasmid pGT5 from P. abyssi strain GE5T was used as a reporter extrachromosomal element to study DNA topology and DNA replication (7, 26).
Many type species of Thermococcales were isolated from either shallow-water hydrothermal regions or deep-sea hydrothermal vents, and in one case an organism was isolated from a terrestrial hot spring; 19 species, including 15 Thermococcus species and 4 Pyrococcus species, are described in the latest edition of Bergey's Manual of Systematic Bacteriology (17). In general, only one species or a small number of species have been found at each site. However, preliminary data have suggested that a high diversity of Thermococcales could be found at a single geographic location. In particular, one Pyrococcus strain and three Thermococcus strains were isolated from shallow-water hydrothermal locations on the island of Vulcano in Italy (reviewed in reference 17), and both Thermococcus and Pyrococcus strains were identified in a collection of 20 isolates obtained from hydrothermal vents located in two different regions of the Pacific Ocean floor (27). The latter collection was used to screen for extrachromosomal elements (3) and to develop genetic tools. In particular, P. abyssi GE9, which lacks pGT5, has been used recently as a recipient for transformation by a vector based on this plasmid (25).
We describe here a molecular analysis of 86 Thermococcales strains isolated from a single hydrothermal field located in the mid-Pacific Ocean ridge (13°N 104°W). This work was undertaken to gain more insight into the genetic diversity of the Thermococcales in a single location and to increase the number of extrachromosomal elements (plasmids and/or viruses) that can be used for future genetic work. These isolates were characterized by the randomly amplified polymorphic DNA (RAPD) method (44) and by 16S rRNA gene sequencing analyses. The RAPD method, as well as pulsed-field gel electrophoresis (21) and amplified fragment length polymorphism analysis (41), are appropriate typing methods for exhaustive analysis of the molecular diversity within and between bacterial species. 16S rRNA gene sequencing allows workers to obtain useful information concerning the phylogenetic positions of unknown isolates. The combination of RAPD typing and 16S rRNA gene sequence analysis is therefore a method that allows workers to identify groups of closely related species and to place them in a more global phylogenetic context. This approach has been used previously to analyze the molecular diversity of bacterial isolates within species and between species of the same genus (4, 8). Here, we used this approach for the first time to characterize thermophilic archaeal isolates. The results show that our collection contains a wide phylogenetic spectrum of Thermococcus and Pyrococcus strains, including many new putative Thermococcus species, which are dispersed among type strains in the 16S rRNA tree. About one-half of the isolates contain extrachromosomal elements whose sizes range from 2 to more than 25 kb. Our work indicates that a single hydrothermal field can contain a wide diversity of members of the Thermococcales and suggests that despite extensive investigation of this archaeal group, a great number of Thermococcus and Pyrococcus species remain to be described.
MATERIALS AND METHODS
Sampling and enrichment cultures.
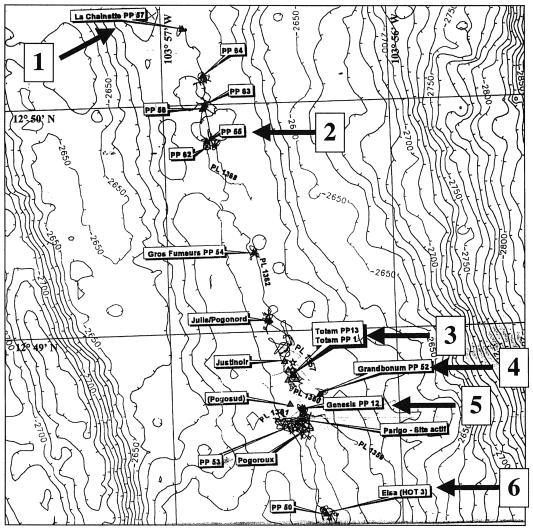
Rock fragments from chimney walls were collected from hydrothermal vent chimneys at a depth of 2,330 m during 18 different dives of the manned submersible Nautile (Ifremer) during the AMISTAD cruise in the 13°N 104°W segment of the East Pacific Ocean ridge. Thirty-four samples were obtained from six different chimneys, including La Chainette PP57 (site 1), Pulsar PP55 (site 2), Totem PP13/1 (site 3), Grandbonum PP52 (site 4), genesis PP12 (site 5), and Elsa (HOT3) (site 6) (Fig. 1), by grinding rock fragments to a fine powder and suspending them in 5 ml of sterilized seawater in Hungate tubes closed with rubber stoppers. These samples were made anaerobic by flushing the tubes with N2, followed by addition of Na2S (0.1 mg/liter) and addition of resazurin (0.001%) as a redox indicator. Some enrichment cultures were prepared on the boat (Atalante [Ifremer]) by inoculating 500-μl samples into Hungate tubes containing 5 ml of YPS medium (7) and incubating the tubes at 90°C for 12 to 48 h. Growth was detected by the presence of turbidity in 28 of the 34 cultures. Immediately after growth became visible, the enrichment cultures were first stored at 0°C (on the boat) and then kept at the ambient temperature (during transport from Mexico to Europe). Other enrichment cultures were grown in Martinsried, Germany, as described above, except that samples were made anaerobic by dropwise addition with a syringe of water saturated with H2S and that YPS medium was replaced by Zillig's broth (ZB). ZB was prepared by mixing 1 volume of solution A (trace elements) and 1 volume of solution B separately sterilized at 120°C. Solution A contained (per 500 ml) 25 g of NaCl, 5 g of MgCl2 · 6H2O, 1 g of CaCl2 · 2H2O, 0.75 g of KCl, 0.5 g of (NH4)SO4, 0.05 g of NaBr · 10H2O, 0.0075 g of SrCl2 · 6H2O, and 0.002 g of FeSO4 · 7H2O, as well as 90 μl of a 1% MnCl2 · 4H2O solution, 224 μl of a 1% Na2B4O7 · 10H2O solution, 11 μl of a 1% ZnSO4 · 7H2O solution, 2.5 μl of a 1% CuCl2 · 2H2O solution, 1.5 μl of a 1% Na2MnO4 · 2H2O solution, 1.5 μl of a 1% VoSO4 · 5H2O solution, 0.5 μl of a 1% CoSO4 · 7H2O solution, and 0.5 μl of a 1% NiSO4 · 7H2O solution. Solution B contained (per 500 ml) 1 g of yeast extract, 4 g of Bacto Tryptone (Difco), 0.2 g of starch, 0.06 g of K2HPO4, 0.06 g of KH2PO4, 10 ml of Tris-HCl (1 M) (pH 7), and 1 ml of resazurin (1 g/liter).
FIG. 1.
Hydrothermal field of the 13°N 104°W segment of the East Pacific Ocean ridge that was sampled during the AMISTAD expedition. The sites were La Chainette PP57 (site 1), Pulsar PP55 (site 2), Totem PP13/1 (site 3), Grandbonum PP52 (site 4), Genesis PP12 (site 5), and Elsa (HOT3) (site 6).
