Abstract
The sensitive detection of cancer biomarkers in urine could revolutionize cancer diagnosis and treatment. Such detectors must be inexpensive, easy to interpret, and sensitive. This report describes a bioaffinity matrix of viruses integrated into PEDOT films for electrochemical sensing of prostate specific membrane antigen (PSMA), a prostate cancer biomarker. High sensitivity to PSMA resulted from synergistic action by two different ligands to PSMA on the same phage particle. One ligand was genetically encoded, and the secondary recognition ligand was chemically synthesized to wrap around the phage. The dual ligands result in a bidentate binder with high copy, dense ligand display for enhanced PSMA detection through a chelate-based, avidity effect. Biosensing with virus-PEDOT films provides a 100 pM limit of detection for PSMA in synthetic urine without requiring enzymatic or other amplification.
INTRODUCTION
More effective biosensors could address a critical need for detecting cancer-associated biomarkers. An estimated 29,000 men in the US will succumb to prostate cancer in 2013.1 Unfortunately, the lack of validated clinical diagnostic markers complicates efforts to develop tests for early prostate cancer detection. For example, a recent report concludes that the Prostate Specific Antigen (PSA) test used for prostate cancer diagnostics is more harmful than beneficial.2 Despite this caveat, PSA remains an important biomarker for detecting recurrent prostate cancer. However, early detection of the disease could enable more effective treatment and prognosis.3 Thus, issues addressable by bioanalytical chemistry include the development of more sensitive measurements of protein concentration and then applying such measurements to identify and validate more effective biomarkers.
Unlike PSA, Prostate Specific Membrane Antigen (PSMA) concentrations in biological fluids appear to offer a more useful metric for prostate cancer diagnosis and prognosis.4 For example, elevated PSMA levels have been observed in prostate cancer patients’ urine.5 The PSMA concentration increases from 0.25 nM to approximately 3.5 nM in prostate cancer patients’ biological fluids including urine.6 PSMA, a 750-residue, 90 kD glycoprotein, is overexpressed on the surface of tumor cells as a non-covalent homodimer in >94.3 and >57.7% of primary and metastatic prostate cancers respectively.7,8 Elevated PSMA levels also correlate with the aggressiveness of tumor growth.9 Thus, PSMA offers an important biomarker for the development of biosensor-based diagnostic devices. This report describes the development of a biosensor capable of detecting clinically relevant concentrations of PSMA (<0.25 nM) in synthetic urine.
In 2003, Petrenko and Vodyanoy demonstrated the use of whole virus particles as a bioaffinity matrix for biosensors.10,11 In an improved generation of biosensors, T7 virus particles with a peptide antigen from the West Nile virus on their surfaces have been incorporated into conducting polymers by Cosnier and coworkers to allow detection of antibodies to the West Nile virus.12 This strategy can offer higher density ligands for biomarker binding, as T7 phage have a high density of peptides displayed on their surface. Improving biosensor sensitivity through increasing the density of ligands on the phage surface inspired in part the approach reported here.
M13 bacteriophage, or more commonly “phage,” serve as receptors for biosensors reported by our laboratories. Viruses that infect only bacteria, the M13 bacteriophage have a readily customized protein coat, which can be tailored to bind to cancer biomarkers.13 The M13 viruses have ssDNA encapsulated by approximately 2700 copies of the major coat protein (P8) and five copies each of the four minor coat proteins. Manipulating the encapsulated DNA can provide peptides and proteins fused to the phage coat proteins, which are displayed on the phage surface.13 Combinatorial engineering of such polypeptides allows molecular evolution to obtain displayed ligands with specific binding affinities and specificities.14,15
For direct electrical detection of biomarkers, M13 bacteriophage have been incorporated into films of an electronically conductive polymer, PEDOT (poly-3,4-ethylenedioxythiophene).16–20 Synthesis of the biosensor film is accomplished by electropolymerizing EDOT on the surface of a planar gold electrode from a solution that contains virus particles. During biosensor measurements, the electrochemical impedance of the virus-PEDOT film increases upon exposure to the biomarker, providing a quantifiable readout for analyte binding.21
Modifications to the biosensing films could further improve the device’s limit of detection (LOD) for translational relevance. In a previous report, our labs described phage-incorporated into PEDOT nanowires, which resulted in biosensors with a >66 nM LOD for PSMA in synthetic urine.22 Conventional phage display results in a low density of genetically encoded ligands displayed on the surface of the phage. Here, we focus on increasing the density of such ligands, as a strategy for more sensitive measurements with higher signal-to-noise ratios. The concept of “phage wrapping” to improve ligand density builds upon our previous reports of wrapping the negatively charged phage surface with positively charged polymers to prevent non-specific binding to the phage.23,24 The approach takes advantage of the presence of negatively charged residues, one Glu and two Asp, on the N-terminus of each P8. Since each phage includes 2700 copies of P8, such carboxylate-bearing residues result in a high negative charge on the outer surface of the virus particle.25 As reported here, additional ligands wrapped onto the phage surface due to this electrostatic interaction, lead to enhanced affinity and selectivity for PSMA.
