Abstract
Silage quality is typically assessed by the measurement of several individual parameters, including pH, lactic acid, acetic acid, bacterial numbers, and protein content. The objective of this study was to use a holistic metabolic fingerprinting approach, combining a high-throughput microtiter plate-based fermentation system with Fourier transform infrared (FT-IR) spectroscopy, to obtain a snapshot of the sample metabolome (typically low-molecular-weight compounds) at a given time. The aim was to study the dynamics of red clover or grass silage fermentations in response to various inoculants incorporating lactic acid bacteria (LAB). The hyperspectral multivariate datasets generated by FT-IR spectroscopy are difficult to interpret visually, so chemometrics methods were used to deconvolute the data. Two-phase principal component-discriminant function analysis allowed discrimination between herbage types and different LAB inoculants and modeling of fermentation dynamics over time. Further analysis of FT-IR spectra by the use of genetic algorithms to identify the underlying biochemical differences between treatments revealed that the amide I and amide II regions (wavenumbers of 1,550 to 1,750 cm−1) of the spectra were most frequently selected (reflecting changes in proteins and free amino acids) in comparisons between control and inoculant-treated fermentations. This corresponds to the known importance of rapid fermentation for the efficient conservation of forage proteins.
Ruminant animal production in the United Kingdom relies mainly on grass (either grazed or conserved as silage and often supplemented with high-protein feed concentrates) (28, 38). A ban on animal protein in livestock feeds has led to renewed interest in using plant-based N sources, for example, high-protein legume crops such as red and white clover (which typically contain 20 to 30% crude protein in the dry matter compared to 15% in grasses) (D. E. Beever, Abstr. Proc. XVII Int. Grassland Cong., p. 535-541, 1993). However, leguminous crops tend to have intrinsically lower sugar content and a high level of buffering capacity compared to grasses (6), which can result in problems during ensilage. Silage is the product of an anaerobic fermentation characterized by rapid acidification due to lactate production by lactic acid bacteria (LAB). This process comprises four main stages, as described in Table 1. The duration of the first (aerobic) phase and the rate of pH decrease in (anaerobic) phase 2 are both critical factors in determining the quality of the silage at feed-out (10). Good fermentation is characterized by a rapid initial decrease in pH. The decrease in pH (compared to that of fresh herbage, with a pH typically ranging from 5.6 to 6.6) to pH 4.2 or less ideally should occur in 24 to 48 h (8). This rapid acidification inhibits spoilage organisms such as clostridia, enterobacteria, yeasts, and molds (7) and the action of intrinsic plant enzymes such as proteases (7, 18, 28), thus preserving the nutrient value of the feed for the ruminant. The acidification should also be stable over time, allowing long-term storage of the silage (8).
TABLE 1.
The four main phases of the ensilage processa
| Phase | Key event | Duration |
|---|---|---|
| Aerobic | Aerobic organisms active | Short lived (hours) |
| Oxygen depletion | ||
| Anaerobic | Competition between anaerobes | Days |
| Acidification | ||
| LAB domination in a good fermentation | ||
| Stable (storage) | Low pH | Months |
| Maturation | Inhibition of spoilage organisms | |
| Decrease in viable microorganisms | ||
| Feedout | Aerobic instability | Days |
| Increase in pH | ||
| Increase in aerobes; spoilage |
See reference 47.
The use of LAB inoculants to improve grass silage fermentation (and, hence, quality) is now widespread (10, 22; M. K. Woolford, Abstr. Proc. Alltech. 14th Annu. Symp., p. 181-200, 1998). Researchers are now applying knowledge gained during the development of inoculants that work well for grass silage to improve those for use in the ensilage of leguminous crops (28).
Near infrared spectroscopy (NIR) has previously been used to derive models for the study of forage composition (36, 37), sward quality (1, 44), and silage composition (42). Wachendorf et al. (44) applied NIR to predict the clover content in mixtures of red and white clover with grass, a value previously estimated by manual separation of the plant material or by visual assessment in the field. NIR covers the spectral region of wavenumbers from 10,000 to 4,000 cm−1. NIR region spectra are based on the overtone and combination-band absorption characteristics of CH, NH, and OH groups. By contrast, the mid-IR (MIR) region (wavenumbers from 4,000 to 400 cm−1) (37) is measured using Fourier transform infrared (FT-IR) spectroscopy and provides much greater chemical information content (17, 39) because it measures the fundamental vibration. In the region of wavenumbers between 4,000 and 1,500 cm−1 there is absorption by various stretching modes of functional groups of molecules, and the region of wavenumbers below 1,500 cm−1 is significant for deformation, bending, and ring vibrations and is frequently referred to as the fingerprint region of the spectrum (39). Reeves (36, 37) compared the use of NIR spectroscopy to that of MIR spectroscopy for the study of forage grasses and concluded that MIR performed at least as well as NIR for modeling grass composition. In a later paper, Reeves (35) reported the possibility of combining both spectral regions but showed that no significant advantage was gained by adding the NIR to the MIR spectra.
Current methods to study silage fermentation rely on the time-consuming measurement of various chemical and biological parameters, such as levels of pH, dry matter (DM), lactic acid concentration, volatile fatty acids, crude protein, ammonia, dry-matter digestibility, and viable microbial counts (6, 46, 48). These data, although informative, only provide selective information about the fermentation characteristics and a limited picture of the actual biochemical changes that occur during fermentation. The application of a model fermentation system and FT-IR spectroscopy provides a high-throughput technique for studying the “silage metabolome” in toto. The metabolome is defined as the total biochemical composition of a sample at a given time (30). Analysis by FT-IR spectroscopy also includes the measurement of proteins and nucleic acids which are not naturally included in the metabolome definition. FT-IR spectroscopy has previously been successfully applied to study bioprocesses (industrial fermentations) with respect to the detection of degenerate variants in solventogenic clostridia (40), the monitoring of α2 interferon production by Escherichia coli (27), and gibberellic acid production by the fungus Gibberella fujikuroi (26). These studies focused on analyzing the actual bacteria or monitoring the production of a specific metabolite. The approach taken here is more accurately defined as metabolic fingerprinting.