Plating and isolation of strains.
For preparation of plates, 8 g of Gelrite gellan gum (Sigma-Aldrich) was dissolved in 500 ml of solution B by heating in a microwave (the gelrite was not dissolved in complete ZB). After complete solubilization of the gelrite, 500 ml of preheated medium A was slowly added at the boiling temperature on a magnetic stirrer, and 20-ml portions of this medium were poured rapidly into petri dishes. After solidification at room temperature, the plates were subjected to reduction by placing them for 1 h into a stainless steel anaerobic jar under CO2(50 kPa)-H2S (5 kPa) as the gas phase. The CO2 and H2S were eliminated by flushing the containers several times with N2, and the plates were removed from the containers in an anaerobic chamber (Coy Laboratory Products, Grass Lake, Mich.). For plating, 0.1-ml dilutions of the enrichment cultures were mixed with 1.5 ml of preheated ZB containing 0.2% gelrite and 0.1 ml of extensively washed colloidal sulfur (Riedel-de Haën; distributed by Sigma-Aldrich). This hot soft-layer-forming mixture was quickly homogenized by vortexing and immediately poured and evenly spread on plates that were preheated to 80°C. After cooling, the plates were placed upside down into airtight containers with a CO2 (50 kPa)-H2 (20 kPa) atmosphere and incubated for 48 h at 80°C (for more details, see references 25 and 46). Colonies were identified by inspection with a binocular microscope in the anaerobic chamber and by the formation of plaque-like halos of sulfur depletion in the opaquely whitish soft layer. The sulfur depletion was induced by colony growth, and a colony was located at the center of each halo. We used dilutions that gave well-separated halos on plates to be sure that each halo contained a single colony. Colonies were picked up with Eppendorf cones (0.1 ml) whose tips were cut, and they were suspended in 0.5 ml of reduced ZB in Eppendorf tubes. The inoculated medium was then transferred into 5-ml portions of reduced ZB in Hungate tubes and incubated for 24 to 48 h at 90°C with shaking in the presence of CO2 (50 kPa) and H2 (20 kPa). The isolates from Daniel Prieur's laboratory from the same cruise were obtained by enrichment at 85°C in Ravot's medium (29). Colonies were picked directly on plates prepared without an overlay and were restreaked on plates three times.
DNA preparation and gel electrophoresis analysis.
Total DNA was prepared from all isolates by a phenol extraction procedure by starting with 5 to 50 ml of a culture in the stationary phase. The cultures were centrifuged, and the pellets were dissolved in 250 μl of TEN (0.01 M Tris-HCl, 0.15 M NaCl, 0.001 M EDTA); this was followed by slow addition of 250 μl of TEN with 1.6% sodium dodecyl sulfate and 0.12% Triton X-100. In some cases, samples were treated successively with RNase (50 μg/ml) and proteinase K (500 μg/ml). After incubation for 20 min at room temperature, the DNA was extracted three times with 1 volume of a solution containing phenol-chloroform (50/50) and isoamyl alcohol (24/1), precipitated with 2 volumes of 100% ethanol or 0.8 volume of isopropanol, and washed with 70% ethanol. After drying, the pellets were dissolved in 20 μl of TE buffer (0.01 M Tris-HCl, 0.001 M EDTA) (pH 7.5). DNA samples were subjected to 1% agarose gel electrophoresis in Tris-borate-EDTA buffer to detect the presence of plasmids and to verify the quality and quantity of DNA for RAPD analysis. For whole-genome restriction analysis, 5 μg of total DNA was cut with 5 U of EcoRI, and the products were separated on a horizontal 0.7% agarose gel in Tris-borate-EDTA buffer for 16 h at 1 V cm−1. Covalently closed circular DNA were extracted by alkaline lysis (32) with the length of the lysis step reduced to 45 s. The relative sizes were roughly determined by using a ladder of supercoiled plasmids (Promega G6231 supercoiled DNA ladder; 2 to 10 kb) as a marker. The gels were electrophoresed in the presence of ethidium bromide (0.1 μg/ml) to positively supercoil plasmids from hyperthermophiles which are normally relaxed or slightly positively supercoiled (7). In this condition, isolated plasmids and marker plasmids can be accurately compared since they have similar superhelical densities.
RAPD conditions, 16S rRNA genes, and 16S-23S rRNA intergenic spacer region (ISR) sequencing.
RAPD fingerprinting conditions that allowed reliable analyses of the data were used as previously described (4, 8, 36) with an annealing temperature of 42°C (28). The conditions used to analyze the diversity of Lactococcus, Leuconostoc, Lactobacillus, and Propionibacterium strains were also suitable and reproducible for Thermococcales isolates. RAPD profiles were determined by using four primers, primers P1 (5′-CGG CCT GGA C-3′), P2 (5′-GGG GCC CTA C-3′), P3 (5′-CGC CCT GCC C-3′), and P4 (5′-GGC GGC GCG G-3′), in separate reactions. The resulting patterns were compared by using the Pearson similarity coefficient and the unweighted pair group method using arithmetic averages included in the GelCompar program (Applied-Maths, Sint Martens-Latem, Belgium). When our RAPD conditions were used, the Pearson similarity coefficient was more than 80% for two profiles of the same strain obtained independently. We decided to propose RAPD groups when the Pearson similarity coefficient for profiles of a group was more than 55%.
The 16S rRNA gene was amplified and sequenced as previously described (8), with the following modifications. A fragment containing the nearly complete 16S rRNA gene and the 16S-23S rRNA ISR was amplified by PCR by using two primers, 16S-P1 (5′-CGG TTG ATC CTG CCG GA-3′; Escherichia coli 16S rRNA gene positions 10 to 28, forward) (5a) and 23S-P1 (5′-CTT TCG GTC GCC CCT ACT-3′; E. coli 23S rRNA gene positions 241 to 258, reverse) (6). The PCR fragment was purified to eliminate salts and excess primers by using a Wizard purification kit (Promega, Charbonnière-les-Bains, France). Three 500- to 600-bp partial and overlapping sequences were obtained by using the purified PCR fragment as the template and the following sequencing primers: 16S-SP1 (5′-GCT TTA GGC CCA ATA ATA G-3′; E. coli positions 558 to 576, reverse), 16S-SP2 (5′-GTG GGT CTC GCT CGT T-3′; E. coli positions 1101 to 1116, reverse), and 16S-SP3 (5′-ATA GGA GGT GAT CGA GC-3′; E. coli positions 1526 to 1542, reverse). Sequences were assembled by using the GCG Wisconsin software package (10). New sequences were compared with the sequences of closely related species of Thermococcales listed in the GenBank database (http://www.ncbi.nlm.nih.gov) by using the GeneBase program (Applied-Maths). This program was used to (i) align the sequences, (ii) calculate phylogenetic distances, (iii) construct distance matrix trees by the neighbor-joining method (31), and (iv) perform a bootstrap analysis (14) with 1,000 data sets. The alignment of 51 16S rRNA gene sequences of members of the Thermococcales was corrected manually before construction of trees. Phylogenetic distances were calculated based on alignment of 1,225 nucleotides of the 16S rRNA gene sequences (E. coli 16S rRNA gene positions 115 to 1365).
