Abstract
Naltrexone, one of four FDA-approved pharmacotherapies for alcohol dependence, has shown moderate efficacy in clinical trials. Pharmacogenetic effects have been reported such that allelic variation at the gene encoding the mu-opioid receptor (OPRM1, rs1799971) predicts naltrexone-induced blunting of the positively reinforcing effects of alcohol. However, naltrexone also binds, albeit to a lesser degree, to kappa and delta opioid receptors in the brain. This alternate binding presents the possibility that single nucleotide polymorphisms (SNPs) in the kappa and delta opioid receptor (OPRK1 and OPRD1) genes may contribute to naltrexone pharmacogenetics. Therefore, the goal of this exploratory study was to re-examine data from a double-blind placebo controlled laboratory trial of naltrexone for pharmacogenetic effects at kappa and delta opioid receptor tag SNPs. Participants were 40 heavy drinkers (12 female) who underwent an intravenous alcohol challenge paradigm after receiving naltrexone (50 mg) or placebo in randomized and crossover fashion. Dependent variables were self-reported alcohol-induced stimulation, sedation, and craving. Multilevel models revealed a significant Naltrexone × OPRK1 Genotype (rs997917) interaction predicting alcohol-induced sedation, such that TT homozygotes reported lower naltrexone-induced alcohol sedation as compared to carriers of the C allele. Moreover, there was a significant Naltrexone × OPRD1 Genotype (rs4654327) interaction predicting alcohol-induced stimulation and craving, such that carriers of the A allele at this locus reported greater naltrexone-induced blunting of alcohol stimulation and alcohol craving compared to GG homozygotes. These findings suggest that additional pharmacogenetic effects in the opioid receptor system may account for individual differences in response to naltrexone in the human laboratory.
Keywords: alcohol administration, naltrexone, OPRM1, OPRK1, OPRD1, pharmacogenetics
1. Introduction
Alcohol abuse and dependence represent a sizeable public health problem in the United States, with about 8.5% of the adult population diagnosed with either alcohol abuse or dependence in a given year (Grant et al., 2004). Treatment strategies for alcohol use disorders (AUDs) include the Food and Drug Administration-approved medication naltrexone, an opioid antagonist that has highest affinity for the mu-opioid receptor (Littleton and Zieglgansberger, 2003). Results from clinical trials have demonstrated moderate efficacy of naltrexone for alcohol dependence. Specifically, findings suggest that naltrexone treatment reduces the occurrence of heavy drinking days (Balldin et al., 2003, Monti et al., 2001, Rubio et al., 2002), increases time to first relapse (Anton et al., 1999, Guardia et al., 2002, Kiefer et al., 2003), and yields lower relapse rates (Heinala et al., 2001, Latt et al., 2002, Volpicelli et al., 1992). Additionally, naltrexone reduces the number of drinking days (O’Malley et al., 1992, Volpicelli et al., 1992) and the number of drinks per drinking episode (Chick et al., 2000, Guardia et al., 2002, Morris et al., 2001, O’Malley et al., 1992). Results from the COMBINE Study, a large multi-site controlled trial, found that naltrexone was an effective treatment for alcohol dependence when delivered in combination with a medically-oriented behavioral intervention (Anton et al., 2006). A few studies however, have not found support for the efficacy of naltrexone (Killeen et al., 2004, Kranzler et al., 2000, Krystal et al., 2001).
Alcohol has a complex pharmacological profile involving affinity for multiple receptor types; the reinforcing effects are thought to be due to the combined release of endogenous opioids like β-endorphin and dopamine (from the midbrain), targeting neurons in the nucleus accumbens (NAc) downstream (Gianoulakis et al., 1996, Volpicelli, 2001). Current models of the effects of naltrexone posit that it acts by diminishing the dopamine response to ethanol in the NAc, which has been supported by pre-clinical research (Benjamin et al., 1993). Human laboratory studies have demonstrated that naltrexone dampens alcohol-induced feelings of stimulation (Drobes et al., 2004, Swift et al., 1994), decreases liking of alcohol (McCaul et al., 2001), and increases alcohol-induced fatigue and confusion (King et al., 1997).
Prior pharmacogenetic studies have focused on the OPRM1 gene, which codes for mu-opioid receptors, for which naltrexone has highest binding affinity (Goldman et al., 2005, Oslin et al., 2003). One of the most widely studied polymorphisms of the OPRM1 gene is the A118G single nucleotide polymorphism (SNP) (rs1799971). Studies of the functional significance of this variant have produced mixed results with some reports suggesting that the 118G variant binds more strongly to β-endorphin than the major allele when expressed in Xenopus oocytes (Bond et al., 1998) while others have not replicated this result for β-endorphin or found differential binding of morphine when assayed in human embryonic kidney cells (Beyer et al., 2004). Additionally, the 118G variant appears to decrease OPRM1 mRNA and protein yield suggesting that this SNP may function more as a loss of function effect (Zhang et al., 2005).
Pharmacogenetic studies of the A118G SNP have found that G-allele carriers exhibit greater naltrexone-induced blunting of alcohol high (Ray and Hutchison, 2007) and lower relapse rates in clinical trials of naltrexone for alcoholism (Anton et al., 2008, Oslin et al., 2003). A recent study in Asian Americans found that G-allele carriers reported greater alcohol-induced sedation, subjective intoxication, and lower alcohol craving while on naltrexone versus placebo, and as compared to A-allele homozygotes (Ray et al., 2012b). Some studies, however, have failed to support this pharmacogenetic effect (Coller et al., 2011, Gelernter et al., 2007, Tidey et al., 2008). These null findings may be due, in part, to the relatively small effect size of naltrexone and its interaction with OPMR1 genotype compared to more robust psychosocial interventions, high type II error, and study design limitations as several did not prospectively genotype participants prior to medication randomization (for review see Ray et. al, 2012a).
While the primary focus of pharmacogenetic studies has been on the mu class of opioid receptors, research has shown that naltrexone also binds to delta and kappa opioid receptors (Takemori et al., 1988, Takemori and Portoghese, 1992), although with lower affinity than for mu. Specifically, preferential binding for mu over kappa receptors has been shown to be in the range of three to ten times greater (Ananthan et al., 1999, Ko et al., 1998). For mu receptors versus delta, the affinity for mu may be as much as 63 times greater (Emmerson et al., 1994).
As naltrexone has activity at multiple opioid receptor classes, it remains unknown where in the brain naltrexone exerts its clinical effects. All three opioid receptor classes (i.e., mu, kappa, delta) are present in the nucleus accumbens and other areas associated with the reinforcing effects of alcohol, like the ventral pallidum (VP) (Mansour et al., 1988), and pre-clinical research has demonstrated that the roles of specific receptor classes in these areas are complex. DAMGO, a selective mu opioid antagonist, microinjected in the VP suppresses voluntary alcohol consumption, while morphine had the opposite effect (Kemppainen et al., 2012). Microinjection of CTOP (mu antagonist) in the nucleus accumbens, however, failed to alter alcohol intake in animals (Hyytia and Kiianmaa, 2001). Likewise, delivery of delta or kappa agonists and antagonists, as well as naltrexone, in the VP had no effect on alcohol intake in rats (Kemppainen et al., 2012). These data stand in contrast to other studies examining delta and kappa effects on drinking. Delta opioid agonists specific to the delta opioid receptor sub-type one administered subcutaneously reduced drinking in mice, while a non-subtype-specific delta opioid agonist had the opposite effect by increasing drinking relative to baseline (van Rijn et al., 2010). Further, the delta antagonist, naltridole, when delivered to the nucleus accumbens reduces drinking (Hyytia and Kiianmaa, 2001).
