Abstract
High abundance proteins in serum and plasma (e.g., albumin) are routinely removed during proteomic sample processing as they can mask lower abundance proteins and peptides of biological/clinical interest. A common method of albumin depletion is based on immunoaffinity capture, and many immunoaffinity devices are designed for multiple uses. In this case, it is critical that the albumin captured on the affinity matrix is stripped from the column prior to regeneration of the matrix and processing of subsequent samples, to ensure no carryover and that maximal binding sites are available for subsequent samples. The current study examines the ability of a manufacturer’s protocol to remove the proteins and peptides captured by an immunoaffinity spin column. The data presented in the current work illustrate the difficulty in completely removing albumin from the immunoaffinity device, and consequently, may explain the variability and decreased efficiency shown for this device in previous studies. In summary, the current data present important considerations for the implementation of multiple-use immunoaffinity devices for processing subsequent clinical samples in a proteomic workflow.
Keywords: Albumin, Antihuman serum albumin column, Serum
1 Introduction
High abundance proteins in serum and plasma (e.g., albumin) are commonly removed during proteomic sample processing as they can mask lower abundance proteins and peptides of biological interest. Albumin depletion, for example, is a routine first step in the processing of serum and plasma for proteomic studies, and its use has been described in numerous publications (for reviews see [1–3]). This need has led to the development of chemical-based [4–6], as well as multiple commercial technologies for the depletion of albumin from serum and plasma samples. The commercially available devices include both dye (e.g., cibacron blue) and immunoaffinity-based methods and are available in a variety of formats including spin columns and on-line LC columns. Importantly, in addition to albumin depletion, these albumin-capture devices are also useful for studying albumin itself, which has been suggested to be modified, both in concentration and metal binding properties, in correlation with myocardial ischemia [7–9]. Other disease states such as diabetes mellitus [10], liver disease [11, 12], and renal dysfunction [13, 14], have also been associated with changes in albumin oxidation states [7, 8, 12, 15, 16]. In addition to changes to the primary sequence of albumin, it is known that albumin binds various ligands including proteins, peptides and small molecules [16–22], and a number of recent studies have discussed the utility of capturing the “albuminome” (albumin and its bound proteins/peptides) as a means to enrich for potential disease biomarkers [23–27]. Examples of interesting potential biomarkers that have been found to be associated with albumin, and thus may be removed during albumin depletion, include BRCA2 peptides, C-reactive protein, cystatin C [23], apoliprotein CIII, lumican, and angiotensinogen [28].
Immunoaffinity devices have been shown to typically be more specific than dye based methods [29–33], and thus are often the method of choice for both albumin-depletion and albumin-capture studies. Due to the expense of the antibody-based devices, they are typically designed for repeated use. Importantly, however, several previous studies have compared the effectiveness of a number of the commercially available affinity based methods and have shown that these affinity based devices are vulnerable to nonspecific binding of proteins/peptides to the ligand and column materials and carryover between experiments in the case of LC columns [30, 32, 34–37]. These studies highlight the importance of validating that the protein captured by the column is completely removed prior to processing subsequent samples, to ensure no carryover and that maximal binding sites are available for subsequent samples. This is critical both for studies that are interested in examining the albumin-depleted sample, as well as those interested in albumin and its bound proteins/peptides.
In proteomic analyses, optimal sample preparation methods must be highly reproducible while minimizing the potential carryover among multiple samples in order for meaningful comparisons, for example differences among disease states, to be interpreted. For these reasons, it is critical that sample preparation methods are validated prior to implementation into the proteomic workflow. Thus, immunoaffinity depletion methods should be examined for their reproducibility, specificity, and potential to introduce sample carryover when used for multiple samples. The specificity (i.e., ability to capture only the desired protein) of albumin depletion using immunoaffinity based methods has been previously examined [29–33]. In particular, the specificity of spin columns [38] and on-line LC formats [38, 39] of chicken IgY based immunoaffinity devices has been examined. In a separate study, the reproducibility of the same chicken IgY-based immunoaffinity spin column was assessed by analyzing the resulting albumin-depleted fraction of serum using SELDI-TOF MS [40]. The data presented showed variability in this approach, with inconsistencies observed across both the MS profiles and the concentration of albumin that remains in the depleted fraction of multiple samples. Furthermore, the efficiency of depletion was shown to decrease with increased uses of the same spin column. These data suggest a loss of binding capacity of the column with increased usage, possibly through the loss of antigen binding or the inaccessibility of the antibody due to incomplete stripping of the bound albumin between subsequent samples.