The 16S-23S ISR was amplified by using the 16S rRNA gene-ISR PCR product as the template and primers 16S-P2 (5′-TAC GGT TGG ATC ACC TC-3′; E. coli 16S rRNA gene positions 1522 to 1538, forward) and 23S-P1. PCR amplification was carried out in a 100-μl mixture containing 20 to 100 ng of DNA, 1.5 mM MgCl2, each primer at a concentration of 600 nM, 200 μM dATP, 200 μM dCTP, 200 μM dGTP, 200 μM dTTP (Roche Diagnostics, Meylan, France), and 2.5 U of Taq DNA polymerase (Q.BIOgene, Illkirch, France) in 10 mM Tris-HCl (pH 9.0). A Perkin-Elmer 2400 thermal cycler was used for 30 cycles of amplification (96°C for 10 s, 50°C for 30 s, and 72°C for 30 s) after a DNA-denaturing step consisting of 96°C for 4 min. Sequencing of the ISR amplified fragment used as the template was carried out by using primer 23S-SP1 (5′-GCT TTT GCT TTC TTT TCC T-3′; E. coli positions 213 to 231, reverse). The sequences were compared by using the GeneBase program. Phylogenetic distances were calculated based on alignment of 189 nucleotides of the 16S-23S ISR sequences.
Nucleotide sequence accession numbers.
The GenBank accession numbers of the 16S rRNA gene and 16S-23S rRNA ISR sequences of the new members of the Thermococcales are shown in Table 1.
TABLE 1.
GenBank accession numbers of the 16S rRNA gene and 16S-23S rRNA ISR sequences of the new Thermococcales isolates
| Taxon | Straina | Accession no.
|
|
|---|---|---|---|
| 16S rRNA gene | 16S-23S rRNA ISR | ||
| Thermococcus sp. | 5-1 | AY099165 | AY099201 |
| Thermococcus sp. | 12-4 | AY099152 | AY099188 |
| Thermococcus sp. | 13-2 | AY099153 | AY099189 |
| Thermococcus sp. | 13-3 | AY099154 | AY099190 |
| Thermococcus sp. | 21-1 | AY099155 | AY099191 |
| Thermococcus sp. | 23-1 | AY099156 | AY099192 |
| Thermococcus sp. | 23-2 | AY206704 | AY206706 |
| Thermococcus sp. | 26-2 | AY099157 | AY099193 |
| Thermococcus sp. | 26-3 | AY099158 | AY099194 |
| Thermococcus sp. | 28-1 | AY099159 | AY099195 |
| Thermococcus sp. | 29-1 | AY099160 | AY099196 |
| Thermococcus sp. | 30-1 | AY099161 | AY099197 |
| Thermococcus sp. | 31-1 | AY099162 | AY099198 |
| Thermococcus sp. | 31-3 | AY099163 | AY099199 |
| Pyrococcus sp. | 32-4 | AY099164 | AY099200 |
| Pyrococcus abyssi | GE23 | AY099166 | AY099202 |
| Pyrococcus abyssi | GE5 | AY099167 | AY099203 |
| Pyrococcus glycovorans | AL585 | AY099168 | AY099204 |
| Pyrococcus horikoshii | DSM12428 | AY099169 | AY099205 |
| Thermococcus acidaminovorans | DSM11906 | AY099170 | AY099206 |
| Thermococcus aegaeus | DSM12767 | AY099171 | AY099207 |
| Thermococcus barophilus | DSM11836 | AY099172 | AY099208 |
| Thermococcus barossii | DSM9535 | AY099173 | AY099209 |
| Thermococcus celer | DSM2476 | AY099174 | AY099210 |
| Thermococcus chitonophagus | DSM10152 | AY099175 | AY099211 |
| Thermococcus fumicolans | ST557 | AY099176 | AY099212 |
| Thermococcus gammatolerans | DSM15229 | AY206705 | AY206707 |
| Thermococcus gorgonarius | DSM10395 | AY099177 | AY099213 |
| Thermococcus guaymasensis | DSM11113 | AY099178 | AY099214 |
| Thermococcus hydrothermalis | AL662 | AY099179 | AY099215 |
| Thermococcus litoralis | DSM5474 | AY099180 | AY099216 |
| Thermococcus mexicalis | GY869 | AY099181 | AY099217 |
| Thermococcus pacificus | DSM10394 | AY099182 | AY099218 |
| Thermococcus peptonophilus | DSM10343 | AY099183 | AY099219 |
| Thermococcus profundus | DSM9503 | AY099184 | AY099220 |
| Thermococcus siculi | DSM12349 | AY099185 | AY099221 |
| Thermococcus stetteri | DSM5262 | AY099186 | AY099222 |
| Thermococcus waiotapuensis | DSM12768 | AY099187 | AY099223 |
DSM, Deutsche Sammlung von Mikroorganismen.
RESULTS
Isolation of heterotrophic anaerobic hyperthermophiles.
Samples of rocks were collected by the submersible Nautile (Ifremer) from six different hydrothermal chimneys located at a depth of 2,330 m in the 13°N 104°W segment of the East Pacific Ocean ridge (Fig. 1). Enrichment cultures were prepared by incubating the samples at 90°C under anaerobic conditions (see Materials and Methods). Isolates were obtained from 18 enrichment cultures by plating dilutions of the cultures in soft layers containing colloidal sulfur on gelrite plates and then incubating the plates for 2 days at 80°C. Colonies were surrounded by plaque-like halos formed by sulfur depletion of the soft layer. Various sizes and morphologies were observed; most colonies were surrounded by either small or large circular halos with either clear-cut or diffuse edges, whereas other colonies were surrounded by irregularly shaped halos. We prepared one plate for each positive enrichment culture and usually observed several different colony shapes on each plate. In an attempt to increase the diversity of our isolates, we picked several colonies per plate, each with a different morphology. This procedure resulted in 70 isolates that were designated according to the number of the enrichment culture and the number of the colony (i.e., isolate 30-1 was first colony picked from the plate inoculated with enrichment culture 30).
Identification of extrachromosomal genetic elements.
DNA was extracted from all 70 isolates to analyze EcoRI restriction patterns by agarose gel electrophoresis in order to detect the presence of multicopy extrachromosomal elements, either plasmids or viral genomes. This method was used previously to isolate plasmids and viruses from Sulfolobus species (46). Different EcoRI restriction patterns were usually obtained from different colonies picked from a single plate, whereas similar patterns were often obtained for colonies with very different shapes, either from the same plate or from different plates. The EcoRI restriction patterns of the 70 isolates exhibited great diversity that could not be correlated simply with the site of sampling or colony morphology (data not shown). This result was strikingly different from the results obtained previously during screening of terrestrial hot springs for Sulfolobus isolates, since all the isolates in the latter analysis exhibited only two types of EcoRI restriction patterns, corresponding to only two Sulfolobus species (46).
