Abstract
We developed and assessed real-time PCR (RTi-PCR) assays for the detection and quantification of the food-borne pathogen Listeria monocytogenes and the closely related nonpathogenic species L. innocua. The target genes were hly and iap for L. monocytogenes and lin02483 for L. innocua. The assays were 100% specific, as determined with 100 Listeria strains and 45 non-Listeria strains, and highly sensitive, with detection limits of one target molecule in 11 to 56% of the reactions with purified DNA and 3 CFU in 56 to 89% of the reactions with bacterial suspensions. Quantification was possible over a 5-log dynamic range, with a limit of 15 target molecules and R2 values of >0.996. There was an excellent correspondence between the predicted and the actual numbers of CFU in the samples (deviations of <23%). The hly-based assay accurately quantified L. monocytogenes in all of the samples tested. The iap-based assay, in contrast, was unsuitable for quantification purposes, underestimating the bacterial counts by 3 to 4 log units in a significant proportion of the samples due to serovar-related target sequence variability. The combination of the two assays enabled us to classify L. monocytogenes isolates into one of the two major phylogenetic divisions of the species, I and II. We also assessed the new AmpliFluor technology for the quantitative detection of L. monocytogenes by RTi-PCR. The performance of this system was similar to that of the TaqMan system, although the former system was slightly less sensitive (detection limit of 15 molecules in 45% of the reactions) and had a higher quantification limit (60 molecules).
Bacteria of the facultative anaerobic gram-positive genus Listeria are widely distributed in the environment, particularly the closely related species Listeria monocytogenes and L. innocua. Both of these Listeria spp. are frequently found in food products, where they can grow over a pH range of 4.39 to 9.40, even at refrigeration temperatures. Ingestion of foods contaminated with L. monocytogenes can result in listeriosis, a severe infectious disease characterized by meningoencephalitis, abortion, septicemia, and a high fatality rate (30%). Listeriosis predominantly affects certain risk groups, including pregnant women, newborns, elderly people, and immunocompromised patients. L. innocua, in contrast, is nonpathogenic, and its presence in foods is no hazard to human health (20, 25, 35, 37, 41). Human listeriosis outbreaks are most often associated with ready-to-eat food products that are consumed without prior cooking (8, 36). To err on the side of caution, food safety regulations have tended to adopt a zero-tolerance attitude for L. monocytogenes in these products (9). However, as clinical cases of listeriosis are usually associated with high loads of L. monocytogenes (6, 8) and as it is difficult to eradicate listeriae from the environment of food-processing plants (11), the International Commission on Microbiological Specification for Foods concluded that 100 CFU of L. monocytogenes per g of food is acceptable for consumers not in risk groups (29, 31). A prerequisite for the general adoption of this less stringent criterion is the availability of appropriate laboratory methods for the differentiation of L. monocytogenes and L. innocua and for the specific and precise quantification of L. monocytogenes in food.
As well as not always being reliable, conventional bacteriological methods for the detection and quantification of L. monocytogenes are laborious and time-consuming and require individual biochemical confirmation of the species in a number of isolated colonies (7). These drawbacks are overcome by PCR-based methods, particularly by the development of real-time PCR (RTi-PCR), which is highly specific and can very accurately quantify target DNA (which is directly related to the size of the bacterial population present in the sample). As this quantification is based on the emission of a fluorescence signal as the specific PCR progresses, no post-PCR manipulations are required. This feature reduces the risk of cross-contamination in the laboratory and permits high throughput and automation (reviewed in reference 22).
A potential problem that can seriously compromise the applicability of the RTi-PCR technique for quantification purposes is the existence of interstrain variability in the target DNA sequence. Although sequences exhibiting a certain degree of divergence can still be detected, primers and probes anneal less efficiently to nonidentical target sequences, resulting in weak signals and underestimation of the amount of DNA in the sample. Most PCR assays for L. monocytogenes are based on the detection of the virulence genes hly and iap, encoding the hemolysin listeriolysin O (27) and the invasion-associated surface protein p60 (23), respectively. A number of RTi-PCR assays based on these targets have been developed (12, 18, 24, 29), but the quantification abilities of these assays were assessed with only one L. monocytogenes isolate. While the hly gene is relatively well conserved in all L. monocytogenes strains, the iap gene is not. Although iap contains conserved portions at the 5′ and 3′ ends, its central region is highly variable and contains sequence polymorphisms even among strains of the same serovar (5, 26, 33).
Here we evaluated the usefulness of the hly and iap genes as targets for the specific quantitative detection of L. monocytogenes by RTi-PCR. Specific, sensitive, and accurate quantification of L. monocytogenes was consistently achieved with the hly-based assay. The iap-based assay, in contrast, yielded heterogeneous results, and reliable quantification was possible only when homologous strains or strains belonging to the same serovar-related phylogenetic branch were tested. We also developed an efficient quantitative PCR assay for L. innocua based on the detection of lin02483 gene sequences. Finally, we assessed the new AmpliFluor system (Intergen Co., Purchase, N.Y.) for the detection of food-borne pathogenic bacteria. In contrast to the widely used TaqMan system, which requires an energy tranfer-labeled probe specific for each PCR assay, the AmpliFluor technology uses a universal energy transfer hairpin primer (UniPrimer) which emits a fluorescence signal when unfolded during its incorporation into an amplification product. The UniPrimer contains a 3′ Z tail sequence that is also present at the 5′ end of one of the target-specific primers so that it anneals to the PCR product and acts as a universal PCR primer. In our experiments, the AmpliFluor and TaqMan technologies performed similarly, with only slight differences in detection and quantification limits.
MATERIALS AND METHODS
Bacterial strains, culture media, and growth conditions.
One hundred Listeria strains (49 L. monocytogenes, 17 L. innocua, 7 L. grayi, 10 L. seeligeri, 5 L. welshimeri, and 12 L. ivanovii strains) and 45 non-Listeria strains were used in this study (Tables 1 and 2). They were maintained at −80°C in Luria-Bertani or MRS (lactic acid bacteria) broth supplemented with 15% (vol/vol) glycerol. Listeria strains were grown in brain heart infusion broth at 37°C, and non-Listeria strains were grown in MRS broth or tryptone soya broth at 30°C. For plate cultures, 1.5% (wt/vol) agar was added to these media. All media were purchased from Oxoid (Hampshire, United Kingdom).
TABLE 1.
