Abstract
Growing axons encounter numerous developmental signals to which they must promptly respond in order to properly form complex neural circuitry. In the axons, these signals are often transduced into a local increase or decrease in protein levels. Contrary to the traditional view that the cell bodies are the exclusive source of axonal proteins, it is becoming increasingly clear not only that de novo protein synthesis takes place in axons, but also that it is required for the axons to respond to certain signals. Here we review the current knowledge of local mRNA translation in developing neurons with a special focus on protein synthesis occurring in axons and growth cones.
INTRODUCTION
Localizing a gene product to a precise subcellular domain at a given time is an incredible feat every cell within developing organisms must perform. This is even more challenging to highly polarized cells like neurons. Developing neurons extend long processes such as axons and dendrites, which receive numerous signals as they grow. The signals received at the distal ends of the neurites are locally processed, and can result in subtle changes in protein localization, which are used as a mode of signal transduction. Accumulating evidence suggests that local mRNA translation is one of the key mechanisms by which precise temporal and spatial control of protein localization is achieved in neurons. Indeed, local mRNA translation appears to play essential roles in many aspects of neuronal development including cell survival, axon guidance, and synaptogenesis.
Based on the early electron microscopy (EM) studies of mature axons1 in which no clear evidence of protein synthesis machinery was found, it had been traditionally believed that the cell body is the exclusive source of axonal proteins. However, a simple calculation reveals an apparent paradox in this model. The rate of the microtubule-based slow axonal transport, which accounts for the transport of cytoskeletal proteins, does not exceed 3 mm/day,2 and an axon of a mammal can be very long—in the case of blue whales, as long as 50 m! In this case, non-stop travel to the end of an axon would take almost 40 years and not many proteins would survive this long trip.3 This extreme example argues for the need of local protein synthesis in axons. In fact, there have been a number of overlooked EM studies in which mRNA translational machinery was found in axons of mature as well as developing neurons.4-6 Furthermore, recent studies have revealed that axonal protein synthesis is upregulated upon nerve injury and may be required for the regeneration of injured axons.7,8 In fact, several studies have shown that active protein synthesis occurs in the axons which were previously shown by EM to possess no translational machinery including mature dorsal root ganglion neurons9 and mature olfactory sensory neurons,10 suggesting that negative EM data should be interpreted with caution.11,12 However, axonal mRNA translation seems to be a more prevalent phenomenon in developing neurons than in mature neurons. Many groups have reported that local protein synthesis is required for axons to properly respond to many developmental signals they receive.13 Here we review the current understanding on the functions and mechanisms of local mRNA translation in developing neurons. Local mRNA translation in dendrites has been covered by many excellent reviews,14 and we focus on the local protein synthesis in growing axons of the developing nervous system.
FUNCTIONS OF LOCAL mRNA TRANSLATION
Several studies using invertebrates and vertebrates have identified a number of axonally localized mRNAs. Considering cellular needs of growing axons, the components of cytoskeleton would be the most likely proteins to be locally synthesized. Indeed, mRNAs encoding cytoskeletal proteins were among the first to be identified.15-17 Likewise mRNAs encoding proteins associated with presynaptic differentiation and lipid biogenesis are also found in growing axons (K. Zivraj and C.E. Holt, personal communication).18 However, these classes of mRNAs are not the only RNAs found in axons. Interestingly, quantitative analyses of mRNAs isolated from cell bodies and axons suggest that they are not even among the most abundant RNAs present in axons.18 mRNAs encoding proteins with diverse functions exist in axons and these include proteins unlikely to be of local use in axons such as transcription factors.19 Here we review some of the axonally localized mRNAs identified so far (Tables 1 and 2).
TABLE 1.
Axonal mRNAs
| Axonal mRNA | Present | RBPs | Function | Cis-element | Regulators | References |
|---|---|---|---|---|---|---|
| Cytoskeletal | ||||||
| β-actin | Squid giant axon | 20 | ||||
| SCG neuron, cortical neuron |
21 | |||||
| Cortical neuron; RGC | ZBP | Axon guidance (GC turning) |
3′UTR | Netrin-1, BDNF, NT-3, cAMP |
22-25 | |
| RhoA | DRG neuron | Axon guidance (GC collapse) |
3′UTR | Sema3A | 26 | |
| Cofilin | SCG neuron | Axon guidance (GC collapse) |
Slit2 | 27 | ||
| ADF | SCG neuron | 28 | ||||
| β-thymosin | Snail CNS neuron | Axon outgrowth | 29 | |||
| α-tubulin | Snail sensory neuron | Synapse formation | 30 | |||
| β-tubulin | Squid giant axon | 20 | ||||
| SCG neuron | 31 | |||||
| MAP-H1 | Squidgiant axon | 32 | ||||
| MAP-tau | Cortical neuron | 33 | ||||
| Neuronal cell line P19 |
HuD | 3′UTR | 34 | |||
| Kinsein | Squid giant axon | 20 | ||||
| Neurofilament | Squid CNS | 35 | ||||
| NF-M | Goldfish Mauthner axon |
36 | ||||
| NF-L, -M, -H | Sciatic nerve | Nerve injury | 37 | |||
| Axon terminal | ||||||
| CDCH | Snail CNS neuron | 38 | ||||
| ELH | Snail CNS neuron | 39 | ||||
| Sensorin A | Snail sensory neuron | 40, 41 | ||||
| Neuropeptide | Snail CNS neuron | 42, 43 | ||||
| Syntaxin | Snail sensory neuron | 44 | ||||
| CGRP | Olfactory sensory neuron |
45 | ||||
| Vasopressin | Hypothalamus | 46 | ||||
| Olfactory marker protein | Sensory neuron | 47, 48 | ||||
| Oxytocin | Hypothalamus | 49 | ||||
| Prodynorphin | Hypothalamus | 50 | ||||
| TRPV1 | Sensory neuron | 51 | ||||
| EphA2 | Commisural neuron | Midline crossing | 3′UTR | 52 | ||
| EphB2 | RGC | 52 | ||||
| NCAM | RGC | 52 | ||||
| Kor | DRG neuron | 5′UTR | Depolarization | 53, 54 | ||
| Sensorin | Aplysia sensory neuron |
Synapse formation | 55 | |||
| Odorant receptors | Olfactory sensory neuron |
10 | ||||
| Nuclear | ||||||
| CREB | DRG neuron | Cell survival | NGF | 19 | ||
| CEBP-1 | C. elegans neuron | DLK-1 | Synapse formation; axon regeneration |
3′UTR | 56 | |
| Importin-β | DRG neuron | Repair | Axotomy | 9 | ||
| Signaling | ||||||
| CamKII-α | Hippocampal neuron | 57 | ||||
| β-catenin | Hippocampal neuron | CPEB1 | NT-3 | 58 | ||
| IMPA-a | SCG neuron | 3′UTR | NGF | 18 | ||
| Hsp70 | Squid photoreceptor | 59 | ||||
| Mitochondrial | ||||||
| Enolase | Squid giant axon | 32 | ||||
| Cox17 | Squid giant axon | 20 | ||||
| Protein synthesis | ||||||
| eEF-α | Snail sensory neuron | 60 | ||||
| Ribosomal proteins |
SCG neuron | 18 | ||||
| Ribosomal RNAs | SCG neuron | 18 | ||||
| Others | ||||||
| L7/pcp2 | Cerebellar Purkinje cell | 61 |
RBP, RNA binding protein; ADF, actin depolymerizing factor; CDCH, caudodorsal cell hormone; ELH, egg-laying hormone; CGRP, calcitonin gene-related peptide; TRPV1, transient receptor potential channel V1; kor, kappa opioid receptor; CREB, CRE-binding protein; CEBP-1, CAAT enhancer binding protein-1; CamKII, calmodulin-dependent kinase II; eEF, eukaryotic elongation factors; RGC, retinal ganglion cells; SCG, superior cervical ganglion; UTR, untranslated region; NGF, nerve growth factor; BDNF, brain-derived neurotrophic factor; HuD, Hu antigen D; MAP, microtubule-associated protein; CNS, central nervous system; NCAM, neural cell adhesion molecule; DRG, dorsal root ganglion; cAMP, cyclic adenosine monophosphate; GC, growth cone.