Analysis of the EcoRI restriction patterns of our isolates suggested that many of them harbor putative extrachromosomal genetic elements, as inferred by the presence of bands above the restriction fragment ladder (46). Additional extrachromosomal genetic elements were detected when unrestricted DNA samples from all isolates were directly analyzed by electrophoresis. Several putative extrachromosomal elements detected by these methods were purified by alkaline lysis from the corresponding isolates and were found to be covalently closed circular DNA. These elements had a wide range of sizes (2 kb to more than 25 kb). Some isolates appeared to contain two or three distinct elements (with different topological forms in each case). One isolate (30-1) contained two plasmids that were induced in the stationary phase (to be described elsewhere). Combining all these data allowed us to identify at least 44 plasmids, including 8 large plasmids (>15 kb), indicating that more than one-half of our isolates harbor extrachromosomal elements (Table 2).
TABLE 2.
Distribution of the 86 Thermococcales isolates in the different RAPD groups
| RAPD group | Isolatesa |
|---|---|
| G1 | 12-1, 21-4, 30-2, 30-3, 30-4, 31-2, 32-1, 32-2, 32-3, 32-4 |
| G2 | 21-2, 23-1, 24-1, 24-4, 27-3, 33-1, 33-2, 33-3, 33-4, 34-1, 34-2, 34-3, 34-4 |
| G3 | 9-3, 23-5, 29-1, 29-2, 29-3, 30-1, 17j, 17Q, 17N, AM17CHA, AM17CHG |
| G4 | 23-2, 29-4 |
| G5 | 13-1, 13-2, 13-3, 13-4 |
| G6 | 15-1, 15-2, 15-3, 15-4, 28-1, 28-2, 28-3, 28-4 |
| G7 | 5-1, 5-2, 5-3, 5-4, 9-1, 9-2, 9-4, 23-3, 23-4, 24-2, 24-3, 27-1, 27-2, 27-4 |
| G8 | 10, 12, 13 |
| G9 | 31-3, 31-4 |
| Unique profiles | 12-2, 12-3, 12-4, 21-1, 21-3, 26-1, 26-2, 26-3, 26-4, 31-1, 03j, 03Q, 03N, AM03CHA, AM03CHG, 15C, 15S, 16.2 |
The following strains were isolated in the laboratory of Daniel Prieur: 17j, 17Q, 17N, AM17CHA, AM17CHG, 10, 12, 13, 03j, 03Q, 03N, AM03CHA, AM03CHG, 15C, 15S, and 16.2. Extrachromosomal elements were detected in strains 21-4, 32-1, 32-2, 32-3, and 32-4 in group G1; in strains 23-1, 33-1, 33-3, 33-4, 34-1, 34-2, 34-3, and 34-4 in group G2; in strains 23-5, 29-1, 29-2, 29-3, and 30-1 in group G3; in strains 23-2 and 29-4 in group G4; in strains 15-2, 15-3, 15-4, 28-1, 28-2, 28-3, and 28-4 in group G6; in strains 5-4, 9-1, 9-4, 23-3, 23-4, 24-2, and 24-3 in group G7; in strain 31-3 in group G9; and in strain 21-1. The 16S rRNA gene sequences were determined for strain 32-4 in group G1; for strain 23-1 in group G2; for strains 29-1 and 30-1 in group G3; for strain 23-2 in group G4; for strains 13-2 and 13-3 in group G5; for strain 28-1 in group G6; for strain 5-1 in group G7; for strains 10 and 12 in group G8; for strain 31-3 in group G9; and for strains 12-4, 21-1, 26-2, 26-3, 31-1, 03j, 15C, 15S, and 16.2.
RAPD grouping of the isolates.
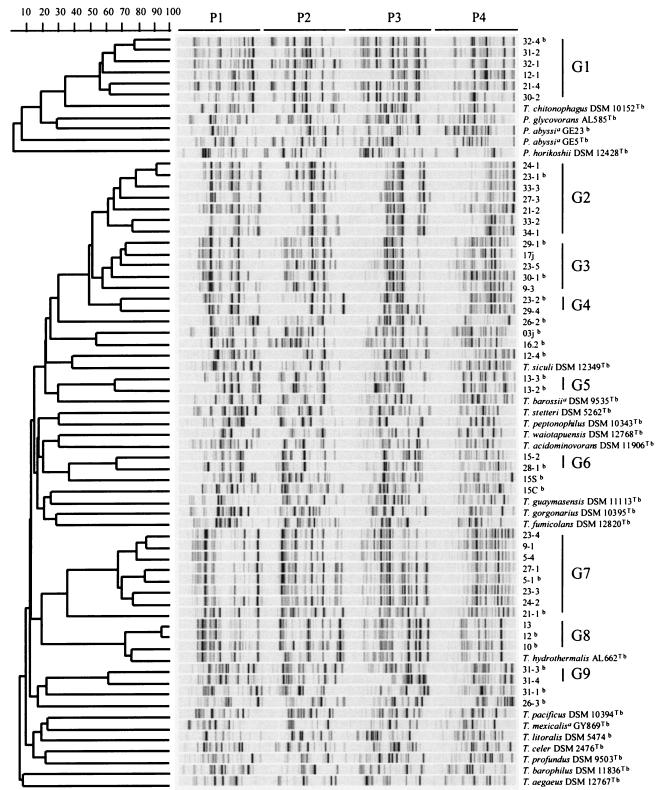
To characterize our isolates at the molecular level, DNA from the 70 isolates were first subjected to RAPD fingerprinting (44). This method detects DNA polymorphism from amplification of random DNA segments by using primers with arbitrary nucleotide sequences. In our experiments, we used four primers in separate reactions. We also analyzed under the same conditions the RAPD profiles of 16 isolates obtained in the laboratory of Daniel Prieur (Plouzané, France) by an independent isolation procedure; these isolates were from the same cruise and location (Table 2). To simplify the presentation of the RAPD results, we selected one representative when several isolates picked from the same petri dish had very similar RAPD profiles, which reduced the number of profiles from 86 to 45 (RAPD profiles were considered similar when the Pearson coefficient was higher than 80%; this value was determined on the basis of our reproducibility tests [see Materials and Methods]). Figure 2 shows a comparison of the 45 selected RAPD profiles, together with the profiles of 22 Thermococcales reference strains (18 Thermococcus strains and 4 Pyrococcus strains). The RAPD profiles of reference strains were unique, including those of the two P. abyssi strains. Nine of the new isolates had unique RAPD profiles, whereas 36 isolates had RAPD profiles that were separated into nine groups (groups G1 to G9) (Pearson coefficients, >55%). We observed no correlation between the grouping of our isolates based on RAPD profiles and the morphology of the colonies. In some cases, isolates picked from the same plate with very similar RAPD profiles exhibited very different morphologies, whereas isolates with very similar atypical colonies (e.g., gelrite eater) belonged to different RAPD groups. The absence of a correlation between colony morphology and grouping of the isolates indicates that colony morphology cannot be used to group members of the Thermococcales.