The current study adds to the assessment of the chicken IgY immunoaffinity spin column for processing multiple samples by specifically examining the ability to remove the proteins and peptides captured by the spin column between samples. By evaluating the ability of the recommended standard operating procedure (SOP) to sufficiently strip the captured proteins from the column, such that there would be no carryover to the next sample, this study aims to complement previously published studies. Assessment of the protein that is left bound to the column, rather than the albumin-depleted fraction, is necessary for validating the use of such spin columns for studies that examine the discrete albuminome of multiple clinical samples. The data presented in the current work illustrate the difficulty in completely removing albumin from the immunoaffinity device, and consequently, may explain the variability and decreased efficiency shown in previous studies [40]. In summary, the current data present important considerations for the implementation of multiple-use immunoaffinity devices for processing subsequent clinical samples in a proteomic workflow.
2 Materials and methods
2.1 Reagents
IgY-anti-HSA spin columns and all kit reagents were kindly provided by Beckman Coulter (Fullerton, CA). Normal human serum (Scantibodies, Santee, CA) was processed using the IgY-anti-HSA column. The same serum sample was used for all spin column experiments. Purified HSA was obtained from Sigma–Aldrich (St. Louis, MO).
2.2 Sample processing using the IgY-anti-HSA spin column
For each experiment, 20 μl serum was used and each experiment was performed in triplicate. As recommended by the manufacturer, the serum was passed through a 0.45 μm spin filter to remove particulates and fibrin clots. Also, prior to the first use of the column, two cyclings of the SOP were performed without the addition of serum in order to remove uncoupled IgY and other chemical components as well as to condition the resin. The SOP as written by the manufacturer was followed for all experiments except where the following components were changed: the concentration of the stripping buffer was either 0.1 M (1×) or 0.25 M (2.5×) glycine-HCl (pH 2.5), the concentration of neutralization buffer was either 0.1 M (1×) or 0.25 M (2.5×) Tris-HCl (pH 8.0), and the number of sample strips ranged from 2 to 6. It is noted that the recommended SOP at the time this work was begun included using two sample strips with 1×stripping buffer, though more recent distributions of the SOP recommend four sample strips with 1×stripping buffer.
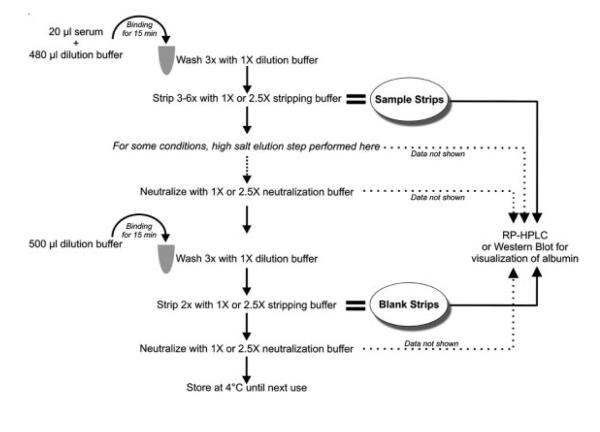
For all experiments, the serum was allowed to bind to the column for 15 min at room temperature with gentle end-over-end rotation. The column was then washed three times with 500 μl dilution buffer (10 mM Tris-HCl, pH 7.4, 150 mM NaCl) followed by 3–6 sample strips with stripping buffer. For some experiments, a high salt elution step after the glycine-HCl stripping step was also performed with either 0.25 or 0.5 M Tris-HCl, pH 8.0, 1 M NaCl, as high salt conditions are often successful in disrupting protein-protein interactions. Immediately following the stripping steps, the column was neutralized with the appropriate concentration of neutralization buffer to match the concentration of the stripping buffer. The column was then stored in dilution buffer until the next cycling. The overview of the method is illustrated in Fig. 1 and the combinations of the various components used to remove the bound protein are summarized in Table 1. Since the previous study by Seam et al. [40] showed decreased efficiency after >.17 cyclings of the spincolumn, we validated our results for each approach using new columns for each new experimental series.
Figure 1.
Schematic illustrating one cycling of the IgY-anti-HSA spin column. First, serum is processed through the column. Secondly, a “blank” sample containing only dilution buffer is processed in order to determine whether any protein left on the column after processing of the first sample would carryover into the subsequent sample.
Table 1.