While the specific dynamics of this circuit have yet to be resolved, these preclinical findings implicate that activity at delta and kappa opioid receptors, in addition to mu, may underlie the effects of naltrexone on alcohol-related phenotypes. This is consistent with human positron emission tomography (PET) studies documenting opioid receptor blockade at the standard therapeutic dose in alcohol dependent human subjects (Weerts et al., 2008). Specifically, while the naltrexone dose (50mg) blocked nearly all (about 94%) of mu opioid receptors with little variability across subjects, delta receptor blockade was lower (about 21%) but also highly variable, potentially explaining individual differences in treatment outcomes (Weerts et al., 2008).
There are several known polymorphisms in this larger family of opioid receptors that may inform naltrexone pharmacogenetics beyond the A118G SNP of the OPRM1 gene. In fact, a clinical study of naltrexone found trend-level main effect for an OPRK1 marker (rs963549) on relapse rates despite not replicating the A118G SNP effect (Gelernter et al., 2007). Therefore, the goal of the present study was to extend the literature by testing for pharmacogenetic effects of kappa and delta opioid receptor SNPs on subjective responses to alcohol and alcohol craving while controlling for the pharmacogenetic effect of the A118G SNP of the OPRM1 gene. This was accomplished by conducting additional sequencing of tag SNPs (tSNPs) in the OPRD1 and OPRK1 genes and completing a re-analysis of a previously published placebo-controlled laboratory study of naltrexone (Ray and Hutchison, 2007). Given that there are no known functional polymorphisms in the OPRD1 and OPRK1 genes, a tSNP approach was used to provide coverage of the two genes of interest. Consistent with this data-driven approach and with the absence of functional data on these markers, no a-priori hypotheses were advanced for the effects of any specific tSNP. Instead, these analyses tested interactions between OPRK1 and OPRD1 genotypes and medication (naltrexone versus placebo) on the stimulant, sedative, and craving effects produced by alcohol administration. This exploratory study seeks to extend naltrexone pharmacogenetics to OPRD1 and OPRK1 genes as they may contribute to the individual differences observed in naltrexone response.
2. Method
2.1 Participants
Participants were recruited from the Boulder, Colorado community using print and online advertisements. Inclusion criteria were the following: (1) a score of 8 or higher on the Alcohol Use Disorders Identification Test (AUDIT) (Allen et al., 1997), indicating a hazardous drinking pattern; (2) self-reported drinking frequency of 3 or more drinks (2 for women) at least twice per week; (3) no history of adverse reactions to needle puncture. All female participants tested negative for pregnancy prior to the alcohol administration and all subjects were required to have a breath alcohol concentration (BrAC) of zero before each session.
The final sample of study completers consisted of forty (12 female) non-treatment seeking heavy drinkers with an average age of 22 (SD = 2.18; Range 21 to 32). The majority of the sample was White (n = 34) with a few participants of Asian (n = 4) and Latino (n = 2) background. The average self-reported number of drinks per occasion in the last year was 4.84 (SD = 2.25), and AUDIT scores reflected, on average, hazardous levels of alcohol use (M = 12.41, SD = 4.23). The study was approved by the University of Colorado Human Research Committee and all participants provided written informed consent after receiving a full explanation of the study.
2.2 Screening and Experimental Procedures
Initial assessment for the inclusion criteria was conducted through a telephone interview, after which eligible participants were invited to the laboratory for an in-person assessment session. Upon arrival at the lab, participants read and signed an informed consent form, provided information about quantity and frequency of drinking episodes over the past year, and provided a saliva sample for DNA analyses. Participants were prospectively genotyped prior to being invited to the physical exam and alcohol challenge sessions in order to oversample for individuals who carry at least one copy of the G allele of the A118G SNP of the OPRM1 gene, thus resulting in exclusion of 64 A allele homozygotes. Genotypes for kappa and delta opioid receptor genes were not considered prospectively. Prior to participating in the ethanol infusion session, participants received a physical examination at the General Clinical Research Center (GCRC) in order to ensure that participants were in good physical health and were medically eligible to take the study medication and to participate in the alcohol challenge. Of the total of 124 participants (39 women) who were screened in the laboratory, 53 completed the physical exam, 7 of whom were ineligible due to a medical reason and 6 of whom decided not to participate in the trial. These procedures resulted in a total of 40 completers.
2.3 Alcohol Administration
In order to control dosage of alcohol during the alcohol challenge, an intravenous administration paradigm was used consistent with previous work (Ray and Hutchison, 2004, Ray et al., 2006). Ethanol infusion sessions took place at the General Clinical Research Center at the University of Colorado at Boulder and were performed by registered nurses under the direct supervision of a physician. Target breath alcohol concentrations (BrACs) at which participants completed study measures were: 0.02, 0.04, and 0.06 g/dL. Upon reaching each of the target BrAC levels, participants’ infusion rates were reduced to half their rate, in order to maintain stable BrAC levels during the assessments. After the alcohol infusion was finished, participants were debriefed, given a meal, and asked to stay in the laboratory until their BrAC was below 0.02 g/dL.
2.4 Medication
Medication (naltrexone 50 mg or matched placebo) was administered in a double-blind and crossover fashion such that each participant completed two infusion visits: one after taking naltrexone and another after taking matched placebo. Participants were asked to take a single dose of medication (either naltrexone or placebo) on each of the two days preceding the infusion visit, with a last dose occurring on the morning of the infusion, totaling three days on the active medication (or placebo). To assess medication compliance, naltrexone and placebo were packed into capsules with 50 mg of riboflavin, which becomes visible in urine samples held under ultraviolet light (Del Boca et al., 1996). Urine samples were collected prior to each infusion session and all samples tested positive for riboflavin content. To assess for the integrity of the medication blind, participants were asked to guess which medication they were currently receiving during each alcohol infusion visit. A total of 79% of the participants correctly guessed in the placebo condition, while 72% guessed correctly while in the naltrexone condition. While higher than chance levels (50%), these percent correct medication guesses did not differ by medication condition in a chi-square test [χ2 (1)= 1.33; p = .25].
2.5 SNP Selection and Sequencing
Bioinformatics resources and results from the International HapMap Project were used to identify tag SNPs (tSNPs) for the OPRK1 and OPRD1 genes. Specifically, Haploview v4.1 (Barrett et al., 2005) was used to conduct a search with the following parameters: (a) haplotype r2 cutoff = 0.8 and (b) minor allele frequency (MAF) of 0.2. Results of this search suggested two tSNPs for the kappa opioid receptor gene (rs997917, rs6985606) and eight tSNPs for the delta opioid receptor gene (rs760588, rs529520, rs2236856, rs499062, rs678849, rs4654327, rs508448, rs581111). Based on this search and the available resources, the following SNPs were assayed in addition to the A118G SNP of the OPRM1 and will be named in the manuscript as follows: OPRK1^1 (rs6985606), OPRK1^2 (rs997917), OPRD1^1 (rs2236856), OPRD1^2 (rs499062), OPRD1^3 (rs678849), OPRD1^4 (rs4654327), and OPRD1^5 (rs508448). For statistical power and data analytic purposes, participants were grouped into two genotype categories where the heterozygote group was combined with the homozygote cell with the lowest frequency (Table 1).
Table 1.