FIG. 2.
RAPD profiles of 67 Thermococcus and Pyrococcus strains obtained by using primers P1, P2, P3, and P4 (as indicated at the top) and deduced dendrogram obtained by the unweighted pair group method using arithmetic averages. The scale bar indicates the correlation values (Pearson's coefficient, ×100). RAPD groups are indicated on the right. T = type strain. A superscript a indicates a species whose name has not been validly published. A superscript b indicates that the 16S rRNA gene sequence was determined.
The RAPD profiles of the isolates were significantly different from those of the 22 reference strains, except for the profiles of Brest isolates 10, 12, and 13 (group G8), which were very similar to the profile of Thermococcus hydrothermalis (Pearson coefficient, >70%), and for the group G1 profiles, which shared bands with the profile of Thermococcus chitonophagus.
Isolates belonging to several RAPD groups were found in each chimney (and up to five of the nine RAPD groups were found in a single chimney), showing that diversity can be detected in a unique location (Table 3). However, the distribution of the groups was different from one chimney to another, and only members of group G7 and one isolate with a unique profile were obtained from the Totem PP13/1 chimney (site 3). This indicates that sampling from multiple chimneys was an important factor for increasing the recovery of biodiversity (Table 3). Several isolates with very similar profiles (Pearson coefficients, >80%) were picked from different chimneys (e.g., isolates 23-1 and 24-1, isolates 5-1 and 27-1, and isolates 12 and 13), indicating that the divergence between these closely related isolates predated the formation of the chimneys. Some RAPD groups may have even diverged before the formation of the hydrothermal field studied, since one group contained the species T. hydrothermalis, which has been isolated from another location (20°N instead of 13°N).
TABLE 3.
Relationship between site of isolation and molecular diversity of the 45 Thermococcales isolates revealed by RAPD analysis
| Site | RAPD groups and isolates with unique profilesa |
|---|---|
| 1 | G2, G6, G7 |
| 2 | G1, G2, G3, G4, G6, 16.2 |
| 3 | G2, G7, 26-1, 26-2, 26-3, 26-4, 15S |
| 4 | G2, G3, G4, G7 |
| 5 | G1, G3, G9, 31-1, AM03CHA, AM03CHG |
| 6 | G1, G2, G5, G7, G8, 12-2, 12-3, 12-4, 21-1, 21-3, 15C |
Strains 16.2, 15S, AM03CHA, AM03CHG, and 15C were isolated in the laboratory of Daniel Prieur.
Molecular identification of isolates based on their 16S rRNA gene sequences.
Twenty-one isolates representative of the molecular diversity observed by RAPD analysis were selected for 16S rRNA gene sequencing (Fig. 2) (at least one isolate for each RAPD group and all isolates with unique profiles were used). The high quality of the sequences obtained by using 16S PCR fragments as templates confirmed that the strain isolation procedure used allowed us to obtain sufficient culture purity. These 16S rRNA gene sequences were compared to those of reference strains whose sequences were available in the GenBank database. To confirm that the 22 reference strains that we used for the RAPD typing were the correct strains, we sequenced their 16S rRNA genes in our laboratory. All sequences obtained were indeed identical to those obtained from the GenBank database for the same strains. In several cases, our sequences allowed us to resolve uncertainties in the previously published sequences (the presence of unknown nucleotides) and/or to increase the length of the sequence. Therefore, all our new sequences have been deposited in the GenBank database (accession numbers are shown in Table 1).
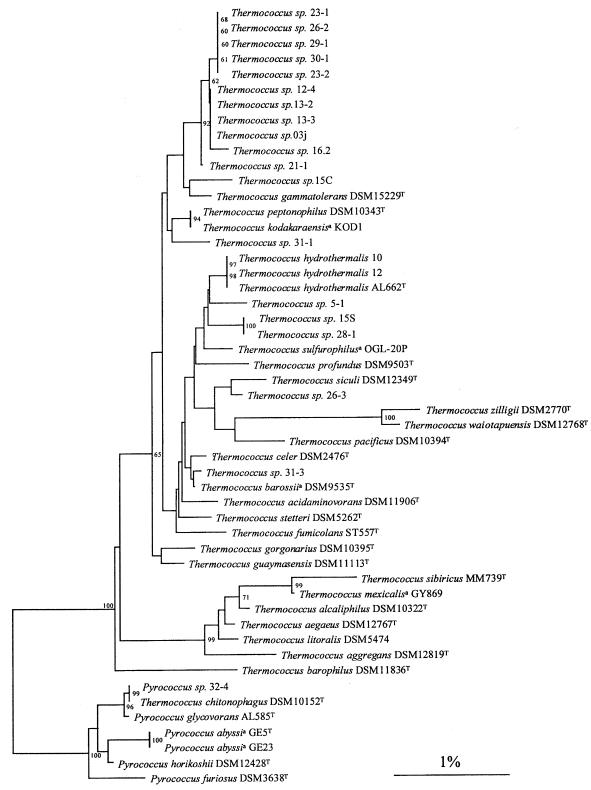
As shown in the distance-based dendrogram in Fig. 3, all our isolates were clearly members of the Thermococcales, as expected from our enrichment procedure. The genera Pyrococcus and Thermococcus are clearly separated on the dendrogram, and all our isolates belong to one of these genera (i.e., we did not obtain isolates that branched between the genera). The six isolates in group G1 (represented by strain 32-4) belong to the genus Pyrococcus, while all the other isolates belong to the genus Thermococcus. The diversity in colony morphology and the diversity in plasmid distribution were similar in the genera Thermococcus and Pyrococcus. Pyrococcus isolates (one group) were present in three of the six chimneys, whereas Thermococcus isolates were present in all of the chimneys. In four cases, Thermococcus and Pyrococcus isolates were obtained from a single plate.
FIG. 3.
Distance tree showing the positions of 21 Thermococcus and Pyrococcus isolates and 30 reference strains. The sequences of T. aggregans (accession no. Y08384), T. alcaliphilus (AB055121), T. kodakaraensis KOD1 (D38650), T. sibiricus (AJ238992), T. sulfurophilus OGL-20P (AF394925), T. zilligii (U76534), and P. furiosus (U20163) were obtained from the GenBank database. The tree was constructed by using the neighbor-joining method (31) included in the GeneBase program (Applied-Maths). Phylogenetic distances were calculated based on 1,225 nucleotides of the 16S rRNA gene sequences (E. coli 16S rRNA gene positions 115 to 1365). Bootstrap values, expressed as percentages of 1,000 data sets, are indicated at the branch points. Only values higher than 60% are shown. Scale bar = 1 nucleotide substitution per 100 nucleotides. A superscript a indicates a species whose name has not been validly published.