RESULTS AND DISCUSSION
Genetically-Encoded, Phage-Displayed Ligands Targeting PSMA
The two forms of PSMA, monomeric and dimeric, offer different targets for ligand binding; the dimeric form is overexpressed by prostate cancer cells, and the monomeric form offers a closely matched negative control for non-specificity, as a protein only found in healthy prostate cells.9 The relative binding affinities of two previously reported phage-displayed ligands,22 phage-1 and phage-2, (sequences and nomenclature in Table 1) for the PSMA isoforms were first examined by ELISA (Figure 1). Phage-2 binds with higher affinity to the PSMA dimer, than phage-1. Neither phage-displayed ligand binds with significant affinity to the PSMA monomer. Thus, the peptide ligands selectively bind the dimeric form of PSMA. The specificity of phage-displayed ligands for the dimeric PSMA is critical for potential clinical applications. Additional negative controls include phage-displayed peptides targeting the blocking agent (bovine serum albumin, BSA) and Stop-4 phage targeting PSMA; the latter phage includes an analogous phagemid packaged into phage without ligands displayed on their surfaces. As expected, the negative controls failed to show any significant binding.
Table 1.
PSMA ligands, sequence and nomenclature.
| X | Amino acid sequence |
Genetically encoded |
Chemically synthesized |
Structure of the chemically synthesized peptide |
|---|---|---|---|---|
| 1 | CALCEFLG | phage – 1 | KCS–1 |  |
| 2 | SECVEVFQNSCDW | phage – 2 | KCS–2 | |
Figure 1.
Phage-based ELISA comparing ligand binding to monomeric and dimeric forms of PSMA. This ELISA includes PSMA monomer; all other reported experiments with PSMA apply the cancer-relevant PSMA dimer. Stop-4 provides a negative control with helper phage packaging the phagemid DNA. Throughout this report, error bars for ELISA data represent standard error (n = 3). All experimental data points with the exception of the negative controls (n = 1) include such error bars, though often these are quite small.
The two PSMA ligands, 1 and 2, provide a starting point for the development of biosensors. For translational relevance, the LOD of the resultant biosensor must be <0.25 nM, and a high-signal-to-noise ratio is essential for definitive diagnosis. In theory, the affinity of the ligands for their target analyte should correlate with their usefulness in biosensor applications. For example, a higher affinity ligand could enhance sensitivity for the analyte. In a previous report, we described using phage-displayed homolog shotgun scanning to improve phage-displayed ligand affinity for PSMA by >100 fold.22 However, this approach requires extensive mutagenesis and selections.