TABLE 2.
Signals regulating axonal mRNA translation
| Signals | References | |||
|---|---|---|---|---|
| Netrin-1 | induces PS | of | β-actin, par3 | 24, 25 |
| via | PI3K, Erk1/2, p38, mTOR | 64 | ||
| requires PS | for | GC turning and collapse; axon outgrowth | 64, 67 | |
| Sema3A | induces PS | of | RhoA | 26 |
| requires PS | for | GC collapse | 26, 64 | |
| Slit2 | induces PS | of | Cofilin | 27 |
| via | MAPK | |||
| requires PS | for | GC collapse | ||
| LPA | does not induce PS | 64 | ||
| S1P | does not induce PS | 66 | ||
| EphrinB/EphB | does not induce PS | 65 | ||
| PACAP | requires PS | for | GC turning | 63 |
| via | PKA, Rho GTPases | |||
| EphA | inhibits PS | via | Tsc2 | 71 |
| En2 | induces PS | of | components of EphrinA5 signaling | 72 |
| requires PS | for | GC turning | 73 | |
| NGF | induces PS | of | CREB | 19 |
| requires PS | for | survival of DRG neurons | ||
| induces PS | of | par3 | 67 | |
| requires PS | for | induced axon outgrowth | ||
| does not induce PS | of | β-actin | 23 | |
| BDNF | induces PS | of | β-actin | 25 |
| via | ZBP | |||
| requires PS | for | GC turning | ||
| NT3 | induces PS | of | β-actin | 23 |
| via | PKA |
LPA, lysophosphatidic acid; PACAP, pituitary adenylate cyclase-activating polypeptide; NGF, nerve growth factor; BDNF, brain-derived neurotrophic factor; mTOR, mammalian target of rapamycin; CREB, CRE-binding protein; ZBP, zipcode binding protein; S1P, sphingosine-1-phosphate;MAPK, mitogen-activated protein kinase; PKA, protein kinase A.
Cytoskeletal Protein-Encoding mRNAs
mRNAs encoding the components of all three major classes of cytoskeleton—actin microfilament, microtubule, and intermediate filament—as well as their associated proteins are found in axons of diverse animals. Many of these mRNAs are locally translated in response to extracellular signals. Molecular mechanisms underlying axonal mRNA translation of cytoskeletal proteins as well as the functional significance are best understood in the context of responses of growing axon tips (i.e., growth cones) to guidance cues.
The components of actin filaments have been found to be particularly important in steering responses of growth cones to guidance cues. Bassell et al. first reported that β-actin mRNA was selectively localized to axons of cultured neurons in contrast to γ-actin mRNA which was restricted to the cell body.22 They later found the cis-element in the 3′-untranslated region (UTR) of β-actin mRNA was responsible for the axonal transport of β-actin mRNA,23 and named it the ‘zipcode’.62 Zipcode binding protein (ZBP) mediates axonal transport of β-actin mRNA by directly binding to the zipcode. Inhibiting ZBP–zipcode interactions impairs growth cone motility.62 In this way, an external signal may regulate growth cone dynamics by sequence-specific axonal transport and local translation of its target mRNAs. Indeed, NT-3 induces axonal transport and local translation of β-actin mRNA in cultured hippocampal neurons, a process which is critical for axon branching.58 Asymmetric translation of β-actin mRNA within a growth cone is required for the correct steering responses to guidance cues such as Netrin-1 and BDNF in Xenopus retinal ganglion cells (RGCs) and spinal neurons in vitro.24,25 Likewise, a neuropeptide pituitary adenylate cyclase-activating polypeptide requires axonal protein synthesis to induce attractive turning responses to growth cones.63 Thus, is local synthesis of cytoskeletal proteins a general mechanism for growth cones to respond to guidance cues? The answer seems to be both yes and no, depending on the guidance cues and the circumstances in which they are applied. For example, repellent cues such as Sema3A and Slit2, which cause the collapse of growth cones when uniformly applied in culture, also require axonal protein synthesis to exert their effects.26,27 Unlike in the case of attractive guidance cues, repulsive cues induce local synthesis of actin-depolymerizing molecules, such as RhoA and Cofilin, to mediate the collapse of growth cones. Similarly, actin depolymerizing factor mRNA is localized to axons and locally translated in chick sympathetic neurons, although the functional significance of this phenomenon was not shown.28 In addition, the actin monomer-sequestering protein β-thymosin mRNA is enriched in the turning points of the axons and decreasing its translation results in an uncontrolled outgrowth of snail axons,29 suggesting that axonal synthesis of actin and actin binding proteins may be a general mechanism neurons use to regulate axon dynamics. However, it does not seem to apply to all guidance cues because some, e.g., lysophosphatidic acid, EphrinB, and sphingosine-1-phosphate, do not require local protein synthesis.64-66
Components of microtubules and intermediate filaments are also locally synthesized in axons. Campenot et al., using a pioneering compartmentalized culture system, found that β-tubulin mRNA as well as β-actin mRNAs were abundant and actively translated in axons.31 They estimated that less than 1% of β-tubulin and β-actin are made in axons compared to the cell bodies and found that blocking around 80% of axonal protein synthesis did not inhibit axonal elongation. They speculated that axonal protein synthesis may be required for axonal maintenance rather than for elongation. This idea has been supported by a later study in which it was shown that blocking protein synthesis or β-actin mRNA translation in axons did not affect axon elongation although it did inhibit steering responses of growth cones to guidance cues.24,64 Interestingly, it has recently been found that axon elongation induced by nerve growth factor (NGF) or Netrin-1 signaling, but not the basal rate of axon elongation, require axonal protein synthesis.67 Jaffrey et al. have shown that stimulated, but not basal, axon elongation requires local synthesis of Par3, a component of the Par complex which is a regulator of cytoskeleton dynamics and enriched in growth cones.67 Microtubule binding protein tau mRNA has also been found in axons.33,34,68 Ginzburg et al. found that the 3′-UTR of tau mRNA contains an axon-localization signal, analogous to the zipcode of β-actin mRNAs, and that the RNA-binding protein HuD mediates kinesin-based anterograde transport of tau mRNA along the axons.34,68 Interestingly, mRNAs encoding kinesin itself, as well as other microtubule-associated proteins, are also found in squid giant axons.20,32 mRNAs encoding neurofilaments have also been found in adult goldfish Mauthner axons and the rat sciatic nerve,36,37 suggesting that axonal synthesis of the components of cytoskeleton may be a conserved mechanism by which axons are maintained.