Composition of reagents used for removing captured albumin from the IgY-anti-HSA spin column
| Condition | Sample stripping buffer | Salt elution after sample strip | Blank stripping buffer |
|---|---|---|---|
| 1 | 0.1 M Glycine-HCl pH 2.5 | – | 0.1 M Glycine-HCl pH 2.5 |
| 2 | 0.1 M Glycine-HCl pH 2.5 | – | 0.25 M Glycine-HCl pH 2.5 |
| 3 | 0.1 M Glycine-HCl pH 2.5 | 0.25 M Tris-HCl, pH 8.0, 1 M NaCl | 0.25 M Glycine-HCl pH 2.5 |
| 4 | 0.1 M Glycine-HCl pH 2.5 | 0.5 M Tris-HCl, pH 8.0, 1 M NaCl | 0.25 M Glycine-HCl pH 2.5 |
| 5 | 0.25 M Glycine-HCl pH 2.5 | – | 0.25 M Glycine-HCl pH 2.5 |
| 6 | 0.25 M Glycine-HCl pH 2.5 | 0.25 M Tris-HCl, pH 8.0, 1 M NaCl | 0.25 M Glycine-HCl pH 2.5 |
| 7 | 0.25 M Glycine-HCl pH 2.5 | 0.5 M Tris-HCl, pH 8.0, 1 M NaCl | 0.25 M Glycine-HCl pH 2.5 |
2.3 Evaluating protein stripping efficiency
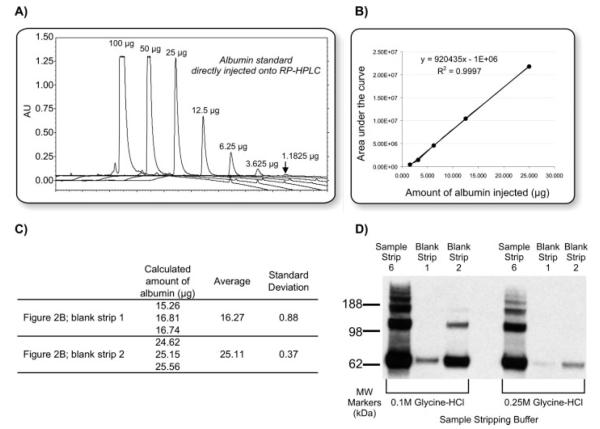
To evaluate whether any albumin was left on the column after processing the serum sample, a “blank” sample containing only dilution buffer was processed in the same manner as the serum sample, except that two sequential incubations with stripping buffer were performed (Fig. 1). The stripping buffer collected after incubation with this “blank” sample is termed a “blank strip,” whereas it is termed a “sample strip” when performed after incubation with the serum sample. Examining the blank strips provided information about the amount, if any, of protein that would carryover after stripping of one sample and potentially be present in subsequent sample strips. All strips were collected and dried in vacuo and resuspended in 0.1% TFA prior to injection onto RP-HPLC. RP-HPLC was chosen as the detection method for this work due to the fact that it provides an easy, antibody-independent method for quantitation that is compatible with the buffers used for washing, stripping, and neutralizing the IgY-anti-HSA spin column. RP-HPLC can be quantitative when measured at 210 nm as the absorbance is relative to the number of peptide bonds present. Other methods, such as the bicinchoninic acid assay, were considered for protein quantitation but not selected because of their incompatibility with the buffer compositions used in processing the spin column. For RP-HPLC, a Jupiter 5 μ C18, 300 Å, 250×4.6 mm2 (Phenomenex, Torrance, CA) column was used on a System Gold (Beckman Coulter) HPLC. The gradient ran 5% B for 10 min, 5–95% B in 25 min, 95% B to 5% B in 5 min, and 5% B for 15 min at 1 mL/min with mobile phase A = 0.1% TFA in water and mobile phase B = 0.1% TFA in ACN. Absorbance was monitored at 210 nm. Duplicate runs of 0.1% TFA only (i.e., no sample) were performed between each sample injection to ensure that any absorbance detected by UV was due to the protein stripped from the IgY-anti-HSA column and not carryover from the RP-HPLC column (data not shown). In addition to the glycine-HCl strips (Figs. 2 and 3), the high salt strips and the neutralization buffers from each experiment were also examined, but no protein was detected in these samples (data not shown). In order to obtain reference absorbencies for comparison to the processed serum samples, various amounts of purified HSA (1.18–100 μg) were directly injected onto the RP-HPLC (Fig. 4A) and a calibration curve (Fig. 4B) was used to calculate the amount of protein observed in the blank strips (Fig. 4C).
