Abstract
We constructed food-grade host-vector and integration systems for Streptococcus thermophilus by using a thymidylate synthase gene (thyA) as the selection marker. Two thyA genes, thyASt and thyALb, were cloned from S. thermophilus and Lactobacillus delbrueckii subsp. bulgaricus, respectively. Thymidine-requiring mutants of S. thermophilus were obtained after successive cultures in the presence of trimethoprim, and one of them, TM1-1, was used as the host. Food-grade vectors were constructed by using either thyASt or thyALb as the selection marker. Transformants of TM1-1 created by using these vectors were selected for thymidine autotrophy as efficiently as for erythromycin resistance. By using the host-vector system developed in this way, a foreign amylase gene (amyA) was expressed in TM1-1 and was also integrated into the chromosome by use of a temperature-sensitive integration vector constructed with thyALb as the selection marker via a double-crossover event. The results obtained show that thyA is an efficient and safe selection marker for S. thermophilus that is suitable for food applications.
Streptococcus thermophilus is one of the important lactic acid bacteria (LAB) and has long been used to produce yogurt and cheeses. It is desirable to improve the industrial characteristics of S. thermophilus by using modern biotechnological methods. Genes of this species encoding cadmium resistance (39), heat shock protein (11), and PepX (2) were proposed as safe food-grade selection markers. Chromosomal gene integration by use of a temperature-sensitive (ts) vector (pG+host5) (3, 23) and an Escherichia coli-derived vector (25) has been reported. For other LAB, especially Lactococcus lactis, food-grade genetic modification systems, chromosomal integration systems (22, 23), and inducible gene expression systems (8, 26, 28, 34) have been reported. For safety considerations, antibiotic resistance markers should be avoided for food applications. Food-grade selection marker genes are classified into two categories (9), dominant genes and complementary genes. Sucrose, xylose, and inulin utilization genes (17, 31), which the host does not naturally express, were reported as being dominant, and the lactose utilization gene (14) and the purine synthesis gene (10) were reported as being complementary. In the case of complementary marker genes, the gene relevant to the host should be inactivated first, which is sometimes difficult to perform by mutation or recombinant DNA techniques. Although spontaneous thyA mutants of some bacteria, including E. coli (27) and Bacillus subtilis (12), have been easily obtained by using antifolates such as trimethoprim (TMP) (19) and aminopterin, only one example of a thyA mutant of Lactobacillus acidophilus (13) has been reported as a host for dairy LAB. Among streptococcal species, thyA mutants of S. pyogenes and S. agalactiae have been reported, but thyA mutants have not been reported for S. sanguis, S. salivarius, S. mitis, or S. pneumoniae (7). The thyA gene of L. lactis has been cloned (32, 33) and was recently used as the target site for the integration of the human interleukin 10 gene (37). Although the clones of L. lactis that lack thyA are now available, they are not spontaneous mutants but were obtained by using recombinant DNA techniques.
In this report, we describe how spontaneous thyA mutants of S. thermophilus were obtained by using high concentrations of TMP. Two thyA genes of S. thermophilus and L. delbrueckii subsp. bulgaricus were successfully used as safe selection markers for transformation. Moreover, a safe chromosomal gene integration system for S. thermophilus was developed by using thyALb. A foreign gene was inserted into the chromosome next to this marker gene, which ensured stabilization of the integrated genes in the host.
The strains and plasmids used in this study are listed in Table 1. S. thermophilus strains were grown at 37°C on skim milk medium (10% skim milk with 0.1% yeast extract) or M17 medium (Difco) containing either 0.5% glucose (GM17) or 0.5% lactose (LM17). E. coli was grown at 37°C on M9 medium (24) supplemented with thymidine (50 μg/ml) when necessary. Erythromycin (Sigma) was used at a concentration of 25 μg/ml for S. thermophilus, and ampicillin (Sigma) was used at a concentration of 50 μg/ml for E. coli.
TABLE 1.