Nature applies evolution-guided affinity maturation, but also relies on another approach for more rapid affinity maturation. The immune system, for example, applies the principle of avidity to boost the apparent affinity of a weaker initial lead. During the initial immune response, the IgM protein presents receptors in a decavalent format, allowing weak initial binders to attain higher apparent affinity through proximity- and chelate-based avidity.26 The large, phage surface with repeatitive structural motifs appears well-suited to this approach, and, indeed avidity effects are often present during phage-based selections and screens.27 This concept could provide a generalized method for expedient improvement of ligand affinity and biosensor sensitivity.
Cycloaddition to Generate the Secondary Recognition Ligand
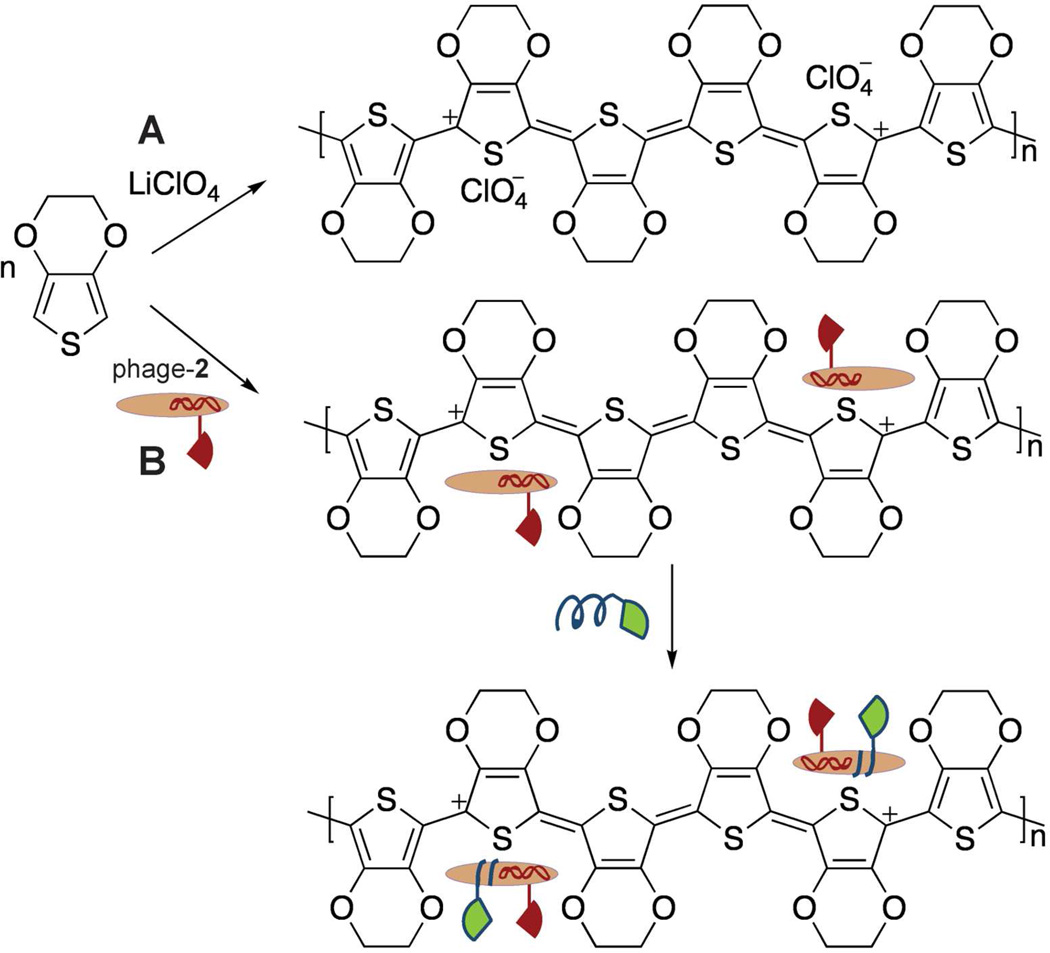
To exploit this avidity effect, phage wrapping was used to boost ligand density, subsequent affinity, and the resultant sensitivity and the signal-to-noise of phage-based biosensors. Each “wrapper” consists of two parts linked together by the CuI-catalyzed azide-alkyne cycloaddition (“click”) reaction (Scheme 1).28 The first component, an oligolysine (K14) peptide provides affinity to the phage surface. For the click reaction, an alkyne (4-pentynoic acid) was coupled to the N-terminus of the K14 peptide. The second component of the wrapper is the peptide ligand to PSMA. In previous studies, the PSMA-binding peptides, 1 and 2, exhibited limited solubility in water. The peptides were therefore synthesized as fusions to the solubilizing peptide sequence K3 on their N-termini. For the click reaction, the N-termini of the peptide ligands were coupled to an azide (4-azidobutanoic acid).
Scheme 1.
The CuI-catalyzed azide-alkyne cycloaddition reaction to provide the secondary recognition ligand, KCS-2.
The two parts of each wrapper were chemically synthesized using solid phase peptide synthesis and purified by reverse-phase HPLC before the cycloaddition reaction. Here, click chemistry offers a convergent synthesis, and the reaction takes place at room temperature and in aqueous solution.28 The resultant secondary recognition ligands thus provide an oligolysine half to wrap the phage (termed KCS for “lysine, chemically synthesized”), and a second component, the PSMA ligand (1 or 2), to bind to the analyte (Figure 2). The products formed by the click reaction (KCS-1 or KCS-2) were characterized by MALDI-TOF MS (Figure S1 in the Supporting Information), and purified by reverse-phase HPLC to an estimated 90% purity.
Figure 2.
Schematic diagram of bidentate binding to PSMA by chemically synthesized (KCS-1, green) and genetically encoded (peptide 2, red) ligands to PSMA (PDB: 1Z8L). The former ligand wraps non-covalently onto the negatively charged P8 proteins found on the phage surface due to conjugation with a positively charged K14 peptide (blue). Simultaneous binding by the two ligands provides higher apparent affinity to PSMA. The inset depicts a simplified version of the schematic appearing in the subsequent figures shown here.