FT-IR spectra are complex multivariate datasets, which can be extremely difficult to analyze by simplistic visual methods; consequently, multivariate mathematical modeling (chemometrics) can be applied to identify patterns within sets of these data (41). Two clustering methods are used here, viz., principal component (PC) analysis (PCA) and discriminant function (DF) analysis (DFA). These methods are used to discriminate between red clover fermentations treated with different inoculants. Since it has been shown that there is discriminatory information within the FT-IR fingerprint, computational search methods (such as genetic algorithms [GA]) can be used to aid in the discovery of important biochemical features in these spectra. These chemometric approaches are described in more detail in the Materials and Methods section.
Here we show (using a model microtiter plate-based fermentation system coupled with FT-IR spectroscopy for high-throughput screening) how a metabolic fingerprinting approach can be applied to studies of model fermentation dynamics in red clover to identify the presence of an inoculant and to discriminate between the effects of different LAB inoculants over time. The results of three experiments studying grass and red clover juice fermentation in response to inoculation with a range of different LAB are presented.
MATERIALS AND METHODS
The model fermentation system.
A model system (devised on the basis of the use of 1-ml “microsilos”) was developed to permit high-throughput screening of the effect of different LAB inoculants on the fermentation of grass and red clover (9; H. E. Johnson, D. Broadhurst, G. W. Griffith, D. B. Kell, M. K. Theodorou, and R. J. Merry, Proc. 19th Gen. Meet. Eur. Grassland Federation, p. 206-208, 2002). The system uses deep 96-well microtiter plates, with each well constituting an individual fermentation combining herbage juice as the substrate and the inoculant in question. Herbage juice was extracted from freshly harvested grass and red clover cultivars with a silage press (Institute of Grassland and Environmental Research, Aberystwyth, United Kingdom). The juice was diluted twofold with sterile distilled H2O prior to loading the wells.
The LAB strains selected for use as inoculants are described in Table 2. These bacteria were grown in MRS broth (Oxoid Ltd, Wade Road, Basingstoke, Hampshire, United Kingdom) at 30°C for 20 h (as described by Winters et al.) (46). The cultures were diluted in sterilized distilled H2O prior to application to give a final concentration after inoculation of 106 CFU ml−1. Each well (1.2 ml in volume) in the deep-well microtiter plate represents a single fermentation. In the control samples, 1 ml of red clover juice was added to each well. In the inoculated samples, 0.99 ml red clover juice plus 10 μl of inoculant suspension was added to each well. The microtiter plates were then sealed with adhesive aluminum foil and incubated at 25°C. At each sampling time point, the pH was measured and the samples were stored in vials (Fisher Scientific Ltd.) at −80°C until further analysis.
TABLE 2.
The LAB strains selected for screening in the microsilo fermentation experiments
| Experiment | Lab straina | Strain isolated from: | Source (code)b |
|---|---|---|---|
| II | Lactobacillus plantarum (Lp01) | Aberelan herbage | IGER (EB2) |
| I + II | Lactobacillus plantarum (Lp02) | Perennial rye grass | IGER (621) |
| II | Lactococcus lactis | Milk powder | IGER (S614) |
| III | Lactobacillus mali | Cider apple juice | NCIMB (10560) |
| III | Lactobacillus delbrueckii subsp. lactis | Unknown (Starter culture for Swiss cheese) | NCIMB (8140) |
| III | Lactobacillus brevis | Silage | NCIMB (947) |
| III | Lactobacillus amylovorus | Cattle waste—corn fermentation | NCIMB (13276) |
All LAB strains were homofermentative (with the exception of L. brevis, which is heterofermentative).
The isolates were obtained from either the National Collections of Industrial and Marine Bacteria (NCIMB) culture collection (http://www.ukncc.co.uk/) or the culture collection at the Institute of Grassland and Environmental Research (IGER), Aberystwyth, United Kingdom.
FT-IR spectroscopy.
Samples were thawed at room temperature and vortexed prior to FT-IR analysis. A total of 5 μl of sample was loaded onto a 400-well bespoke aluminum plate. Each sample was loaded in triplicate (“machine replicates”). The plate was then dried at 50°C for 45 min. The aluminum plate was loaded onto a motorized xy stage of an adapted reflectance thin layer chromatography accessory connected to an IFS28 FT-IR spectrometer (Bruker Spectrospin, Coventry, United Kingdom) equipped with a mercury-cadmium-telluride detector cooled with liquid N2 (16). The spectra were collected over a wavenumber range of 4,000 to 600 cm−1. A total of 256 spectra were coadded and averaged per sample to improve the signal-to-noise ratio. Further instrument and methodological details are given in Winson et al. (45) and Goodacre et al. (15).
The spectrum for each sample contained 882 data points (wavenumbers ranging from 4,000 to 600 cm−1), each representing an absorbance value at a particular wavelength. These data were imported into Matlab software (version 6.2). Unless otherwise stated, all further analyses were performed using Matlab. The whole data set was used in the analysis after the characteristic CO2 peaks at 683 to 656 and 2,403 to 2,272 cm−1 were removed and replaced with a trend line (2).
Cluster analysis.
Grouping of analytical data is possible either by means of clustering methods or by projecting the high-dimensional data onto lower-dimensional space. There are many clustering methods available (12), but it has been shown for spectroscopic data that a multistage analysis method in the form of a projection algorithm (PCA) followed by a clustering algorithm (DFA) is robust and reliable (15, 43).
PCA is an unsupervised clustering method requiring no a priori knowledge of the data set under analysis and acts to reduce the dimensionality of multivariate data while preserving most of the variance within; hence, it is termed a data compression method (5, 21). In contrast to PCA, DFA is a supervised method. A priori information about the class membership of the samples is used to produce measures of within-group variance and between-group variance. These data are then used to define DFs that optimally separate the a priori classes (i.e., biological replicates rather than machine replicates). The functions can then be used as a coordinate system to visualize the DFA scores. The fact that replicates must be within the same class is used to define the classes accordingly (32).
In this study the number of PCs used by the DFA was optimized by cross-validation, which involves forming the model on a training data set and then projecting a previously unseen set of data (the test set) onto the model, as detailed previously (33). By adjusting the number of PCs used in the model and then employing cross-validation an optimal configuration could be found. Each FT-IR data set was divided by biological replicate into two groups (the training set and the test set) in a ratio of 2:1.
GA.
A GA is an optimization method based on the principles of Darwinian selection (14) in which, iteratively, a set of parameter values for a given problem evolves until an optimal, or near-optimal, solution is found.