Bacterial strains and plasmids used in this studya
| Strains and plasmids | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| S. thermophilus | ||
| ATCC 19258 | Type strain of S. thermophilus | ATCC |
| TM1-1 | Spontaneous thyA mutant of ATCC 19258 | This study |
| OLL1131 | Industrial strain for yogurt production | Meiji Dairies Corporation |
| OILL3074 | Industrial strain for yogurt production | Meiji Dairies Corporation |
| L. delbrueckii subsp. bulgaricus M-878 | Industrial strain for yogurt production | Meiji Dairies Corporation |
| E. coli TGthy2 | Spontaneous thyA mutant of E. coli TG1 | This study |
| Plasmids | ||
| pBUL1 | Endogenous plasmid (7.9 kb) of L. delbrueckii subsp. bulgaricus M-878 | 20 |
| pBSt1 | 8.9 kb; Thy+; pBUL1 + thyASt | This study |
| pBLb1 | 8.9 kb; Thy+; pBUL1 + thyALb | This study |
| pX3 | 9.1 kb; Emr; pBUL1 + ermA | 20 |
| pXTLb | 10.1 kb; Emr Thy+; pX3 + thyALb | This study |
| pX4041 | 10.1 kb; Emr Thy+; pX3 + thyASt | This study |
| pSY1 | Endogenous plasmid (2.9 kb) of L. lactis M-128C (Meiji Dairies Corporation) | 21 |
| pSLb1 | 4.0 kb; Thy+; pSY1 + thyALb | This study |
| pSLbA1 | 7.0 kb; Thy+ Amy+; pSLb1 + amyA | This study |
| pSYE2 | 3.9 kb; Emr; pSY1 + ermA | 21 |
| pSYTLb | 5.2 kb; Emr Thy+; pSYE2 + thyALb | This study |
| pSG+E2 | 3.9 kb; ts replication, Emr; ori of pSYE2 replaced with ts ori of pG+host5 (4) | Unpublished results |
| pSintA1 | 8.0 kb; Thy+ Amy+; ts replication, pSG+E2 + thyALb + amyA + two lacZ fragments (ermA deleted) | This study |
Thy, thymidine; Em, erythromycin.
E. coli was transformed by the CaCl2 method (6). S. thermophilus was transformed according to the method described by Holo and Nes (16), with some modifications. Competent cells were prepared as follows. An overnight bacterial culture was inoculated (2%, vol/vol) and incubated at 42°C on M17 medium (adjusted at pH 5.5 with HCl) containing 1% lactose, 0.175 M sucrose, and 0.6% glycine. Cells were harvested by centrifugation at an optical density at 600 nm of 0.08 to 0.1. The cells were washed twice with ice-cold buffer (0.5 M sucrose, 10% glycerol), concentrated in the same buffer (final optical density at 600 nm of 50), and stored at −80°C until use. Forty microliters of the competent cells was given a single electric pulse (2.0 kV, 25 μF) and incubated for 2 h at 37°C in 1 ml of expression medium (M17 medium containing 1% glucose, 0.5 M sucrose, 20 mM MgCl2, 2 mM CaCl2, and 20 μg of thymidine/ml). For the construction of a ts integration vector, the expression was done at 28°C for 3 h. The cells were centrifuged and washed twice with sterilized water to remove thymidine, when necessary. Transformants were selected on LM17 agar plates with or without erythromycin.
The recombinant techniques used were done according to the standard methods (24). The chromosomal DNA of S. thermophilus ATCC 19258 or L. bulgaricus subsp. delbrueckii M-878 was digested with HindIII or BamHI, respectively. Fragments of each digested DNA ranging from 2 to 6 kb were ligated at the HindIII or BamHI site of pBR322. After transformation of E. coli TGthy2 with a ligation mixture, colonies were obtained on an M9 agar plate supplemented with ampicillin and 2 μg of Casamino Acids (Difco)/ml. The chromosomal DNA fragments inserted into pBR322, which complemented the thyA mutation of E. coli TGthy2, were subcloned into pUC118, and the nucleotide sequences were determined.
In order to isolate thyA mutants, the S. thermophilus strains were cultivated on LM17 supplemented with thymidine (20 μg/ml) and TMP (0.3 to 1.5 mg/ml, depending on the strain). After about 50 and 100 generations of cultivation, each culture broth was spread on LM17 agar plates containing thymidine. A hundred TMP-resistant single-colony isolates of each strain were examined by growth on LM17 agar both with and without added thymidine. Those colonies that grew in the presence of thymidine (20 μg/ml) for 1 day but that did not grow after 1 week in the absence of thymidine were classified as Thy− mutants.
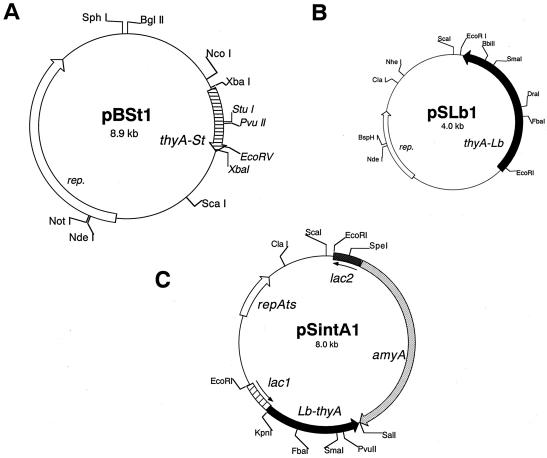
The following novel vectors were constructed by transformation of S. thermophilus TM1-1 after selection for Thy+ on LM17 agar plates. A PCR fragment containing thyASt or thyALb was obtained by using primers 84 and 86 or primers 89 and 90, respectively (Table 2). Each DNA fragment was ligated to the XbaI site of pBUL1, and a ligation mixture was used to transform TM1-1, resulting in two vectors, pBSt1 (Fig. 1A) and pBLb1 (data not shown), respectively. A PCR fragment containing thyALb was inserted at the EcoRI site of pSY1, which resulted in a novel vector, pSLb1 (Fig. 1B). The 2.95-kb PCR fragment containing the amyA gene from Streptococcus bovis 148 was obtained by using primers 150 and 151 and pSAES5 (35). This PCR fragment was digested with EcoRI and was blunt-end ligated to the BbiII site of pSLb1 (Fig. 1B), resulting in an Amy+ plasmid, pSLbA1. Moreover, in order to compare selection efficiencies between thyA and ermA marker genes, a PCR fragment containing either thyALb or thyASt was inserted into pX3 and pSYE2. Three recombinant plasmids were constructed, two from pX3, namely, pXTLb and pX4041, harboring ermA together with thyALb or thyASt, respectively, and one from pSYE2, namely, pSYTLb, harboring ermA and thyALb.