Observed allele frequencies for study sample
| Gene/SNP | Observed Minor Allele Frequency | Minor | Heterozygous | Major | HWE χ2 value |
|---|---|---|---|---|---|
| OPRM1 | G = 20% | GG | GA | AA | |
| rs1799971 | 1 | 14 | 25 | 0.35 | |
| OPRK1 ^1 | T = 50% | TT | TC | CC | |
| rs6985606 | 10 | 18 | 10 | 0.11 | |
| OPRK1 ^2 | C = 29.5% | CC | CT | TT | |
| rs997917 | 2 | 19 | 18 | 1.15 | |
| OPRD1^1 | A = 21.8% | AA | AG | GG | |
| rs2236856 | 1 | 15 | 23 | 0.64 | |
| OPRD1^2 | C = 16.25% | CC | CT | TT | |
| rs499062 | 0 | 13 | 27 | 1.50 | |
| OPRD1^3 | T = 65.4% | TT | TC | CC | |
| rs678849 | 16 | 19 | 4 | 0.23 | |
| OPRD1^4 | G = 57.7% | GG | GA | AA | |
| rs4654327 | 10 | 25 | 4 | 3.82* | |
| OPRD1^5 | A = 48.7% | AA | AG | GG | |
| rs508448 | 10 | 18 | 11 | 0.23 |
p = 0.051
For sequencing purposes, genomic DNA was collected and isolated from buccal cells following published procedures (Freeman et al., 1997, Lench et al., 1988, Walker et al., 1999). An ABI PRISM 7500 instrument (Applied Biosystems, Inc. Foster City, CA) was used to conduct 5′-nuclease (TaqMan) assays of the opioid receptor SNPs using assays commercially available from Applied Biosystems.
2.6 Measures
During the in-person assessment session, participants completed a battery of individual difference measures including demographics and alcohol/drug use questionnaires. During the ethanol infusion, the following measures of subjective responses to alcohol and alcohol craving were administered at baseline and at each target BrAC (i.e., 0.02, 0.04, and 0.06 g/dL): (a) The Biphasic Alcohol Effects Scale (BAES) was used to assess feelings of alcohol-induced stimulation and sedation, with each subscale consisting of seven items answered on a 0–10 scale. The BAES has been shown to be reliable and valid in alcohol administration studies (Martin et al., 1993); and (b) The Alcohol urge questionnaire (AUQ) was used to assess for degree of alcohol craving (Bohn et al., 1995, MacKillop, 2006). The dependent variable was average score for all of the eight AUQ items (range 0–6).
2.7 Statistical Analysis
A series of mixed models (PROC MIXED) in SAS (V9.2 Cary, NC) were conducted for each tSNP on the three dependent variables of interest: alcohol-induced sedation, stimulation, and craving. Specifically, Medication was modeled as a two-level within subjects factor (i.e., Placebo vs. Naltrexone, coded 0 and 1 respectively), Genotype as a two-level between subjects factor (e.g., OPRD1^4: GG = 0 vs. AG/AA = 1 and OPRK^2: TT = 0 vs. CT/CC = 1), and BrAC as a four-level within subjects factor (i.e., BrAC = 0.00, 0.02, 0.04, and 0.06 g/dl, coded 0 to 3). The model also included interactions between genotype and medication, as well as between BrAC and medication. If a non-significant interaction was found (e.g., BrAC × medication), the model was re-run without the interaction term. In addition, models producing significant genotype × medication interactions were followed up by a simple effects analysis. An asterisk (*) in the figures indicates a significant simple effect (p < 0.05). Finally, OPRM1 genotype and the interaction between OPRM1 genotype and medication were used as covariates in subsequent models. This added step controlled for the prospective genotyping based on the A118G SNP of the OPRM1 gene and verifies that any novel pharmacogenetic effects persist over and above OPRM1 results.
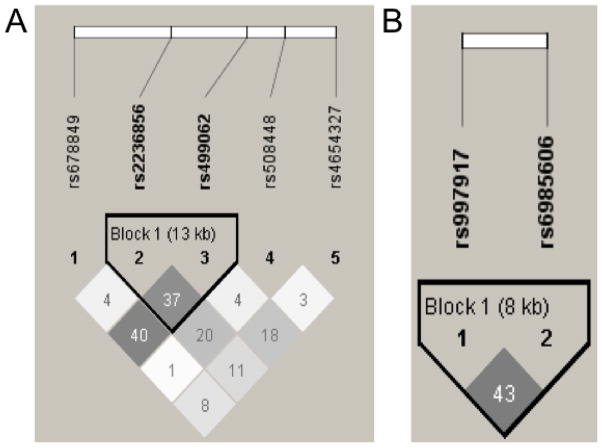
In order to account for multiple comparisons, the significance threshold for pharmacogenetic effects was divided by the number of independent comparisons. Specifically, linkage disequilibrium (LD) across the tSNPs sampled in the OPRK1 and OPRD1 genes were used to determine dependence (i.e., overlap) between the tSNPs of interest. Thus, accounting for one haplotype block for the OPRD1 tSNPs and one for the OPRK1 tSNPs (see Figure 1), a total of 5 independent comparisons were conducted. Therefore, the critical p-value (p = .05) was divided by 5 resulting in a corrected critical p-value of p = 0.01 for the pharmacogenetic hypotheses. Lastly, given that the genes coding for mu, kappa, and delta receptors are located on different chromosomes (chromosomes 6, 8, and 1, respectively), there was no concern about LD across the three genes.
Figure 1.
LD plot from Haploview 4.1 for EA subjects based on HapMap (phase II) data for individuals of European Ancestry for the (A) OPRD1 and (B) OPRK1 tSNPs examined in this study. Pair-wise SNP |D′| values (×100) of linkage are shown along with 2 haplotype blocks identified using the four-gamete rule. Darkened blocks indicate SNP pairs without evidence of extensive recombination (i.e., 4-gamete rule for haplotype block characterization with at least one 2-SNP haplotype having a frequency < 0.02).
3. Results
The participants reported drinking an average of 4.9 (SD = 2.4) standard drinks per drinking episode at a frequency of twice per week (6 = twice a week, M = 6.18; SD = 1.38) during the past year. Allele frequencies for the OPRM1 and tSNPS for OPRK1 and OPRD1 are presented in Table 1. Assessment for violation of Hardy-Weinberg Equilibrium (HWE) resulted in no significant chi-square values (ps>0.05, Table 1), with the exception of OPRD1^4 which reached marginal significance (p=0.051). HWE was then re-estimated among Whites only, which suggested no violation for OPRD1^4 (χ2 = 1.91, p = 0.16).