Initially a random set of N objects, P1 = (p11, p12,…, p1N), each containing a string of binary digits representing the parameters of the problem to be optimized, is created. Each string is then tested against a function, f(pi), which returns a value defining the quality of the parameters represented by that string (referred to as its fitness value). Once all N fitness values have been assigned, a new set of binary strings is created, P2. For P2 to be made fitter than P1, principles analogous to genetic sexual and asexual reproduction within the initial set, P1, are applied (3, 13, 31, 34). The aim of this process is to create new strings which contain the best sections of two parent strings with high fitness values, thus potentially increasing the new string's fitness. At the same time the reproduction process also facilitates a certain amount of stochastic searching of parameter space for new parameter values by allowing not-so-fit parent strings to also reproduce and by including a very small probability of random mutation of each new binary string (by randomly changing binary values from 0 to 1 or from 1 to 0). The probability of a particular parent string being selected for reproduction is proportional to its fitness, so strings with a high fitness value have a greater chance of selection. The process of selection followed by reproduction followed by mutation is repeated until N new strings (i.e., a new set, P2, to replace the old, P1) are created. The fitness value is then evaluated for each of the new strings (p21, p22,…, p2N), and the whole process repeats itself. The algorithm continues until a predefined stopping criterion (such as arrival at a given optimal fitness value, evaluation of a certain number of parameter sets, or convergence of the top n% of the strings in a given set to similar intrinsic parameter values) is reached.
In this study we used the GA to determine the subset of n wavenumbers (taken from the data matrix) which, when applied to a discriminant multiple linear regression (D-MLR) model (4), optimally distinguished between two selected treatments. D-MLR GA analysis for all the possible combinations of the four treatments (i.e., six pairwise combinations) was performed in experiment 2 after 96 h of fermentation. All calculations were performed using in-house software written in C++ running in a Microsoft Windows NT operating system on an IBM-compatible personal computer. Full details of GA-MLR were given previously (4), and a previous application using this technique to discriminate between control and salt-treated tomato fruit was described previously by Johnson et al. (20). Optimization is achieved by minimizing the residual mean square error of prediction for RMSEPTEST, the internal validation set (given that it is less than the residual mean square error of prediction for RMSEPTRAIN, the training set).
In C code:
If (RMSEPTEST < RMSEPTRAIN)
{
fitness_function = 1/RMSEPTEST;
}
else
{
fitness_function = 1/(RMSEPTRAIN+1000);
}
The fitness rule described above restrains the GA from selecting subsets of variables whose MLR models over-train on the validation data set, thus producing a more robust model.
The GA uses two-point crossover with mutation (14), operating on a set of binary-encoded strings, with each string representing v candidate wavelengths (4); v can be set to any integer value (between 2 and the total number of wavelengths used) prior to the execution of any single GA run. The selection of parent strings for the next generation is carried out using a rank-based scheme (D. Whitley, Abstr. Proc. Third Int. Conf. Genet. Algorithms, p. 434-439, 1989). No two identical candidates were allowed in a given set of strings. A total of 50 independent GA runs was performed for each treatment combination. For a given GA run the optimization process was terminated when the top 10% of the stings in a given set were identical for over more than 20 generations.
Spectrum characterization was used to reduce the dimensionality of the GA search space by focusing on key spectral features as follows. Salient features (i.e., subsets of important variables) were found by initially locating local maxima, minima, and points of inflection along a spectrum representative of the total data set [in this case, the mean spectrum of f(v), the total data set, was used], and then points of maximum gradient either side of these stationary points were identified to fully define the shape of the representative spectrum [i.e., points on f(v) where 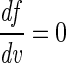 and
and 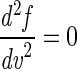 ]. The preprocessing step reduced the dimensionality of the data from 882 variables per sample to 61.
]. The preprocessing step reduced the dimensionality of the data from 882 variables per sample to 61.
RESULTS AND DISCUSSION
Experiment 1: a comparison between the ensiling characteristics of grass and red clover in microsilos.
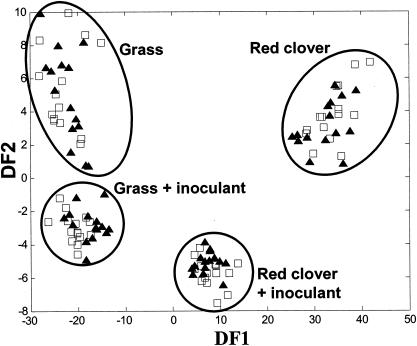
The feasibility of applying the model microsilo system followed by FT-IR spectroscopy for high-throughput screening of silage fermentations was first investigated through a comparison of grass and red clover herbage juices with and without the addition of the bacterial inoculant Lactobacillus plantarum (isolate Lp02) (described in Table 2) after incubation for 6 days at 25°C. Two-phase PC-DF analysis of the FT-IR spectral data enabled clear discrimination between the results seen with grass and red clovers with and without the addition of the inoculant (H. E. Johnson, D. Broadhurst, G. W. Griffith, D. B. Kell, M. K. Theodorou, and R. J. Merry, Abstr. Proc. XIIIth Int. Silage Conf., p. 378-380, 2002) (Fig. 1). The ability of PC-DF analysis to identify herbage type and the presence of an inoculant shows that the FT-IR spectral fingerprints contained important biochemical data characteristic of each type of fermentation.
FIG. 1.
Discrimination between grass and red clover herbage juices fermented for 6 days in the microsilo system with and without an L. plantarum inoculant (isolate Lp02). A cross-validated DFA plot (using 12 PCs accounting for 99.75% of the total variance within the data set) is shown. Both the training data (▴) and the test data (□) are represented. A clear discrimination can be seen between the four treatments (grass and red clover with and without an inoculant). This spectral heterogeneity was not reflected in the pH values measured. The circles around the data points are to improve clarity and are not of any mathematical significance.
Experiment 2: monitoring fermentation dynamics and discrimination between different inoculants in microsilos.
In this experiment, four different treatments were investigated, namely, red clover juice alone and inoculated with Lactococcus lactis, L. plantarum isolate Lp01, or L. plantarum isolate Lp02 (Table 2). A total of 30 replicate fermentations were prepared for each of the four treatments, giving a total of 120 samples. This enabled destructive sampling at eight time points, with three biological replicates for each treatment at each time. The time points selected were 12, 14, 16, 18, 20, 22, 24, 48, 72, and 96 h after inoculation.