Phage Wrapping to Maximize Ligand Density
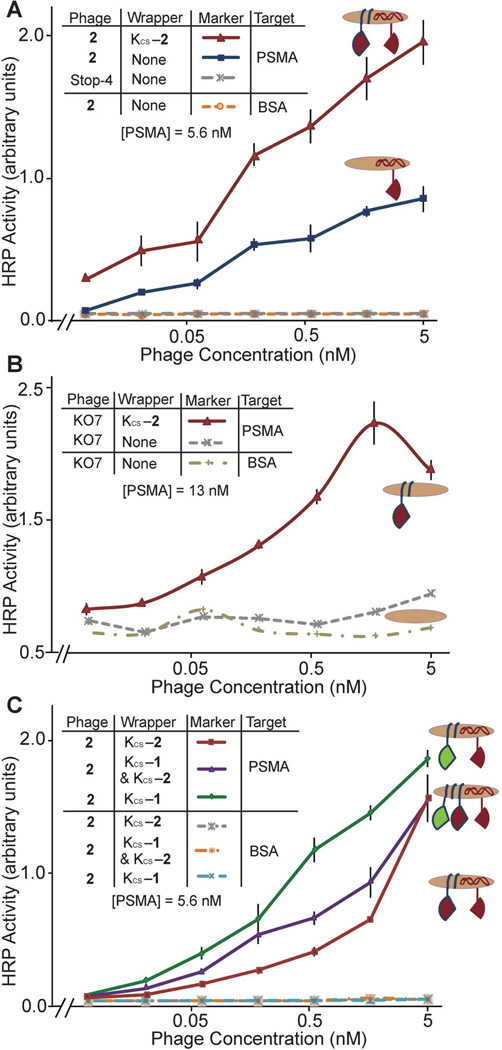
Wrapping the phage with chemically synthesized PSMA ligands described above clearly enhances binding affinity to PSMA (Figure 3). The phage-displayed ligand (phage-2) was wrapped with the secondary recognition ligands (KCS-1, KCS-2 or a mixture of the two) to generate a phage surface displaying two PSMA ligands. The wrapped phage were then assayed for binding to the PSMA dimer. Negative controls, which resulted in no detectable binding, included the PSMA ligands targeting BSA and Stop-4 targeting PSMA. Phage-2 wrapped with KCS-2 exhibits ≈50 times higher affinity for PSMA than phage-2 without the ligand wrapper (Figure 3A). Additional optimization examined the concentration of the wrapper (Figure S2 in Supporting Information). As a result, the wrapper concentration can maximize the ligand density on the phage surface. The effectiveness of wrapping for improved binding affinity is dramatically demonstrated by comparing the binding affinities of the helper phage (KO7) versus KO7 wrapped with KCS-2 (Figure 3B and Figure S3 in the Supporting Information). Lacking a displayed ligand, KO7 phage displays no significant binding to PSMA, but KO7 phage wrapped with KCS-2 binds with significant affinity to PSMA. Taken together, the results demonstrate that the wrapping strategy achieves much higher affinity for the target and the wrapped ligands remain functional.
Figure 3.
Phage-based ELISAs demonstrating the effectiveness of ligand wrapping. (A) Phage with chemically and genetically encoded ligands bind with much higher apparent affinity to the targeted PSMA. (B) The chemically synthesized KCS-2 wrapper converts helper phage, KO7, lacking a genetically encoded PSMA ligand, into a high affinity binding partner to PSMA. The decrease in apparent binding affinity at the highest phage concentration could be attributed to steric effects. (C) ELISA comparing different ligand wrappers. The wrapper combination of KCS-2 & KCS-1 indicates a 1:1 (w/w) ratio of ligand wrappers.
Primary and Secondary Recognition Ligands on Phage for Bidentate Binding
The arrangement and density of the primary, genetically encoded ligand and the secondary, chemically synthesized ligand determines the affinity of the wrapped phage for PSMA. For example, wrapping phage-2 with KCS-1 results in phage with an apparent 4-fold higher affinity for PSMA than phage-2 wrapped with KCS-2 (Figure 3C). Peptide 1, however, has a much lower apparent affinity for PSMA than peptide 2, as shown in Figure 1. Thus, the increased binding affinity of phage-2 wrapped with KCS-1, suggests that the two ligands target different sites on the surface of PSMA. Furthermore, a 1:1 mixture of KCS-1 and KCS-2 wrapped on the surface of phage-2, offers intermediate affinity between neat KCS-1 and neat KCS-2. The results demonstrate that phage-2 wrapped with KCS-1 results in improved affinity due to a bidentate binding interaction. Conversely, phage-2 wrapped with KCS-2 fails to access this bidentate binding mode. Thus, the two ligands displayed on phage, for phage-2 wrapped with KCS-1, can result in a chelate-based avidity effect, which enhances binding affinity beyond the gains achieved purely by maximization of ligand density.
In practice, chelate-based avidity effects can be challenging to design, as geometry and sterics must be satisfied to allow both ligands to reach an optimal interaction with the receptor. In fragment-based drug discovery efforts, for example, development of linkers with appropriate configuration is a non-trivial problem.29 Phage wrapping provides a more expedient solution to this problem. The second ligand, presented by a non-covalently bound wrapper, can equilibrate on the phage surface until finding a satisfactory geometry to allow simultaneous binding for synergistic effect.