Presynaptic Protein-Encoding mRNAs
Synapse formation is a dynamic process that occurs in both developing and mature nervous systems, and it is established that local protein synthesis in dendrites plays an essential role for synaptic plasticity of adult rodents.14 There is now evidence indicating that local mRNA translation may also play important roles in the opposite side of the synapse—the axonal presynaptic terminal.69 For example, inhibiting local protein synthesis decreases the releasable pool of synaptic vesicles in hippocampal neurons. This is likely to result from the decreased local synthesis of calmodulin-dependent kinase II (CamKII) in the presynaptic terminal considering the very short half-life of this protein, although the localization of CamKII mRNA in the presynaptic terminal has yet to be demonstrated.57 It has also been noted for long that the posterior pituitary gland of adult rodents, which are the axon terminals of hypothalamic neurons of the supraoptic and paraventricular nuclei, contains mRNAs encoding oxytocin and vasopressin, the neuropeptides which are locally synthesized and secreted.46,49,50 Other neuropeptides or hormones of invertebrates such as caudodorsal cell hormone, egg-laying hormone, and sensorin A are also locally synthesized at the presynaptic nerve terminals39-41,55,70 suggesting that local mRNA translation may be used as a mechanism for forming and maintaining presynaptic terminal in vivo.
Sensory neurons also synthesize proteins in axons. The axons of olfactory sensory neurons contain mRNAs encoding olfactory receptors, olfactory marker proteins, and calcitonin gene-related peptide,45,47,48 and the evidence showing these mRNAs are locally translated in nerve terminal was recently provided by Trembleau et al.10 Peripheral nociceptive (pain-sensing) neurons, whose cell bodies reside in dorsal root ganglia, send out the peripheral branch of their long axons to the receptive fields. These peripheral nerve endings contain multiple receptors that sense noxious stimuli. The mRNA encoding transient receptor potential channel V1 (TRPV1), a receptor responsible for thermal pain, is found in the axons of these neurons.51 Furthermore, the local inflammation induced by a tissue injury promotes the axonal transport of TRPV1 mRNAs to the injury site, suggesting the increased local synthesis of TRPV1 may be involved in pain hypersensitivity during the inflammatory response.51
The in vitro studies in which local protein synthesis was shown to regulate growth cone steering responses suggest that axonal mRNA translation may be used as a regulatory mechanism to control axon guidance in vivo.18,24-26 Evidence supporting this idea was provided by Flanagan et al.52 Pathfinding axons receive multiple guidance cues simultaneously and therefore must quickly integrate these signals in order to properly respond to their changing environments. The surface expression of the receptors for these signals is therefore tightly controlled. For example, a subset of chick commissural axons begin to express EphA2 receptors after they cross the midline and the expression of these receptors remains restricted in the distal segments of the axons. Flanagan et al. showed that EphA mRNAs are localized to the axons of these neurons and translated only in the distal segments of the axons. Furthermore, they found a cis-element in the 3′-UTR of EphA2 mRNA responsible for this spatiotemporally controlled translation. Although it was not examined what signals and RNA binding proteins (RBPs) regulate this specific translation of EphA2 mRNA, this study showed that the local protein synthesis might be used as a mechanism to control axon guidance in vivo, just as many in vitro studies have suggested. The presence of mRNAs encoding diverse receptors and neuropeptides for the specialized function of presynaptic terminals together with the evidence of axonal translation of some of these mRNAs strongly suggests that regulated axonal protein synthesis plays important roles in the formation and maintenance of presynaptic axon terminals.
Nuclear Protein-Encoding mRNAs
Jaffrey et al. have constructed a cDNA library using mRNAs isolated from the axons of cultured embryonic DRG neurons in a compartmentalized culture system.19 Among the axonal cDNAs, they found an unexpected cDNA-CRE-binding protein (CREB), a transcription factor mediating NGF-induced survival of these neurons. They went further to show that NGF treated to the axons, but not to the cell bodies, stimulated local translation of CREB mRNAs in the axons, and the axonal synthesis of CREB is required for NGF-mediated cell survival effects. Surprisingly, the axonally synthesized CREB was transported retrogradely to the nucleus to exert its survival role. Nuclear CREB could not support the cell survival, suggesting that the CREB synthesized in axons has different characteristics to nuclear CREB although the nature of this difference is unclear. Local synthesis of a nuclear protein in axons had already been observed in the axons of mature DRG neurons. Using a rat model of sciatic nerve injury, Fainzilber et al. found that importin–β, a protein regulating nuclear import of other proteins, was locally synthesized in the injured axons and the new importin–β proteins were transported retrogradely to the cell bodies in DRG.9 Using the same injury model in mice, Fainzilber et al. further showed that Vimentin, an intermediate filament, is also locally synthesized in injured axons and that newly synthesized Vimentin directly associates with phosphorylated Erk1/2 (pErks). The direct binding of Vimentin to Importin–β then mediates retrograde transport of pErks by the dynein motor, resulting in Elk1 activation in cell bodies and neuronal regeneration.74 In line with these studies, Jin et al. have recently shown that mRNA encoding the transcription factor CAAT enhancer binding protein-1 (CEBP-1) is present in axons of Caenorhabditis elegans and the stability of CEBP-1 mRNA in axons is regulated by signals known to influence synaptogenesis and axon growth.56 Moreover, blocking signaling pathways that regulated local translation of CEBP-1 inhibited synapse formation and maintenance as well as regeneration of mature axons, suggesting that local mRNA translation of nuclear proteins may be a conserved mechanism that an axon terminal employs to inform its distant cell body of its synaptic environments. It should be noted that Riccio et al. failed to detect CREB mRNAs in the axons of sympathetic neurons. In this study, they isolated axonal mRNAs from superior cervical ganglion (SCG) neurons cultured in a compartmentalized chamber. They performed a comparative analysis of axonal and soma RNAs and made an intriguing observation that the most abundant axonal RNA is IMPA-1 mRNA, an enzyme responsible for biogenesis of myo-inositol, which is widely used as a precursor for important signaling molecules. This study showed that NGF stimulates anterograde transport of IMPA-1 mRNA which is then translated locally in the axons. Blocking axonal transport or translation of IMPA-1 mRNA results in degeneration of the axons. These studies suggest that different cell types have different repertoires of axonally localized mRNAs.18
Others
Riccio et al. have shown in their recent study that the composition of axonal RNAs is distinct from that of somal RNAs, suggesting that a selective pool of RNAs is actively transported to the axons.18 Unexpectedly, mRNAs encoding cytoskeletal proteins are not the most abundant in axons. Surprisingly, the most abundant classes of RNAs found in the axons of SCG neurons encode proteins involved in the functions of mitochondria and ribosomes. In particular, mRNAs encoding most ribosomal proteins as well as ribosomal RNAs are found in axons, suggesting an intriguing possibility of axonal ribogenesis or ribosome repair.18,75,76 Indeed, Twiss et al. had reported that translation of preexisting mRNAs encoding L4, a ribosomal protein, is required for the rapid neurite regeneration of PC12 cells.77 Although the subcellular localization of l4 mRNA translation was not examined, this study demonstrated that de novo synthesis of ribosomal proteins is required for neurite regeneration which indicates that a transcription-independent control of gene expression is required for rapid responses to changing environments and that the same mechanism may be used to maintain the translational machinery itself. Indeed, mRNAs encoding proteins of the translational machinery are also enriched in axons of Aplysia, suggesting that local genesis and/or repair of ribosomes may occur across species to support axonal mRNA translation during development.30
MECHANISMS OF LOCAL mRNA TRANSLATION
An increasing body of evidence has suggested that the controlled translation of axonally localized mRNAs takes part in diverse cellular functions. The repertoires of axonal mRNAs seem to be distinct in different neurons (e.g., DRG neurons, SCG neurons, and RGCs),18,26 and the same neurons seem to have developmental stage-specific changes in axonally translated mRNAs. Furthermore, translation of many axonal mRNAs is triggered by external cues, making it very difficult to propose a general model using current knowledge to describe how axonal mRNA translation is regulated in developing axons.