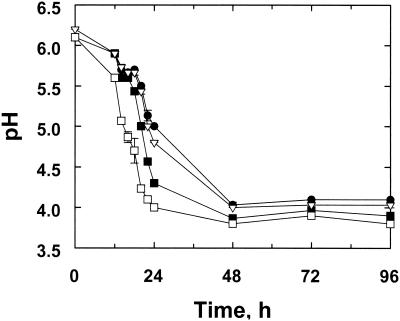
The separate addition of the three different bacterial inoculants to clover juice in microsilos caused a more rapid decrease in pH over a 96-h period compared to the results seen with an uninoculated control (Fig. 2). In terms of pH decline, the effect of inoculation with L. plantarum isolate Lp02 relative to that seen with the other inoculant treatments was most apparent after 12 to 18 h of fermentation, when the initial rate of pH decline, which is critical to the maximization of silage quality (47), was faster. L. plantarum isolate Lp01 also produced a lower pH relative to the results seen with the uninoculated control; however, this difference was not observed until 18 to 24 h of fermentation. Samples inoculated with either L. plantarum isolate consistently had a lower pH relative to the results seen with the other treatments. The addition of an L. lactis inoculant appeared to have little effect on the pH of the fermentation compared to the results seen with an uninoculated control. These data clearly illustrate the various effects of different bacterial inoculants on the fermentation of red clover juice.
FIG. 2.
Fermentation of red clover juice in the microsilo system, as indicated by pH measurement with respect to four different treatments: herbage juice only (control) (•), L. lactis (▿), L. plantarum isolate Lp01 (▪), and L. plantarum isolate Lp02 □). The data shown represent mean values and standard deviations of experiments (n = 3).
A biochemical fingerprint was obtained using FT-IR spectroscopy for samples at each time point. The aim was to use a combination of the cluster analysis techniques (PCA and DFA) to study the effect of different inoculants on the fermentation dynamics of red clover juice. PCA was first applied to reduce the dimensionality of the data set. The DFA model was then formed using a selected number of PCs, the number of which was optimized by cross-validation as described in Materials and Methods. The figures presented here all show the resultant PC-DFA models, with the optimized number of PCs used indicated in the legends.
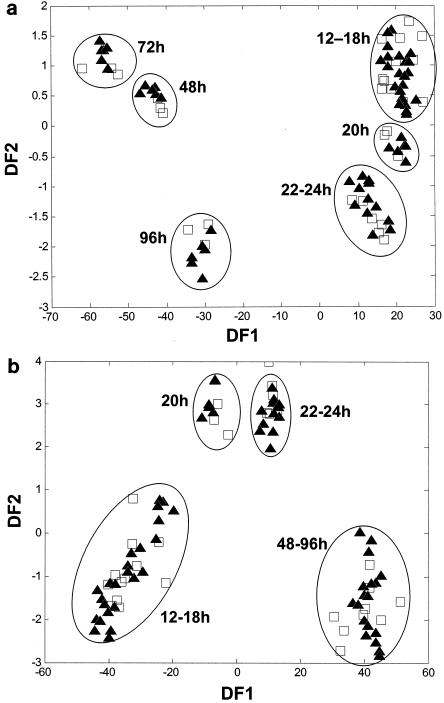
This two-phase PC-DF analysis was performed individually for each treatment over the 96-h incubation period. The aim was to study the discrimination between samples over time and to enable comparisons to be made between the fermentation dynamics results for each treatment. For all the treatments the results seen with the replicate samples in the DFA model grouped together, with discrete clusters corresponding to different time intervals during the incubation being observed; however, the time intervals to which these clusters corresponded differed between treatments. This is illustrated in Fig. 3a and 3b, which show the DFA models for the control and Lp02 fermentations, respectively. These plots were selected as they show very distinct fermentation patterns. The DFA model for the Lp01 fermentation was very similar to that observed with Lp02 (Fig. 3b), and treatment with L. lactis produced fermentation dynamics comparable to that of the control.
FIG. 3.
The dynamics of fermentation of red clover juice fermented at 25°C over a 96-h period without (control) (a) and with (b) an L. plantarum Lp02 inoculant; samples were taken for analysis at 12, 14, 16, 18, 20, 22, 24, 48, 72, and 96 h after inoculation. Two cross-validated DFA models (each using 20 PCs) accounted for 99.99% of the total variance within the data set and used a priori knowledge of class structure. Numbers on the plot indicate fermentation times in hours. Solid triangles (▴) represent training data; open squares (□) represent test data. The circles around the data points are to improve clarity and are not of any mathematical significance.
In Fig. 3a there are three closely associated groups shown, corresponding to samples taken at 12 to 18 h, 20 h, and 22 to 24 h of incubation. A similar trend of three discrete clusters was observed with the addition of the L. plantarum Lp02 inoculant (Fig. 3b). Although clustering was tighter in the control treatment, in both data sets there were no discrete clusters over the first 12 to 18 h of incubation (Fig. 3). In contrast to the control treatment (Fig. 3a), however, clusters in the inoculant treatment showed greater separation at 20 h and 22 to 24 h of incubation (Fig. 3b), indicating marked differences in the biochemistry characteristics of the corresponding samples over this time period. These marked differences are reflected in the pH measurements taken at sampling; fermentations inoculated with L. plantarum Lp02 showed the most rapid initial change relative to the results seen with uninoculated control samples (as depicted by the uppermost and lowermost curves in Fig. 2). Further differences were observed as the fermentation progressed (Fig. 3). In the results seen with the control fermentation (Fig. 3a) there is clear discrimination between samples fermented for 48, 72, and 96 h. In contrast, the results for Lp02-inoculated samples taken at these time points cluster together and no such discrimination is observed (Fig. 3b).
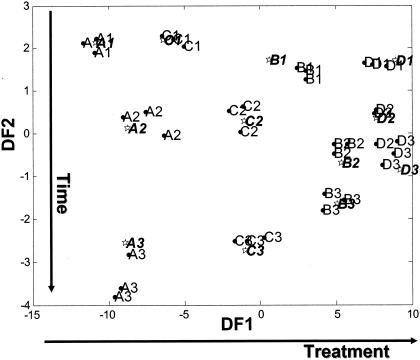
The data presented in Fig. 3 are representative of the data collected for all four treatments and show that it was possible to discriminate between different time points within one treatment. It was also possible to discriminate between the four different treatments at a specific time point (for example, after 96 h of incubation), as shown in Fig. 4. The different inoculant treatments are separated by DF 1 (DF1) and the incubation time on DF2. Figure 2 shows clear differences between the four treatments with respect to pH over the first 24 h of incubation. Beyond this time point, the pH values differed by only 0.3 pH units but (despite this small difference) clear discrimination between the treatment results can be seen (Fig. 4).