Listeria strains used in this studya
| Species | Strain | Other designation(s) | Serovar | Source | PCR result forb:
|
||
|---|---|---|---|---|---|---|---|
| hly | iap | lin02483 | |||||
| L. monocytogenes | CECT 911c | 1/2c | Collection | + | + | − | |
| CECT 932c | 1/2a | Collection | + | + | − | ||
| CECT 934c | 4a | Collection | + | + | − | ||
| CECT 935c | 4b | Collection | + | + | − | ||
| CECT 936c | 1/2b | Collection | + | + | − | ||
| CECT 937c | 3b | Collection | + | + | − | ||
| CECT 938c | 3c | Collection | + | + | − | ||
| CECT 940c | 4d | Collection | + | + | − | ||
| CECT 4031c | ATCC 15313T | 1/2a | Collection | + | + | − | |
| CECT 4032c | 4b | Collection | + | + | − | ||
| UdG 1010c | CTC 1010 | 1/2c | Food plant, meat | + | + | − | |
| UdG 1011c | CTC 1011 | 1/2c | Food plant, meat | + | + | − | |
| UdG 1034c | CTC 1034 | 4b | Food plant, meat | + | + | − | |
| NCMi-3 | 3b | Cheese | + | + | − | ||
| NCFe-2 | Chicken | + | + | − | |||
| NCFe-4 | Paté | + | + | − | |||
| NCFi-2 | 1/2b | Trout | + | + | − | ||
| NCFi-4 | Smoked salmon | + | + | − | |||
| NCU-3 | Environment | + | + | − | |||
| NCU-4 | Environment | + | + | − | |||
| NCMe-3 | Clinical, human | + | + | − | |||
| NCMe-4 | Clinical, human | + | + | − | |||
| NCVe-1 | Clinical, human | + | + | − | |||
| NCVe-3 | Milk | + | + | − | |||
| NCRe-3 | ATCC 5577 | 1/2c | Collection | + | + | − | |
| NCRe-9 | NCTC 11994 | 4b | Collection | + | + | − | |
| NCRe-10 | ATCC 5579 | 4c | Collection | + | + | − | |
| PAM 35 | NCTC 7973, SLCC 2371 | 1/2a | Collection | + | + | − | |
| PAM 484 | SLCC 2755 | 1/2b | Collection | + | + | − | |
| PAM 485 | NCTC 5348, SLCC 2373 | 1/2c | Collection | + | + | − | |
| PAM 486 | ATCC 19113, SLCC 2373 | 3a | Collection | + | + | − | |
| PAM 487 | SLCC 2540 | 3b | Collection | + | + | − | |
| PAM 489 | NCTC 5214, SLCC 2374 | 4a | Collection | + | + | − | |
| PAM 491 | NCTC 10527, SLCC 2375 | 4b | Collection | + | + | − | |
| PAM 493 | ATCC 19116, SLCC 2376 | 4c | Collection | + | + | − | |
| PAM 494 | NCTC 10888, SLCC 2377 | 4d | Collection | + | + | − | |
| PAM 495 | SLCC 2482 | 7 | Collection | + | + | − | |
| PAM 358 | EGD-e | 1/2a | Collection | + | + | − | |
| PAM 61 | 1/2a | Cheese | + | + | − | ||
| PAM 62 | 1/2b | Cheese | + | + | − | ||
| PAM 70 | 4b | Cheese | + | + | − | ||
| PAM 75 | 3b | Cheese | + | + | − | ||
| PAM 68 | 1/2c | Environment | + | + | − | ||
| PAM 80 | 3c | Environment | + | + | − | ||
| PAM 9 | 4b | Clinical, ovine | + | + | − | ||
| PAM 51 | 1/2c | Clinical, human | + | + | − | ||
| PAM 348 | 1/2b | Clinical, human | + | + | − | ||
| PAM 349 | 4b | Clinical, human | + | + | − | ||
| PAM 602 | 1/2a | + | + | − | |||
| L. innocua | CECT 4030 | Collection | − | − | + | ||
| UdG 1012 | CTC 1012 | Food plant, meat | − | − | + | ||
| UdG 1014 | CTC 1014 | Food plant, meat | − | − | + | ||
| NCIN-1 | Shrimp | − | − | + | |||
| NCIN-2 | Ham | − | − | + | |||
| NCIN-12 | ATCC 5578 | Collection | − | − | + | ||
| NCIN-17 | Cheese | − | − | + | |||
| NCIN-19 | DSM 20649 | 6a | Collection | − | − | + | |
| PAM 152 | ATCC 33091, SLCC 3423 | 6b | Collection | − | − | + | |
| PAM 153 | ATCC 33090 | 6a | Collection | − | − | + | |
| PAM 154 | SLCC 3379 | 6a | Collection | − | − | + | |
| PAM 443 | − | − | + | ||||
| PAM 490 | NCTC 10528, SLCC 4951 | 4ab | Collection | − | − | + | |
| PAM 550 | 6b | − | − | + | |||
| PAM 569 | 6b | Meat | − | − | + | ||
| PAM 583 | 6b | Milk | − | − | + | ||
| CECT 910 | 6a | Collection | − | − | + | ||
| L. grayi | CECT 931 | Collection | − | − | − | ||
| CECT 942 | Collection | − | − | − | |||
| CECT 4181 | Collection | − | − | − | |||
| NCGR-1 | Milk | − | − | − | |||
| NCGR-3 | DSM 20601 | Collection | − | − | − | ||
| PAM 450 | SLCC 3322 | Collection | − | − | − | ||
| PAM 466 | SLCC 4425 | Collection | − | − | − | ||
| L. seeligeri | CECT 600 | Collection | − | − | − | ||
| CECT 917 | 1/2b | Collection | − | − | − | ||
| CECT 939d | Collection | − | − | − | |||
| CECT 941d | Collection | − | − | − | |||
| NCSE-1 | Salad | − | − | − | |||
| NCSE-3 | DSM 20751 | 1/2b | Collection | − | − | − | |
| PAM 498 | SLCC 5921 | 1/2b | Collection | − | − | − | |
| PAM 499 | SLCC 3954, CIP 100100T | 1/2b | Collection | − | − | − | |
| PAM 606 | 1/2b | − | − | − | |||
| UdG 1024 | CTC 1024 | Food plant, meat | − | − | − | ||
| L. welshimeri | PAM 497 | SLCC 5334, CIP 8149T | 6a | Collection | − | − | − |
| CECT 919 | 6a | Collection | − | − | − | ||
| UdG 1013 | CTC 1013 | Food plant, meat | − | − | − | ||
| NCWe-1 | DSM 20650 | Collection | − | − | − | ||
| NCWe-3 | Salami | − | − | − | |||
| L. ivanovii | PAM 424 | ATCC 19119T | 5 | Collection | − | − | − |
| PAM 55 | 5 | Clinical, ovine | − | − | − | ||
| CECT 913 | 5 | Collection | − | − | − | ||
| UdG 2001 | 5 | Clinical, caprine | − | − | − | ||
| UdG 2002 | 5 | Clinical, caprine | − | − | − | ||
| UdG 2003 | 5 | Clinical, caprine | − | − | − | ||
| UdG 2004 | 5 | Clinical, ovine | − | − | − | ||
| UdG 2005 | 5 | Clinical, ovine | − | − | − | ||
| UdG 2006 | 5 | Clinical, ovine | − | − | − | ||
| UdG 2007 | 5 | Clinical, ovine | − | − | − | ||
| NCIv-1 | 5 | Milk | − | − | − | ||
| NCIv-3 | DSM 20750 | 5 | Collection | − | − | − | |
CECT, Spanish Type Culture Collection, Valencia, Spain; UdG, collection of Food Microbiology Department, University of Girona, Girona, Spain; NC, kindly provided by Nigel Cook, Central Science Laboratory, Sand Hutton, York, United Kingdom; PAM, collection of Microbial Pathogenesis Group, Veterinary Molecular Microbiology Section, University of Bristol, Bristol, United Kingdom; NCTC, National Collection of Type Cultures, Central Public Health Laboratory, London, United Kingdom; SLCC, H. P. R. Seeliger's Special Listeria Culture Collection; ATCC, American Type Culture Collection; DSM, German Collection of Microorganisms; CIP, collection of the Pasteur Institute; CTC, Centre de Tecnologia de la Carn, iRTA, Monells, Spain.
Qualitative results of conventional PCR and RTi-PCR: +, positive; −, negative.
Used for target gene sequencing (see Fig. 2B).
Originally deposited as L. monocytogenes but recently assigned to the species L. seeligeri.
TABLE 2.