The members of groups G2, G3, and G4, which were clustered by their RAPD profiles, had very similar 16S rRNA gene sequences. They formed a large cluster that included group G5 and several isolates with unique RAPD profiles but no type strain. Other groups and isolates with unique RAPD profiles were also clearly separated from type strains, with the two exceptions detected by RAPD analysis. The 16S rRNA analysis confirmed that isolates 10 and 12 (group G8) can be assigned to T. hydrothermalis and isolate 32-4 (group G1) can be assigned to T. chitonophagus.
Based on the 16S rRNA dendrogram, T. chitonophagus DSM10152T (18) is clearly misclassified at the genus level and should be renamed Pyrococcus chitonophagus.
We also amplified by PCR and sequenced the 16S-23S rRNA ISR of all 21 representative isolates and of the 22 reference species in an attempt to gain better insight into the phylogenetic relationships among members of our collection. However, the 16S-23S rRNA ISR also turned out to be highly conserved in the Thermococcales (the levels of identity were between 80 and 100%). Nevertheless, a dendrogram based on the 16S-23S rRNA ISR sequence comparison confirmed the delimitation between the Pyrococcus and Thermococcus strains observed when the 16S rRNA gene sequences were used (data not shown). This analysis also confirmed the clustering of groups G2, G3, G4, and G5, the assignment of strain DSM10152T (T. chitonophagus) to the genus Pyrococcus, the close relationship of group G1 to P. chitonophagus and Pyrococcus glycovorans, and the identity of isolate 10 and 12 sequences with the sequence of T. hydrothermalis.
DISCUSSION
In the last 10 years, molecular studies of environmental samples performed by using PCR amplification of 16S rRNA genes have shown that cultivable species account for only a minority of the biosphere in all environments analyzed. In particular, recent studies have shown the great 16S rRNA gene diversity of archaeal organisms in deep oceanic regions (24, 37, 38, 39). In contrast, there was no previous study to estimate the biodiversity of cultivable hyperthermophiles belonging to a particular order in a given environment. We focused on the Thermococcales by analyzing many isolates obtained by plating at a high temperature on solid medium. We chose members of the Thermococcales since they are the best-studied hyperthermophilic anaerobes and a large number of species have been already described in both the genus Thermococcus and the genus Pyrococcus.
In this work, we characterized by molecular techniques (RAPD fingerprinting and rRNA analyses) 86 Thermococcales isolates obtained from six different chimneys located in the same area of the East Pacific Ocean ridge. The molecular diversity of these isolates was determined first by using the RAPD typing method. Typing methods are now widely used to study the molecular diversity of bacteria (35). In particular, this technique was used previously to evaluate the redundancy and to detect rare representatives in a collection of bacterial isolates with low or high G+C contents (4, 5, 8, 28, 36). We showed here that this method can also be used to study the diversity of hyperthermophilic archaea since a unique protocol was applied to archaea whose G+C contents ranged from 37 mol% (Thermococcus barophilus) to 60 mol% (Thermococcus barossii).
We were able to define nine RAPD groups and nine unique RAPD isolates. There was no correlation between this grouping and colony morphology or sampling site. The ability to recover known species from the environment by RAPD typing is exemplified by the fact that three isolates (isolates 10, 12, and 13 in RAPD group G8) had RAPD profiles that were very similar to the profile of the type strain of T. hydrothermalis. The 83 other isolates had RAPD profiles different from those of the 22 reference strains, although group G1 can be clearly affiliated with T. chitonophagus (Fig. 2). The analysis of the 16S rRNA gene sequences allowed us to classify unambiguously our new isolates in the genus Pyrococcus or the genus Thermococcus (Fig. 3) and confirmed the grouping suggested by the RAPD analysis. In addition, the 16S rRNA tree showed that the type strain of T. chitonophagus should be reclassified as P. chitonophagus. These results were confirmed by a comparative analysis of the 16S-23S rRNA spacer region sequences. The data from the two dendrograms should help us to perform quantitative DNA-DNA hybridization experiments required to characterize the molecular taxonomic positions of new isolates (33).
How many new species are present among our isolates? New species can be predicted with confidence only when they exhibit levels of 16S rRNA gene sequence similarity of less than 97% with other species (33). However, the divergence between 16S rRNA gene sequences of previously described type strains of Thermococcales species was often much less than 3% (Fig. 3). In such a case, DNA-DNA hybridization experiments are required to rigorously describe new species. However, since different Thermococcales species could have identical 16S rRNA gene sequences (e.g., Thermococcus kodakaraensis and Thermococcus peptonophilus), one can tentatively conclude that strains which do not have identical 16S rRNA gene sequences belong to different species. If this criterion is used, our collection contains at least 11 new species on the basis of 16S rRNA gene sequences and RAPD profile comparisons (Table 4 and Fig. 3). Since about 20 different Thermococcales species have been described, this means that we were able to increase the biodiversity of this taxon by about 50% by using a rather limited number of enrichment cultures obtained with samples collected at a single hydrothermal field. This result was somewhat surprising since, using a similar enrichment and isolation procedure to isolate Sulfolobales strains from terrestrial hot springs, two groups of workers reported that all of the isolates which they obtained belonged to either a single species, Sulfolobus islandicus (43), or two species, S. islandicus and Sulfolobus solfataricus (46); the reason for this difference is unclear and merits further investigation. However, our results are in agreement with the previous finding of three different Thermococcus species—Thermococcus celer, Thermococcus alcaliphilus, and Thermococcus litoralis—in shallow-water hot springs on the island of Vulcano (17). In any case, our collection could be a good starting point for further studies on the biogeography of marine hyperthermophiles.
TABLE 4.
Potential new Thermococcales species and representative isolates
| Representative isolate | RAPD group | No. of isolates |
|---|---|---|
| 23-1 | G2 | 7 |
| 29-1 or 30-1 | G3 | 5 |
| 23-2 | G4 | 2 |
| 13-2 or 13-3 | G5 | 2 |
| 03ja | Unique profile | |
| 12-4 | Unique profile | |
| 28-1 | G6 | 2 |
| 15Sa | Unique profile | |
| 5-1 | G7 | 7 |
| 15Ca | Unique profile | |
| 16.2a | Unique profile | |
| 21-1 | Unique profile | |
| 26-2 | Unique profile | |
| 26-3 | Unique profile | |
| 31-1 | Unique profile | |
| 31-3 | Unique profile |
Strain isolated in the laboratory of Daniel Prieur
Several authors have described great genetic diversity of closely related bacterial isolates (usually belonging to a single species) in various environments (2, 4, 5, 8, 12). Our results indicate that such diversity also exists in deep-sea hydrothermal vents (i.e., in an environment with strong selection pressure). The diversity in our collection is especially striking for the genus Thermococcus. Interestingly, the Thermococcus isolates in our collection are as dispersed in the Thermococcus tree as the type strains themselves are. This suggests that the diversity of Thermococcus species detected by culturing many isolates from a single hydrothermal vent is similar to the diversity previously observed by culturing single isolates or a few isolates from various terrestrial, marine, and submarine hot springs located all around the world (Atlantic Ocean, Pacific Ocean, Mediterranean Sea, New Zealand hot springs). The diversity of cultivable organisms in one genus is thus greatly underestimated when only type species are considered. A more exhaustive search for new isolates could have increased even more the present biodiversity of the Thermococcales, possibly confusing the border between currently recognized species. In contrast, the absence of intermediates between Pyrococcus and Thermococcus species in our collection raises the question of which specific feature(s) is responsible for the clear-cut distinction at the genus level.