FIG. 4.
Discrimination (with respect to inoculants and time points) between the results seen with four fermentations of red clover juice. The results of cross-validated DFA using 15 PCs accounting for 99.97% total variance and a priori knowledge of the sample class structure, for which A (control), B (L. plantarum Lp01), C (L. lactis), and D (L. plantarum Lp02) represent the results seen after fermentation for 48 h (A1, B1, C1, and D1), 72 h (A2, B2, C2, and D2), and 96 h (A3, B3, C3, and D3); circles and lightface characters indicate training data, and stars and boldface characters indicate test data. The points shown represent the mean DFA scores calculated from three machine replicates taken for each sample.
Experiment 3: discrimination of homo- and heterofermentative LAB inoculants.
All plant material contains epiphytic LAB populations with a predominance of heterofermentative species and hence undergoes mixed-acid fermentation under ensilage conditions (7). In comparison to homofermentative species (and due to the pK of lactic acid), heterolactic species are less effective at reducing pH in the silo; hence, the quality of the resultant silage is very much dependent on the initial bacterial population (18; Woolford, Abstr. Proc. Alltech. 14th Annu. Symp., p. 181-200, 1998). In the previous experiments homolactic-fermenting LAB were used (producing two lactic acid molecules from one molecule of glucose, with a net gain of 2 ATP) (25). Here we wanted to investigate whether FT-IR could be used to identify heterolactic bacterial fermentation in a range of homolactic species. Heterofermentative LAB produce lactic acid, ethanol, and CO2 (rather than lactic acid alone) as well as smaller amounts of acetic acid, formic acid, and glycerol, during fermentation.
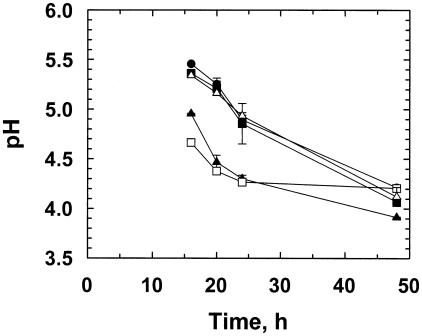
This final experiment, comparing four different LAB strains (three homofermentative strains and one heterofermentative strain [Lactobacillus brevis 947]), as detailed in Table 2, followed the same methodology as used previously. Although some of these LAB strains are not typically used as silage inoculants, all strains have been identified in lactic-acidic-type fermentations (Table 2) (http://www.ukncc.co.uk/). At inoculation, the pH of the diluted red clover juice was 5.6; addition of 10 μl of inoculant to 990 μl of juice had no marked effect on this starting pH. The microsilos were incubated and sampled in triplicate at 16, 20, 24, and 48 h after incubation. The heterofermentative L. brevis strain rapidly decreased the pH over the first 16 h to a value of (on average) 4.7 compared to 5.6 for the control. A similar pH decline was shown after inoculation with Lactobacillus mali, with a mean pH value of 4.9 after 16 h (Fig. 5). After 24 h of incubation, cultures inoculated with L. brevis and L. mali both had a pH value of 4.3 compared to 4.9 in microsilos inoculated with Lactobacillus amylovorus, Lactobacillus delbrueckii, and the control. Over the next 24 h, however, the pH of all treatments (with the exception of that of L. brevis, which reached a plateau at 24 h) decreased steadily, reaching a range of pH 4.2 to 3.9 at 48 h.
FIG. 5.
The fermentation of red clover juice over 48 h of incubation measured by pH with respect to five different treatments: herbage juice only (•), Lactobacillus mali 10560 (▴), L. delbrueckii subsp. lactis 8140 (▪), L. brevis 947 (□), and L. amylovorus 13276 (▵). At h 0, the red clover juice had a pH value of 5.6. Data shown are mean and standard deviation values for the experiments performed (n = 3).
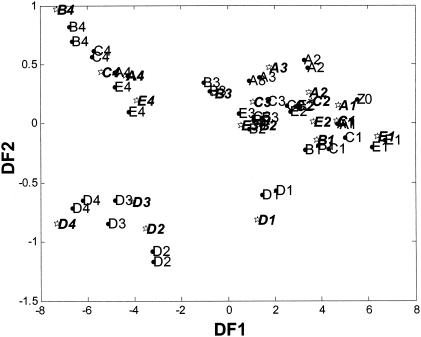
The DFA model (Fig. 6) shows the clear discrimination between the two types of fermentation over time. It can be seen that L. brevis clusters in a different area of the DFA model than the three homofermentative inoculants and the control. After 48 h the pH values measured for all treatments were very similar, ranging from 4.3 (L. brevis and the control) to 3.9 (L. mali). However, despite these similarities there is still clear discrimination between the two types of fermentation. These results (although determined with only a few LAB inoculants) highlight the potential of using FT-IR for the rapid identification and detection of different fermentation types.
FIG. 6.
Cross-validated DFA model showing the fermentation of red clover over a 48-h period in response to different LAB inoculants. The DFA used 25 PCs, which accounted for 99.98% of the total variance within the data set, and knowledge of class structure, where A (control), B (L. mali), C (L. delbrueckii), D (L. brevis), and E (L. amylovorus) represent the results seen after incubation for 0 h (A0, B0, C0, D0, and E0), 16 h (A1, B1, C1, D1, and E1), 20 h (A2, B2, C2, D2, and E2), 24 h (A3, B3, C3, D3, and E3), and 48 h (A4, B4, C4, D4, and E4); circles and lightface characters indicate training data, and stars and boldface characters indicate test data. The points shown are the mean DFA scores calculated from three machine replicates taken for each sample.
Application of GA to identify potentially discriminatory regions in the FT-IR spectra.
GA are members of a class of optimization methods which permit the identification of potentially key regions for discrimination between different treatment pairs (4). Here we have applied the technique to try to identify the key spectral regions for discrimination between all treatment combinations (four treatments [A, B, C, and D]; six possible pair combinations) at 96 h, as described for experiment 2. Successful discrimination for a given variable subset was defined as being achieved when both the RMSEPTRAIN percentage and the RMSEPTEST percentage were less than 20% for the D-MLR model in question.