Non-Listeria strains used in this studya
| Species | Strainb | Other des- ignation | Source |
|---|---|---|---|
| Bacillus subtilis | PAM 870 | NCTC 10400 | Collection |
| Bacillus cereus | PAM 871 | NCTC 7464 | Collection |
| Brochothrix thermosphacta | UdG 1510 | CTC 1510 | Food plant, meat |
| Brochothrix thermosphacta | PAM 873 | Collection | |
| Citrobacter freundii | PAM 878 | ATCC 8090 | Collection |
| Enterobacter aerogenes | PAM 863 | NCTC 10006 | Collection |
| Enterococcus faecalis | UdG 2708 | CTC 2708 | Food plant, meat |
| Enterococcus faecalis | PAM 872 | NCTC 775 | Collection |
| Enterococcus faecium | UdG 492 | CTC 492 | Food plant, meat |
| Enterococcus malodoratus | UdG 7007 | Food plant, meat | |
| Enterococcus malodoratus | UdG 7008 | Food plant, meat | |
| Enterococcus malodoratus | UdG 7009 | Food plant, meat | |
| Klebsiella aerogenes | PAM 862 | NCTC 9528 | Collection |
| Kurthia gibsonii | PAM 876 | Collection | |
| Kurthia zopfii | PAM 875 | ATCC 6900 | Collection |
| Lactobacillus curvatus | UdG 742 | CTC 742 | Food plant, meat |
| Lactobacillus curvatus | UdG 759 | CTC 759 | Food plant, meat |
| Lactobacillus curvatus | UdG 1174 | CTC 1174 | Food plant, meat |
| Lactobacillus murinus | UdG 7004 | Food plant, meat | |
| Lactobacillus murinus | UdG 7005 | Food plant, meat | |
| Lactobacillus murinus | UdG 7006 | Food plant, meat | |
| Lactobacillus plantarum | UdG 305 | CTC 305 | Food plant, meat |
| Lactobacillus reuteri | UdG 7010 | Food plant, meat | |
| Lactobacillus reuteri | UdG 7011 | Food plant, meat | |
| Lactobacillus reuteri | UdG 7012 | Food plant, meat | |
| Lactobacillus reuteri | UdG 7013 | Food plant, meat | |
| Lactobacillus sakei | UdG 746 | CTC 746 | Food plant, meat |
| Lactobacillus sakei | UdG 748 | CTC 748 | Food plant, meat |
| Lactobacillus sakei | UdG 756 | CTC 756 | Food plant, meat |
| Lactobacillus sakei | UdG 757 | CTC 757 | Food plant, meat |
| Lactococcus garvieae | UdG 7001 | Food plant, meat | |
| Lactococcus garvieae | UdG 7002 | Food plant, meat | |
| Lactococcus garvieae | UdG 7003 | Food plant, meat | |
| Lactococcus lactis | UdG 122 | CTC 122 | Food plant, meat |
| Leuconostoc carnosum | UdG 747 | CTC 747 | Food plant, meat |
| Pediococcus pentosaceus | UdG 745 | CTC 745 | Food plant, meat |
| Pediococcus acidolactici | UdG 771 | CTC 771 | Food plant, meat |
| Pseudomonas aeruginosa | PAM 860 | Collection | |
| Rhodococcus equi | CECT 555T | Collection | |
| Staphylococcus aureus | CECT 4520T | Collection | |
| Staphylococcus aureus | PAM 868 | Collection | |
| Staphylococcus epidermidis | PAM 869 | Collection | |
| Streptococcus faecalis | PAM 879 | Collection | |
| Streptococcus pyogenes | PAM 880 | Collection |
All the strains were negative in the PCR assays.
See Table 1, footnote a.
DNA isolation and quantification.
Bacterial genomic DNA was isolated from planktonic overnight cultures by using a Wizard genomic DNA purification kit (Promega, Madison, Wis.) according to the manufacturer's recommendations. DNA concentrations were determined by using PicoGreen (Molecular Probes, Inc., Eugene, Oreg.) and luminescence spectrometer LS50B (Perkin-Elmer Corp., Norwalk, Conn.). Concentrations were further checked by agarose gel electrophoresis and ethidium bromide staining. UV fluorescence emission was recorded and quantified by using Quantity One software (Bio-Rad Laboratories Inc., Hercules, Calif.).
Oligonucleotides.
Primer Express, version 2.0, software (Applied Biosystems Division, Perkin-Elmer Corp., Foster City, Calif.) was used to design oligonucleotides targeting the L. monocytogenes hly gene (GenBank accession no. M24199) (27) and iap gene (GenBank accession no. X52268) (23) and the L. innocua lin02483 gene (http://genolist.pasteur.fr/ListiList/). The oligonucleotides were purchased from MWG-Biotech AG (Ebensburg, Germany).
PCR.
TaqMan RTi-PCR assays were performed and evaluated essentially as described by Hernández et al. (13) with TaqMan PCR core reagents (Applied Biosystems-Roche Molecular Systems Inc., Branchburg, N.J.) and a 20-μl reaction volume containing 1× PCR TaqMan buffer A (including 5-carboxy-X-rhodamine [ROX] as a passive reference dye); 4.5 mM (iap reactions) or 6 mM (hly and lin02483 reactions) MgCl2; 200 μM each dATP, dCTP, and dGTP; 400 μM dUTP; 50 nM primers; 100 nM probe; 1 U of AmpliTaq Gold DNA polymerase; 0.2 U of AmpErase uracil N-glycosylase; and 1 μl of the target DNA solution. Reactions were run on an ABI Prism 7700 apparatus (Applied Biosystems Division, Perkin-Elmer) with the following program: 2 min at 50°C, 10 min at 95°C, and 50 cycles of 15 s at 95°C and 1 min at 63°C. AmpliFluor RTi-PCR assays were performed with a 20-μl reaction volume containing 1× Ex Taq buffer (TaKaRa Bio, Inc., Shiga, Japan), 1.5 mM MgCl2, 250 μM each deoxynucleoside triphosphate, 50 nM hlyZ primer, 500 nM hlyQR primer, 500 nM UniPrimer (Intergen Co., Purchase, N.Y.), 1 U of TaKaRa Ex Taq polymerase, and 1 μl of the target DNA solution. The conditions for AmpliFluor RTi-PCR assays were 4 min at 95°C and 45 cycles of 15 s at 95°C, 20 s at 55°C, and 40 s at 72°C. Fluorescence was measured only at the melting point.
TaqMan and AmpliFluor RTi-PCR assays were evaluated by using sequence detection system software, version 1.7 (Applied Biosystems Division, Perkin-Elmer). Quantification was performed by interpolation in a standard regression curve of threshold cycle (CT) values generated from samples at known concentrations. Negative values or a lack of amplification for RTi-PCR was set at a CT value of >50 or >45 for the TaqMan or the AmpliFluor system, respectively. Unless otherwise stated, all reactions were performed in triplicate. The 95% confidence interval was calculated for every serial dilution. The calculations were performed according to a binomial distribution (21) by using the SAS statistical software system for Windows, version 8.0 (SAS Institute Inc., Cary, N.C.).
Conventional PCR assays were performed under the same conditions as those used for TaqMan RTi-PCR assays, except that PCR buffer II was used instead of PCR TaqMan buffer A. PCR products were detected by ethidium bromide staining after electrophoresis in 3% agarose gels.
Sequencing of hly and iap gene fragments.
L. monocytogenes genomic DNA was PCR amplified with primers hlyF and hlyR (512-bp fragment) and primers iapF and iapR (687-bp fragment) (Table 3) in 50-μl reaction mixtures containing 1× PCR buffer II, 1.5 mM MgCl2, 200 μM each deoxynucleoside triphosphate, 0.9 μM primers, and 1 U of AmpliTaq Gold DNA polymerase. Reactions were performed by using GeneAmp PCR system 9600 (Applied Biosystems Division, Perkin-Elmer) and the following program: 10 min at 95°C; 40 cycles of 20 s at 95°C, 30 s at 56°C (hly) or 53°C (iap), and 1 min at 72°C; and a final extension of 7 min at 72°C. The PCR products were purified by using a QIAEXII gel extraction kit (Qiagen, Hilden, Germany) and sequenced on both strands with the same primers by using an ABI Prism Big Dye Terminator, version 3.0, cycle sequencing kit and an ABI Prism 377 DNA sequencer (Applied Biosystems Division, Perkin-Elmer).