Biosensing with Virus-PEDOT Films
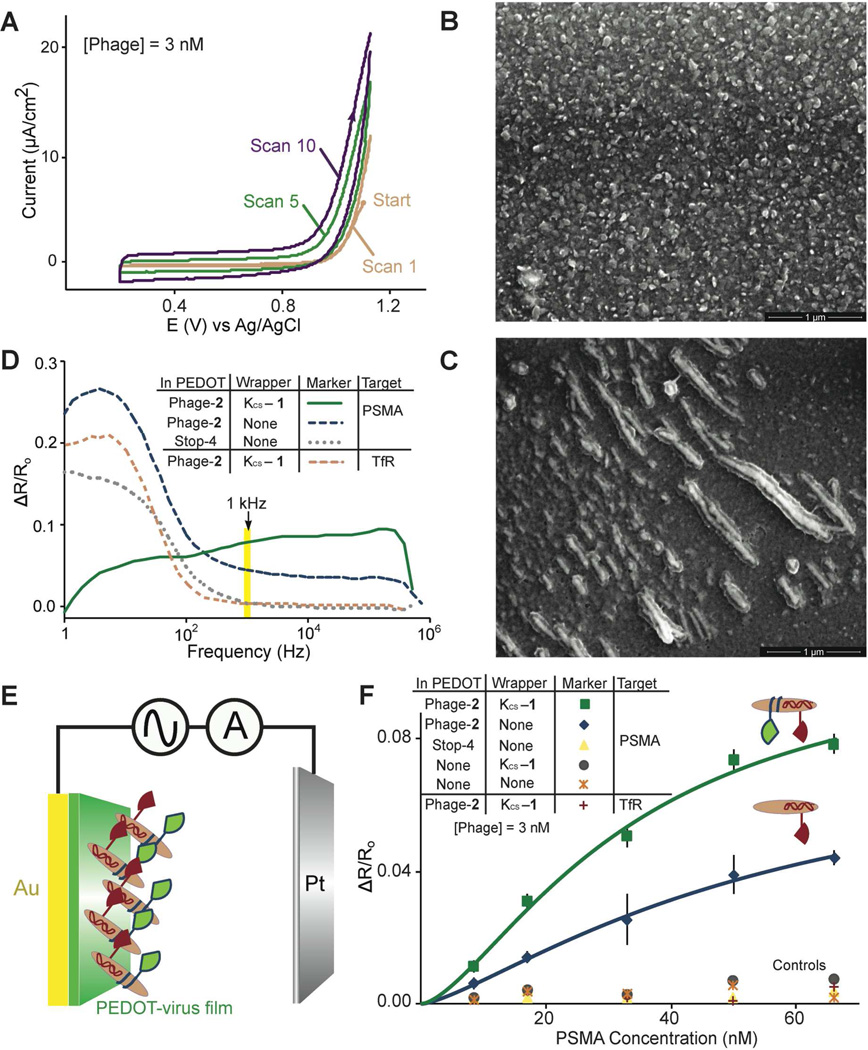
Can these wrapped virus particles be exploited to create virus-electrode biosensors with a higher sensitivity for PSMA? To explore this question, films of PEDOT were prepared on gold electrodes by electropolymerization in the presence of phage-2 (Scheme 2). The PEDOT formed in solution during electropolymerization, associate with the negatively charged perchlorate ions from the electrolyte solution as it is deposited onto the gold electrode.30 Polymerization of EDOT in the presence of negatively charged phage particles leads to incorporation of virus particles into the polymeric film as counter-ion dopants due to electrostatic interactions.21 The cyclic voltammogram acquired during electrodeposition of the virus-PEDOT bioaffinity matrix indicates that the maximum current increases with every deposition cycle, consistent with the expected increase in the surface area of the film during growth (Figure 4A). SEM imaging confirms incorporation of phage into the bioaffinity matrix; we observe both filament-like and less-extended features having dimensions consistent with phage integrated as rope-like bundles into the polymer at various angles to the film (Figure 4B, 4C). The KCS-1 wrapper was then applied in vitro simply by exposing the resultant phage-2 film for a short time to an aqueous solution of the wrapper. For the biosensing measurements, only the highest affinity ligand combination of phage-2 wrapped with KCS-1 was used, and studied in comparison to phage-2 films.
Scheme 2.
Polymerization of EDOT in the presence of (A) LiClO4 or (B) phage-2, followed by wrapping with KCS-1 (green and blue).
Figure 4.
Biosensing with virus-PEDOT films. (A) Cyclic voltammogram for depositing virus-PEDOT films on gold electrodes. (B) SEM image of a PEDOT film. (C) SEM image of a virus-PEDOT film prepared under the same conditions. (D) Relative change in resistance, ΔR/Ro, versus frequency for phage-displayed ligands targeting PSMA. Data collected at 1 kHz (highlighted), were used for the analysis of PSMA binding. (E) Schematic diagram of the biosensing experiment. (F) ΔR/Ro of the film increases with the PSMA concentration. Throughout this report, error bars for the biosensing data represent standard error (n = 5). Data was fit to the indicated lines using the Hill equation, resulting in an R2 value of >0.99.