TABLE 2.
Primers used in this study
| Primer | Relevant gene or plasmid | Nucleotide sequence |
|---|---|---|
| 06 | thyALb | 5′-AAATGTGGTTTTTATGTTCAAGCAAG-3′ |
| 37 | pSY1 plasmid | 5′-CTTGTTATTATATTAATTTTTA-3′ |
| 68 | thyALb | 5′-CTGCCAGCGGATTTTGGATGATG-3′ |
| 84 | thyASt | 5′-GGTCTAGAGCAACATTATTTACAGCTATA-3′ |
| 86 | thyASt | 5′-GGTCTAGAGGGCTTAAATCATAAGCATTT-3′ |
| 89 | thyALb | 5′-GGTCTAGACCTGTCCGGGAAAAAAGG-3′ |
| 90 | thyALb | 5′-GGTCTAGAGGTCTAATTTCCTGGTGG-3′ |
| 98 | lacZ | 5′-GCGAGCTCG GTGATGCATCTGTTAATGG-3′ |
| 99 | lacZ | 5′-CGGGTACCGATATTCCATGAAGGTTCGC-3′ |
| 114 | lacZ | 5′-GCTATTCCAAAAGTTCACGTTC-3′ |
| 115 | lacZ | 5′-CTAGCTAGGAAAGTATAAGCAG-3′ |
| 135 | lacZ | 5′-CTAATTTAGTGGTTCAATCA-3′ |
| 150 | amyA | 5′-GCGAATTCATCGGTACAATTGATATTA-3′ |
| 151 | amyA | 5′-GCGAATTCGTACAAAAAGAAGTAGCC-3′ |
FIG. 1.
Maps of constructed food-grade vectors. Relevant restriction enzyme sites are shown. (A) pBSt1 was constructed with pBUL1 by using thyASt as a selection marker. White arrow, replication (rep.) protein gene of pBUL1. (B) pSLb1 was constructed with pSY1 by using thyALb as a selection marker. White arrow, replication protein gene of pSY1. (C) A food-grade integration vector, pSintA1, which has a ts ori (repAts), thyALb as a selection marker (black arrow), and amyA as a foreign gene (dotted arrow). thyALb and amyA are inserted between two DNA fragments, lac-1 (striped box with a superimposed arrow) and lac-2 (stippled box with a superimposed arrow), derived from the lacZ gene of S. thermophilus. The lacZ gene was selected as an integration target site. Relevant restriction enzyme sites are shown.
A novel ts integration vector was constructed as follows. First, the PCR fragment (targeted with primers 89 and 90) containing thyALb was inserted at the SmaI site of pSG+E2, a ts replication vector, in the same direction as ermA gene transcription. Second, the resultant plasmid, designated pS+ETLb2, received a 413-bp DNA fragment (lac-2St; Fig. 2A) of a β-galactosidase gene (lacZ) (36) from S. thermophilus ATCC 19258 at its BamHI site. A DNA fragment of lac-2St had been obtained by NdeI digestion of the 1,343-bp PCR fragment (targeted with primers 114 and 115) of lacZ and was blunt-end ligated with the BamHI blunt-end-treated pS+ETLb2. Third, a 1.1-kb DNA fragment containing ermA was removed from this new plasmid by two restriction enzyme digestions, with PstI and ScaI, followed by a blunt-end self-ligation, which resulted in pS+iTLb22. Fourth, the other DNA fragment (lac-1St [459 bp]) (Fig. 2A) of lacZ was inserted at the KpnI and SacI sites of pS+iTLb22. A DNA fragment of lac-1St had been amplified by PCR with primers 98 and 99 and was digested by the same enzymes. And finally, as a foreign model gene, amyA was inserted at the BbiII site (47 bp downstream of the stop codon of thyALb) of this plasmid, resulting in pSintA1 (Fig. 1C).
FIG. 2.