3.1 Kappa Opioid Receptor tSNPs
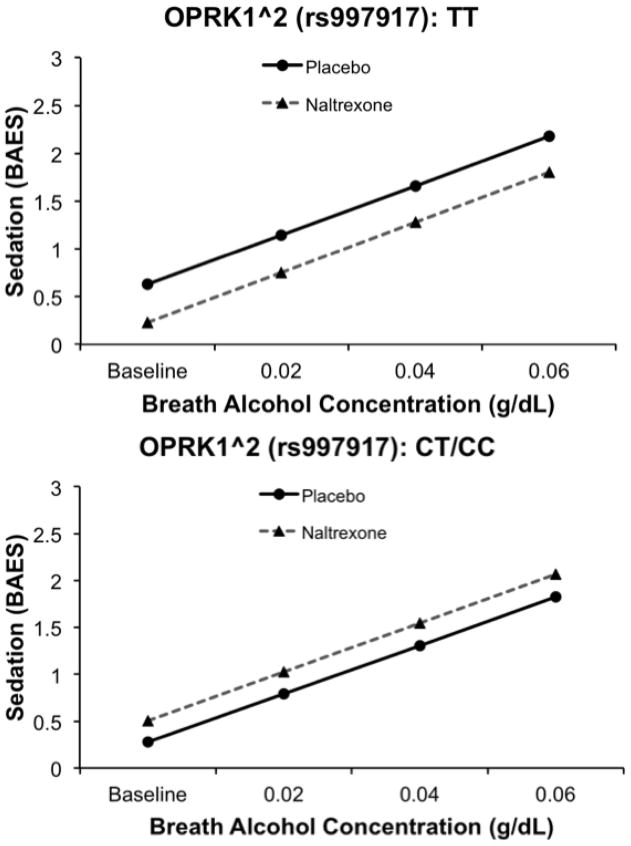
Of the two OPRK1 tSNPs examined, one marker (OPRK1^2, rs997917) showed a significant effect on alcohol-induced sedation. Specifically, there was no significant effect of OPRK1^2 genotype (β = −0.36, SE = 0.21, t = −1.62, p > .05), an effect of medication (β = −0.39, SE = 0.13, t = −2.90, p < .01), and an OPRK1^2 × medication interaction (β = 0.63, SE = 0.182, t = 3.43, p < .001). There was also an effect of rising BrAC (β = 0.52, SE = 0.08, t = 6.46, p < 0.0001). There was no interaction between BrAC and medication (p > 0.1) and this interaction term was not included in the model reported above. As shown in Figure 2, individuals who were homozygous for TT reported reduced feelings of alcohol sedation on naltrexone versus placebo, and as compared to C allele carriers. Controlling for the OPRM1 A118G SNP did not alter the significance of the pharmacogenetic effect reported above. However, when the model was reexamined among Whites only, the OPRK1^2 × medication interaction was reduced to a trend level of significance (β = 0.26, SE = 0.5, t = 1.74, p =0.083).
Figure 2.
Effect of OPRK1^2 genotype on sedation while on naltrexone versus placebo. Data presented are predicted values based on betas from mixed-model regression analyses. TT homozygotes showed reduced sedation while on naltrexone.
There were no significant pharmacogenetic effects for OPRK1^2 on stimulation (β = 0.071, SE = 0.203, t = 0.35, p = 0.73) or craving (β = 0.046, SE = 0.147, t = 0.31, p = 0.76). For the other kappa marker (OPRK1^1), there was a trend-level pharmacogenetic effect on sedation (β = −0.41, SE = 0.215, t = −1.92, p = 0.055), but no such effects on stimulation (β = −0.0007, SE = 0.232, t = −0.00, p = 0.99), or craving (β = 0.22, SE = 0.169, t = 1.30, p = 0.196).
3.2 Delta Opioid Receptor tSNPs
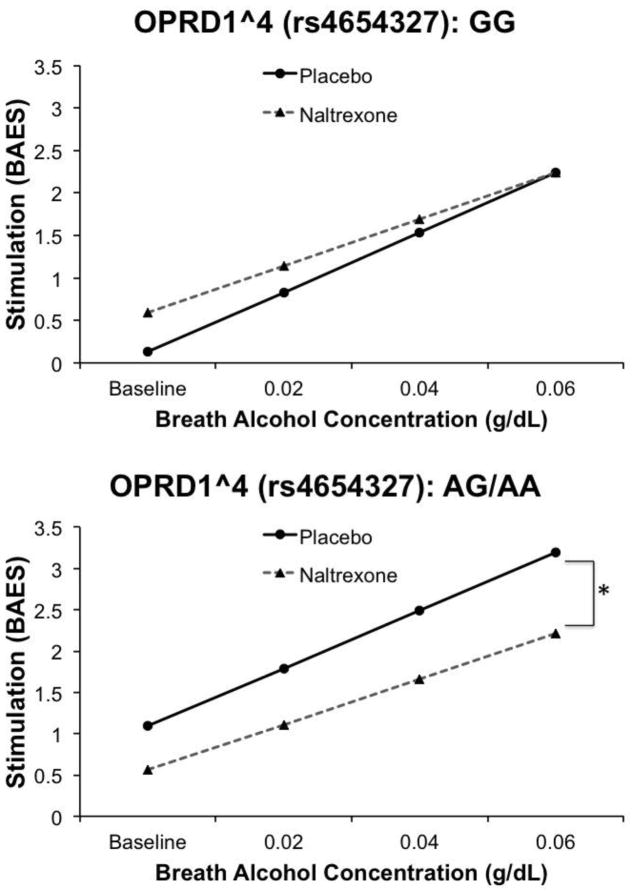
Analysis for OPRD1 receptor tSNPs revealed that of the five tSNPs tested, one marker (OPRD1^4, rs4654327) reached corrected p-value significance in the model assessing stimulation scores. Specifically, there was a significant effect of OPRD1^4 genotype on stimulation (β = 0.96, SE = 0.31, t = 3.07, p < .01), an effect of medication (β = 0.46, SE = 0.231, t = 2.0, p < 0.05), and a significant OPRD1^4 × medication interaction (β = −0.99, SE = 0.222, t = −4.46, p < .0001). This model also showed a significant effect of BrAC (β = 0.70, SE = 0.09, t = 7.69, p < .0001) and a trend-level BrAC × medication interaction (β = −0.15, SE = 0.087, t = −1.74, p = 0.08). As shown in Figure 3, the pharmacogenetic effect was such that carriers of the A allele at this locus reported greater naltrexone-induced blunting of alcohol stimulation. Controlling for OPRM1 genotype and removing non-Whites from analysis did not alter the significance of the medication × OPRD1^4 interaction.
Figure 3.
Effect of OPRD10^4 genotype on alcohol-induced stimulation while on naltrexone versus placebo. Data presented are predicted values based on betas from mixed-model regression analyses. A-allele carriers showed naltrexone-induced blunting of alcohol stimulation. An asterisk (** p < 0.01) indicates a significant effect of medication within a genotype group across the infusion session
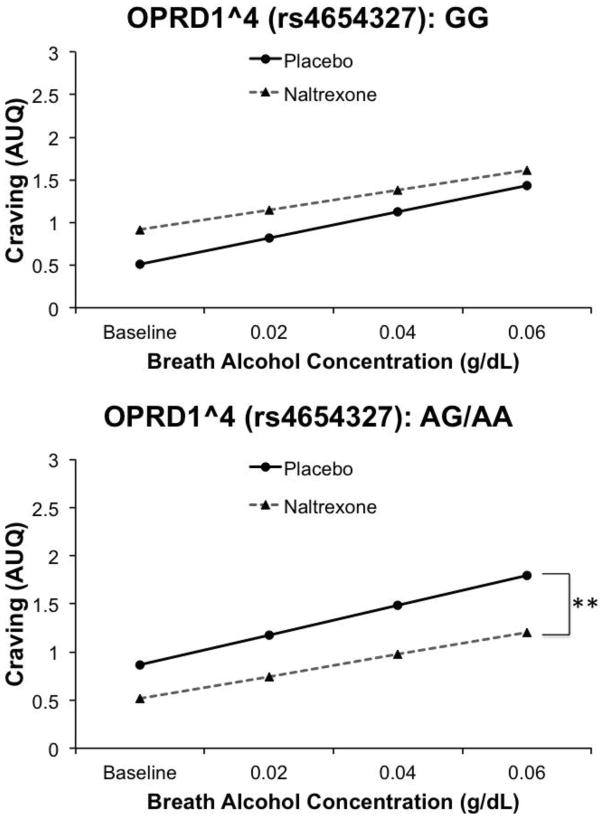
A similar pharmacogenetic effect of OPRD1^4 was observed for alcohol craving (Figure 4). In this model, there was no significant effect of OPRD1^4 (β = 0.36, SE = 0.32, t = 1.1, p > .01), but an effect of medication (β = 0.29, SE = 0.14, t = 2.11, p < 0.05), and a significant OPRD1^4 × medication interaction (β = −0.76, SE = 0.16, t = −4.76, p < 0.0001). Finally, there was an effect of BrAC (β = 0.27, SE = 0.045, t = 5.95, p < 0.001). There was no BrAC × medication interaction (p > 0.1), and thus this interaction term was not included in the above model. For this tSNP, A-allele carriers showed greater naltrexone-induced blunting of alcohol craving as compared to G-allele homozygotes. Controlling for the OPRM1 A118G SNP or restricting the sample to Whites only did not alter the significance of the OPRD1^4 pharmacogenetic effect. For OPRD1^4, there was no significant pharmacogenetic effect on sedation (β = 0.39, SE = 0.212, t = 1.84, p = 0.066). Models testing the other OPRD1 tSNPs (^1, ^2, ^3, ^5) yielded no significant pharmacogenetic effects on either subjective intoxication measure or craving (all ps > 0.01).