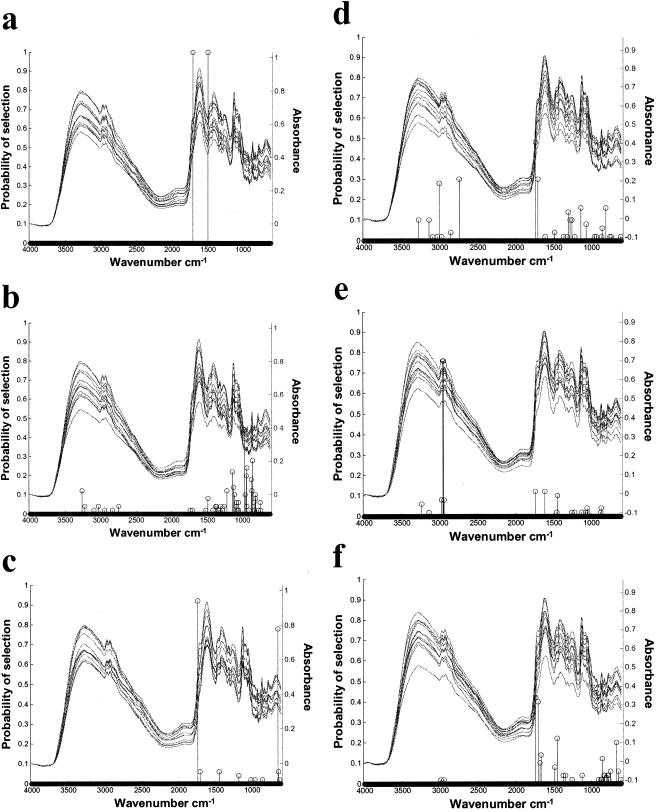
Due to the stochastic nature of GA, each analysis consisted of 50 independent runs of identically set-up D-MLR GA. The optimal model from each of the 50 GA runs was recorded. Then the numbers of times each variable (spectral feature) was used in each of these models were summed. These data were plotted in the form of a frequency stem plot overlaid on a plot of the total FT-IR input data set. The results obtained for each pairwise experiment are shown in Fig. 7. The GA was able to discriminate all treatments by treatment type for both the training and the test data with 100% accuracy. The results obtained are shown in Fig. 7.
FIG. 7.
Identification (using GA) of discriminatory regions in the FT-IR spectra for discrimination between treatments after 96 h of microsilo fermentation. All pairwise treatment combinations were analyzed; with four treatments, this resulted in six possible combinations. Due to the stochastic nature of GA, each analysis consisted of 50 independent runs of identically set-up D-MLR GA. Successful discrimination for a given variable subset was defined as being achieved when both the RMSEPTRAIN percentage and the RMSEPTEST percentage were less than 20% for the D-MLR model in question. The optimal model from each of the 50 GA runs was recorded. Then the numbers of times each variable (spectral feature) was used in each of these models were summed. These data were plotted in the form of a frequency stem plot overlaid on a plot of the total FT-IR input data set. The results of the six pairwise GA results are shown in panels a to f: (a) control and L. plantarum Lp01; (b) control and L. lactis; (c) control and L. plantarum Lp02; (d) L. plantarum Lp01 and L. lactis; (e) L. plantarum Lp01 and L. plantarum Lp02; (f) L. lactis and L. plantarum Lp02.
In all models a maximum of five and a minimum of two variables out of 61 possible spectral features were needed to discriminate between treatment pairs. Figure 7a shows that only two wavenumbers (at 1,706 and 1,500 cm−1) were consistently chosen to discriminate between the control and Lp01 samples. These regions correspond to the amide I region, a strong C = O stretch at 1,706 cm−1 (identified using IR mentor software; Bio-Rad Laboratories), and to the amide II region (around 1,550 cm−1) (11). Variables within the amide I region, which ranges from 1,740 to 1,670 cm−1, were also selected for the discrimination between control and isolate Lp02 (Fig. 7c), isolate Lp01 and L. lactis (Fig. 7d), and isolate Lp02 and L. lactis (Fig. 7f). Figure 7c, 7d, and 7f show that at least one variable within the amide I region was used by the GA in 49, 48, and 50 out of 50 GA runs, respectively.
In contrast to these results, the GA consistently chose a different region at 2,950 to 2,965 cm−1 to discriminate the fermentations produced with the two L. plantarum strains (Fig. 7e). This region is termed the fatty acid region, corresponding to strong C-H vibrations. Although classification between the other pairs of treatment combinations was 100% accurate, many different combinations of variables were used (with a maximum of 5 in any model and 50 models produced in total). Figure 7b is distinct again in that the amide I and amide II regions were not selected by the GA for the classification fermentations with the control and with L. lactis; instead, variables were selected in the low-frequency region ranging from 974 to 931 cm−1 (on the edge of an region attributed to vibrations corresponding to polysaccharides). The GA did not select the same region to discriminate between all the additive combinations, indicating differences between samples with respect to their biochemical fingerprints. Initial investigations indicated that amide I and amide II were the regions of the FT-IR spectra most frequently selected for discrimination between treatments, potentially reflecting changes in proteins and free amino acids occurring in the fermentations (8). During ensiling, extensive proteolysis occurs; the rapid decrease in pH during the first few days of fermentation through the addition of homofermentative LAB inhibits the protease activity, thus reducing proteolysis (19, 25). The application of both L. plantarum strains resulted in a marked decrease in pH during the first 24 h of the fermentation compared to the results seen with the control and L. lactis (Fig. 2). When the GA was used to discriminate either of the L. plantarum fermentations from the results seen with control or L. lactis treatments, the amide region of the FT-IR spectra was consistently selected. Studies by Heron et al. (19) and Davies et al. (8) both investigated the effect of application of a L. plantarum-containing inoculant on the degree of proteolysis in ryegrass silage. The data from these independent experiments led to the conclusion that the addition of such an inoculant, which rapidly reduced the pH of the fermentation through the efficient production of lactic acid, reduced the extent of proteolysis. These results confirmed the contention by Muck (29) that a combination of good silo preparation and rapid pH decrease reduces proteolysis. From this data, therefore, we can hypothesize that the GA-selected variables are related to the degree of proteolysis occurring during ensilage, a factor known to play a role in determining the nutritional value of silage to ruminants (28).