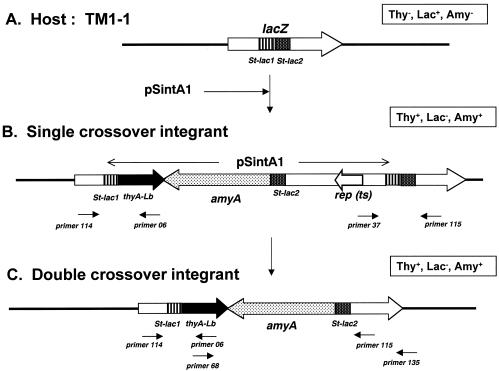
Chromosome structures of the host (TM1-1) (A), the single-crossover integrant (B), and the double-crossover integrant (C). Integrants were obtained after cultivation of a transformant harboring the pSintA1 plasmid on LM17 at 37°C. The chromosome structures were estimated by the PCR experiment data shown in Table 4. The phenotype of each clone is shown in an open square. Relevant PCR primers used in this study are shown.
A strain was judged to be Thy− or Thy+ based on its requirement for thymidine (20 μg/ml) on LM17 for normal growth. A strain was determined to be Amy+ if a clear halo around the colony was detected on GM17 agar plates containing 0.5% soluble starch after exposure to iodine vapor. A strain was judged to be Lac+ or Lac− by the color of colonies grown on a GM17 agar plate supplemented with a 0.2% (vol/vol) concentration of 4% X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) solution in dimethylformamide.
Characterization of thyA genes of yogurt starter strains.
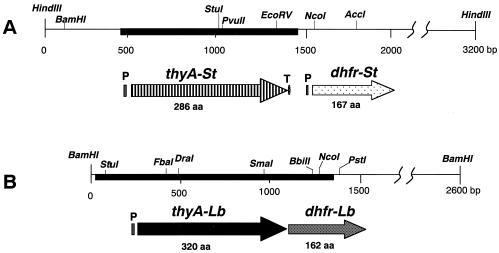
A 3.2-kb HindIII DNA fragment and a 2.6-kb BamHI fragment from S. thermophilus ATCC 19258 and L. delbrueckii subsp. bulgaricus M-878, respectively, were selected for complementation of Thy− in E. coli. Also, the thyA genes thyASt and thyALb and the dihydrofolate reductase genes dhfrSt and dhfrLb were deduced as shown in Fig. 3. Homology search analyses revealed that the amino acid sequence deduced from thyASt had 97.8, 85.3, 82.2, 81.1, and 75.6% homologies with those of S. thermophilus CHOC2136 (S. Moia, M. B. Pedersen, and K. Soerensen, submitted for publication), S. pneumoniae (18), S. agalactiae (38), S. mutans (1), and L. lactis (5), respectively. Also, the amino acid sequence deduced from thyALb had 68.7% homology with that of Lactobacillus casei (30). The sequence homology between thyASt and thyALb products was relatively low (35.2%).
FIG. 3.
(A) Restriction map of the 3.2-kb HindIII DNA fragment of the S. thermophilus ATCC 19258 chromosome. The thymidylate synthase gene (thyA-St) and the dihydrofolate reductase gene (dhfr-St) were deduced from the nucleotide sequence. A heavy line indicates a DNA fragment amplified by PCR with primers 84 and 86 containing the thyASt gene. (B) Restriction map of the 2.6-kb BamHI DNA fragment of the L. delbrueckii subsp. bulgaricus M-878 chromosome and the deduced thyALb and dhfrLb products. A heavy line indicates a DNA fragment amplified by PCR with primers 89 and 90 containing thyALb. A potential promoter (P), a potential terminator (T), and relevant restriction enzyme sites are indicated. aa, amino acids.
Our initial attempts to isolate thyA mutants were unsuccessful because S. thermophilus strains grew well in the presence of 50 μg of TMP/ml. Spontaneous thyA mutants were obtained after about 100 generations of cultivation in LM17 broth supplemented with 20 μg of thymidine/ml and higher concentrations of TMP, i.e., 1,500 μg/ml for strain ATCC 19258 and 300 μg/ml for strains OLL1131 and OLL3074. One of the stable thyA mutants obtained from ATCC 19258 was designated TM1-1 and was used as the host in this study. We found two mutations in the thyA region of the TM1-1 chromosome, a deletion of G at nucleotide 319 and one base exchange (from A to T) at nucleotide 323, which indicated that the thyA gene had been inactivated in this mutant. No changes were detected in the nucleotide sequence of dhfrSt in TM1-1 compared to that of the wild-type strain, in contrast to the mutation of dhfr in S. pneumoniae which resulted in TMP resistance (29). TM1-1 grew as well as the wild-type strain, ATCC 19258, on LM17 or skim milk medium only if supplemented with more than 10 μg of thymidine/ml. The Thy− phenotype of TM1-1 was stable, since no Thy+ revertants appeared even after 50 generations of cultivation on LM17 containing 20 μg of thymidine/ml (data not shown). Thy+ revertants were estimated to number fewer than 1 out of 1011 colonies in an experiment in which a concentrated TM1-1 competent cell suspension was spread and cultivated on LM17 agar plates.