Figure 4.
Effect of OPRD1^4 genotype on alcohol craving while on naltrexone versus placebo. Data presented are predicted values based on betas from mixed-model regression analyses. A-allele carriers showed naltrexone-induced attenuation of alcohol craving. An asterisk (* p < 0.05) indicates a significant effect of medication within a genotype group across the infusion session.
4. Discussion
The goal of this study was to assess for interactions between tag single nucleotide polymorphisms (tSNPs) in the genes encoding kappa and delta opioid receptors and naltrexone on subjective responses to alcohol and craving in the laboratory. Results revealed a significant interaction between OPRK1 genotype at one locus (OPRK1^2, rs997917) and medication (naltrexone vs. placebo), demonstrating differential naltrexone-induced changes in alcohol sedation. Specifically, among TT homozygotes, naltrexone dampened feelings of sedation compared to placebo, while among C-allele carriers naltrexone enhanced alcohol-induced sedation. OPRK1^2 has been previously tested for association with alcohol, opiate, or other drug dependence, but was not a significant predictor of drug or alcohol problems (Zhang et al., 2008). Interestingly, a nearby marker in OPRK1 (rs963549) showed marginal significance in predicting relapse rates and time to relapse in a naltrexone treatment study (Gelernter et al, 2007). However, this marginal effect was limited to a main effect of genotype in that study, as this marker did not significantly interact with medication treatment (naltrexone vs. placebo). As ascertained through publicly available data from HapMap analyzed through Haploview, these two OPRK1 SNPs are about 10kb apart and do share some variance in the general population (r2 = 0.38), although not enough to suggest major overlap.
Pharmacogenetic effects were also observed for a tSNP in the OPRD1 gene (^4, rs4654327) on dampening self-reported alcohol stimulation and craving for alcohol in the laboratory. Specifically, GG homozygotes showed no effect of naltrexone, versus placebo, on alcohol-induced stimulation and alcohol craving, while for A carriers, naltrexone blunted alcohol’s stimulant effects and attenuated alcohol craving. These results may be clinically meaningful as naltrexone’s purported mechanisms of action include dampening the rewarding effects of alcohol and reductions in craving (Anton et al., 2004, King et al., 1997, Swift et al., 1994). Taken together, these findings suggest that variation in delta and kappa opioid receptor genes may further explain variation in the effects of naltrexone on subjective responses to alcohol and alcohol craving. Further, these results suggest that the biobehavioral mechanisms underlying the treatment efficacy of naltrexone for alcohol dependence may extend beyond mu opioid receptors.
As noted elsewhere (Arias et al., 2006, Ray et al., 2012a), individuals from different ethnic backgrounds show varying allele frequencies for the A118G SNP of the OPRM1 gene, which in turn may impact the application of personalized medicine approaches to optimizing naltrexone treatment for alcoholism (Tate and Goldstein, 2004). To that end, based on publicly available bioinformatics resources (HapMap in particular), the kappa and delta tSNPs that showed pharmacogenetic effects in this study also vary in minor allele frequency by ethnicity. Specifically, while the G allele of OPRD1^4 is present in approximately 43% of Whites, it is much more frequently observed in people of African ancestry (about 73%). For OPRK1^2, the C allele is present in about 28% of those with European ancestry, but it is more frequent among those of African descent (about 65%) and among Han Chinese (about 43%). Therefore it stands to reason that these preliminary findings, if supported, may be useful in addressing naltrexone personalized medicine in ethnic minorities.
Our data may also complement the literature examining the role of endogenous opiates in the reinforcing effects of alcohol. Recent imaging data have supported that alcohol induces endogenous opioid release in the nucleus accumbens (Mitchell et al., 2012), a target of the mesolimbic dopamine projection from the ventral tegmental area. Some pre-clinical studies, however, suggest that some of the reinforcing effects of alcohol may in fact be independent of this dopamine pathway and are better explained by opioidergic effects in this system. Specifically, after toxic lesion of the mesolimbic dopamine pathway, rats are able to acquire and continue to seek ethanol in an operant paradigm (Koistinen et al., 2001, Shoemaker et al., 2002). When delivered systemically, naloxone (Shoemaker et al., 2002), an opioid antagonist with high affinity for the mu receptor, and naltrexone (Koistinen et al., 2001), reduce this voluntary drinking in rats with dopaminergic damage. However, mice lacking D2 receptors do show reduced preference for ethanol as well (Phillips et al., 1998), indicating that dopamine binding at these sites remains an important component in ethanol self-administration behavior.
The findings of this exploratory study must be weighed with regard to the strengths and limitations of the study. First, population stratification was a possibility as participants were not exclusively from a homogenous ethnic background. To that end, controlling for population stratification by screening out non-White participants reduced the significance of the OPRK1^2 × medication interaction to a trend-level relationship. Thus, larger sample sizes affording greater statistical power are needed to confirm and extend these initial findings. Second, all participants received alcohol infusions only and there was no placebo (saline) infusion condition to control for alcohol expectancies. Third, in line with the purpose of the original study (Ray and Hutchison, 2007), participants were prospectively genotyped at the A118G variant of OPRM1. Thus, allele frequencies for OPRK1 and OPRD1 SNPs may not reflect those that would be observed if sampling from the population at random. However, as OPRM1 lies on chromosome 6, OPRK1 on chromosome 8, and OPRD1 on chromosome 1, it is unlikely that oversampling for 118G carriers biased the sample for the OPRK1 and OPRD1 markers. Next, given that participants guessed their current medication (naltrexone or placebo) above chance levels, it is possible that medication expectancies may have influenced results. Finally, the tSNPs examined were selected to provide sufficient coverage of the genes of interest, and it remains unknown where the true genetic “signal” lies within these genes as well as their mode of inheritance.
The statistical control for OPRM1 A118G genotype in all models represented a significant strength of this study, and doing so did not diminish the significance of the novel pharmacogenetic effects reported. Second, this trial of naltrexone was double-blinded, placebo-controlled, and consisted of a crossover design such that all participants received both naltrexone and placebo, enhancing the reliability and power of our findings. Third, alcohol was administered intravenously rather than orally, which resulted in tightly controlled blood alcohol concentrations. Lastly, a proper p-value correction for multiple comparisons was implemented, reducing the likelihood of false positives in this exploratory analysis.