TABLE 3.
Oligonucleotides used in RTi-PCR assays for L. monocytogenes and L. innocua and target gene sequencing
| Use | Target gene | Name | Type | Sequence |
|---|---|---|---|---|
| TaqMan RTi-PCR | hly | hlyQFa | Forward primer | 5′-CAT GGC ACC ACC AGC ATC T-3′ |
| hlyQRa | Reverse primer | 5′-ATC CGC GTG TTT CTT TTC GA-3′ | ||
| hlyQP | TaqMan probe | 5′-FAM-CGC CTG CAA GTC CTA AGA CGC CA-TAMRA-3′ | ||
| iap | iapQFa | Forward primer | 5′-AAT CTG TTA GCG CAA CTT GGT TAA-3′ | |
| iapQRa | Reverse primer | 5′-CAC CTT TGA TGG ACG TAA TAA TAC TGT T-3′ | ||
| iapQP | TaqMan probe | 5′-FAM-CAA CAC CAG CGC CAC TAC GGA CG-TAMRA-3′ | ||
| lin02483 | lipHQFa | Forward primer | 5′-AAC CGG GCC GCT TAT GA-3′ | |
| lipHQRa | Reverse primer | 5′-CGA ACG CAA TTG GTC ACG-3′ | ||
| lipHQP | TaqMan probe | 5′-FAM-TTC GAA TTG CTA GCG GCA CAC CAG T-TAMRA-3′ | ||
| AmpliFluor RTi-PCR | hly | hlyZ | AmpliFluor primer | 5′-act gaa cct gac cgt aca CAT GGC ACC ACC AGC ATC T-3′b |
| hlyQR | Reverse primer | 5′-ATC CGC GTG TTT CTT TTC GA-3′ | ||
| Sequencing | hly | hly-F | Forward primer | 5′-TAA CGA CGA TAA AGG GAC AGC AGG ACT A-3′ |
| hly-R | Reverse primer | 5′-AAT GAA TCA CGT TTT ACA GGG AGA A-3′ | ||
| iap | P60-F | Forward primer | 5′-TAA AGG GAC TAC TGT TGA CG-3′ | |
| P60-R | Reverse primer | 5′-GCT TCT GTT GGT GCT TTA GGT GCT GTT TG-3′ |
Also used for conventional PCR.
The sequence in lowercase type corresponds to the Z tail sequence of the AmpliFluor primer.
Nucleotide sequence accession numbers.
The partial hly and iap DNA sequences from 13 different L. monocytogenes strains (Table 1) can be found in the GenBank database under accession numbers AY174657 to AY174669 (hly) and AY174670 to AY17682 (iap).
RESULTS
Design and optimization of L. monocytogenes- and L. innocua-specific RTi-PCR assays.
Regions suitable for the design of L. monocytogenes-specific PCR primers and probes were identified by aligning all hly and iap sequences deposited in public databases by using the CLUSTALW multiple-alignment tool (European Bioinformatics Institute, EMBL; www.ebi.ac.uk). L. innocua-specific oligonucleotides were designed on the basis of the lin02483 gene (10). The BLAST-N tool (National Center for Biotechnology Information; www.ncbi.nlm.nih.gov) was used to confirm that none of the selected oligonucleotides (Table 3) recognized any registered DNA sequence other than the target sequence. Primer pair hlyQF-hlyQR amplified a 64-bp fragment from the L. monocytogenes hly gene (positions 113 to 177). Primer pair iapQF-iapQR amplified a 77-bp fragment within the 5′ conserved region of the L. monocytogenes iap gene (nucleotides 242 to 319). Primer pair lipHQF-lipHQR amplified a 62-bp fragment from the L. innocua lin02483 gene (positions 265 to 327). All of these amplicons are of optimal sizes for RTi-PCR and can be detected in agarose gels.
Primer, TaqMan probe, and MgCl2 concentrations were optimized for TaqMan RTi-PCR assays by using as a template 1 ng of DNA from L. monocytogenes strain UdG 1010 or L. innocua strain CECT 910. Optimal conditions (described in Materials and Methods) were the minimum primer and probe concentrations giving the lowest CT value and the highest fluorescence intensity for a normalized reporter value (Perkin-Elmer Applied Biosystems User Bulletin 2 [ABI Prism 7700 sequence detection system], 1997). These conditions yielded the largest quantity of amplification product from the corresponding target DNA in conventional PCR assays.
Specificity of the assays.
The capacity of our PCR assays to discriminate between target and nontarget bacteria was tested by using as a template 1 ng of genomic DNA (≈3 × 105 CFU) from 100 Listeria strains and 45 non-Listeria strains. Only the target species were detected by both RTi-PCR and conventional PCR (Tables 1 and 2). Tests were also performed with a representative set of strains (13 L. monocytogenes, 4 L. innocua, 21 other Listeria, and 33 non-Listeria strains) by using as a template either 1 μl of an overnight liquid culture or a colony from an agar plate. The results were the same as those obtained with purified genomic DNA. These data indicated that the PCR assays were specific for L. monocytogenes and L. innocua.
Sensitivity and quantification range of the assays.
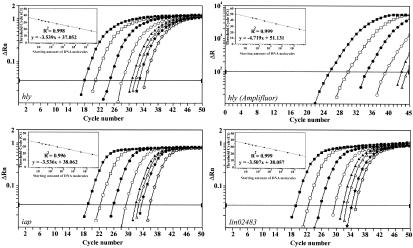
The detection and quantification limits of the PCR assays were determined by using genomic DNA isolated from overnight cultures of L. monocytogenes strain UdG 1010 and L. innocua strain CECT 910. Amplification reactions were performed with a range of DNA concentrations equivalent to approximately 3 × 105, 3 × 104, 3 × 103, 3 × 102, 60, 30, 15, 8, 4, and 1 target molecules. On the basis of the sizes of the L. monocytogenes and L. innocua genomes (10), one molecule of genomic DNA corresponds to 2.94 and 3.01 fg of DNA, respectively. Figure 1 illustrates the amplification profiles and the regression curves obtained with each RTi-PCR assay; Table 4 shows the mean CT values for a total of nine replicates in three independent experiments. The RTi-PCR assays yielded similar results in terms of absolute detection values. Positive amplification in all nine replicates of each DNA dilution was achieved when 8 or more target molecules were present (15 for lin02483), and as few as 1 target molecule could be detected with 33 to 55% probability (11% for lin02483) (Table 4). Conventional reactions consistently detected the target molecules when at least 60 target molecules were present and could detect 15 target molecules with a 44% probability.
FIG. 1.
RTi-PCR detection and amplification of the hly, iap, and lin02483 sequences. Representative amplification plots are shown. Serial dilutions of L. monocytogenes or L. innocua genomic DNA, equivalent to 3 × 105 (▪), 3 × 104 (□), 3 × 103 (•), 3 × 102 (○), 60 (▴), 30 (▵), 15 (★), and 8 (⋆) target molecules per reaction, were used. Note that the AmpliFluor reactions could not detect 15 and 8 target molecules. Insets show representative standard curves generated from the amplification data. ΔRn, normalized reporter value (with ROX). ΔR, reporter value (without ROX).
TABLE 4.