Transformation by using novel food-grade vectors.
Three novel vectors for S. thermophilus were constructed by using pBUL1 or pSY1; pBSt1 (Fig. 1A), pBLb1, and pSLb1 (Fig. 1B). For unknown reasons, we have never been able to insert the DNA fragment containing thyASt into pSY1. We regard these vectors as being food grade since they are constituted of DNA sequences derived only from LAB for manufacturing dairy products. Transformants of TM1-1 were reproducibly obtained by using either one of these vectors, although transformation efficiencies per microgram of DNA were lower with pBLb1 (4 × 102) and pBSt1 (3 × 102) than with pSLb1 (4 × 104), presumably depending on the replicons used. The transformation efficiencies of TM1-1 with the thyA gene were compared with those of an Emr gene (ermA) by using pXTLb, pX4041, and pSYTLb (Table 3). The results were almost the same under both selection conditions. Fifty colonies randomly selected for Thy+ in each experiment were all Emr and vice versa. These results indicate that thyALb and thyASt worked as efficiently as ermA for the selection of genetic transformants of S. thermophilus.
TABLE 3.
Comparisons of transformation efficiencies using thyA and ermA
| Plasmid | Selection marker | Selection mediuma | No. of transformants/ μg of plasmid DNA |
|---|---|---|---|
| pSYTLb | thyALb | LM17 | 6.4 × 104 |
| ermA | LM17 + Em + Thy | 6.2 × 104 | |
| pXTLb | thyALb | LM17 | 2.8 × 102 |
| ermA | LM17 + Em + Thy | 3.0 × 102 | |
| pX4041 | thyASt | LM17 | 2.0 × 102 |
| ermA | LM17 + Em + Thy | 2.3 × 102 |
Erythromycin (Em) and thymidine (Thy) were added at 25 and 20 μg/ml, respectively.
Expression of a foreign gene using the food-grade host- vector system.
An extracellular α-amylase gene (amyA) of S. bovis 148 was inserted into pSLb1, resulting in a novel plasmid, pSLbA1. This plasmid was used to transform TM1-1, and the transformants were selected on LM17 agar plates supplemented with 0.5% starch. All colonies selected for Thy+ formed a clear α-amylase-positive halo around each colony after exposure to iodine vapor (data not shown). This result showed that pSLb1 can be used as a vector for a foreign gene expression in a spontaneous mutant, TM1-1, which indicates that the novel food-grade host-vector system for S. thermophilus is useful for industrial applications.
Chromosomal gene integration with thyA as a selection marker.
Finally, the thyA gene was evaluated as a selection marker for chromosomal gene integration in S. thermophilus. The transformation efficiency of ATCC 19258 with pG+host5 (4) was slightly low, and we constructed another ts vector using pSYE2, which showed a higher transformation efficiency. The ts mutation of pG+host5 was introduced in vitro into the ori of pSYE2, which resulted in a ts vector, pSG+E2 (unpublished results). In order to avoid recombination at the thyA locus of S. thermophilus, thyALb, which has a low sequence homology with thyASt, was used. The thyALb gene and a model foreign gene, amyA, were inserted into pSG+E2, resulting in a ts integration plasmid, pSintA1 (Fig. 1C). This plasmid was used to transform TM1-1. Transformants were selected for Thy+ on LM17 at 28°C. All colonies obtained on LM17 plates showed the Amy+ phenotype (data not shown). Twelve of the colonies, randomly selected, were verified as transformants by the presence of pSintA1 DNA. To integrate the plasmid into the chromosome, one of the transformants was incubated on GM17 broth at 28°C for 16 h, and then 0.1% of the culture was inoculated and cultivated for 15 generations in fresh GM17 broth at 37°C (nonpermissive temperature for the replication of pSintA1). Aliquots of the culture were spread and grown on GM17 plates containing 0.5% soluble starch and X-Gal. All colonies obtained were white (Lac−), indicating that pSintA1 was inserted at either one of the target sites, lac-1St or lac-2St, and hence, lacZ was inactivated. Forty-eight colonies randomly selected on the plates were Amy+. Their chromosomal structure was further examined by PCR with the following four sets of primers: primers 114 and 06, 37 and 115, 114 and 115, and 68 and 135 (Fig. 2C). As shown in Table 4, the results of these PCR experiments suggest that one copy of pSintA1 was inserted at one of the target sites, lac-1St, in 21 out of 48 colonies, presumably via a single-crossover event (single-crossover integrant) (Fig. 2B). However, the other 27 colonies gave different PCR results (Table 4), which suggested that only the thyALb and amyA genes were retained in the lacZ gene and that no DNA fragments of the vector pSG+E2 remained. We believe that these 27 colonies were obtained via a double-crossover event as shown in Fig. 2C.
TABLE 4.