At the beginning of this work, we were especially interested in isolating new members of the Thermococcales containing extrachromosomal elements. Extrachromosomal elements are indeed widespread in our isolates, confirming and extending previous work by Benbouzid-Rollet et al. (3). This collection of Thermococcales isolates could also be useful for extending preliminary studies on gene transfer and the mobility of genetic elements in the Thermococcales (11, 22, 48), developing genetic tools for hyperthermophiles, and searching for novel replication proteins (23).
Acknowledgments
We thank Daniel Prieur for inviting Evelyne Marguet and Wolfram Zillig to participate in the AMISTAD expedition in May 1999 and Christian Jeanthon, who was the responsible scientist for this expedition. We thank Francisco Canganella, Rob Daniel, Frank Robb, Michael Thomm, Georges Barbier, Marie-Anne Cambon-Bonavita, and Edmond Jolivet for providing type strains and Jocelyne DiRuggiero for correcting the manuscript. We are grateful to Benjamin Kottler, Julien Bouchoux, and Nicolas Soler for help with growing the isolates and performing some DNA extractions.
REFERENCES
- 1.Adams, M. W. 1993. Enzymes and proteins from organisms that grow near and above 100°C. Annu. Rev. Microbiol. 47:627-658. [DOI] [PubMed] [Google Scholar]
- 2.Baldy-Chudzik, K., J. Niedbach, and M. Stosik. 2003. REP-PCR fingerprinting as a tool for the analysis of genomic diversity in Escherichia coli strains isolated from aqueous/freshwater environment. Cell. Mol. Biol. Lett. 8:793-798. [PubMed] [Google Scholar]
- 3.Benbouzid-Rollet, N., P. Lopez-Garcia, L. Watrin, G. Erauso, D. Prieur, and P. Forterre. 1997. Isolation of new plasmids from hyperthermophilic archaea of the order Thermococcales. Res. Microbiol. 148:767-777. [DOI] [PubMed] [Google Scholar]
- 4.Ben Omar, N., F. Ampe, M. Raimbault, J.-P. Guyot, and P. Tailliez. 2000. Molecular diversity of lactic acid bacteria from cassava sour starch (Colombia). Syst. Appl. Microbiol. 23:285-291. [DOI] [PubMed] [Google Scholar]
- 5.Bringel, F., P. Quénée, and P. Tailliez. 2001. Polyphasic investigation of the diversity within Lactobacillus plantarum related strains revealed two L. plantarum subgroups. Syst. Appl. Microbiol. 24:561-571. [DOI] [PubMed] [Google Scholar]
- 5a.Brosius, J., M. L. Palmer, P. J. Kennedy, and H. F. Noller. 1978. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc. Natl. Acad. Sci. 75:4801-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brosius, J., T. J. Dull, and H. F. Noller. 1980. Complete nucleotide sequence of a 23S ribosomal RNA gene from Escherichia coli. Proc. Natl. Acad. Sci. 77:201-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charbonnier, F., G. Erauso, T. Barbeyron, D. Prieur, and P. Forterre. 1992. Evidence that a plasmid from a hyperthermophilic archaebacterium is relaxed at physiological temperatures. J. Bacteriol. 174:6103-6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cibik, R., E. Lepage, and P. Tailliez. 2000. Molecular diversity of Leuconostoc mesenteroides and Leuconostoc citreum isolated from traditional French cheeses as revealed by RAPD fingerprinting, 16S rDNA sequencing and 16S rDNA fragment amplification. Syst. Appl. Microbiol. 23:267-278. [DOI] [PubMed] [Google Scholar]
- 9.Cohen, G., V. Barde, D. Flament, M. D., Galperin, R. Heilig, O. Lecompte, O. Poch, D. Prieur, J. Querellou, R. Ripp, J. C. Thierry, J. Van Der Oost, J. Weissenbach, Y. Zivanovic, and P. Forterre. 2003. An integrated analysis of the genome of the hyperthermophilic archaeon Pyrococcus abyssi: chromosome plasticity and key metabolic features. Mol. Microbiol. 47:1495-1512. [DOI] [PubMed] [Google Scholar]
- 10.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DiRuggiero, J., D. Dunn, D. L. Maeder, R. Holley-Shanks, J. Chatard, R. Horlacher, F. T. Robb, W. Boos, and R. B. Weiss. 2001. Evidence of recent lateral gene transfer among hyperthermophilic archaea. Mol. Microbiol. 38:684-693. [DOI] [PubMed] [Google Scholar]
- 12.Dogan, B., and K. J. Boor. 2003. Genetic diversity and spoilage potentials among Pseudomonas spp. isolated from fluid milk products and dairy processing plants. Appl. Environ. Microbiol. 69:130-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erauso, G., S. Marsin, N. Benbouzid-Rollet, M. F. Baucher, T. Barbeyron, Y. Zivanovic, D. Prieur, and P. Forterre. 1996. Sequence of plasmid pGT5 from the archaeon Pyrococcus abyssi: evidence for rolling-circle replication. J. Bacteriol. 178:3232-3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 15.Fiala, G., and K. O. Stetter. 1986. Pyrococcus furiosus sp. nov. represents a novel genus of marine heterotrophic archaebacteria growing optimally at 100°C. Arch. Microbiol. 145:56-61. [Google Scholar]
- 16.Galtier, N., N. Tourasse, and M. Gouy. 1999. A nonhyperthermophilic common ancestor to extant life forms. Science 283:220-221. [DOI] [PubMed] [Google Scholar]
- 17.Garrity, G. M., and J. G. Holt. 2002. Phylum All. Euryarchaeota phy. nov., p. 211-355. In R. D. Boone and R. W. Castenholz (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 1. Springer, New York, N.Y.