Detection and quantification limits of RTi-PCR assays with genomic DNA from standard curve strains L. monocytogenes UdG 1010 and L. innocua CECT 910
| Gene | Approx no. of template molecules | Confidence interval limita
|
Signal ratio (positive signals/ 9 reactions) | CT value
|
||
|---|---|---|---|---|---|---|
| Lower | Upper | Mean | SD | |||
| hly | 3 × 105 | 298,928 | 301,073 | 9 | 18.16 | 0.05 |
| 3 × 104 | 29,661 | 30,340 | 9 | 21.27 | 0.05 | |
| 3 × 103 | 2,893 | 3,108 | 9 | 24.87 | 0.09 | |
| 3 × 102 | 267 | 334 | 9 | 28.43 | 0.15 | |
| 60 | 45 | 76 | 9 | 30.75 | 0.15 | |
| 30 | 20 | 41 | 9 | 32.17 | 0.19 | |
| 15 | 8 | 23 | 9 | 33.42 | 0.34 | |
| 8 | 3 | 13 | 9 | 34.99 | 0.78 | |
| 4 | 1 | 8 | 8 | 37.23 | 4.81 | |
| 1 | 0 | 3 | 3 | 45.12 | 7.32 | |
| hly (AmpliFluor) | 3 × 105 | 298,928 | 301,073 | 9 | 24.67 | 0.18 |
| 3 × 104 | 29,661 | 30,340 | 9 | 29.14 | 0.14 | |
| 3 × 103 | 2,893 | 3,108 | 9 | 34.17 | 0.04 | |
| 3 × 102 | 267 | 334 | 9 | 39.29 | 0.08 | |
| 60 | 45 | 76 | 9 | 43.81 | 0.21 | |
| 30 | 20 | 41 | 7 | 44.53 | 0.37 | |
| 15 | 8 | 23 | 4 | 44.77 | 0.40 | |
| 8 | 3 | 13 | 0 | 45.00 | 0.00 | |
| 4 | 1 | 8 | 0 | 45.00 | 0.00 | |
| 1 | 0 | 3 | 0 | 45.00 | 0.00 | |
| iap | 3 × 105 | 298,928 | 301,073 | 9 | 18.97 | 0.09 |
| 3 × 104 | 29,661 | 30,340 | 9 | 22.20 | 0.20 | |
| 3 × 103 | 2,893 | 3,108 | 9 | 25.94 | 0.33 | |
| 3 × 102 | 267 | 334 | 9 | 29.41 | 0.23 | |
| 60 | 45 | 76 | 9 | 31.95 | 0.31 | |
| 30 | 20 | 41 | 9 | 32.99 | 0.52 | |
| 15 | 8 | 23 | 9 | 34.31 | 0.60 | |
| 8 | 3 | 13 | 9 | 36.38 | 1.18 | |
| 4 | 1 | 8 | 7 | 39.46 | 6.01 | |
| 1 | 0 | 3 | 5 | 42.34 | 7.36 | |
| lin02483 | 3 × 105 | 298,928 | 301,073 | 9 | 17.25 | 0.19 |
| 3 × 104 | 29,661 | 30,340 | 9 | 20.93 | 0.18 | |
| 3 × 103 | 2,893 | 3,108 | 9 | 24.60 | 0.26 | |
| 3 × 102 | 267 | 334 | 9 | 28.52 | 0.26 | |
| 60 | 45 | 76 | 9 | 30.55 | 0.20 | |
| 30 | 20 | 41 | 9 | 32.11 | 0.44 | |
| 15 | 8 | 23 | 9 | 34.10 | 0.70 | |
| 8 | 3 | 13 | 7 | 37.87 | 6.88 | |
| 4 | 1 | 8 | 4 | 41.80 | 7.89 | |
| 1 | 0 | 3 | 1 | 48.36 | 4.93 | |
Calculated for the expected number of template molecules at each dilution at the 95% confidence level.
The slopes of the linear regression curves calculated over a 5-log range were similar to the theoretical optimum of −3.32 (17) and showed that the amplification rates were very efficient (−3.49, hly; −3.52, iap; and −3.67, lin02483). Moreover, R2 values were above 0.996, indicating that the RTi-PCR systems were highly linear. The confidence intervals based on the standard deviations of CT values did not overlap each other down to 15 target molecules, indicating that reliable quantification was possible above this limit. Experimental results were also statistically significant (P < 0.05), taking into consideration the error associated with the serial dilutions.
The sensitivity of the RTi-PCR assays was investigated by using intact bacterial cells instead of DNA. Tenfold dilutions of overnight cultures of L. monocytogenes UdG 1010 and L. innocua CECT 910 were used as templates in the RTi-PCR assays and were plated in parallel to count the bacterial CFU. The overall detection limit for the RTi-PCR assays was 30 CFU, although just 3 CFU were detected in 55.55% (lin02483), 66.66% (iap), and 88.89% (hly) of the replicates (Table 5). Linear regression analysis of CT values and bacterial numbers in the reactions yielded R2 values (above 0.996) and slopes (Table 5) similar to those obtained with purified genomic DNA, indicating that our RTi-PCR assays potentially can be used to quantify accurately the L. monocytogenes or L. innocua populations present in a sample.
TABLE 5.
Detection and quantification limits of RTi-PCR assays with suspensions of standard curve strains L. monocytogenes UdG 1010 and L. innocua strain CECT 910a
| Species | Gene | Slope | R2 | Mean ± SD CFU in the reaction (3 replicates) | Signal ratio (positive signals/9 reactions) | CT values
|
|
|---|---|---|---|---|---|---|---|
| Mean | SD | ||||||
| L. monocytogenes | hly | −3.81 | 0.998 | 291,250 ± 13,150 | 9 | 18.01 | 0.11 |
| 29,125 ± 1,315.0 | 9 | 21.33 | 0.08 | ||||
| 2,912 ± 131.50 | 9 | 25.00 | 0.08 | ||||
| 291 ± 13.15 | 9 | 29.33 | 0.10 | ||||
| 29 ± 1.31 | 9 | 33.10 | 0.39 | ||||
| 3 ± 0.13 | 8 | 37.54 | 4.71 | ||||
| hly (AmpliFluor) | −3.84 | 0.996 | 291,250 ± 13,150 | 9 | 27.54 | 0.09 | |
| 29,125 ± 1,315.0 | 9 | 30.85 | 0.30 | ||||
| 2,912 ± 131.50 | 9 | 35.38 | 0.36 | ||||
| 291 ± 13.15 | 9 | 38.83 | 0.34 | ||||
| 29 ± 1.31 | 2 | 44.30 | 1.40 | ||||
| 3 ± 0.13 | 0 | 45.00 | 0.00 | ||||
| iap | −3.71 | 0.996 | 291,250 ± 13,150 | 9 | 17.72 | 0.19 | |
| 29,125 ± 1,315.0 | 9 | 20.91 | 0.32 | ||||
| 2,912 ± 131.50 | 9 | 24.24 | 0.23 | ||||
| 291 ± 13.15 | 9 | 28.84 | 0.12 | ||||
| 29 ± 1.31 | 9 | 32.29 | 0.46 | ||||
| 3 ± 0.13 | 6 | 40.20 | 7.40 | ||||
| L. innocua | lin02483 | −3.85 | 0.997 | 231,661 ± 13,714 | 9 | 17.75 | 0.14 |
| 23,166 ± 1,371.4 | 9 | 20.93 | 0.17 | ||||
| 2,317 ± 137.14 | 9 | 24.71 | 0.23 | ||||
| 232 ± 13.71 | 9 | 29.21 | 0.25 | ||||
| 23 ± 1.38 | 9 | 32.95 | 0.48 | ||||
| 2 ± 0.14 | 5 | 42.81 | 6.88 | ||||
Regression curves were calculated from approximately 3 × 105 to 30 template molecules.
Relative accuracy of quantification.