Previous work has demonstrated that translational machinery exists in growth cones and axons of developing neurons as well as of mature neurons in certain circumstances.6,64 For example, mature ribosomes as well as ribosomal proteins are found in growth cones. Eukaryotic initiation factor-4E (eIF-4E), which binds the 5′ cap structure of an mRNA and regulates the rate-limiting step of cap-dependent mRNA translation, as well as its regulator eIF-4E binding protein (4E-BP) are found in growth cones. Under basal conditions, 4E-BP exists in a hypophosphorylated form, which binds and sequesters eIF-4E from the translation initiation complex. Signaling cascades that activate phosphorylation of 4EBP relieve this repression and thus activate cap-dependent translational initiation and general translation.78 Different guidance cues such as Netrin-1 and Sema3A have been shown to increase phosphorylation of 4EBP via mammalian target of rapamycin (mTOR), MAP kinases, and/or Mnk-1.27,79 Interestingly, EphrinA, which acts through EphA receptor and in general does not require local protein synthesis to signal, was recently found to decrease mTOR activity.71 mTOR activity is positively regulated by the small GTPase Ras homolog enriched in brain (Rheb). Tuberous sclerosis protein 2 (Tsc2), the GTPase activating protein of Rheb, is negatively regulated by Erk1/2 MAP kinase, and thus Erk1/2 is a positive regulator of mTOR pathway. Sahin et al. have shown that EphrinA increases Tsc2-mediated mTOR inhibition by decreasing Erk1/2 activity.71 These studies suggest an interesting model in which developing axons regulate spatiotemporal translational activity within axons by integrating diverse signals from their environments that increase or decrease local mRNA translation and this mechanism may play an important role in axon guidance and/or topographic mapping (Figure 1). Recently, Flanagan et al. reported a new link between Netrin-1 and translation initiation. Under basal condition, DCC, a transmembrane receptor of Netrin-1, forms a complex with multiple components of translational machinery inhibiting translation. Netrin-1 binding then releases translational machinery from DCC and promotes active local translation.80 This study suggests an interesting model that external signals may regulate local translation by directly associating with translational machinery, and it will be interesting to see if signals known to decrease translation (e.g., EphA) also directly associate with translational machinery.
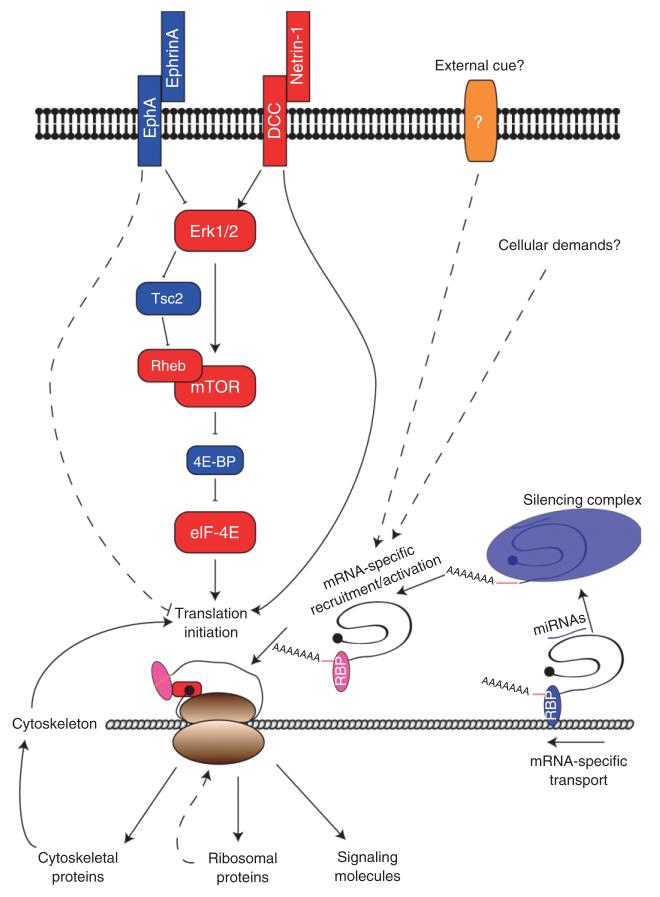
FIGURE 1.
Regulation of local mRNA translation in growth cones. Growth cones integrate multiple external signals that increase or decrease mRNA translation activity. In general, mRNA translation is regulated at the level of eIF-4E, which activates cap-dependent translation initiation. Activation of mammalian target of rapamycin (mTOR) induces phosphorylation of 4E-BP, an inhibitor of eIF-4E, and subsequently relieves repression of translation initiation. mTOR is regulated by the small GTPase Rheb, and can be inhibited by signals that activate Tsc2, the Rheb GTPase activating protein (GAP). Tsc2 is negatively regulated by Erk1/2 MAP kinase and therefore activation of Erk1/2 results in activation of mTOR. Erk1/2 can also directly phosphorylate 4E-BP. For example, Netrin-1 increases mRNA translation by activating Erk1/2, whereas EphrinA decreases mRNA translation by inhibiting Erk1/2. Alternatively, binding of Netrin-1 to DCC may directly activate translation by regulating the direct association between DCC and translation machinery. It remains to be seen whether any external signals negatively regulate translation also by directly associating with translational machinery (e.g., EphA). Translation machinery is in physical contact with cytoskeletal networks where translation occurs. Local translation of cytoskeletal proteins regulates cytoskeletal dynamics, and the changes in cytoskeleton can also regulate the degree of local translation. Local translation of ribosomal proteins may occur to replenish or repair ribosomes. A subset of mRNAs are transported along cytoskeleton to growth cones by associating with RNA binding proteins (RBPs), which then bind to motor proteins. Specific cis-elements in the 5′- or 3′-untranslated regions (UTRs) are recognized by RBPs. Most mRNAs in growth cones are translationally repressed and signals may regulate release of certain mRNAs from repression. MicroRNAs may participate sequence-specific repression and derepression of mRNA translation.
Studies in cultured cells have revealed a novel role of the cytoskeleton in regulating mRNA translation. Actin filaments, microtubules, and intermediate filaments not only bind to the components of translational machinery including eIFs, eukaryotic elongation factors, cap-binding protein, and aminoacyl-tRNA synthase, by serving as a scaffold, but also actively participate in signaling pathways that regulate protein synthesis.81 For example, when epithelial cells increase protein synthesis during the repair in response to tissue injury, Keratin 17, an intermediate filament protein, signals by directly binding to the adaptor protein 14-3-3σ to increase mTOR activity and global protein synthesis.82 Considering dynamic changes in the cytoskeletal structure of pathfinding axons, it is conceivable that local protein synthesis is being constantly regulated by the changes in cytoskeleton of axons and growth cones as protein synthesis of cytoskeletal proteins regulates cytoskeletal dynamics.