The data presented above demonstrate the potential of using a combination of microsilos and FT-IR spectroscopy for the high-throughput screening of different LAB isolates as potential silage inoculants. They also provide a means of modeling fermentation dynamics and of detecting inoculant and fermentation types. Here we used simple clustering techniques and GA to deconvolve the multivariate spectral data, which enabled clear discriminations between the effects of treatments and time and allowed us to postulate the biochemical basis for the effects. This is an inductive experimental approach in which hypotheses are derived as the output rather than constituting the input, providing a direction for future research (23, 24).
Acknowledgments
We thank the BBSRC (grant 2D/13533) for financial support.
We acknowledge Roy Goodacre and members of the Molecular Spectroscopic and Systematics Group for their technical advice. We are also grateful to Eleanor Blakewell (IGER) for her skilled technical assistance with the LAB cultures.
REFERENCES
- 1.Alomar, D., R. Montero, and R. Fuchslocher. 1999. Effect of freezing and grinding method on near-infrared reflectance (NIR) spectra variation and chemical composition of fresh silage. Anim. Feed Sci. 78:57-63. [Google Scholar]
- 2.Alsberg, B. K., W. G. Wade, and R. Goodacre. 1998. Chemometric analysis of diffuse reflectance-absorbance Fourier transform infrared spectra using rule induction methods: application to the classification of Eubacterium species. Appl. Spectrosc. 52:823-832. [Google Scholar]
- 3.Back, T., D. B. Fogel, and Z. Michalewicz. 1997. Handbook of evolutionary computation. IOP Publishing—Oxford University Press, Oxford, United Kingdom.
- 4.Broadhurst, D., R. Goodacre, A. Jones, J. J. Rowland, and D. B. Kell. 1997. Genetic algorithms as a method for variable selection in multiple linear regression and partial least squares regression, with applications to pyrolysis mass spectrometry. Anal. Chim. Acta 348:71-86. [Google Scholar]
- 5.Causton, D. R. 1987. A biologist's advanced mathematics. Allen & Unwin, London, United Kingdom.
- 6.Cussen, R. F., R. J. Merry, A. P. Williams, and J. K. S. Tweed. 1995. The effect of additives on the ensilage of forage of differing perennial ryegrass and white clover content. Grass Forage Sci. 50:249-258. [Google Scholar]
- 7.Daeschel, M. A., R. E. Andersson, and H. P. Fleming. 1987. Microbial ecology of fermenting plant materials. FEMS Microbiol. Rev. 46:357-367. [Google Scholar]
- 8.Davies, D. R., R. J. Merry, A. P. Williams, E. L. Bakewell, D. Leemans, and J. K. S. Tweed. 1998. Proteolysis during ensilage of forages varying in soluble sugar content. J. Dairy Sci. 81:444-453. [DOI] [PubMed] [Google Scholar]
- 9.Davies, Z. S. 2000. Manipulation of silage fermentation using biological additives and genetic algorithms. Ph.D. thesis. University of Wales, Aberystwyth.
- 10.Done, D. L. 1986. Silage inoculants—a review of experimental work. Res. Dev. Agric. 3:83-87. [Google Scholar]
- 11.Ellis, D. I., D. Broadhurst, D. B. Kell, J. J. Rowland, and R. Goodacre. 2002. Rapid and quantitative detection of the microbial spoilage of meat by Fourier transform infrared spectroscopy and machine learning. Appl. Environ. Microbiol. 68:2822-2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Everitt, B. S. 1993. Cluster analysis. Edward Arnold, London, United Kingdom.
- 13.Goldberg, D. E. 2002. The design of innovation: lessons from and for competent genetic algorithms. Kluwer Academic Publishers, Boston, Mass.
- 14.Goldberg, D. E. 1989. Genetic algorithms in search, optimization and machine learning. Addison-Wesley, Reading, Mass.
- 15.Goodacre, R., E. M. Timmins, R. Burton, N. Kaderbhai, A. M. Woodward, D. B. Kell, and P. J. Rooney. 1998. Rapid identification of urinary tract infection bacteria using hyperspectral whole-organism fingerprinting and artificial neural networks. Microbiology (United Kingdom) 144:1157-1170. [DOI] [PubMed] [Google Scholar]
- 16.Goodacre, R., E. M. Timmins, P. J. Rooney, J. J. Rowland, and D. B. Kell. 1996. Rapid identification of Streptococcus and Enterococcus species using diffuse reflectance-absorbance Fourier transform infrared spectroscopy and artificial neural networks. FEMS Microbiol. Lett. 140:233-239. [DOI] [PubMed] [Google Scholar]
- 17.Griffiths, P. R., and J. A. de Haseth. 1986. Fourier transform infrared spectrometry. John Wiley & Sons, New York, N.Y.
- 18.Henderson, N. 1993. Silage additives. Anim. Feed Sci. Technol. 45:35-56. [Google Scholar]
- 19.Heron, S. J. E., R. A. Edwards, and P. McDonald. 1988. The effects of inoculation, addition of glucose and mincing on fermentation and proteolysis in ryegrass ensiled in laboratory silos. Anim. Feed Sci. Technol. 19:85-96. [Google Scholar]
- 20.Johnson, H. E., D. Broadhurst, R. Goodacre, and A. R. Smith. 2003. Metabolic fingerprinting of salt-stressed tomatoes. Phytochemistry 62:919-928. [DOI] [PubMed] [Google Scholar]
- 21.Jolliffe, I. T. 1986. Principal component analysis. Springer, New York, N.Y.
- 22.Jones, R. 1995. Role of biological additives in crop conservation, p. 465-482. In T. P. Lyons and K. A. Jacques (ed.), Proceedings of Alltech’s 11th Annual Symposium. Nottingham University Press, Nottingham, United Kingdom.
- 23.Kell, D. B. 2002. Genotype-phenotype mapping: genes as computer programs. Trends Genet. 18:555-559. [DOI] [PubMed] [Google Scholar]
- 24.Kell, D. B., and S. G. Oliver. Here is the evidence, now what is the hypothesis? The complementary roles of inductive and hypothesis-driven science in the post-genomic era. BioEssays, in press. [DOI] [PubMed]
- 25.McDonald, P., N. Henderson, and S. Heron. 1991. The biochemistry of silage, 2nd ed. Chalcombe Publications, Marlow, Buckinghamshire, United Kingdom.