4.1 Conclusions
In summary, the present study re-analyzed a previously published dataset for pharmacogenetic effects beyond the mu opioid receptor gene on subjective responses to alcohol and craving. One kappa receptor tSNP (rs997917) and one delta tSNP (rs4654327) moderated alcohol-induced sedation and alcohol-induced stimulation and craving, respectively. These findings extend the literature by indicating that allelic variation at or near these sites may further explain the variability in the biobehavioral effects of naltrexone. Replication of these preliminary findings in larger samples is warranted, particularly with regard to ethnic groups that differ in terms of minor allele frequencies at these loci. Likewise, molecular studies identifying functional polymorphisms in these genes are needed to afford a more focused pharmacogenetic investigation.
Highlights.
Novel pharmacogenetic findings for naltrexone are demonstrated
Tag SNPs in kappa and delta opioid receptor genes are examined
OPRK1 SNP predicts sedative response to alcohol while on naltrexone
OPRD1 SNP predicts stimulation and craving responses to alcohol while on naltrexone
Acknowledgments
This research was supported by grants from the National Institute on Alcoholism and Alcohol Abuse (AA014847) and by a grant from the General Clinical Research Center Program of the National Center for Research Resources, National Institutes of Health (M01-RR00051). The authors would like to thank the members of the UCLA Addictions Research Laboratory for their thoughtful feedback on earlier drafts of this manuscript. The authors also wish to thank the staff at the General Clinical Research Center at the University of Colorado, at Boulder.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allen JP, Litten RZ, Fertig JB, Babor T. A review of research on the Alcohol Use Disorders Identification Test (AUDIT) Alcohol Clin Exp Res. 1997;21:613–9. [PubMed] [Google Scholar]
- Ananthan S, Kezar HS, 3rd, Carter RL, Saini SK, Rice KC, Wells JL, et al. Synthesis, opioid receptor binding, and biological activities of naltrexone-derived pyrido- and pyrimidomorphinans. Journal of medicinal chemistry. 1999;42:3527–38. doi: 10.1021/jm990039i. [DOI] [PubMed] [Google Scholar]
- Anton RF, Drobes DJ, Voronin K, Durazo-Avizu R, Moak D. Naltrexone effects on alcohol consumption in a clinical laboratory paradigm: temporal effects of drinking. Psychopharmacology (Berl) 2004;173:32–40. doi: 10.1007/s00213-003-1720-7. [DOI] [PubMed] [Google Scholar]
- Anton RF, Moak DH, Waid LR, Latham PK, Malcolm RJ, Dias JK. Naltrexone and cognitive behavioral therapy for the treatment of outpatient alcoholics: results of a placebo-controlled trial. Am J Psychiatry. 1999;156:1758–64. doi: 10.1176/ajp.156.11.1758. [DOI] [PubMed] [Google Scholar]
- Anton RF, O’Malley SS, Ciraulo DA, Cisler RA, Couper D, Donovan DM, et al. Combined pharmacotherapies and behavioral interventions for alcohol dependence: the COMBINE study: a randomized controlled trial. JAMA. 2006;295:2003–17. doi: 10.1001/jama.295.17.2003. [DOI] [PubMed] [Google Scholar]
- Anton RF, Oroszi G, O’Malley S, Couper D, Swift R, Pettinati H, et al. An evaluation of mu-opioid receptor (OPRM1) as a predictor of naltrexone response in the treatment of alcohol dependence: results from the Combined Pharmacotherapies and Behavioral Interventions for Alcohol Dependence (COMBINE) study. Arch Gen Psychiatry. 2008;65:135–44. doi: 10.1001/archpsyc.65.2.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias A, Feinn R, Kranzler HR. Association of an Asn40Asp (A118G) polymorphism in the mu-opioid receptor gene with substance dependence: a meta-analysis. Drug Alcohol Depend. 2006;83:262–8. doi: 10.1016/j.drugalcdep.2005.11.024. [DOI] [PubMed] [Google Scholar]
- Balldin J, Berglund M, Borg S, Mansson M, Bendtsen P, Franck J, et al. A 6-month controlled naltrexone study: combined effect with cognitive behavioral therapy in outpatient treatment of alcohol dependence. Alcohol Clin Exp Res. 2003;27:1142–9. doi: 10.1097/01.ALC.0000075548.83053.A9. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Benjamin D, Grant ER, Pohorecky LA. Naltrexone reverses ethanol-induced dopamine release in the nucleus accumbens in awake, freely moving rats. Brain Res. 1993;621:137–40. doi: 10.1016/0006-8993(93)90309-b. [DOI] [PubMed] [Google Scholar]
- Beyer A, Koch T, Schroder H, Schulz S, Hollt V. Effect of the A118G polymorphism on binding affinity, potency and agonist-mediated endocytosis, desensitization, and resensitization of the human mu-opioid receptor. J Neurochem. 2004;89:553–60. doi: 10.1111/j.1471-4159.2004.02340.x. [DOI] [PubMed] [Google Scholar]
- Bohn MJ, Krahn DD, Staehler BA. Development and initial validation of a measure of drinking urges in abstinent alcoholics. Alcoholism, clinical and experimental research. 1995;19:600–6. doi: 10.1111/j.1530-0277.1995.tb01554.x. [DOI] [PubMed] [Google Scholar]
- Bond C, LaForge KS, Tian M, Melia D, Zhang S, Borg L, et al. Single-nucleotide polymorphism in the human mu opioid receptor gene alters beta-endorphin binding and activity: possible implications for opiate addiction. Proc Natl Acad Sci U S A. 1998;95:9608–13. doi: 10.1073/pnas.95.16.9608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chick J, Anton R, Checinski K, Croop R, Drummond DC, Farmer R, et al. A multicentre, randomized, double-blind, placebo-controlled trial of naltrexone in the treatment of alcohol dependence or abuse. Alcohol Alcohol. 2000;35:587–93. doi: 10.1093/alcalc/35.6.587. [DOI] [PubMed] [Google Scholar]
- Coller JK, Cahill S, Edmonds C, Farquharson AL, Longo M, Minniti R, et al. OPRM1 A118G genotype fails to predict the effectiveness of naltrexone treatment for alcohol dependence. Pharmacogenet Genomics. 2011;21:902–5. doi: 10.1097/FPC.0b013e32834c5445. [DOI] [PubMed] [Google Scholar]
- Del Boca FK, Kranzler HR, Brown J, Korner PF. Assessment of medication compliance in alcoholics through UV light detection of a riboflavin tracer. Alcohol Clin Exp Res. 1996;20:1412–7. doi: 10.1111/j.1530-0277.1996.tb01142.x. [DOI] [PubMed] [Google Scholar]
- Drobes DJ, Anton RF, Thomas SE, Voronin K. Effects of naltrexone and nalmefene on subjective response to alcohol among non-treatment-seeking alcoholics and social drinkers. Alcohol Clin Exp Res. 2004;28:1362–70. doi: 10.1097/01.alc.0000139704.88862.01. [DOI] [PubMed] [Google Scholar]