PCR DNA amplification patterns of 48 colonies examined
| Primers used for targeting | Detectiona | Length of PCR fragment (kb) detected inb,c:
|
|
|---|---|---|---|
| Group A | Group B | ||
| 114 and 06 | 48/48 | 1.1 (21) | 1.1 (27) |
| 37 and 115 | 21/48 | 1.5 (21) | ND (0)d |
| 114 and 115 | 48/48 | 9.3 (21) | 5.7 (27) |
| 68 and 135 | 48/48 | 9.1 (21) | 5.5 (27) |
Number of colonies in which DNA amplification was detected by PCR/total number of colonies.
Forty-eight colonies were divided into two groups of 21 and 27 colonies, according to their PCR amplification patterns.
Numbers in parentheses indicate numbers of colonies showing PCR amplification in group A and B.
ND, no DNA amplification was detected.
The host-vector systems of S. thermophilus reported here are composed of genetic materials derived solely from LAB used in dairy production. Therefore, they are regarded as food grade and fulfill the following conditions that are preferable in industrial applications. First, the selection marker genes thyALb and thyASt, derived from yogurt starter organisms, are relatively small and easy to handle in the construction of recombinant plasmids. Second, the host cell (thyA mutant) of industrial strains of S. thermophilus can be easily obtained through spontaneous mutation simply by cultivation with higher concentrations of TMP. Third, transformants of S. thermophilus can be selected on normal growth media such as skim milk and M17, and the selection efficiency is almost the same as that of an antibiotic resistance marker gene, ermA.
In our experience, stable thyA mutants of S. thermophilus were obtained after about 100 generations of cultivation on LM17 broth supplemented with 20 μg of thymidine/ml and higher concentrations (300 to 1,500 μg/ml) of TMP. We tried to apply higher concentrations of TMP to obtain thyA mutants of L. delbrueckii subsp. bulgaricus and L. lactis, but the strains we tested grew well even in the presence of 2.0 mg of TMP/ml, which is nearly the saturation concentration of this drug in water. This explains why no Thy− mutants of these LAB have so far been obtained by using TMP.
We also report here a novel food-grade integration system in which both the gene of interest (amyA) and the selection marker gene (thyA) are connected and inserted at a target site of the chromosome via a double-crossover event. We planned to insert pSintA1 (Fig. 1C) at the lacZ locus of S. thermophilus (15) as an integration target site since the desired integrants could be easily detected by the presence of white colonies on agar plates containing X-Gal. Other integration target sites can be selected according to the objectives of experiments or applications. When a ts integration plasmid of pSintA1 was used, about half of the integrants obtained after cultivation at a high temperature (37°C) were double-crossover integrants, as shown in Fig. 2C. Theoretically, both single- and double-crossover integrants may be obtained because the selection pressure is thymidine autotrophy and no erythromycin is added (Fig. 2B and C). This finding explains why one-step cultivation at 37°C was sufficient to obtain the objective gene integrants which appeared after the second homologous recombination event, concomitant with a loop-out of the vector DNA. An advantage of this novel system is the ability to obtain objective integrants after a double-crossover event because the desired gene is directly connected with the marker gene (thyA), which is essential for growth. This result is in contrast to that of other methods, in which a marker gene is not inserted next to the gene of interest, and, therefore, wild-type revertants easily appear after a double-crossover event. It is clear that the integrants do not lose the gene of interest after cultivation for a long period of time, because the selection pressure remains unless thymidine is added to the medium. The integrants' genetic stability ensured in this way is favorable for industrial applications.
It is clear from the data shown here that the thymidylate synthase gene (thyA) of yogurt starter strains can be used as a safe selection marker in molecular breeding of S. thermophilus.
Nucleotide sequence accession numbers.
The DNA sequences of the thyA gene of S. thermophilus ATCC 19258 and L. delbrueckii subsp. bulgaricus M-878 were registered in the GenBank nucleotide sequence data library under accession numbers E12779 and E12778, respectively.
Acknowledgments
We appreciate E. Maguin for supplying us with the pG+host5 plasmid. We thank Mariko Takeda and Yoshiko Honme for their technical assistance. We also thank R. Walton for reading the manuscript.