EIS to Quantify PSMA Binding
As we have observed in our prior work,21 the electrochemical impedance of the virus-PEDOT film increases as PSMA selectively binds to the phage-displayed peptide ligands (Figure 4D). The real component of the impedance, R, in particular, increases upon PSMA binding. In previous work, we demonstrated that the increase in R, ΔR, normalized by the initial resistance, Ro, (ΔR/Ro) can be correlated with the concentration of a target molecule.21 Here, impedance data were acquired in PBF (phosphate buffered fluoride)-Tween buffer, spanning a frequency range from 0.1 Hz to 1 MHz in an electrochemical cell with a Pt counter electrode, and virus-PEDOT film electroplated on a planar gold working electrode (Figure 4E). The films incorporating phage-2 wrapped with KCS-1 provide higher sensitivity for PSMA detection than films incorporating phage-2 (Figure 4F). For example, the relative impedance change, ΔR/Ro, at each concentration of PSMA is three-fold higher. The noise present in this measurement (estimated as the standard deviation for five impedance measurements) is unchanged resulting in a much higher signal-to-noise ratio for phage-2 wrapped with KCS-1 relative to unwrapped phage-2. A series of negative controls validate the data obtained. The PEDOT films incorporating Stop-4 phage, PEDOT films lacking PSMA binding ligands, and PEDOT films incubated with KCS-1 result in no significant binding to PSMA, as expected. The specificity of PSMA binding was investigated by using an alternative target, transferrin receptor (TfR), which has 54% sequence similarity to PSMA. No significant binding affinity to TfR was observed, as expected. This experiment illustrates the negligible change in impedance caused by the wrapper due to the small size of the wrapper.
Calculating the Hill Coefficient
The biosensing data acquired for phage-2, and phage-2 wrapped with KCS-1 targeting PSMA, follows a Langmurian adsorption model. The data acquired was fit to the following Hill equation:
where Y is ΔR/Ro, L is the ligand concentration, and n is the Hill coefficient.31 Consequently, the dissociation constant, Kd and n were determined for phage-2 (Kd = 54 nM, n = 1.3, LOD = 6 nM), and phage-2 wrapped with KCS-1 (Kd = 33 nM, n = 1.5, LOD = 3.1 nM). Here, the two LODs, defined as 3x over background signal, were calculated from line fits to the data shown in Figure 4F. The response obtained for phage-2 wrapped with KCS-1 displays a much higher signal-to-noise ratio, compared to films incorporating only phage-2. Such sensitivity can play a crucial role in low concentration analyte detection. The n-values obtained are >1, indicating the presence of multiple binding sites and a cooperative binding effect. Phage-2 can access cooperative binding due to the avidity effect of multi-copy phage-displayed ligands. Wrapping phage with additional ligands leads to a further increase in cooperativity for phage-2 wrapped with KCS-1. Again, the synergism of the two ligands leads to higher PSMA binding affinity for phage-2 wrapped with KCS-1.
PSMA Detection in Synthetic Urine
To further demonstrate the usefulness of the approach for potential clinical applications, biosensing data was next acquired in synthetic urine. This complex solution includes water, nitric acid, urea, sodium sulfate, potassium chloride, sodium dihydrogen phosphate, sodium chloride, ammonium chloride and 10 other components; the resultant solution has a high salt concentration (a calculated osmolality of 516.2 mOsm/kg and a pH of 5.8).32,33 The solution provides a good model for the clinical challenge of identifying cancer biomarkers found in urine samples. Substituting synthetic urine for PBF, impedance measurements with virus-PEDOT films were acquired as before (Figure 5A). Having already established phage-2 wrapped with KCS-1 as the most effective ligand combination for detecting PSMA in terms of sensitivity, specificity and signal-to-noise ratio, our experiments focused on this ligand combination for synthetic urine-based biosensing. In the presence of PSMA, the impedance scans for virus-PEDOT films of phage-2 wrapped with KCS-1 follow similar trends in both PBF and synthetic urine. However, the lower frequency ranges differ dramatically for the negative controls. For example, the negative control with Stop-4 phage in PBF displayed a much higher ΔR/Ro; this response at low frequencies was suppressed in synthetic urine. Thus, the measurements in synthetic urine resulted in higher specificity for the PSMA-ligand interaction.
Figure 5.
Detection of PSMA in synthetic urine using virus-PEDOT film biosensors. (A) ΔR/Ro versus frequency for the detection, in synthetic urine, of PSMA. (B) ΔR/Ro versus PSMA concentration. The inset expands the low PSMA concentration region. Data was fit to the indicated lines using the Hill equation, resulting in an R2 value of >0.99.