- Emmerson PJ, Liu MR, Woods JH, Medzihradsky F. Binding affinity and selectivity of opioids at mu, delta and kappa receptors in monkey brain membranes. The Journal of pharmacology and experimental therapeutics. 1994;271:1630–7. [PubMed] [Google Scholar]
- Freeman B, Powell J, Ball D, Hill L, Craig I, Plomin R. DNA by mail: an inexpensive and noninvasive method for collecting DNA samples from widely dispersed populations. Behav Genet. 1997;27:251–7. doi: 10.1023/a:1025614231190. [DOI] [PubMed] [Google Scholar]
- Gelernter J, Gueorguieva R, Kranzler HR, Zhang H, Cramer J, Rosenheck R, et al. Opioid receptor gene (OPRM1, OPRK1, and OPRD1) variants and response to naltrexone treatment for alcohol dependence: results from the VA Cooperative Study. Alcoholism, clinical and experimental research. 2007;31:555–63. doi: 10.1111/j.1530-0277.2007.00339.x. [DOI] [PubMed] [Google Scholar]
- Gianoulakis C, Krishnan B, Thavundayil J. Enhanced sensitivity of pituitary beta-endorphin to ethanol in subjects at high risk of alcoholism. Arch Gen Psychiatry. 1996;53:250–7. doi: 10.1001/archpsyc.1996.01830030072011. [DOI] [PubMed] [Google Scholar]
- Goldman D, Oroszi G, O’Malley S, Anton R. COMBINE genetics study: the pharmacogenetics of alcoholism treatment response: genes and mechanisms. J Stud Alcohol Suppl. 2005:56–64. doi: 10.15288/jsas.2005.s15.56. discussion 33. [DOI] [PubMed] [Google Scholar]
- Grant BF, Dawson DA, Stinson FS, Chou SP, Dufour MC, Pickering RP. The 12-month prevalence and trends in DSM-IV alcohol abuse and dependence: United States, 1991–1992 and 2001–2002. Drug Alcohol Depend. 2004;74:223–34. doi: 10.1016/j.drugalcdep.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Guardia J, Caso C, Arias F, Gual A, Sanahuja J, Ramirez M, et al. A double-blind, placebo-controlled study of naltrexone in the treatment of alcohol-dependence disorder: results from a multicenter clinical trial. Alcohol Clin Exp Res. 2002;26:1381–7. doi: 10.1097/01.ALC.0000030561.15921.A9. [DOI] [PubMed] [Google Scholar]
- Heinala P, Alho H, Kiianmaa K, Lonnqvist J, Kuoppasalmi K, Sinclair JD. Targeted use of naltrexone without prior detoxification in the treatment of alcohol dependence: a factorial double-blind, placebo-controlled trial. J Clin Psychopharmacol. 2001;21:287–92. doi: 10.1097/00004714-200106000-00006. [DOI] [PubMed] [Google Scholar]
- Hyytia P, Kiianmaa K. Suppression of ethanol responding by centrally administered CTOP and naltrindole in AA and Wistar rats. Alcohol Clin Exp Res. 2001;25:25–33. doi: 10.1111/j.1530-0277.2001.tb02123.x. [DOI] [PubMed] [Google Scholar]
- Kemppainen H, Raivio N, Suo-Yrjo V, Kiianmaa K. Opioidergic modulation of ethanol self-administration in the ventral pallidum. Alcohol Clin Exp Res. 2012;36:286–93. doi: 10.1111/j.1530-0277.2011.01611.x. [DOI] [PubMed] [Google Scholar]
- Kiefer F, Jahn H, Tarnaske T, Helwig H, Briken P, Holzbach R, et al. Comparing and combining naltrexone and acamprosate in relapse prevention of alcoholism: a double-blind, placebo-controlled study. Arch Gen Psychiatry. 2003;60:92–9. doi: 10.1001/archpsyc.60.1.92. [DOI] [PubMed] [Google Scholar]
- Killeen TK, Brady KT, Gold PB, Simpson KN, Faldowski RA, Tyson C, et al. Effectiveness of naltrexone in a community treatment program. Alcohol Clin Exp Res. 2004;28:1710–7. doi: 10.1097/01.alc.0000145688.30448.2c. [DOI] [PubMed] [Google Scholar]
- King AC, Volpicelli JR, Frazer A, O’Brien CP. Effect of naltrexone on subjective alcohol response in subjects at high and low risk for future alcohol dependence. Psychopharmacology (Berl) 1997;129:15–22. doi: 10.1007/s002130050156. [DOI] [PubMed] [Google Scholar]
- Ko MC, Butelman ER, Traynor JR, Woods JH. Differentiation of kappa opioid agonist-induced antinociception by naltrexone apparent pA2 analysis in rhesus monkeys. The Journal of pharmacology and experimental therapeutics. 1998;285:518–26. [PMC free article] [PubMed] [Google Scholar]
- Koistinen M, Tuomainen P, Hyytia P, Kiianmaa K. Naltrexone suppresses ethanol intake in 6-hydroxydopamine-treated rats. Alcohol Clin Exp Res. 2001;25:1605–12. [PubMed] [Google Scholar]
- Kranzler HR, Modesto-Lowe V, Van Kirk J. Naltrexone vs. nefazodone for treatment of alcohol dependence. A placebo-controlled trial. Neuropsychopharmacology. 2000;22:493–503. doi: 10.1016/S0893-133X(99)00135-9. [DOI] [PubMed] [Google Scholar]
- Krystal JH, Cramer JA, Krol WF, Kirk GF, Rosenheck RA. Naltrexone in the treatment of alcohol dependence. N Engl J Med. 2001;345:1734–9. doi: 10.1056/NEJMoa011127. [DOI] [PubMed] [Google Scholar]
- Latt NC, Jurd S, Houseman J, Wutzke SE. Naltrexone in alcohol dependence: a randomised controlled trial of effectiveness in a standard clinical setting. Med J Aust. 2002;176:530–4. doi: 10.5694/j.1326-5377.2002.tb04550.x. [DOI] [PubMed] [Google Scholar]
- Lee MC, Wagner HN, Jr, Tanada S, Frost JJ, Bice AN, Dannals RF. Duration of occupancy of opiate receptors by naltrexone. J Nucl Med. 1988;29:1207–11. [PubMed] [Google Scholar]
- Lench N, Stanier P, Williamson R. Simple non-invasive method to obtain DNA for gene analysis. Lancet. 1988;1:1356–8. doi: 10.1016/s0140-6736(88)92178-2. [DOI] [PubMed] [Google Scholar]
- Littleton J, Zieglgansberger W. Pharmacological mechanisms of naltrexone and acamprosate in the prevention of relapse in alcohol dependence. Am J Addict. 2003;12 (Suppl 1):S3–11. doi: 10.1111/j.1521-0391.2003.tb00492.x. [DOI] [PubMed] [Google Scholar]
- MacKillop J. Factor structure of the alcohol urge questionnaire under neutral conditions and during a cue-elicited urge state. Alcoholism, clinical and experimental research. 2006;30:1315–21. doi: 10.1111/j.1530-0277.2006.00159.x. [DOI] [PubMed] [Google Scholar]
- Mansour A, Khachaturian H, Lewis ME, Akil H, Watson SJ. Anatomy of CNS opioid receptors. Trends Neurosci. 1988;11:308–14. doi: 10.1016/0166-2236(88)90093-8. [DOI] [PubMed] [Google Scholar]
- Martin CS, Earleywine M, Musty RE, Perrine MW, Swift RM. Development and validation of the Biphasic Alcohol Effects Scale. Alcohol Clin Exp Res. 1993;17:140–6. doi: 10.1111/j.1530-0277.1993.tb00739.x. [DOI] [PubMed] [Google Scholar]
- McCaul ME, Wand GS, Stauffer R, Lee SM, Rohde CA. Naltrexone dampens ethanol-induced cardiovascular and hypothalamic- pituitary-adrenal axis activation. Neuropsychopharmacology. 2001;25:537–47. doi: 10.1016/S0893-133X(01)00241-X. [DOI] [PubMed] [Google Scholar]