One remaining questing is how mRNAs to be translated are selected? One way to control mRNA-specific translation would be to control the local mRNA repertoire by transporting specific mRNAs to axons. Several cis-elements residing in the 5′- and/or 3′-UTR have been implicated in axonal transport of mRNAs. These include the ‘zipcode’ of β-actin mRNA as well as the cis-elements found in the 3′-UTRs of RhoA, EphA2, and IMPA-1 mRNAs which can even induce axonal transport of otherwise soma restricted mRNAs.18,22,52 Trans-elements recognize these motifs, form ribonucleoprotein (RNP) complexes, and transport the mRNAs to axons while repressing translation during the transport. Some evidence suggests that the movement of these complexes can be regulated by external signals. For example, a gradient of guidance cues such as brain-derived neurotrophic factor (BDNF) and Netrin-1 applied to a growth cone induces asymmetric accumulation of Vg1RBP, the Xenopus of homolog of ZBP, in the near side of the gradient, and also results in an increased local synthesis of β-Actin protein.24,25 The release from translationally silent RNP complexes can also be the step of translational regulation.83 For example, Netrin-1 increases axonal translation of kappa opioid receptor mRNA by releasing growth factor receptor-bound protein 7, an RBP that inhibits translation, from the 5′-UTR by activation of focal adhesion kinase-mediated phosphorylation of this protein.84 Fragile X mental retardation protein, an RBP known to be a regulator of local translation in dendrites, is also localized to developing axons suggesting a possible role in controlling axonal mRNA translation (H. Svoboda and C. E. Holt, personal communication).26 Another candidate regulator is cytoplasmic polyadenylation element binding protein (CPEB), an RBP that regulates the length of the poly A tails of target mRNAs thereby controlling translation.85 CPEB binds to the cytoplasmic polyadenylation element (CPE) in the 3′ UTRs of several mRNAs including EphA2 receptors, which localize to developing axons,52 and the expression of dominant negative CPEB1 inhibits translation-dependent chemotropic responses in vitro and axon outgrowth in vivo.86
Finally, microRNAs may participate in mRNA-specific regulation of local translation. In dendrites, for example, the microRNA miR-134 keeps its target lim kinase 1 mRNA from being translated until signals such as BDNF release this inhibition.87 Jaffrey et al. have shown that small interfering RNA specifically delivered to axons could degrade target mRNAs locally in the axons indicating that functional RNA interference machinery is present in axons.26 Because axons and growth cones have diverse miRNAs (M. L. Baudet and C. E. Holt, personal communication), it is plausible that similar mechanisms may take part in regulating mRNA-specific axonal translation in axons.
CONCLUSION
The progress made in recent years increased our understanding of local mRNA translation in developing neurons. A selective pool of mRNAs is actively transported to axons and dendrites to make cell type- and developmental stage-specific repertoires of local mRNAs. These mRNAs encode proteins of diverse functions including cytoskeleton, cell signaling, and even transcription and translation. RBPs recognizing axon- or dendrite-localizing elements in the 5′- and 3′-UTR of the target mRNAs are involved in transporting mRNAs to axons and dendrites. Developmental signals such as guidance cues and morphogens modulate global translational activity in axons and dendrites and mRNA-specific regulation may be mediated by specificity of RNB–mRNA interactions. Local protein synthesis regulates various cellular processes such as axon guidance and cell survival. With such understanding of local mRNA translation, we can now ask new questions. Firstly, most studies used neurons cultured in vitro and it should be noted that Letourneau et al. have reported that neither axon outgrowth nor growth cone turning required axonal protein synthesis in some circumstances.88,89 They showed, using a novel ex vivo preparation of the chick brainstem spinal cord projection, that protein synthesis at distal axons is not required for axon elongation88 in good accordance with previous in vitro studies which reported that growth cone chemotropic responses but not axon extension require axonal protein synthesis.8,31,64 However, in a separate study, they observed no difference even in growth cone responses to guidance cues in cultured chick neurons.89 Differences in the conditions of in vitro culture protocols may be responsible for this apparent discrepancy. Therefore, developing methods to inactivate axonal protein synthesis in vivo would be required to test the relevance of the results obtained by in vitro studies. In this sense, examining the in vivo repertoire of axonal mRNAs is of equal importance and results will shed light on the functions played by local protein synthesis. Secondly, most locally synthesized proteins are already present and it is not clear how it is advantageous to use local protein synthesis as a signaling mechanism. One may speculate that a more rapid control of protein levels can be achieved by regulating local protein synthesis independent of the soma control. Furthermore, mRNA localization may provide a mechanism for highly precise control over where a protein is made and localized because multiple localization signals can be added to a protein without altering its function by using different UTRs, whereas adding localization signals to a protein may change its structure and function.90 Also newly synthesized proteins may have novel properties distinct from the same preexisting proteins, which may serve as a useful signaling mechanism. Thirdly, the mechanisms responsible for transport of certain mRNAs are far from clear. Several studies have identified many mRNAs localized in the same subcellular compartments, but there were no clear similarities in the 5′- and 3′-UTRs at least at the primary sequence level. Fourthly, it is not clear how only a certain mRNAs are translated at a given time from a pool of diverse mRNAs. MicroRNAs may participate in mRNA selection, and in this sense the dicer knockout mouse will be a very useful tool to test if local translation is controled by microRNAs. Fifthly, many mRNAs encoding the components of protein synthesis machinery are found in the axons of different neurons.18 It will be very interesting to see if these mRNAs are locally translated, and if so, if this is a mechanism to support the ongoing needs of translational machinery by making or repairing ribosomes within axons. Finally, some guidance cues are known to signal by activating protein degradation by ubiquitin-proteasome system (UPS) and/or local protein synthesis.64 For example, Netrin-1 requires both protein synthesis and degradation to induce chemotropic turning responses of growth cones suggesting that protein synthesis and protein degradation work in serial rather than in parallel. The proteins whose UPS-mediated degradation is activated by Netrin-1 include the regulators of protein synthesis, such as the tumor suppressor phosphatase and tensin homolog (PTEN). Since PTEN is a negative regulator of the PI3 kinase pathway which activates mTOR and mRNA translation, it will be intriguing to see whether protein synthesis and protein degradation are functionally linked.
ACKNOWLEDGEMENTS
We thank Louis Leung for helpful suggestions and critical reading of the manuscript.