REFERENCES
- 1.Ajdic, D., W. M. McShan, R. E. McLaughlin, G. Savic, J. Chang, M. B. Carson, C. Primeaux, R. Tian, S. Kenton, H. Jia, S. Lin, Y. Qian, S. Li, H. Zhu, F. Najar, H. Lai, J. White, B. A. Roe, and J. J. Ferrett. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. USA 99:14434-14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anastasiou, R., M. Papadelli, M. D. Georgalaki, G. Kalantzopoulos, and E. Tsakalidou. 2002. Cloning and sequencing of the gene encoding X-prolyl-dipeptidyl aminopeptidase (PepX) from Streptococcus thermophilus strain ACA-DC 4. J. Appl. Microbiol. 93:52-59. [DOI] [PubMed] [Google Scholar]
- 3.Baccigalupi, L., G. Naclerio, M. D. Felice, and E. Ricca. 2000. Efficient insertional mutagenesis in Streptococcus thermophilus. Gene 258:9-14. [DOI] [PubMed] [Google Scholar]
- 4.Biswas, I., A. Gruss, S. D. Ehrlich, and E. Maguin. 1993. High-efficiency gene inactivation and replacement system for gram-positive bacteria. J. Bacteriol. 175:3628-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11:731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen, S. N., A. C. Y. Chang, and L. Hsu. 1972. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc. Natl. Acad. Sci. USA 69:2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coll, P. F., V. R. Ausina, J. V. Vernis, B. O. Mirelis, and G. P. Prats. 1984. Exogenous thymidine and reversal of the inhibitory effect of sulfamethoxazole-trimethoprim on streptococci. Eur. J. Microbiol. 3:424-426. [DOI] [PubMed] [Google Scholar]
- 8.de Ruyter, P. G. G. A., O. P. Kuipers, and W. M. de Vos. 1996. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 62:3662-3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Vos, W. M. 1999. Safe and sustainable systems for food-grade fermentations by genetically modified lactic acid bacteria. Int. Dairy J. 9:3-10. [Google Scholar]
- 10.Dickely, F., D. Nilsson, E. B. Hansen, and E. Johansen. 1995. Isolation of Lactococcus lactis nonsense suppressors and construction of a food-grade cloning vector. Mol. Microbiol. 15:839-847. [DOI] [PubMed] [Google Scholar]
- 11.El Demerdash, H. A. M., K. J. Heller, and A. Geis. 2003. Application of the shsp gene, encoding a small heat shock protein, as a food-grade selection marker for lactic acid bacteria. Appl. Environ. Microbiol. 69:4408-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farmer, J. L., and F. Rothman. 1965. Transformable thymine-requiring mutant of Bacillus subtilis. J. Bacteriol. 89:262-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu, X., and J.-G. Xu. 2000. Development of a chromosome-plasmid balanced lethal system for Lactobacillus acidophilus with thyA gene as selective marker. Microbiol. Immunol. 44:551-556. [DOI] [PubMed] [Google Scholar]
- 14.Hashiba, H., R. Takiguchi, K. Jyoho, and K. Aoyama. 1992. Establishment of a host-vector system in Lactobacillus helveticus with beta-galactosidase activity as a selection marker. Biosci. Biotechnol. Biochem. 56:190-194. [DOI] [PubMed] [Google Scholar]
- 15.Herman, R. E., and L. L. McKay. 1986. Cloning and expression of the β-d-galactosidase gene from Streptococcus thermophilus in Escherichia coli. Appl. Environ. Microbiol. 52:45-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holo, H., and I. F. Nes. 1989. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl. Environ. Microbiol. 55:3119-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hols, P., T. Ferain, D. Garmyn, N. Bernard, and J. Delcour. 1994. Use of homologous expression-secretion signals and vector-free stable integration in engineering of Lactobacillus plantarum for α-amylase and levanase expression. Appl. Environ. Microbiol. 60:1401-1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoskins, J., W. E. Alborn, Jr., J. Arnold, L. C. Blaszczak, S. Burgett, B. S. DeHoff, S. T. Estrem, L. Fritz, D.-J. Fu, W. Fuller, C. Geringer, R. Gilmour, J. S. Glass, H. Khoja, A. R. Kraft, R. E. Lagace, D. J. LeBlanc, L. N. Lee, E. J. Lefkowitz, J. Lu, P. Matsushima, S. M. McAhren, M. McHenney, K. McLeaster, C. W. Mundy, T. I. Nicas, F. H. Norris, M. O'Gara, R. B. Peery, G. T. Robertson, P. Rockey, P.-M. Sun, M. E. Winkler, Y. Yang, M. Young-Bellido, G. Zhao, C. A. Zook, R. H. Baltz, S. R. Jaskunas, P. R. Rosteck, Jr., P. L. Skatrud, and J. I. Glass. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 183:5709-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huovinen, P. 1987. Trimethoprim resistance. Antimicrob. Agents Chemother. 31:1451-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito, Y., Y. Sasaki, and T. Sasaki. June 1991. Novel plasmid pBUL1 derived from Lactobacillus and derivative thereof. Japanese patent P8103275.
- 21.Ito, Y., Y. Sasaki, and T. Sasaki. December 1991. Novel plasmid pSY1 derived from Lactococcus. Japanese patent P5176776.