- 26.McGovern, A. C., D. Broadhurst, J. Taylor, N. Kaderbhai, M. K. Winson, D. A. Small, J. J. Rowland, D. B. Kell, and R. Goodacre. 2002. Monitoring of complex industrial bioprocesses for metabolite concentrations using modern spectroscopies and machine learning: application to gibberellic acid production. Biotechnol. Bioeng. 78:527-538. [DOI] [PubMed] [Google Scholar]
- 27.McGovern, A. C., R. Ernill, B. V. Kara, D. B. Kell, and R. Goodacre. 1999. Rapid analysis of the expression of heterogous proteins in Escherichia coli using pyrolysis mass spectrometry and Fourier transform infrared spectroscopy with chemometrics: application to α2-interferon production. J. Biotechnol. 72:157-167. [DOI] [PubMed] [Google Scholar]
- 28.Merry, R. J., R. Jones, and M. K. Theodorou. 2001. Alternative forages—back to the future. Biologist 48:30-34. [PubMed] [Google Scholar]
- 29.Muck, R. E. 1988. Factors influencing silage quality and their implications for management. J. Dairy Sci. 71:2992-3002. [Google Scholar]
- 30.Oliver, S. G., M. K. Winson, D. B. Kell, and F. Baganaz. 1998. Systemic functional analysis of the yeast genome. Trends Biotechnol. 16:373-378. [DOI] [PubMed] [Google Scholar]
- 31.Pham, D. T., and D. Karaboga. 2000. Intelligent optimisation techniques: genetic algorithms, Tabu search, simulated annealing and neural networks. Springer-Verlag, Berlin, Germany.
- 32.Raamsdonk, L. M., B. Teusink, D. Broadhurst, N. Zhang, A. Hayes, M. Walsh, J. A. Berden, K. M. Brindle, D. B. Kell, J. J. Rowland, H. V. Westerhoff, K. van Dam, and S. G. Oliver. 2001. A functional genomics strategy that uses metabolome data to reveal the phenotype of silent mutations. Nat. Biotechnol. 19:45-50. [DOI] [PubMed] [Google Scholar]
- 33.Radovic, B. S., R. Goodacre, and E. Anklam. 2001. Contribution of pyrolysis mass spectrometry (Py-MS) to authenticity testing of honey. J. Anal. Appl. Pyrolysis 60:79-87. [Google Scholar]
- 34.Reeves, C. R. 2002. Genetic algorithms—principles and perspectives: a guide to GA theory. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 35.Reeves, J. B. 1997. Concatenation of near- and mid-infrared spectra to improve calibrations for determining forage composition. J. Agric. Food Chem. 45:1711-1714. [Google Scholar]
- 36.Reeves, J. B. 1994. Near- versus mid-infrared diffuse reflectance spectroscopy for the quantitative determination of the composition of forages and by-products. J. Near Infrared Spectrosc. 2:49-57. [Google Scholar]
- 37.Reeves, J. B. 1994. Near versus mid-infrared spectroscopy for quantitative analysis of chlorite treated forages and by-products. Near Infrared Spectrosc. 2:153-162. [Google Scholar]
- 38.Salawu, M. B., E. H. Warren, and A. T. Adesogan. 2001. Fermentation characteristics, aerobic stability and ruminal degradation of ensiled pea/wheat bi-crop forages treated with two microbial inoculants, formic acid or quebracho tannins. J. Sci. Food Agric. 81:1263-1268. [Google Scholar]
- 39.Schmitt, J., and H. Flemming. 1998. FTIR-spectroscopy in microbial and material analysis. Intl. Biodeterior. Biodegrad. 41:1-11. [Google Scholar]
- 40.Schuster, K. C., R. Goodacre, J. R. Grapes, and M. Young. 2001. Degeneration of solventogenic Clostridium strains monitored by Fourier transform infrared spectroscopy of bacterial cells. J. Ind. Microbiol. Biotechnol. 27:314-321. [DOI] [PubMed] [Google Scholar]
- 41.Shaw, A. D., M. K. Winson, A. M. Woodward, A. C. McGovern, H. M. Davey, N. Kaderbhai, D. Broadhurst, R. J. Gilbert, J. Taylor, E. M. Timmins, R. Goodacre, and D. B. Kell. 1999. Rapid analysis of high-dimensional bioprocesses using multivariate spectroscopies and advanced chemometrics. Adv. Biochem. Eng. Biotechnol. 66:84-113. [DOI] [PubMed] [Google Scholar]
- 42.Steen, R., W. J., F. J. Gordon, L. E. R. Dawson, R. S. Park, C. S. Mayne, R. E. Agnew, D. J. Kilpatrick, and M. G. Porter. 1998. Factors affecting the intake of grass silage by cattle and prediction of silage intake. Anim. Sci. 66:115-127. [Google Scholar]
- 43.Timmins, E. M., S. A. Howell, B. K. Alsberg, W. C. Noble, and R. Goodacre. 1998. Rapid differentiation of closely related Candida species and strains by pyrolysis-mass spectrometry and Fourier transform-infrared spectroscopy. J. Clin. Microbiol. 36:367-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wachendorf, M., B. Ingwersen, and F. Taube. 1999. Prediction of the clover content of red clover- and white clover-grass mixtures by near-infrared reflectance spectroscopy. Grass Forage Sci. 54:87-90. [Google Scholar]
- 45.Winson, M. K., R. Goodacre, É. M. Timmins, A. Jones, B. K. Alsberg, A. M. Woodward, J. J. Rowland, and D. B. Kell. 1997. Diffuse reflectance absorbance spectroscopy taking in chemometrics (DRASTIC). A hyperspectral FT-IR-based approach to rapid screening for metabolite overproduction. Anal. Chim. Acta 348:273-282. [Google Scholar]
- 46.Winters, A. L., R. J. Merry, M. Muller, D. R. Davies, G. Pahlow, and T. Muller. 1998. Degradation of fructans by epiphytic and inoculant lactic acid bacteria during ensilage of grass. J. Appl. Microbiol. 84:304-312. [Google Scholar]
- 47.Woolford, M. K. 1984. The Silage Fermentation, vol. 14. Marcel Dekker Inc., New York, N.Y.
- 48.Zhu, W.-Y., M. K. Theodorou, A. C. Longland, B. B. Nielsen, J. Dijkstra, and A. P. J. Trinci. 1996. Growth and survival of anaerobic fungi in batch and continuous-flow cultures. Anaerobe 2:29-37. [Google Scholar]