Turning next to the measurement of PSMA concentration, the synthetic urine solution also appeared to enhance measurement sensitivity. The calibration curves in PBF and synthetic urine were overlaid for comparison (Figure 5B, green and purple, respectively). At high analyte concentrations, measurements in PBF and synthetic urine are superimposable. Both conditions reach saturation at high PSMA concentrations, hence the overlap in device response. At the lowest PSMA concentrations, the higher ΔR/Ro response in synthetic urine can be attributed to higher sensitivity and selectivity for PSMA under the high-salt conditions. Phage-2 films wrapped with KCS-1 targeting PSMA in synthetic urine, yields a 100 pM experimentally observed LOD. Applying the calculation to determine LOD described above yields a 10 pM LOD for the detection of PSMA in synthetic urine. Furthermore, this sensitivity requires no signal or enzymatic amplification. As before no significant change in impedance was observed for the negative controls.
The high salt concentration in synthetic urine appears to prevent non-specific charge-charge interactions. Binding of the biomarker to the virus-PEDOT film generates a positive ΔR/Ro, whereas a negligible ΔR/Ro for the negative controls indicates a non-significant extent of non-specific binding by the analyte. The resultant specificity increase in synthetic urine boosts the apparent sensitivity of the device by decreasing background binding. This effect also enhances the sensitivity by unmasking a higher concentration of ligands for analyte detection, which would otherwise be occluded through non-specific binding. Thus, a dramatic improvement in specificity and sensitivity can be obtained through the decreased non-specific interactions. The results suggest a general strategy for improving biosensor performance through focusing on decreased non-specific binding.
CONCLUSION
The 100 pM experimentally measured LOD obtained for phage-2 wrapped with KCS-1 is a significant improvement over our previous effort, which had a >66 nM LOD22 for PSMA. This detection sensitivity satisfies a key requirement for clinical applications. The chelate-based avidity effect and high specificity obtained due to the bidentate binding mode distinguishes this work from our previous efforts, and also accounts for the greatly improved signal-to-noise ratio and LOD obtained without enzymatic amplification. Furthermore, inclusion of synthetic urine as an analyte solution expands the applicability of the technique. However, the reported biosensing films have not been optimized. Further improvements could be made to achieve an increased response, without amplification, for lower PSMA concentrations, as demonstrated by earlier experiments with similar biosensing films.34
In conclusion, wrapping phage with additional ligands can increase the target affinity due to a chelate-based synergistic effect. Phage-2 wrapped with KCS-1 binds to PSMA with a 33 nM Kd and a 3.1 nM LOD in PBF, and a 100 pM experimentally measured LOD in synthetic urine. In the future, the biosensors will be optimized for improved PSMA detection.
Supplementary Material
ACKNOWLEDGMENT
R.M.P. acknowledges the financial support of this work from the National Science Foundation (CHE-0956524), and G.A.W. acknowledges support from the NAID (1 R43 AI074163) and the NCI (R01 CA133592-01) of the National Institutes of Health.
Footnotes
ASSOCIATED CONTENT
Additional materials and methods are available in the supporting information. This information is available free of charge via the internet at http://pubs.acs.org.
REFERENCES
- 1.Siegel R, Naishadham D, Jemal A. CA-Cancer J. Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Moyer VA. Ann. Intern. Med. 2012;157:120–134. doi: 10.7326/0003-4819-157-2-201207170-00459. [DOI] [PubMed] [Google Scholar]
- 3.Madu CO, Lu YL. J. Cancer. 2010;1:150–177. doi: 10.7150/jca.1.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murphy GP, Kenny GM, Ragde H, Wolfert RL, Boynton AL, Holmes EH, Misrock SL, Bartsch G, Klocker H, Pointner J, Reissigl A, McLeod DG, Douglas T, Morgan T, Gilbaugh J. Urology. 1998;51:89–97. doi: 10.1016/s0090-4295(98)00082-x. [DOI] [PubMed] [Google Scholar]
- 5.Sokoloff RL, Norton KC, Gasior CL, Marker KM, Grauer LS. Prostate. 2000;43:150–157. doi: 10.1002/(sici)1097-0045(20000501)43:2<150::aid-pros10>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 6.Xiao Z, Adam B-L, Cazares LH, Clements MA, Davis JW, Schellhammer PF, Dalmasso EA, Wright GL. Cancer Res. 2001;61:6029–6033. [PubMed] [Google Scholar]