- 22.Leenhouts, K., A. Bolhuis, G. Venema, and J. Kok. 1998. Construction of a food-grade multiple-copy integration system for Lactococcus lactis. Appl. Microbiol. Biotechnol. 49:417-423. [DOI] [PubMed] [Google Scholar]
- 23.Maguin, E., H. Prévost, S. D. Ehrlich, and A. Gruss. 1996. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J. Bacteriol. 178:931-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 25.Mollet, B., J. Knol, B. Poolman, O. Marciset, and M. Delley. 1993. Directed genenomic integration, gene replacement, and integrative gene expression in Streptococcus thermophilus. J. Bacteriol. 175:4315-4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nauta, A., D. van Sinderen, H. Karsens, E. Smit, G. Venema, and J. Kok. 1996. Inducible gene expression mediated by a repressor-operator system isolated from Lactococcus lactis bacteriophage rlt. Mol. Microbiol. 19:1331-1341. [DOI] [PubMed] [Google Scholar]
- 27.Okada, T., K. Yanagisawa, and F. J. Ryan. 1960. Elective production of thymine-less mutants. Nature 188:340-341. [DOI] [PubMed] [Google Scholar]
- 28.O'Sullivan, D., S. A. Walker, S. G. West, and T. R. Klaenhammer. 1996. Development of an expression strategy using a lytic phage to trigger explosive plasmid amplification and gene expression. Bio/Technology 14:82-87. [DOI] [PubMed] [Google Scholar]
- 29.Pikis, A., J. A. Donkersloot, W. J. Rodriguez, and J. M. Keith. 1998. A conservative amino acid mutation in the chromosome-encoded dihydrofolate reductase confers trimethoprim resistance. J. Infect. Dis. 178:700-706. [DOI] [PubMed] [Google Scholar]
- 30.Pinter, K., V. J. Davisson, and D. V. Santi. 1988. Cloning, sequencing, and expression of the Lactobacillus casei thymidylate synthase gene. DNA 7:235-241. [DOI] [PubMed] [Google Scholar]
- 31.Posno, M., P. T. H. M. Heuvelmans, M. J. F. van Giezen, B. C. Lokman, R. J. Leer, and P. H. Pouwels. 1991. Complementation of the inability of Lactobacillus strains to utilize d-xylose with d-xylose catabolism-encoding genes of Lactobacillus pentosus. Appl. Environ. Microbiol. 57:2764-2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ross, P., F. O'Gara, and S. Condon. 1990. Cloning and characterization of the thymidylate synthase gene from Lactococcus lactis subsp. lactis. Appl. Environ. Microbiol. 56:2156-2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ross, P., F. O'Gara, and S. Condon. 1990. Thymidylate synthetase gene from Lactococcus lactis as a genetic marker: an alternative to antibiotic resistance genes. Appl. Environ. Microbiol. 56:2164-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanders, J. W., G. Venema, and J. Kok. 1997. A chloride-inducible gene expression cassette and its use in induced lysis of Lactococcus lactis. Appl. Environ. Microbiol. 63:4877-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Satoh, E., Y. Ito, Y. Sasaki, and T. Sasaki. 1997. Application of the extracellular α-amylase gene from Streptococcus bovis 148 to construction of a secretion vector for yogurt starter strains. Appl. Environ. Microbiol. 63:4593-4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schroeder, C. J., C. Robert, G. Lenzen, L. L. McKay, and A. Mercenier. 1991. Analysis of the lacZ sequences from two Streptococcus thermophilus strains: comparison with the Escherichia coli and Lactobacillus bulgaricus beta-galactosidase sequences. J. Gen. Microbiol. 137:369-380. [DOI] [PubMed] [Google Scholar]
- 37.Steidler, L., S. Neirynck, N. Huyghebaert, V. Snoeck, A. Vermeire, B. Goddeeris, E. Cox, J. P. Remon, and E. Remaut. 2003. Biological containment of genetically modified Lactococcus lactis for intestinal delivery of human interleukin 10. Nat. Biotechnol. 21:785-789. [DOI] [PubMed] [Google Scholar]
- 38.Tettelin, H., V. Masignani, M. J. Cieslewicz, J. A. Eisen, S. Peterson, M. R. Wessels, I. T. Paulsen, K. E. Nelson, I. Margarit, T. D. Read, L. C. Madoff, A. M. Wolf, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. T. DeBoy, S. Durkin, J. F. Kolonay, L. A. Umayam, R. Madupu, M. R. Lewis, D. Radune, N. B Fedorova, D. Scanlan, H. Khouri, S. Mulligan, H. A. Carty, R. T. Cline, J. Gill, M. Scarselli, M. Mora, E. T. Iacobini, C. Brettoni, G. Galli, M. Mariani, F. Vegni, D. Maione, D. Rinaudo, R. Rappuoli, J. L. Telford, D. L. Kasper, G. Grandi, and C. M. Fraser. 2002. Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. Proc. Natl. Acad. Sci. USA 99:12391-12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wong, W. Y., P. Su, G. E. Allison, C.-Q. Liu, and N. W. Dunn. 2003. A potential food-grade cloning vector for Streptococcus thermophilus that uses cadmium resistance as the selectable marker. Appl. Environ. Microbiol. 69:5767-5771. [DOI] [PMC free article] [PubMed] [Google Scholar]