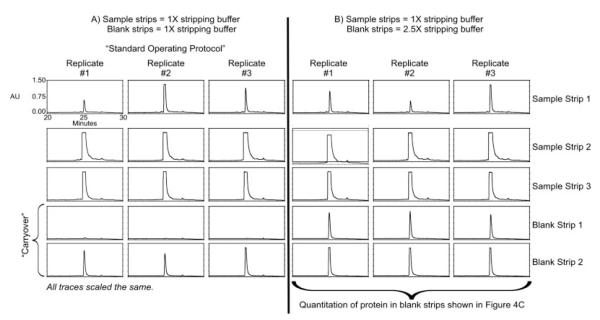
Figure 2.
RP-HPLC chromatograms illustrating protein removed using three sample strips. Panel A shows results when 1X stripping buffer was used for both sample strips and blank strips. Panel B shows results when 1X stripping buffer was used for sample strips and 2.5X stripping buffer was used for blank strips.
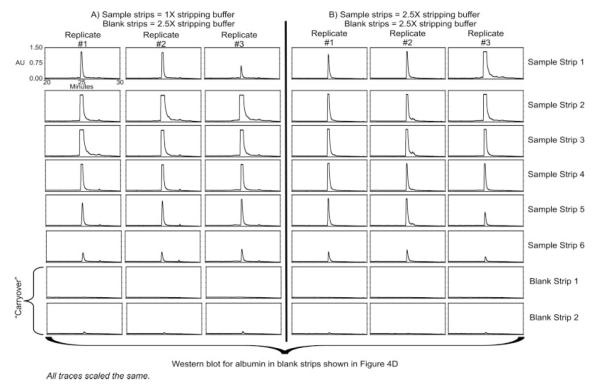
Figure 3.
RP-HPLC chromatograms illustrating protein removed using six sample strips. Panel A shows results when 1X stripping buffer was used for sample strips and 2.5X stripping buffer was used for blank strips. Panel B shows results when 2.5X stripping buffer was used for both sample strips and blank strips
Figure 4.
Quantification of albumin standard and protein stripped from the immunoaffinity column. (A) RP-HPLC chromatograms overlaid illustrating reference absorbencies observed for various amounts of an albumin standard directly injected onto the RP-HPLC. (B) Corresponding calibration curve for the albumin standard concentrations shown in (A). Concentrations which saturate the detector (.50 μg) were not included in the calibration curve. This calibration curve and resulting equation was then used to calculate the amounts of albumin removed in three replicate blank strips whose chromatograms are shown in Fig. 2B. The result of these calculations are listed in (C). (D) Western blot probing for albumin after six sample strips with 2.5×stripping buffer (correlates to blank strips in Fig. 3).
Additionally, for selected conditions, a Western blot was used to determine the presence of albumin. Dried IgY column elements were solubilized in 40 μl ddH2O. One microliter of that solution was separated on a 4–12% NuPage Bis– Tris gel (Invitrogen, Carlsbad, CA), transferred to a NC membrane (GE Water & Process Technologies, Trevose, PA) then blocked overnight at 4°C with Western Blocking Reagent (Roche Diagnostics, Indianapolis, IN) diluted 1:10 in TBS-T (20 mM Tris, pH 7.4, 150 mM NaCl, and 0.1% Tween-20). The immunoblots were probed with 1:10 000 mouse monoclonal antihuman albumin antibody (clone HSA-11, Sigma, St. Louis, MO) as primary and 1:10 000 goat antimouse IgG conjugated to alkaline alkaline phosphatase (AP; Jackson Immunologicals, West Grove, PA) as secondary antibody. Blots were developed by Immun-Star AP Substrate Pack (BioRad, Hercules, CA) according to the manufacturer’s protocol. For the gel, various different sample dilutions were attempted (1:1–1:20), though due to the high concentration of glycine-HCl, dilutions less than 1:8 resulted in poorly resolved protein bands (data not shown). For the Western blot, less than 5 s exposure time was required to detect the antigen.