- Mitchell JM, O’Neil JP, Janabi M, Marks SM, Jagust WJ, Fields HL. Alcohol consumption induces endogenous opioid release in the human orbitofrontal cortex and nucleus accumbens. Sci Transl Med. 2012;4:116ra6. doi: 10.1126/scitranslmed.3002902. [DOI] [PubMed] [Google Scholar]
- Monti PM, Rohsenow DJ, Swift RM, Gulliver SB, Colby SM, Mueller TI, et al. Naltrexone and cue exposure with coping and communication skills training for alcoholics: treatment process and 1-year outcomes. Alcohol Clin Exp Res. 2001;25:1634–47. [PubMed] [Google Scholar]
- Morris PL, Hopwood M, Whelan G, Gardiner J, Drummond E. Naltrexone for alcohol dependence: a randomized controlled trial. Addiction. 2001;96:1565–73. doi: 10.1046/j.1360-0443.2001.961115654.x. [DOI] [PubMed] [Google Scholar]
- O’Malley SS, Jaffe AJ, Chang G, Schottenfeld RS, Meyer RE, Rounsaville B. Naltrexone and coping skills therapy for alcohol dependence. A controlled study. Arch Gen Psychiatry. 1992;49:881–7. doi: 10.1001/archpsyc.1992.01820110045007. [DOI] [PubMed] [Google Scholar]
- Oslin DW, Berrettini W, Kranzler HR, Pettinati H, Gelernter J, Volpicelli JR, et al. A functional polymorphism of the mu-opioid receptor gene is associated with naltrexone response in alcohol-dependent patients. Neuropsychopharmacology. 2003;28:1546–52. doi: 10.1038/sj.npp.1300219. [DOI] [PubMed] [Google Scholar]
- Phillips TJ, Brown KJ, Burkhart-Kasch S, Wenger CD, Kelly MA, Rubinstein M, et al. Alcohol preference and sensitivity are markedly reduced in mice lacking dopamine D2 receptors. Nature neuroscience. 1998;1:610–5. doi: 10.1038/2843. [DOI] [PubMed] [Google Scholar]
- Ray LA, Barr CS, Blendy JA, Oslin D, Goldman D, Anton RF. The Role of the Asn40Asp Polymorphism of the Mu Opioid Receptor Gene (OPRM1) on Alcoholism Etiology and Treatment: A Critical Review. Alcohol Clin Exp Res. 2012a;36:385–94. doi: 10.1111/j.1530-0277.2011.01633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray LA, Bujarski S, Chin PF, Miotto K. Pharmacogenetics of naltrexone in asian americans: a randomized placebo-controlled laboratory study. Neuropsychopharmacology. 2012b;37:445–55. doi: 10.1038/npp.2011.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray LA, Hutchison KE. A polymorphism of the mu-opioid receptor gene (OPRM1) and sensitivity to the effects of alcohol in humans. Alcohol Clin Exp Res. 2004;28:1789–95. doi: 10.1097/01.alc.0000148114.34000.b9. [DOI] [PubMed] [Google Scholar]
- Ray LA, Hutchison KE. Effects of naltrexone on alcohol sensitivity and genetic moderators of medication response: a double-blind placebo-controlled study. Arch Gen Psychiatry. 2007;64:1069–77. doi: 10.1001/archpsyc.64.9.1069. [DOI] [PubMed] [Google Scholar]
- Ray LA, McGeary J, Marshall E, Hutchison KE. Risk factors for alcohol misuse: examining heart rate reactivity to alcohol, alcohol sensitivity, and personality constructs. Addict Behav. 2006;31:1959–73. doi: 10.1016/j.addbeh.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Rubio G, Manzanares J, Lopez-Munoz F, Alamo C, Ponce G, Jimenez-Arriero MA, et al. Naltrexone improves outcome of a controlled drinking program. J Subst Abuse Treat. 2002;23:361–6. doi: 10.1016/s0740-5472(02)00296-9. [DOI] [PubMed] [Google Scholar]
- Shoemaker WJ, Vavrousek-Jakuba E, Arons CD, Kwok FC. The acquisition and maintenance of voluntary ethanol drinking in the rat: effects of dopaminergic lesions and naloxone. Behav Brain Res. 2002;137:139–48. doi: 10.1016/s0166-4328(02)00290-5. [DOI] [PubMed] [Google Scholar]
- Swift RM, Whelihan W, Kuznetsov O, Buongiorno G, Hsuing H. Naltrexone-induced alterations in human ethanol intoxication. Am J Psychiatry. 1994;151:1463–7. doi: 10.1176/ajp.151.10.1463. [DOI] [PubMed] [Google Scholar]
- Takemori AE, Ho BY, Naeseth JS, Portoghese PS. Nor-binaltorphimine, a highly selective kappa-opioid antagonist in analgesic and receptor binding assays. J Pharmacol Exp Ther. 1988;246:255–8. [PubMed] [Google Scholar]
- Takemori AE, Portoghese PS. Selective naltrexone-derived opioid receptor antagonists. Annu Rev Pharmacol Toxicol. 1992;32:239–69. doi: 10.1146/annurev.pa.32.040192.001323. [DOI] [PubMed] [Google Scholar]
- Tate SK, Goldstein DB. Will tomorrow’s medicines work for everyone? Nat Genet. 2004;36:S34–42. doi: 10.1038/ng1437. [DOI] [PubMed] [Google Scholar]
- Tidey JW, Monti PM, Rohsenow DJ, Gwaltney CJ, Miranda R, Jr, McGeary JE, et al. Moderators of naltrexone’s effects on drinking, urge, and alcohol effects in non-treatment-seeking heavy drinkers in the natural environment. Alcoholism, clinical and experimental research. 2008;32:58–66. doi: 10.1111/j.1530-0277.2007.00545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rijn RM, Brissett DI, Whistler JL. Dual efficacy of delta opioid receptor-selective ligands for ethanol drinking and anxiety. J Pharmacol Exp Ther. 2010;335:133–9. doi: 10.1124/jpet.110.170969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpicelli JR. Alcohol abuse and alcoholism: an overview. J Clin Psychiatry. 2001;62 (Suppl 20):4–10. [PubMed] [Google Scholar]
- Volpicelli JR, Alterman AI, Hayashida M, O’Brien CP. Naltrexone in the treatment of alcohol dependence. Arch Gen Psychiatry. 1992;49:876–80. doi: 10.1001/archpsyc.1992.01820110040006. [DOI] [PubMed] [Google Scholar]
- Walker AH, Najarian D, White DL, Jaffe JF, Kanetsky PA, Rebbeck TR. Collection of genomic DNA by buccal swabs for polymerase chain reaction-based biomarker assays. Environ Health Perspect. 1999;107:517–20. doi: 10.1289/ehp.99107517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerts EM, Kim YK, Wand GS, Dannals RF, Lee JS, Frost JJ, et al. Differences in delta- and mu-opioid receptor blockade measured by positron emission tomography in naltrexone-treated recently abstinent alcohol-dependent subjects. Neuropsychopharmacology. 2008;33:653–65. doi: 10.1038/sj.npp.1301440. [DOI] [PubMed] [Google Scholar]
- Zhang H, Kranzler HR, Yang BZ, Luo X, Gelernter J. The OPRD1 and OPRK1 loci in alcohol or drug dependence: OPRD1 variation modulates substance dependence risk. Mol Psychiatry. 2008;13:531–43. doi: 10.1038/sj.mp.4002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang D, Johnson AD, Papp AC, Sadee W. Allelic expression imbalance of human mu opioid receptor (OPRM1) caused by variant A118G. J Biol Chem. 2005;280:32618–24. doi: 10.1074/jbc.M504942200. [DOI] [PubMed] [Google Scholar]