REFERENCES
- 1.Lasek RJ, Dabrowski C, Nordlander R. Analysis of axoplasmic RNA from invertebrate giant axons. Nat New Biol. 1973;244:162–165. doi: 10.1038/newbio244162a0. [DOI] [PubMed] [Google Scholar]
- 2.Campenot RB, Eng H. Protein synthesis in axons and its possible functions. J Neurocytol. 2000;29:793–798. doi: 10.1023/a:1010939307434. [DOI] [PubMed] [Google Scholar]
- 3.Smith DH. Stretch growth of integrated axon tracts: extremes and exploitations. Prog Neurobiol. 2009;89:231–239. doi: 10.1016/j.pneurobio.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tennyson VM. The fine structure of the axon and growth cone of the dorsal root neuroblast of the rabbit embryo. J Cell Biol. 1970;44:62–79. doi: 10.1083/jcb.44.1.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamada KM, Spooner BS, Wessells NK. Ultrastructure and function of growth cones and axons of cultured nerve cells. J Cell Biol. 1971;49:614–635. doi: 10.1083/jcb.49.3.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zelena J. Ribosome-like particles in myelinated axons of the rat. Brain Res. 1970;24:359–363. doi: 10.1016/0006-8993(70)90120-4. [DOI] [PubMed] [Google Scholar]
- 7.Twiss JL, van Minnen J. New insights into neuronal regeneration: the role of axonal protein synthesis in pathfinding and axonal extension. J Neurotrauma. 2006;23:295–308. doi: 10.1089/neu.2006.23.295. [DOI] [PubMed] [Google Scholar]
- 8.Verma P, Chierzi S, Codd AM, Campbell DS, Meyer RL, Holt CE, Fawcett JW. Axonal protein synthesis and degradation are necessary for efficient growth cone regeneration. J Neurosci. 2005;25:331–342. doi: 10.1523/JNEUROSCI.3073-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanz S, Perlson E, Willis D, Zheng JQ, Massarwa R, Huerta JJ, Koltzenburg M, Kohler M, van-Minnen J, Twiss JL, et al. Axoplasmic importins enable retro-grade injury signaling in lesioned nerve. Neuron. 2003;40:1095–1104. doi: 10.1016/s0896-6273(03)00770-0. [DOI] [PubMed] [Google Scholar]
- 10.Dubacq C, Jamet S, Trembleau A. Evidence for developmentally regulated local translation of odorant receptor mRNAs in the axons of olfactory sensory neurons. J Neurosci. 2009;29:10184–10190. doi: 10.1523/JNEUROSCI.2443-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koenig E, Martin R, Titmus M, Sotelo-Silveira JR. Cryptic peripheral ribosomal domains distributed intermittently along mammalian myelinated axons. J Neurosci. 2000;20:8390–8400. doi: 10.1523/JNEUROSCI.20-22-08390.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nedelec S, Dubacq C, Trembleau A. Morphological and molecular features of the mammalian olfactory sensory neuron axons: what makes these axons so special? J Neurocytol. 2005;34:49–64. doi: 10.1007/s11068-005-5047-7. [DOI] [PubMed] [Google Scholar]
- 13.Hengst U, Jaffrey SR. Function and translational regulation of mRNA in developing axons. Semin Cell Dev Biol. 2007;18:209–215. doi: 10.1016/j.semcdb.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sutton MA, Schuman EM. Dendritic protein synthesis, synaptic plasticity, and memory. Cell. 2006;127:49–58. doi: 10.1016/j.cell.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 15.Koenig E. Cycloheximide-sensitive [35S]methionine labeling of proteins in goldfish retinal ganglion cell axons in vitro. Brain Res. 1989;481:119–123. doi: 10.1016/0006-8993(89)90491-5. [DOI] [PubMed] [Google Scholar]
- 16.Koenig E, Adams P. Local protein synthesizing activity in axonal fields regenerating in vitro. J Neurochem. 1982;39:386–400. doi: 10.1111/j.1471-4159.1982.tb03960.x. [DOI] [PubMed] [Google Scholar]
- 17.Kaplan BB, Gioio AE, Capano CP, Crispino M, Giuditta A. beta-Actin and beta-Tubulin are components of a heterogeneous mRNA population present in the squid giant axon. Mol Cell Neurosci. 1992;3:133–144. doi: 10.1016/1044-7431(92)90017-v. [DOI] [PubMed] [Google Scholar]
- 18.Andreassi C, Zimmermann C, Mitter R, Fusco S, Devita S, Saiardi A, Riccio A. An NGF-responsive element targets myo-inositol monophosphatase-1 mRNA to sympathetic neuron axons. Nat Neurosci. 2010;13:291–301. doi: 10.1038/nn.2486. [DOI] [PubMed] [Google Scholar]
- 19.Cox LJ, Hengst U, Gurskaya NG, Lukyanov KA, Jaffrey SR. Intra-axonal translation and retrograde trafficking of CREB promotes neuronal survival. Nat Cell Biol. 2008;10:149–159. doi: 10.1038/ncb1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gioio AE, Chun JT, Crispino M, Capano CP, Giuditta A, Kaplan BB. Kinesin mRNA is present in the squid giant axon. J Neurochem. 1994;63:13–18. doi: 10.1046/j.1471-4159.1994.63010013.x. [DOI] [PubMed] [Google Scholar]
- 21.Olink-Coux M, Hollenbeck PJ. Localization and active transport of mRNA in axons of sympathetic neurons in culture. J Neurosci. 1996;16:1346–1358. doi: 10.1523/JNEUROSCI.16-04-01346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bassell GJ, Zhang H, Byrd AL, Femino AM, Singer RH, Taneja KL, Lifshitz LM, Herman IM, Kosik KS. Sorting of beta-actin mRNA and protein to neurites and growth cones in culture. J Neurosci. 1998;18:251–265. doi: 10.1523/JNEUROSCI.18-01-00251.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang HL, Singer RH, Bassell GJ. Neurotrophin regulation of beta-actin mRNA and protein localization within growth cones. J Cell Biol. 1999;147:59–70. doi: 10.1083/jcb.147.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leung KM, van Horck FP, Lin AC, Allison R, Standart N, Holt CE. Asymmetrical beta-actin mRNA translation in growth cones mediates attractive turning to netrin-1. Nat Neurosci. 2006;9:1247–1256. doi: 10.1038/nn1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yao J, Sasaki Y, Wen Z, Bassell GJ, Zheng JQ. An essential role for beta-actin mRNA localization and translation in Ca2+-dependent growth cone guidance. Nat Neurosci. 2006;9:1265–1273. doi: 10.1038/nn1773. [DOI] [PubMed] [Google Scholar]
- 26.Wu KY, Hengst U, Cox LJ, Macosko EZ, Jeromin A, Urquhart ER, Jaffrey SR. Local translation of RhoA regulates growth cone collapse. Nature. 2005;436:1020–1024. doi: 10.1038/nature03885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piper M, Anderson R, Dwivedy A, Weinl C, van Horck F, Leung KM, Cogill E, Holt C. Signaling mechanisms underlying Slit2-induced collapse of Xenopus retinal growth cones. Neuron. 2006;49:215–228. doi: 10.1016/j.neuron.2005.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee SK, Hollenbeck PJ. Organization and translation of mRNA in sympathetic axons. J Cell Sci. 2003;116:4467–4478. doi: 10.1242/jcs.00745. [DOI] [PubMed] [Google Scholar]
- 29.van Kesteren RE, Carter C, Dissel HM, van Minnen J, Gouwenberg Y, Syed NI, Spencer GE, Smit AB. Local synthesis of actin-binding protein beta-thymosin regulates neurite outgrowth. J Neurosci. 2006;26:152–157. doi: 10.1523/JNEUROSCI.4164-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moccia R, Chen D, Lyles V, Kapuya E, E Y, Kalachikov S, Spahn CM, Frank J, Kandel ER, Barad M, Martin KC. An unbiased cDNA library prepared from isolated Aplysia sensory neuron processes is enriched for cytoskeletal and translational mRNAs. J Neurosci. 2003;23:9409–9417. doi: 10.1523/JNEUROSCI.23-28-09409.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eng H, Lund K, Campenot RB. Synthesis of beta-tubulin, actin, and other proteins in axons of sympathetic neurons in compartmented cultures. J Neurosci. 1999;19:1–9. doi: 10.1523/JNEUROSCI.19-01-00001.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chun JT, Gioio AE, Crispino M, Giuditta A, Kaplan BB. Differential compartmentalization of mRNAs in squid giant axon. J Neurochem. 1996;67:1806–1812. doi: 10.1046/j.1471-4159.1996.67051806.x. [DOI] [PubMed] [Google Scholar]