3 Results
Three sample strips with 1×stripping buffer (condition 1, Table 1; see Fig. 1 for schema) is the recommended SOP, but it was not sufficient to completely remove the protein bound to the spin column, as shown by the elution of albumin in the subsequent three blank strips (Fig. 2A). This is further confirmed in Fig. 2B, where even more albumin was detected in the blank strips when a higher than recommended concentration of stripping buffer (2.5×vs. 1×) was used for the blank strips (condition 2, Table 1). The albumin detected in each blank strip with the 2.5×stripping buffer was quantified and results are shown in Fig. 4C. In summary, more than 40 μg of protein remains on the column after three strips with 1×stripping buffer. These results were not anticipated, and it is important to reiterate that the initial goal of this study was not to determine the optimum conditions for removing all of the bound protein, but rather to validate the manufacturer’s SOP for use in future examinations of the albuminome in clinical samples. Any changes to the SOP for the purpose of improving the efficiency of albumin removal were made in such a way to minimize the potential adverse affects on the antibody, as drastic changes would require investigation of the impact of such conditions on the lifetime of the antibody.
The number of sample strips was increased from 3 to 4– 6, and only after six strips with 1×stripping buffer, did it appear by UV, that nearly all of the protein was successfully removed (Fig. 3A). To determine if it would be possible to observe the same results with fewer strips, the same process was also examined using the 2.5×stripping buffer, but again it appeared that the minimum number of sample strips required to observe a blank strip at baseline UV levels was six sample strips (Fig. 3B). The marginal difference in the amount of albumin eluted in the blank strips after using six sample strips with 1×or 2.5×stripping buffer, compared to the removal due to increasing the number of strips from 3 to 6, suggests that the volume of stripping buffer that the column is incubated with is perhaps more critical than the stripping buffer concentration, at least under these conditions. This could, however, be due to the longer incubation time that the stripping buffer is in contact with the column rather than the number of strips, per say. To test this, a longer stripping time (6 min) with 2.5×stripping buffer (compared to the 3 min with 2.5×stripping buffer) was als otested. In this case, six total strips were still required in order to observe a baseline chromatogram in the blank strip (data not shown). Taken together, the amount of sample dilution which increases with each sequential incubation of stripping buffer appears to be more critical for the removal of protein bound to the column rather than the total amount of incubation time.
It was considered that increasing the concentration and number of strips alone may not be sufficient to completely remove the captured albumin from the column. For this reason, several experiments used a high salt elution step after the sample strip step (see Table 1 for conditions). However, no protein was detected in these high salt elutions (data not shown), even though subsequent blank strips after this high salt treatment did reveal detectable peaks. Thus it appeared that the high salt elution was also unable to remove the bound albumin. We therefore concluded that given the apparent lack of improvement in stripping efficiency and potential incompatibilities with downstream analysis (e.g., MS examination), the high salt elution was deemed to be not beneficial in this case.
As shown in Fig. 4A, the detection limit of the UV detector on the RP-HPLC is close to 1 μg. Therefore, anti-albumin Western blots were carried out on the improved conditions (6 strips with 2.5×stripping buffer) to determine whether any albumin was present in the blank strips at a level below the UV detection limit. It is clear that albumin still remains on the column even after six sample strips with 2.5×stripping buffer and this represents the potential carryover to the subsequent samples (Fig. 4D). There is less carryover potential, however, with 2.5×versus 1×stripping buffer.
Finally, it is important to point out that the first strip following both the sample and blank loading conditions contained less protein (Figs. 2–4) than subsequent strips. This may seem counterintuitive, as it would be expected that the majority of albumin would be removed in the first strip. However, this phenomenon is reproducible, as it is observed in every experimental series for both the data shown here and data not shown. Therefore, the fact that the first strips contain less protein than subsequent strips is attributed to the fact that the stripping buffer in the first strip is somewhat diluted by any remaining dilution buffer (i.e., void volume) from the preceding washing step, thus reducing the effectiveness of stripping buffer.
4 Discussion
It is critical for proteomics analyses that multiple-use devices are implemented in a manner that minimizes the carryover between samples, regardless of whether the investigator is interested in the proteins and peptides that are captured by the column or rather what remains after depletion. The consequences of incomplete stripping can include both decreased binding capacity as well as carryover to subsequent samples, and thus impede efforts to find proteins and peptides that change with disease. Results presented here clearly demonstrate that the manufacturer’s SOP does not completely remove the protein bound to the column and these results are consistent with previous observations [40] that the binding efficiency of the spin column is variable with increased usage. While the current study has focused on a single spin-column device for removing albumin, the importance of such an assessment is applicable to other types of multiple-use immunoaffinity devices.