- 18.Huber, R., J. Stöhr, S. Hohenhaus, R. Rachel, S. Burggraf, H. Jannash, and K. O. Stetter. 1995. Thermococcus chitonophagus sp. nov., a novel, chitin-degrading, hyperthermophilic archaeum from deep-sea hydrothermal vent environment. Arch. Microbiol. 164:255-264. [Google Scholar]
- 19.Huber, R., H. Huber, and K. O. Stetter. 2000. Towards the ecology of hyperthermophiles: biotopes, new isolation strategies and novel metabolic properties. FEMS Microbiol. Rev. 24:615-623. [DOI] [PubMed] [Google Scholar]
- 20.Kawarabayasi, Y., M. Sawada, H. Horikawa, Y. Haikawa, Y. Hino, S. Yamamoto, M. Sekine, S. Baba, H. Kosugi, A. Hosoyama, Y. Nagai, M. Sakai, K. Ogura, R. Otsuka, H. Nakazawa, M. Takamiya, Y. Ohfuku, T. Funahashi, T. Tanaka, Y. Kudoh, J. Yamazaki, N. Kushida, A. Oguchi, K. Aoki, and H. Kikuchi. 1998. Complete sequence and gene organization of the genome of a hyperthermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 5:147-155. [DOI] [PubMed] [Google Scholar]
- 21.Lai, E., B. W. Birren, S. M. Clarck, M. I. Simon, and L. Hood. 1989. Pulsed-field gel electrophoresis. BioTechniques 7:34-42. [PubMed] [Google Scholar]
- 22.Lecompte, O., R. Ripp, V. Puzos-Barbe, S. Duprat, R. Heilig, J. Dietrich, J.-C. Thierry, and O. Poch. 2001. Genome evolution at the genus level: comparison of three complete genomes of hyperthermophilic archaea. Genome Res. 11:981-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lipps, G., S. Rother, C. Hart, and G. Krauss. 2003. A novel type of replicative enzyme harbouring ATPase, primase and DNA polymerase activity. EMBO J. 22:2516-2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.López-Garcia, P., D. Moreira, A. López-López, and F. Rodríguez-Valera. 2001. A novel haloarchaeal-related lineage is widely distributed in deep oceanic regions. Environ. Microbiol. 3:72-78. [DOI] [PubMed] [Google Scholar]
- 25.Lucas, S., L. Toffin, Y. Zivanovic, D. Charlier, H. Moussard, P. Forterre, D. Prieur, and G. Erauso. 2002. Construction of a shuttle vector for, and spheroplast transformation of, the hyperthermophilic archaeon Pyrococcus abyssi. Appl. Environ. Microbiol. 68:5528-5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marsin, S., and P. Forterre. 2001. pGT5 replication initiator protein Rep75 from Pyrococcus abyssi. Methods Enzymol. 334:193-204. [DOI] [PubMed] [Google Scholar]
- 27.Marteinsson, V. T., L. Watrin, D. Prieur, J. C. Caprais, G. Ragenes, and G. Erauso. 1995. Phenotypic characterization, DNA similarities, and protein profiles of twenty sulfur-metabolizing hyperthermophilic anaerobic archaea isolated from hydrothermal vents in the southwestern Pacific Ocean. Int. J. Syst. Microbiol. 45:623-632. [Google Scholar]
- 28.Matte-Tailliez, O., E. Lepage, M. Tenenhaus, and P. Tailliez. 2002. Use of predictive modeling for Propionibacterium strain classification. Syst. Appl. Microbiol. 25:386-395. [DOI] [PubMed] [Google Scholar]
- 29.Ravot, G., M. Magot, M. L. Fardeau, B. K. Patel, G. Prensier, A. Egan, J. L. Garcia, and B. Ollivier. 1995. Thermotoga elfii sp. nov., a novel thermophilic bacterium from an African oil-producing well. Int J. Syst Bacteriol. 45:308-314. [DOI] [PubMed] [Google Scholar]
- 30.Robb, F. T., D. L. Maeder, J. R. Brown, J. DiRuggiero, M. D. Stump, R. K. Yeh, R. B. Weiss, and D. M. Dunn. 2001. Genomic sequence of hyperthermophile, Pyrococcus furiosus: implications for physiology and enzymology. Methods Enzymol. 330:134-157. [DOI] [PubMed] [Google Scholar]
- 31.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Stackebrandt, E., and B. M. Goebel. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 34.Stetter, K. O. 1999. Extremophiles and their adaptation to hot environments. FEBS Lett. 452:22-25. [DOI] [PubMed] [Google Scholar]
- 35.Tailliez, P., D. Beaud, and J.-C. Ogier. 2002. Molecular approaches for microbial classification and ecology. Sci. Aliments 22:5-21. [Google Scholar]
- 36.Tailliez, P., J. Tremblay, S. D. Ehrlich, and A. Chopin. 1998. Molecular diversity and relationship within Lactococcus lactis, as revealed by randomly amplified polymorphic DNA (RAPD). Syst. Appl. Microbiol. 21:530-538. [DOI] [PubMed] [Google Scholar]
- 37.Takai, K., and K. Horikoshi. 1999. Genetic diversity of Archaea in deep-sea hydrothermal vent environments. Genetics 152:1285-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takai, K., T. Komatsu, F. Inagaki, and K. Horikoshi. 2001. Distribution of archaea in a black smoker chimney structure. Appl. Environ. Microbiol. 67:3618-3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vetriani, C., H. W. Jannasch, B. J. MacGregor, D. A. Stahl, and A.-L. Reysenbach. 1999. Population structure and phylogenetic characterization of marine benthic Archaea in deep-sea sediments. Appl. Environ. Microbiol. 65:4375-4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vieille, C., and G. J. Zeikus. 2001. Hyperthermophilic enzymes: sources, uses, and molecular mechanisms for thermostability. Microbiol. Mol. Biol. Rev. 65:1-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vos, P., R. Hogers, M. Bleeker, M. Reijans, T. Van De Lee, M. Hornes, A. Frijters, J. Pot, J. Peleman, M. Kuiper, and M. Zabeau. 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23:4407-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ward, D. E., I. M. Revet, R. Nandakumar, J. H. Tuttle, W. M. de Vos, J. van der Oost, and J. DiRuggiero. 2002. Characterization of plasmid pRT1 from Pyrococcus sp. strain JT1. J. Bacteriol. 184:2561-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whitaker, R. J., D. W. Grogan, and J. W. Taylor. 2003. Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science 301:976-978. [DOI] [PubMed] [Google Scholar]
- 44.Williams, J. G. K., A. R. Kubelik, K. J. Livak, J. A. Rafalski, and S. V. Tingey. 1990. DNA polymorphisms amplified by arbitrary primers are useful as genetics markers. Nucleic Acids Res. 18:6531-6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Woese, C. R., O. Kandler, and M. L. Wheelis. 1990. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. 87:4576-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zillig, W., A. Kletzin, C. Schleper, I. Holz, D. Janekovic, J. Hain, M. Lanzendörfer, and J. K. Kristjansson. 1994., Screening for Sulfolobales, their plasmids and their viruses in Icelandic Solfataras. Syst. Appl. Microbiol. 16:609-628. [Google Scholar]
- 47.Zillig, W., I. Holz, D. Janekovic, W. Schäfer, and W. D. Reiter. 1983. The archaebacterium Thermococcus celer represents a novel genus within the thermophilic branch of the archaebacteria. Syst. Appl. Microbiol. 4:88-94. [DOI] [PubMed] [Google Scholar]
- 48.Zivanovic, Y., P. Lopez, H. Philippe, and P. Forterre. 2002. Pyrococcus genome comparison evidences chromosome shuffling-driven evolution. Nucleic Acids Res. 30:1902-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]