- 33.Litman P, Barg J, Rindzoonski L, Ginzburg I. Subcellular localization of tau mRNA in differentiating neuronal cell culture: implications for neuronal polarity. Neuron. 1993;10:627–638. doi: 10.1016/0896-6273(93)90165-n. [DOI] [PubMed] [Google Scholar]
- 34.Aronov S, Aranda G, Behar L, Ginzburg I. Axonal tau mRNA localization coincides with tau protein in living neuronal cells and depends on axonal targeting signal. J Neurosci. 2001;21:6577–6587. doi: 10.1523/JNEUROSCI.21-17-06577.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giuditta A, Menichini E, Perrone Capano C, Langella M, Martin R, Castigli E, Kaplan BB. Active polysomes in the axoplasm of the squid giant axon. J Neurosci Res. 1991;28:18–28. doi: 10.1002/jnr.490280103. [DOI] [PubMed] [Google Scholar]
- 36.Weiner OD, Zorn AM, Krieg PA, Bittner GD. Medium weight neurofilament mRNA in goldfish Mauthner axoplasm. Neurosci Lett. 1996;213:83–86. doi: 10.1016/0304-3940(96)12860-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sotelo-Silveira JR, Calliari A, Kun A, Benech JC, Sanguinetti C, Chalar C, Sotelo JR. Neurofilament mRNAs are present and translated in the normal and severed sciatic nerve. J Neurosci Res. 2000;62:65–74. doi: 10.1002/1097-4547(20001001)62:1<65::AID-JNR7>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 38.Dirks RW, van Dorp AG, van Minnen J, Fransen JA, van der Ploeg M, Raap AK. Ultrastructural evidence for the axonal localization of caudodorsal cell hormone mRNA in the central nervous system of the mollusc Lymnaea stagnalis. Microsc Res Tech. 1993;25:12–18. doi: 10.1002/jemt.1070250104. [DOI] [PubMed] [Google Scholar]
- 39.Lee W, Jones AM, Ono JK, Wayne NL. Regional differences in processing of locally translated prohormone in peptidergic neurons of Aplysia californica. J Neurochem. 2002;83:1423–1430. doi: 10.1046/j.1471-4159.2002.01252.x. [DOI] [PubMed] [Google Scholar]
- 40.Hu JY, Meng X, Schacher S. Target interaction regulates distribution and stability of specific mRNAs. J Neurosci. 2002;22:2669–2678. doi: 10.1523/JNEUROSCI.22-07-02669.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schacher S, Wu F, Panyko JD, Sun ZY, Wang D. Expression and branch-specific export of mRNA are regulated by synapse formation and interaction with specific postsynaptic targets. J Neurosci. 1999;19:6338–6347. doi: 10.1523/JNEUROSCI.19-15-06338.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Landry C, Crine P, DesGroseillers L. Differential expression of neuropeptide gene mRNA within the LUQ cells of Aplysia californica. J Neurobiol. 1992;23:89–101. doi: 10.1002/neu.480230109. [DOI] [PubMed] [Google Scholar]
- 43.van Minnen J. Axonal localization of neuropeptide-encoding mRNA in identified neurons of the snail Lymnaea stagnalis. Cell Tissue Res. 1994;276:155–161. doi: 10.1007/BF00354795. [DOI] [PubMed] [Google Scholar]
- 44.Hu JY, Meng X, Schacher S. Redistribution of syntaxin mRNA in neuronal cell bodies regulates protein expression and transport during synapse formation and long-term synaptic plasticity. J Neurosci. 2003;23:1804–1815. doi: 10.1523/JNEUROSCI.23-05-01804.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Denis-Donini S, Branduardi P, Campiglio S, Carnevali MD. Localization of calcitonin gene-related peptide mRNA in developing olfactory axons. Cell Tissue Res. 1998;294:81–91. doi: 10.1007/s004410051158. [DOI] [PubMed] [Google Scholar]
- 46.Mohr E, Fehr S, Richter D. Axonal transport of neuropeptide encoding mRNAs within the hypothalamo-hypophyseal tract of rats. EMBO J. 1991;10:2419–2424. doi: 10.1002/j.1460-2075.1991.tb07781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vassar R, Chao SK, Sitcheran R, Nunez JM, Vosshall LB, Axel R. Topographic organization of sensory projections to the olfactory bulb. Cell. 1994;79:981–991. doi: 10.1016/0092-8674(94)90029-9. [DOI] [PubMed] [Google Scholar]
- 48.Wensley CH, Stone DM, Baker H, Kauer JS, Margolis FL, Chikaraishi DM. Olfactory marker protein mRNA is found in axons of olfactory receptor neurons. J Neurosci. 1995;15:4827–4837. doi: 10.1523/JNEUROSCI.15-07-04827.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jirikowski GF, Sanna PP, Bloom FE. mRNA coding for oxytocin is present in axons of the hypothalamo-neurohypophysial tract. Proc Natl Acad Sci U S A. 1990;87:7400–7404. doi: 10.1073/pnas.87.19.7400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mohr E, Richter D. Diversity of mRNAs in the axonal compartment of peptidergic neurons in the rat. Eur J Neurosci. 1992;4:870–876. doi: 10.1111/j.1460-9568.1992.tb00197.x. [DOI] [PubMed] [Google Scholar]
- 51.Tohda C, Sasaki M, Konemura T, Sasamura T, Itoh M, Kuraishi Y. Axonal transport of VR1 capsaicin receptor mRNA in primary afferents and its participation in inflammation-induced increase in capsaicin sensitivity. J Neurochem. 2001;76:1628–1635. doi: 10.1046/j.1471-4159.2001.00193.x. [DOI] [PubMed] [Google Scholar]
- 52.Brittis PA, Lu Q, Flanagan JG. Axonal protein synthesis provides a mechanism for localized regulation at an intermediate target. Cell. 2002;110:223–235. doi: 10.1016/s0092-8674(02)00813-9. [DOI] [PubMed] [Google Scholar]
- 53.Bi J, Hu X, Loh HH, Wei LN. Mouse kappa-opioid receptor mRNA differential transport in neurons. Mol Pharmacol. 2003;64:594–599. doi: 10.1124/mol.64.3.594. [DOI] [PubMed] [Google Scholar]
- 54.Bi J, Tsai NP, Lin YP, Loh HH, Wei LN. Axonal mRNA transport and localized translational regulation of kappa-opioid receptor in primary neurons of dorsal root ganglia. Proc Natl Acad Sci U S A. 2006;103:19919–19924. doi: 10.1073/pnas.0607394104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lyles V, Zhao Y, Martin KC. Synapse formation and mRNA localization in cultured Aplysia neurons. Neuron. 2006;49:349–356. doi: 10.1016/j.neuron.2005.12.029. [DOI] [PubMed] [Google Scholar]
- 56.Yan D, Wu Z, Chisholm AD, Jin Y. The DLK-1 kinase promotes mRNA stability and local translation in C. elegans synapses and axon regeneration. Cell. 2009;138:1005–1018. doi: 10.1016/j.cell.2009.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sebeo J, Hsiao K, Bozdagi O, Dumitriu D, Ge Y, Zhou Q, Benson DL. Requirement for protein synthesis at developing synapses. J Neurosci. 2009;29:9778–9793. doi: 10.1523/JNEUROSCI.2613-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 58.Kundel M, Jones KJ, Shin CY, Wells DG. Cytoplasmic polyadenylation element-binding protein regulates neurotrophin-3-dependent beta-catenin mRNA translation in developing hippocampal neurons. J Neurosci. 2009;29:13630–13639. doi: 10.1523/JNEUROSCI.2910-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gioio AE, Eyman M, Zhang H, Lavina ZS, Giuditta A, Kaplan BB. Local synthesis of nuclear-encoded mitochondrial proteins in the presynaptic nerve terminal. J Neurosci Res. 2001;64:447–453. doi: 10.1002/jnr.1096. [DOI] [PubMed] [Google Scholar]