The current study does not attempt to find the optimum conditions for completely removing albumin from the IgY-anti-HSA spin column, but aims rather to illustrate the difficulty in removing albumin captured from normal human serum using the manufacturer’s SOP and several variations thereof. While the methods used here do present an improvement in the removal of albumin compared to the manufacturer’s SOP, the results indicate that it may indeed be difficult to find optimal conditions that would allow for complete removal of albumin and associated proteins, while preserving the antibody. The affect on the antibody lifetime and binding capacity under any new stripping conditions is important for determining the applicability of any protocol. Additionally, optimum stripping conditions should also allow for analysis of albumin and any albumin-bound proteins or peptides in typical downstream proteomic analyses, such as MS. In this case, salt, detergents, and other components that are incompatible with MS should be avoided or kept to a minimum. Also, sample handling and any opportunity for sample degradation should be kept to a minimum to preserve integrity of any bound proteins/peptides, which may be present in low abundance. Furthermore, differences in the sample, such as lipid content and pH will affect the column effectiveness either by plugging the column/filter or hindering the initial antigen-antibody binding, thus reducing the binding capacity of the column. In these cases, the sample would need to be delipidated and neutralized prior to affinity chromatography. Consequently, until the optimum method is developed, the percentage of binding sites that are occupied between samples and/or the amount of carryover that is acceptable for a particular proteomic analysis is for the investigator to determine, and will likely depend on the downstream analysis methods and biological questions. As the results shown here illustrate, it is critical that the potential for sample carryover on multiple-use devices is acknowledged prior to implementation of any method into the proteomic workflow. Finally, the current study suggests that an alternative composition for the stripping buffer, or alternative protocol, which would allow for more efficient removal of the albumin from the column, while also allowing for regeneration of the column (i.e., not prove detrimental for the antibody), would be beneficial to the proteomics community.
Supplementary Material
Acknowledgments
This research was supported in part by funding from the NHLBI Proteomics Innovation Contract N01-HV-28180 (RLG, IT, JEV). MYW is a CJ Martin Postdoctoral Research Fellow supported by The National Health and Medical Research Foundation (464905) of Australia. Special thanks to Dr. Qin Fu for careful review of this manuscript.
Abbreviation
- SOP
standard operating procedure
Footnotes
The authors have declared no conflict of interest.
5 References
- [1].Kim MR, Kim CW. Human blood plasma preparation for two-dimensional gel electrophoresis. J. Chromatogr. B Anal. Technol. Biomed. Life. Sci. 2007;849:203–210. doi: 10.1016/j.jchromb.2006.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Luque-Garcia JL, Neubert TA. Sample preparation for serum/plasma profiling and biomarker identification by mass spectrometry. J. Chromatogr. A. 2007;1153:259–276. doi: 10.1016/j.chroma.2006.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Jacobs JM, Adkins JN, Qian WJ, Liu T, et al. Utilizing human blood plasma for proteomic biomarker discovery. J. Proteome Res. 2005;4:1073–1085. doi: 10.1021/pr0500657. [DOI] [PubMed] [Google Scholar]
- [4].Fu Q, Garnham CP, Elliott ST, Bovenkamp DE, Van Eyk JE. A robust, streamlined, and reproducible method for proteomic analysis of serum by delipidation, albumin and IgG depletion, and two-dimensional gel electrophoresis. Proteomics. 2005;5:2656–2664. doi: 10.1002/pmic.200402048. [DOI] [PubMed] [Google Scholar]
- [5].Chen YY, Lin SY, Yeh YY, Hsiao HH, et al. A modified protein precipitation procedure for efficient removal of albumin from serum. Electrophoresis. 2005;26:2117–2127. doi: 10.1002/elps.200410381. [DOI] [PubMed] [Google Scholar]
- [6].Cohn EJ, Strong LE, Hughes WL, Mulford DJ, et al. Preparation and properties of serum and plasma proteins. IV. A system for the separation into fractions of the protein and lipoprotein components of biological tissues and fluids. J. Am. Chem. Soc. 1946;68:459–475. doi: 10.1021/ja01207a034. [DOI] [PubMed] [Google Scholar]
- [7].Bar-Or D, Lau E, Winkler JV. A novel assay for cobalt-albumin binding and its potential as a marker for myocardial ischemia—A preliminary report. J. Emerg. Med. 2000;19:311–315. doi: 10.1016/s0736-4679(00)00255-9. [DOI] [PubMed] [Google Scholar]
- [8].Bar-Or D, Curtis G, Rao N, Bampos N, Lau E. Characterization of the Co(21) and Ni(21) binding amino-acid residues of the N-terminus of human albumin. An insight into the mechanism of a new assay for myocardial ischemia. Eur. J. Biochem. 2001;268:42–47. doi: 10.1046/j.1432-1327.2001.01846.x. [DOI] [PubMed] [Google Scholar]
- [9].Bar-Or D, Edward L, Winkler JV. PCT Int, US. 2004.