A series of experiments were conducted with the same bacterial cultures to determine the exact degree of correspondence between the quantitative data obtained by the RTi-PCR assays and those obtained by the standard plate count technique (1), used as a reference method (i.e., the relative accuracy, defined as the closeness of the agreement between a test result and the accepted reference value [2]). The CT values determined with serial 10-fold dilutions of overnight cultures of our reference strains, L. monocytogenes UdG 1010 and L. innocua CECT 910, were extrapolated to the corresponding standard regression curves, previously calculated experimentally, and the resulting theoretical CFU numbers were compared to those obtained by plate counting (Table 6). The values differed by less than 23% over a 5-log range, indicating excellent relative accuracy.
TABLE 6.
Quantification accuracy of RTi-PCR assays for L. monocytogenes and L. innocuaa
| Strain | Mean ± SD CFU in the reaction (3 replicates) | Theoretical CFU (% of actual CFU) determined with the following gene:
|
|||
|---|---|---|---|---|---|
| hly | hly (AmpliFluor) | iap | lin02483 | ||
| L. monocytogenes UdG 1010 | 291,250 ± 13,150 | 268,389 (92) | 265,836 (91) | 216,679 (74) | |
| 29,125 ± 1,315 | 3,391 (116) | 36,634 (126) | 29,335 (101) | ||
| 2,912 ± 131.50 | 3,453 (119) | 2,419 (83) | 3,646 (125) | ||
| 291 ± 13.15 | 234 (80) | 305 (105) | 204 (70) | ||
| 29 ± 1.31 | 22 (77) | + | 24 (81) | ||
| L. monocytogenes CECT 935 | 199,667 ± 11,060 | 168,518 (84) | 2,658 (1.3) | ||
| 19,967 ± 1,106 | 15,039 (75) | 60 (0.3) | |||
| 1,997 ± 110.60 | 2,429 (121) | + | |||
| 200 ± 11.06 | 218 (109) | − | |||
| 20 ± 1.11 | 18 (88) | − | |||
| L. innocua CECT 910 | 231,661 ± 13,714 | 190,956 (82) | |||
| 23,166 ± 1,371.4 | 26,443 (114) | ||||
| 2,317 ± 137.14 | 2,989 (129) | ||||
| 232 ± 13.71 | 203 (88) | ||||
| 23 ± 1.38 | 22 (94) | ||||
CT values determined with bacterial cultures were extrapolated to regression curves previously obtained with bacterial cell suspensions to calculate the theoretical CFU per reaction. The actual CFU were determined in parallel by standard plate counting (reference method), and the deviation with respect to the theoretical value was expressed as a percentage (i.e., relative accuracy [2]). The assays were carried out with standard curve strains and L. monocytogenes CECT 935 to illustrate the different performances of the hly- and iap-based assays, depending on the homology group to which the test strain belonged. Note the lack of quantifiability of strain CECT 935, belonging to serovar 4b (division I), i.e., from a homology group different from that of standard curve strain UdG 1010, of serovar 1/2c (division II), with the iap-based RTi-PCR. +, positive reaction below the limit of quantification; −, amplification not detectable.
Ribotyping and virulence gene polymorphism studies have shown that most L. monocytogenes isolates belong to two main serovar-related evolutionary branches: division I, comprising serovars 1/2b, 3b, and 4b (and some serovar 3c and 4c isolates), and division II, comprising serovars 1/2a, 1/2c, and 3a. A more distant subbranch, division III, comprises serovar 4a and most serovar 4c isolates (4, 19, 28, 32-34, 42, 44). The strain that we used to construct the standard curve, UdG 1010, belongs to serovar 1/2c, as does strain L028 (27), used to design the hlyQF and hlyQR primers. Virtually no sequence polymorphisms exist between serovar 1/2c and serovar 1/2a, which includes strain EGD-e (23), used to design the iapQF and iapQR primers. We therefore tested a strain from another homology group, CECT 935, of serovar 4b (division I). The quantification accuracy was unaffected in the hly-based RTi-PCR assay; however, the iap-based RTi-PCR assay underestimated the real bacterial load by 3 to 4 log units (Table 6).
Impact of target gene sequence polymorphisms on the quantitative performance of the L. monocytogenes-specific RTi-PCR.
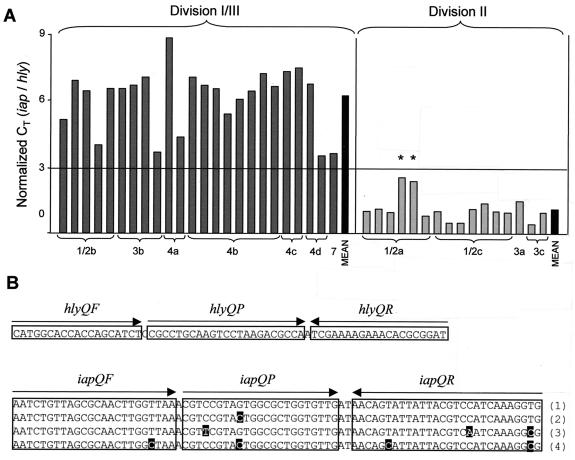
The above results prompted us to investigate in more detail the effect of interstrain hly and iap target sequence polymorphisms on our RTi-PCR assays. The hly- and iap-based RTi-PCR assays were carried out by using 1 ng of purified DNA from 40 strains representing most of the known L. monocytogenes serovars (Table 1), and the mean CT values were calculated from four independent experiments. For hly-targeted reactions, the mean CT values were all within a range of 1 cycle and had an overall standard deviation of 0.46. Conversely, the CT values obtained with the iap-based RTi-PCR assay had strong deviations, up to 7.8 cycles, depending on the L. monocytogenes strain tested. Interestingly, the CT values were distributed into two categories that coincided with the serotype-related phylogenetic divisions of L. monocytogenes (see above). Thus, strains of division II serovars had CT values (mean and standard deviation) of 19.24 ± 0.79, whereas strains of division I or III serovars had significantly higher (P < 0.05) CT values (23.80 ± 1.30). The results of these analyses are represented in Fig. 2A, in which the iap CT values were normalized to those of the hly reaction (i.e., iap CT − hly CT) as a reference to control for possible variability due to differences in DNA quantity or quality. According to our data, L. monocytogenes strains belonging to division II have an iap/hly CT value of <3 (mean value, 1.09), and those belonging to divisions I and III have an iap/hly CT value of >3 (mean value, 5.56) (Fig. 2A). Interestingly, our assay identified serovars 4d and 7 as belonging to division I or III.
FIG. 2.
Effect of target gene sequence polymorphisms on the performance of the iap RTi-PCR assay. (A) Normalized CT values (iap/hly; see the text) obtained with 40 L. monocytogenes strains of different serovars (indicated below the graphs). The results for each strain (mean of four replicates) are represented by bars. Note that the strains can be ascribed to two statistically significant groups (P < 0.05) which coincide with serovar-related phylogenetic division I or III (serovars 1/2b, 3b, 4a, 4b, 4c, 4d, and 7) and division II (serovars 1/2a, 1/2c, 3a, and 3c) of L. monocytogenes delimited by an iap/hly CT value of 3. Note also that two strains of serovar 1/2a have higher iap/hly CT values (indicated by an asterisk); these strains harbor a mismatch in the target sequence of the probe (see panel B, sequence 2, black shading). (B) Nucleotide sequences of hly and iap fragments targeted by PCR primers and probes (boxed, with arrows indicating the 5′→3′ orientation) (Table 3). The bottom panel shows strains CECT 911, CECT 938, UdG 1010, and UdG 1011 (sequence 1); strains CECT 932 and CECT 4031 (sequence 2); strains CECT 935, CECT 936, CECT 937, CECT 940, CECT 4032, and UdG 1034 (sequence 3); and strain CECT 934 (sequence 4) (Table 1). Mismatches are represented by black shading.