- 60.Giustetto M, Hegde AN, Si K, Casadio A, Inokuchi K, Pei W, Kandel ER, Schwartz JH. Axonal transport of eukaryotic translation elongation factor 1alpha mRNA couples transcription in the nucleus to long-term facilitation at the synapse. Proc Natl Acad Sci U S A. 2003;100:13680–13685. doi: 10.1073/pnas.1835674100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wanner I, Baader SL, Brich M, Oberdick J, Schilling K. Subcellular localization of specific mRNAs and their protein products in Purkinje cells by combined fluorescence in situ hybridization and immunocytochemistry. Histochem Cell Biol. 1997;108:345–357. doi: 10.1007/s004180050175. [DOI] [PubMed] [Google Scholar]
- 62.Zhang HL, Eom T, Oleynikov Y, Shenoy SM, Liebelt DA, Dictenberg JB, Singer RH, Bassell GJ. Neurotrophin-induced transport of a beta-actin mRNP complex increases beta-actin levels and stimulates growth cone motility. Neuron. 2001;31:261–275. doi: 10.1016/s0896-6273(01)00357-9. [DOI] [PubMed] [Google Scholar]
- 63.Guirland C, Buck KB, Gibney JA, DiCicco-Bloom E, Zheng JQ. Direct cAMP signaling through G-protein-coupled receptors mediates growth cone attraction induced by pituitary adenylate cyclase-activating polypeptide. J Neurosci. 2003;23:2274–2283. doi: 10.1523/JNEUROSCI.23-06-02274.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Campbell DS, Holt CE. Chemotropic responses of retinal growth cones mediated by rapid local protein synthesis and degradation. Neuron. 2001;32:1013–1026. doi: 10.1016/s0896-6273(01)00551-7. [DOI] [PubMed] [Google Scholar]
- 65.Mann F, Miranda E, Weinl C, Harmer E, Holt CE. B-type Eph receptors and ephrins induce growth cone collapse through distinct intracellular pathways. J Neurobiol. 2003;57:323–336. doi: 10.1002/neu.10303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Strochlic L, Dwivedy A, van Horck FP, Falk J, Holt CE. A role for S1P signalling in axon guidance in the Xenopus visual system. Development. 2008;135:333–342. doi: 10.1242/dev.009563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hengst U, Deglincerti A, Kim HJ, Jeon NL, Jaffrey SR. Axonal elongation triggered by stimulus-induced local translation of a polarity complex protein. Nat Cell Biol. 2009;11:1024–1030. doi: 10.1038/ncb1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aronov S, Aranda G, Behar L, Ginzburg I. Visualization of translated tau protein in the axons of neuronal P19 cells and characterization of tau RNP granules. J Cell Sci. 2002;115:3817–3827. doi: 10.1242/jcs.00058. [DOI] [PubMed] [Google Scholar]
- 69.Zhang X, Poo MM. Localized synaptic potentiation by BDNF requires local protein synthesis in the developing axon. Neuron. 2002;36:675–688. doi: 10.1016/s0896-6273(02)01023-1. [DOI] [PubMed] [Google Scholar]
- 70.Liu K, Hu JY, Wang D, Schacher S. Protein synthesis at synapse versus cell body: enhanced but transient expression of long-term facilitation at isolated synapses. J Neurobiol. 2003;56:275–286. doi: 10.1002/neu.10242. [DOI] [PubMed] [Google Scholar]
- 71.Nie D, Di Nardo A, Han JM, Baharanyi H, Kramvis I, Huynh T, Dabora S, Codeluppi S, Pandolfi PP, Pasquale EB, et al. Tsc2-Rheb signaling regulates EphA-mediated axon guidance. Nat Neurosci. 2010;13:163–172. doi: 10.1038/nn.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wizenmann A, Brunet I, Lam JS, Sonnier L, Beurdeley M, Zarbalis K, Vogt D, Weinl C, Dwivedy A, Joliot A, et al. Extracellular Engrailed participates in the topographic guidance of retinal axons in vivo. Neuron. 2009;64:355–366. doi: 10.1016/j.neuron.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brunet I, Weinl C, Piper M, Trembleau A, Volovitch M, Harris W, Prochiantz A, Holt C. The transcription factor Engrailed-2 guides retinal axons. Nature. 2005;438:94–98. doi: 10.1038/nature04110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Perlson E, Hanz S, Ben-Yaakov K, Segal-Ruder Y, Seger R, Fainzilber M. Vimentin-dependent spatial translocation of an activated MAP kinase in injured nerve. Neuron. 2005;45:715–726. doi: 10.1016/j.neuron.2005.01.023. [DOI] [PubMed] [Google Scholar]
- 75.Willis DE, van Niekerk EA, Sasaki Y, Mesngon M, Merianda TT, Williams GG, Kendall M, Smith DS, Bassell GJ, Twiss JL. Extracellular stimuli specifically regulate localized levels of individual neuronal mRNAs. J Cell Biol. 2007;178:965–980. doi: 10.1083/jcb.200703209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Taylor AM, Berchtold NC, Perreau VM, Tu CH, Li Jeon N, Cotman CW. Axonal mRNA in uninjured and regenerating cortical mammalian axons. J Neurosci. 2009;29:4697–4707. doi: 10.1523/JNEUROSCI.6130-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Twiss JL, Smith DS, Chang B, Shooter EM. Translational control of ribosomal protein L4 mRNA is required for rapid neurite regeneration. Neurobiol Dis. 2000;7:416–428. doi: 10.1006/nbdi.2000.0293. [DOI] [PubMed] [Google Scholar]
- 78.Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Campbell DS, Holt CE. Apoptotic pathway and MAPKs differentially regulate chemotropic responses of retinal growth cones. Neuron. 2003;37:939–952. doi: 10.1016/s0896-6273(03)00158-2. [DOI] [PubMed] [Google Scholar]
- 80.Tcherkezian J, Brittis PA, Thomas F, Roux PP, Flanagan JG. Transmembrane receptor DCC associates with protein synthesis machinery and regulates translation. Cell. 2010;141:632–644. doi: 10.1016/j.cell.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim S, Coulombe PA. Emerging role for the cytoskeleton as an organizer and regulator of translation. Nat Rev Mol Cell Biol. 2010;11:75–81. doi: 10.1038/nrm2818. [DOI] [PubMed] [Google Scholar]
- 82.Kim S, Wong P, Coulombe PA. A keratin cytoskeletal protein regulates protein synthesis and epithelial cell growth. Nature. 2006;441:362–365. doi: 10.1038/nature04659. [DOI] [PubMed] [Google Scholar]
- 83.Krichevsky AM, Kosik KS. Neuronal RNA granules: a link between RNA localization and stimulation-dependent translation. Neuron. 2001;32:683–696. doi: 10.1016/s0896-6273(01)00508-6. [DOI] [PubMed] [Google Scholar]
- 84.Tsai NP, Bi J, Wei LN. The adaptor Grb7 links netrin-1 signaling to regulation of mRNA translation. EMBO J. 2007;26:1522–1531. doi: 10.1038/sj.emboj.7601598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Richter JD. CPEB: a life in translation. Trends Biochem Sci. 2007;32:279–285. doi: 10.1016/j.tibs.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 86.Lin AC, Tan CL, Lin CL, Strochlic L, Huang YS, Richter JD, Holt CE. Cytoplasmic polyadenylation and cytoplasmic polyadenylation element-dependent mRNA regulation are involved in Xenopus retinal axon development. Neural Dev. 2009;4:8. doi: 10.1186/1749-8104-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 88.Blackmore M, Letourneau PC. Protein synthesis in distal axons is not required for axon growth in the embryonic spinal cord. Dev Neurobiol. 2007;67:976–986. doi: 10.1002/dneu.20395. [DOI] [PubMed] [Google Scholar]
- 89.Roche FK, Marsick BM, Letourneau PC. Protein synthesis in distal axons is not required for growth cone responses to guidance cues. J Neurosci. 2009;29:638–652. doi: 10.1523/JNEUROSCI.3845-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Holt CE, Bullock SL. Subcellular mRNA localization in animal cells and why it matters. Science. 2009;326:1212–1216. doi: 10.1126/science.1176488. [DOI] [PMC free article] [PubMed] [Google Scholar]