- [10].Suzuki E, Yasuda K, Takeda N, Sakata S, et al. Increased oxidized form of human serum albumin in patients with diabetes mellitus. Diabetes Res. Clin. Pract. 1992;18:153–158. doi: 10.1016/0168-8227(92)90140-m. [DOI] [PubMed] [Google Scholar]
- [11].Sogami M, Era S, Nagaoka S, Kuwata K, et al. High-performance liquid chromatographic studies on nonmercapt in equilibrium with mercapt conversion of human serum albumin. II. J. Chromatogr. 1985;332:19–27. doi: 10.1016/s0021-9673(01)83283-0. [DOI] [PubMed] [Google Scholar]
- [12].Oettl K, Stadlbauer V, Petter F, Greilberger J, et al. Oxidative damage of albumin in advanced liver disease. Biochim. Biophys. Acta. 2008;1782:469–473. doi: 10.1016/j.bbadis.2008.04.002. [DOI] [PubMed] [Google Scholar]
- [13].Soejima A, Matsuzawa N, Hayashi T, Kimura R, et al. Alteration of redox state of human serum albumin before and after hemodialysis. Blood Purif. 2004;22:525–529. doi: 10.1159/000082524. [DOI] [PubMed] [Google Scholar]
- [14].Terawaki H, Yoshimura K, Hasegawa T, Matsuyama Y, et al. Oxidative stress is enhanced in correlation with renal dysfunction: Examination with the redox state of albumin. Kidney Int. 2004;66:1988–1993. doi: 10.1111/j.1523-1755.2004.00969.x. [DOI] [PubMed] [Google Scholar]
- [15].Oettl K, Stauber RE. Physiological and pathological changes in the redox state of human serum albumin critically influence its binding properties. Br. J. Pharmacol. 2007;151:580–590. doi: 10.1038/sj.bjp.0707251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Peters T., Jr. All About Albumin. Academic Press; San Diego: 1996. [Google Scholar]
- [17].Baczynskyj L, Bronson GE, Kubiak TM. Application of thermally assisted electrospray ionization mass spectrometry for detection of noncovalent complexes of bovine serum albumin with growth hormone releasing factor and other biologically active peptides. Rapid Commun. Mass Spectrom. 1994;8:280–286. doi: 10.1002/rcm.1290080311. [DOI] [PubMed] [Google Scholar]
- [18].Sjobring U, Bjorck L, Kastern W. Streptococcal protein G. Gene structure and protein binding properties. J. Biol. Chem. 1991;266:399–405. [PubMed] [Google Scholar]
- [19].Carter WA. Binding of human interferons to immobilized albumin. Methods Enzymol. 1981;78:576–582. doi: 10.1016/0076-6879(81)78171-0. [DOI] [PubMed] [Google Scholar]
- [20].Carter DC, Ho JX. Structure of serum albumin. Adv. Protein Chem. 1994;45:153–203. doi: 10.1016/s0065-3233(08)60640-3. [DOI] [PubMed] [Google Scholar]
- [21].Peters T., Jr. Serum albumin. Adv. Protein Chem. 1985;37:161–245. doi: 10.1016/s0065-3233(08)60065-0. [DOI] [PubMed] [Google Scholar]
- [22].Fasano M, Curry S, Terreno E, Galliano M, et al. The extraordinary ligand binding properties of human serum albumin. IUBMB Life. 2005;57:787–796. doi: 10.1080/15216540500404093. [DOI] [PubMed] [Google Scholar]
- [23].Lowenthal MS, Mehta AI, Frogale K, Bandle RW, et al. Analysis of albumin-associated peptides and proteins from ovarian cancer patients. Clin. Chem. 2005;51:1933–1945. doi: 10.1373/clinchem.2005.052944. [DOI] [PubMed] [Google Scholar]
- [24].Zhou M, Lucas DA, Chan KC, Issaq HJ, et al. An investigation into the human serum “interactome”. Electrophoresis. 2004;25:1289–1298. doi: 10.1002/elps.200405866. [DOI] [PubMed] [Google Scholar]