We sequenced the iap target region from a sample of 13 representative L. monocytogenes strains to identify the polymorphisms underlying the observed variability in quantitative RTi-PCR performance. The hly target region was also sequenced as a control. No mismatches were detected in any of the sequences corresponding to the hly-specific primers and probes (Fig. 2B). Multiple alignments of the sequenced 512-bp hly fragment, including that derived from strain EGD-e, of serovar 1/2a (10), showed a high degree of interstrain sequence conservation, with only minor differences between serovars in division II and division I or III (96.7 to 100% identity) (nucleotides −143 to +369 of the sequence in GenBank accession no. M24199). The iap target sequences from strains of division II serovars (iap/hly CT value, <3), which were perfectly quantifiable with our iap-based assay, were also identical to the primer and probe oligonucleotides (with the exception of a single mismatch within the probe target sequence in a serovar 1/2a strain). In contrast, target sequences from strains of division I or III serovars (iap/hly CT value, >3), which showed poor quantification results, exhibited three or four mismatches compared to primer and probe oligonucleotides (Fig. 2B). The sequence of the 687-bp iap fragment analyzed (positions +148 to +835 of the sequence in GenBank accession no. X52268) confirmed that the overall degree of divergence in the iap gene was high (pair distances as low as 89.5%) and included transitions, transversions, and deletions, with clear differences between division II and division I or III serovars.
Assessment of AmpliFluor technology.
We also carried out a series of experiments to assess the performance of AmpliFluor technology for the quantitative detection of L. monocytogenes by RTi-PCR. We used the same hlyQF and hlyQR primers, the former including a Z tail sequence (hlyZ), and the optimal conditions described in Materials and Methods. For the same samples, AmpliFluor assays were as specific as TaqMan assays, although the detection limit of the former was slightly higher (Table 4), equivalent to that of conventional PCR assays. The AmpliFluor reactions were efficient and showed a linear and accurate relationship between the CT value and the initial DNA concentration over a 5-log range (slope of the linear regression curve, −4.67; R2, 0.999). Quantification was reliable when 60 or more target molecules were present. The AmpliFluor assays also performed adequately when L. monocytogenes cells were used as a template (Table 5), allowing reliable quantification of 300 target CFU and detection of 30 CFU in 22% of the replicates.
DISCUSSION
In this study, we developed and compared quantitative RTi-PCR assays directed against the listerial virulence genes hly and iap, two targets commonly used in PCR-based methods for detection of the food-borne pathogen L. monocytogenes (12, 18, 24, 29). We also developed a quantitative RTi-PCR assay targeting the lin02483 gene of L. innocua, a nonpathogenic species closely related to L. monocytogenes that is also frequently found in foods but is harmless to the consumer (8, 11, 41). The availability of rapid molecular assays able to distinguish among these Listeria spp. would be of great value for assessing the risks of food-borne listeriosis and preventing the unnecessary recall and destruction of valuable food products. As these Listeria spp. are genetically and ecologically similar (10, 40, 41), the detection and quantification of L. innocua could also be a useful indicator of potential contamination by L. monocytogenes in food plants. Our RTi-PCR assays unequivocally distinguished target samples from nontarget samples, demonstrating their absolute specificity. Moreover, they performed perfectly not only with purified DNA but also directly with broth- or agar-grown bacterial cultures, meaning that it should be possible to use them for the specific identification of L. monocytogenes and L. innocua under routine laboratory conditions.
Perhaps one of the most challenging aspects of designing methods for the detection of food-borne pathogens is achievement of a low detection limit. This goal is of particular interest for L. monocytogenes, as it is often present in low numbers in food products (7, 8). The RTi-PCR assays that we developed could detect approximately one target genome in at least 11% of the experiments, 3 CFU in 55 to 89% of replicates, and 30 CFU in all cases. Detection limits were similar to those previously reported for other RTi-PCR assays targeting single-copy genes (14, 15, 16, 29). Limits of detection of 9 (18), 6 to 60 (29), and 500 (24) target molecules were previously reported for hly, and six genome copies were reported for iap (12).
The quantification accuracy of our assays was excellent compared to that of the reference microbiological method (relative quantification accuracy) (2) under optimal conditions; i.e., the RTi-PCR results were very similar to the quantitative results obtained by the plate count method over a wide range of CFU for the standard curve strains. However, the performance of RTi-PCR and, thus, its detection limit and quantification accuracy, depend on the degree of conservation of the target sequence(s) of the particular microorganism being analyzed. Strains of L. monocytogenes have been divided into three serovar-related homology groups or divisions on the basis of gene sequence diversity (19, 26, 28, 33, 42-44). To ensure that our hly- and iap-based RTi-PCR assays were appropriate for the precise quantification of an L. monocytogenes strain regardless of its genetic background, we extensively evaluated our oligonucleotides by using a large panel of isolates of serovars representative of the three phylogenetic divisions of the species. The hly-based RTi-PCR assay always yielded perfectly homogeneous results with all of the strains tested and had excellent quantification accuracy for bacteria from serovars belonging to different homology groups (i.e., divisions I, II, and III), indicating that this assay can be applied to the entire species L. monocytogenes. Preliminary results obtained with different L. innocua strains indicated that the species-specific lin02483 RTi-PCR assay behaves similarly.
The situation was completely different with the iap-based RTi-PCR assay. Although this assay targets the 5′ conserved portion of the iap gene, our results demonstrated that it was reliable only for L. monocytogenes strains from the same homology group as the strain used to design the primers and probe (division II). Although strains from different homology groups (divisions I and III) yielded positive amplification signals, the efficiencies of the reactions were too low, and quantification was not possible. These results indicate that the iap-based assay cannot be routinely used for the quantitative detection of L. monocytogenes under field conditions. These results also highlight the need to assess primers and probes for quantification purposes carefully and exhaustively, by using a large and comprehensive collection of isolates representative of the biodiversity within the target species, before an RTi-PCR assay is adopted for routine quantitative testing. Finally, they show that target sequences prone to genetic variability must be avoided even when a conserved region is selected. This issue is not trivial, because a number of RTi-PCR assays targeting iap were recently described for the quantitative detection of L. monocytogenes (12, 24). The frequency of spontaneous mutations in prokaryotes, aggravated by the relative abundance of hypermutation phenotypes among certain pathogenic bacteria (3, 30), therefore appears to be a potential limitation of RTi-PCR-based methods for the quantitative detection of bacterial pathogens in natural samples.
Analysis of the target sequences in a sample of isolates representative of the three homology divisions of L. monocytogenes showed that the number of mismatches in the primer and probe sequences is directly correlated with the efficiency of the RTi-PCR. Strains with target sequences containing three or four mismatches in the iap-specific primers and probe (division I or III serovars) were inefficiently amplified and had CT values about 7 cycles higher than those obtained in the hly-based RTi-PCR (Fig. 2). In contrast, strains with target sequences identical to the iap-specific primers and probe (division II serovars) were very efficiently amplified and had CT values close to those obtained in the hly-based assay. It must be noted that two strains of serovar 1/2a (division II) exhibited a single mismatch in the probe sequence which slightly increased the CT value (Fig. 2). The observed correlation allowed us to establish a cutoff for iap-based RTi-PCR CT values (3 cycles after normalization to hly-based CT values) that divided the L. monocytogenes strains into two statistically different groups corresponding to division II and to division I or III (Fig. 2A). An example is strain PAM 602; this strain was originally classified as L. seeligeri but, according to its CT value, corresponded to a division I L. monocytogenes strain. Reidentification and serotype determination confirmed that PAM 602 was indeed L. monocytogenes serovar 1/2a (Table 1), suggesting that the iap-based RTi-PCR assay could be used in conjunction with the “universal” hly-based assay for rapid discrimination between division II and division I or III L. monocytogenes strains. This application is interesting because most clinical isolates of L. monocytogenes belong to division I (primarily serovar 4b, followed by serovar 1/2b), whereas division II isolates are less frequently associated with disease (in particular, serovar 1/2c, which is frequently found in food but rarely found in listeriosis patients) (4, 28, 33, 38, 42, 44, 45). Within division II, the iap-based assay discriminated strains of serovar 1/2a harboring point mutations in the probe sequence (serovar 1/2a has been shown to split into two sequence homology groups) (39) (Fig. 2A).