- 7.Schülke N, Varlamova OA, Donovan GP, Ma D, Gardner JP, Morrissey DM, Arrigale RR, Zhan C, Chodera AJ, Surowitz KG, Maddon PJ, Heston WDW, Olson WC. Proc. Natl. Acad. Sci. USA. 2003;100:12590–12595. doi: 10.1073/pnas.1735443100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chuang A-Y, DeMarzo AM, Veltri RW, Sharma RB, Bieberich CJ, Epstein JI. Am. J. Surg. Pathol. 2007;31:1246–1255. doi: 10.1097/PAS.0b013e31802f5d33. [DOI] [PubMed] [Google Scholar]
- 9.Su SL, Huang IP, Fair WR, Powell CT, Heston WD. Cancer Res. 1995;55:1441–1443. [PubMed] [Google Scholar]
- 10.Petrenko VA, Vodyanoy VJ. J. Microbiol. Methods. 2003;53:253–262. doi: 10.1016/s0167-7012(03)00029-0. [DOI] [PubMed] [Google Scholar]
- 11.Nanduri V, Sorokulova IB, Samoylov AM, Simonian AL, Petrenko VA, Vodyanoy V. Biosens. Bioelectron. 2007;22:986–992. doi: 10.1016/j.bios.2006.03.025. [DOI] [PubMed] [Google Scholar]
- 12.Ionescu RE, Cosnier S, Herrmann S, Marks RS. Anal. Chem. 2007;79:8662–8668. doi: 10.1021/ac0707129. [DOI] [PubMed] [Google Scholar]
- 13.Smith GP. Science. 1985;228:1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 14.Kehoe JW, Kay BK. Chem. Rev. 2005;105:4056–4072. doi: 10.1021/cr000261r. [DOI] [PubMed] [Google Scholar]
- 15.Levin AM, Weiss GA. Mol. Biosyst. 2006;2:49–57. doi: 10.1039/b511782h. [DOI] [PubMed] [Google Scholar]
- 16.Weiss GA, Penner RM. Anal. Chem. 2008;80:3082–3089. doi: 10.1021/ac086009h. [DOI] [PubMed] [Google Scholar]
- 17.Diaz JE, Yang L-MC, Lamboy JA, Penner RM, Weiss GA. Methods Mol. Biol. 2008;504:255–274. doi: 10.1007/978-1-60327-569-9_16. [DOI] [PubMed] [Google Scholar]
- 18.Yang L-MC, Diaz JE, McIntire TM, Weiss GA, Penner RM. Anal. Chem. 2008;80:5695–5705. doi: 10.1021/ac8008109. [DOI] [PubMed] [Google Scholar]
- 19.Yang L-MC, Tam PY, Murray BJ, McIntire TM, Overstreet CM, Weiss GA, Penner RM. Anal. Chem. 2006;78:3265–3270. doi: 10.1021/ac052287u. [DOI] [PubMed] [Google Scholar]
- 20.Arter JA, Taggart DK, McIntire TM, Penner RM, Weiss GA. Nano Lett. 2010;10:4858–4862. doi: 10.1021/nl1025826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Donavan KC, Arter JA, Pilolli R, Cioffi N, Weiss GA, Penner RM. Anal. Chem. 2011;83:2420–2424. doi: 10.1021/ac2000835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arter JA, Diaz JE, Donavan KC, Yuan T, Penner RM, Weiss GA. Anal. Chem. 2012;84:2776–2783. doi: 10.1021/ac203143y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lamboy JA, Arter JA, Knopp KA, Der D, Overstreet CM, Palermo EF, Urakami H, Yu T-B, Tezgel O, Tew GN, Guan Z, Kuroda K, Weiss GA. J. Am. Chem. Soc. 2009;131:16454–16460. doi: 10.1021/ja9050873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamboy JA, Tam PY, Lee LS, Jackson PJ, Avrantinis SK, Lee HJ, Corn RM, Weiss GA. ChemBioChem. 2008;9:2846–2852. doi: 10.1002/cbic.200800366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Welsh LC, Symmons MF, Sturtevant JM, Marvin DA, Perham RN. J. Mol. Biol. 1998;283:155–177. doi: 10.1006/jmbi.1998.2081. [DOI] [PubMed] [Google Scholar]
- 26.Paul W, editor. Fundamental Immunology. 6th ed. Lippincott Williams & Wilkins; 2008. p. 164. [Google Scholar]
- 27.Murase K, Morrison KL, Tam PY, Stafford RL, Jurnak F, Weiss GA. Chem. Biol. 2003;10:161–168. doi: 10.1016/s1074-5521(03)00025-5. [DOI] [PubMed] [Google Scholar]
- 28.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. Angew. Chem. Int. Ed. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 29.Erlanson DA. Top. Curr. Chem. 2012;317:1–32. doi: 10.1007/128_2011_180. [DOI] [PubMed] [Google Scholar]
- 30.Sharma PS, Pietrzyk-Le A, D’Souza F, Kutner W. Anal. Bioanal. Chem. 2012;402:3177–3204. doi: 10.1007/s00216-011-5696-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hill AV. J. Physiol. (Lond) 1910;40:4–7. [Google Scholar]
- 32.Mccurdy D, Lin Z, Inn KGW, Iii RB, Wagner S, Efurd DW, Steiner R, Duffy C, Hamilton TF, Brown TA, Marchetti AA. J. Radioanal. Nucl. Chem. 2005;263:447–455. [Google Scholar]
- 33.McClatchey KD, editor. Clinical Laboratory Medicine. 2nd ed. Lippincott Williams & Wilkins; 2002. p. 381. [Google Scholar]
- 34.Donavan KC, Arter JA, Weiss GA, Penner RM. Langmuir. 2012;28:12581–12587. doi: 10.1021/la302473j. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.