- [25].Gundry R, Fu Q, Jelinek C, Van Eyk J, Cotter R. Investigation of an albumin-enriched fraction of human serum and its albuminome. Proteomics Clin. Appl. 2007;1:73–88. doi: 10.1002/prca.200600276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Mehta AI, Ross S, Lowenthal MS, Fusaro V, et al. Biomarker amplification by serum carrier protein binding. Dis. Markers. 2003;19:1–10. doi: 10.1155/2003/104879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Gundry RL, Cotter RJ. In: Clinical Proteomics: From Diagnosis to Therapy. Dunn MJ, Van Eyk JE, editors. Wiley; London: 2007. p. 263. [Google Scholar]
- [28].Gundry RL, Fu Q, Jelinek CA, Van Eyk JE, Cotter RJ. Investigation of an albumin-enriched fraction of human serum and its albuminome. Proteomics Clin. Appl. 2007;1:73–88. doi: 10.1002/prca.200600276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ahmed N, Barker G, Oliva K, Garfin D, et al. An approach to remove albumin for the proteomic analysis of low abundance biomarkers in human serum. Proteomics. 2003;3:1980–1987. doi: 10.1002/pmic.200300465. [DOI] [PubMed] [Google Scholar]
- [30].Steel LF, Trotter MG, Nakajima PB, Mattu TS, et al. Efficient and specific removal of albumin from human serum samples. Mol. Cell. Proteomics. 2003;2:262–270. doi: 10.1074/mcp.M300026-MCP200. [DOI] [PubMed] [Google Scholar]
- [31].Subramanian S. Dye-ligand affinity chromatography: The interaction of Cibacron Blue F3GA with proteins and enzymes. CRC Crit. Rev. Biochem. 1984;16:169–205. doi: 10.3109/10409238409102302. [DOI] [PubMed] [Google Scholar]
- [32].Björhall K, Miliotis T, Davidsson P. Comparison of different depletion strategies for improved resolution in proteomic analysis of human serum samples. Proteomics. 2005;5:307–317. doi: 10.1002/pmic.200400900. [DOI] [PubMed] [Google Scholar]
- [33].Govorukhina NI, Keizer-Gunnink A, van der Zee AG, de Jong S, et al. Sample preparation of human serum for the analysis of tumor markers. Comparison of different approaches for albumin and gamma-globulin depletion. J. Chromatogr. A. 2003;1009:171–178. doi: 10.1016/s0021-9673(03)00921-x. [DOI] [PubMed] [Google Scholar]
- [34].Chromy BA, Gonzales AD, Perkins J, Choi MW, et al. Proteomic analysis of human serum by two-dimensional differential gel electrophoresis after depletion of high-abundant proteins. J. Proteome Res. 2004;3:1120–1127. doi: 10.1021/pr049921p. [DOI] [PubMed] [Google Scholar]
- [35].Colantonio DA, Dunkinson C, Bovenkamp DE, Van Eyk JE. Effective removal of albumin from serum. Proteomics. 2005;5:3831–3835. doi: 10.1002/pmic.200401235. [DOI] [PubMed] [Google Scholar]
- [36].Zolotarjova N, Martosella J, Nicol G, Bailey J, et al. Differences among techniques for high-abundant protein depletion. Proteomics. 2005;5:3304–3313. doi: 10.1002/pmic.200402021. [DOI] [PubMed] [Google Scholar]
- [37].Stanley BA, Gundry RL, Cotter RJ, Van Eyk JE. Heart disease, clinical proteomics and mass spectrometry. Dis. Markers. 2004;20:167–178. doi: 10.1155/2004/965261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Huang L, Harvie G, Feitelson JS, Gramatikoff K, et al. Immunoaffinity separation of plasma proteins by IgY microbeads: Meeting the needs of proteomic sample preparation and analysis. Proteomics. 2005;5:3314–3328. doi: 10.1002/pmic.200401277. [DOI] [PubMed] [Google Scholar]
- [39].Liu T, Qian WJ, Mottaz HM, Gritsenko MA, et al. Evaluation of multiprotein immunoaffinity subtraction for plasma proteomics and candidate biomarker discovery using mass spectrometry. Mol. Cell. Proteomics. 2006;5:2167–2174. doi: 10.1074/mcp.T600039-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Seam N, Gonzales DA, Kern SJ, Hortin GL, et al. Quality control of serum albumin depletion for proteomic analysis. Clin. Chem. 2007;53:1915–1920. doi: 10.1373/clinchem.2007.091736. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.