Finally, we also assessed AmpliFluor technology for the quantification of L. monocytogenes by using primers targeting the hly gene. We compared the efficiency of this technique with that of the TaqMan system. The AmpliFluor assay was, like the conventional PCR assay, slightly less sensitive, as the detection limit was 15 target molecules. The quantification limit was also slightly less favorable with the AmpliFluor assay (60 target molecules versus 15 with the TaqMan assay). Otherwise, the accuracies were high with both techniques (R2 values of >0.996), and the linearity persisted up to 3 × 106 molecules (data not shown). Although in our experiments the AmpliFluor technique was slightly less efficient than the TaqMan technique, it offers the theoretical advantage of being less sensitive to mutations in the target DNA because it does not require a third (probe) target sequence. It is also more cost-effective when different genes are to be targeted because of the use of the universal fluorigenic UniPrimer and is easy to adapt on the basis of previously established conventional PCR systems. This is the first time that a universal RTi-PCR probe (UniPrimer, based on a Z tail sequence, which is claimed to be nonexistent in nature) has been used for the specific detection and quantification of a food-borne pathogen. We show experimentally that it does not cross-react with a number of bacterial species (Tables 1 and 2).
In conclusion, we report highly specific, sensitive, and reliable RTi-PCR assays for the quantitative detection of L. monocytogenes and L. innocua. The assays show excellent quantification characteristics in terms of both linear dynamic range and relative accuracy with respect to the standard plate count technique, thus offering a promising alternative to traditional microbiological methods. We also warn about the dangers of using genes prone to genetic variability as targets for RTi-PCR-based detection of food-borne pathogens and emphasize the necessity of extensively checking assays with the broadest possible panel of representative strains, as interstrain sequence polymorphisms affecting target sequences can result in an underestimation of the bacterial load present in a sample by several orders of magnitude.
Acknowledgments
This work was supported by the Spanish Ministerio de Ciencia y Tecnología (grants INIA CAL01-058-C2-2 and AGL2002-03496). D.R.-L. received a studentship from the University of Girona (BR01-08), and M.H. was supported by the EU (project QLRT-1999-01301).
We thank M. Hugas and N. Cook for providing bacterial strains; H. Monzó for help with DNA extractions; and C. Jacquet, J. Rocourt, and P. Martin (Listeria Reference Laboratory, Institut Pasteur, Paris, France) for serotyping PAM isolates.
REFERENCES
- 1.Anonymous. 1998. Microbiology of food and animal feeding stuffs—horizontal method for the detection and enumeration of Listeria monocytogenes. Part 2. Enumeration method. ISO 11290-2. International Organization for Standardization, Geneva, Switzerland.
- 2.Anonymous. 2002. Microbiology of food and animal feeding stuffs—protocol for the validation of alternative method ISO/DIS 16140. International Organization for Standardization, Geneva, Switzerland.
- 3.Bayliss, C. D., and E. R. Moxon. 2002. Hypermutation and bacterial adaptation. ASM News 68:549-555. [Google Scholar]
- 4.Boerlin, P., F. Boerlin-Petzold, E. Bannerman, J. Bille, and T. Jemmi. 1997. Typing Listeria monocytogenes isolates from fish products and human listeriosis cases. Appl. Environ. Microbiol. 63:1338-1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bubert, A., S. Köhler, and W. Goebel. 1992. The homologous and heterologous regions within the iap gene allow genus- and species-specific identification of Listeria spp. by polymerase chain reaction. Appl. Environ. Microbiol. 58:2625-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dalton, C. B., C. C. Austin, J. Sobel, P. S. Hayes, W. F. Bibb., L. M. Graves, B. Swaminathan, M. E. Proctor, and P. M. Griffin. 1997. An outbreak of gastroenteritis and fever due to Listeria monocytogenes in milk. N. Engl. J. Med. 336:100-105. [DOI] [PubMed] [Google Scholar]
- 7.Donnelly, C. W. 1999. Conventional methods to detect and isolate Listeria monocytogenes, p. 225-260. In E. T. Ryser and E. H. Marth (ed.), Listeria, listeriosis, and food safety, 2nd ed. Marcel Dekker, Inc., New York, N.Y.
- 8.Farber, J. M., and P. I. Peterkin. 1991. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 55:476-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gallagher, L., E. D. Ebel, and J. R. Kause. 2003. Draft FSIS risk assessment for Listeria in ready-to-eat meat and poultry products. Food Safety and Inspection Service, Washington, D.C.
- 10.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Domínguez-Bernal, E. Duchaud, L. Durant, T. O. Dussurge, K. D. Entian, H. Fsihi, F. G. Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gómez-López, T. Hain, J. Huf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Pérez-Díaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vázquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 11.Gravani, R. 1999. Incidence and control of Listeria in food-processing facilities, p. 657-709. In E. T. Ryser and E. H. Marth (ed.), Listeria, listeriosis, and food safety, 2nd ed. Marcel Dekker, Inc., New York, N.Y.
- 12.Hein, I., D. Klein, A. Lehner, A. Bubert, E. Brandl, and M. Wagner. 2001. Detection and quantification of the iap gene of Listeria monocytogenes and Listeria innocua by a new real-time quantitative PCR assay. Res. Microbiol. 152:37-46. [DOI] [PubMed] [Google Scholar]
- 13.Hernández, M., A. Rio, T. Esteve, S. Prat, and M. Pla. 2001. A rapeseed-specific gene, acetyl-CoA carboxylase, can be used as a reference for qualitative and real-time quantitative PCR detection of transgenes from mixed food samples. J. Agric. Food Chem. 49:3622-3627. [DOI] [PubMed] [Google Scholar]
- 14.Hernández, M., M. Pla, T. Esteve, S. Prat, P. Puigdomènech, and A. Ferrando. 2003. A specific real-time quantitative PCR detection system for event MON810 in maize YieldGard based on the 3-transgene integration sequence. Transgenic Res. 12:179-189. [DOI] [PubMed] [Google Scholar]
- 15.Hernández, M., A. Ferrando, T. Esteve, P. Puigdomènech, S. Prat, and M. Pla. 2003. Real-time and conventional PCR systems based on the metallocarboxypeptidase inhibitor gene for specific detection and quantification of potato and tomato in processed food. J. Food Prot. 66:1063-1070. [DOI] [PubMed] [Google Scholar]
- 16.Hernández, M., T. Esteve, S. Prat, and M. Pla. 2004. Development of real-time PCR systems based on SYBR Green I, Amplifluor and TaqMan technologies for specific quantitative detection of the transgenic maize event GA21. J. Cereal Sci. 39:99-107. [Google Scholar]
- 17.Higuchi, R., C. Fockler, G. Dollinger, and R. Watson. 1993. Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Bio/Technology 11:1026-1030. [DOI] [PubMed] [Google Scholar]
- 18.Hough, A. J., S. A. Harbison, M. G. Savill, L. D. Melton, and G. Fletcher. 2002. Rapid enumeration of Listeria monocytogenes in artificially contaminated cabbage using real-time polymerase chain reaction. J. Food Prot. 65:1329-1332. [DOI] [PubMed] [Google Scholar]
- 19.Jeffers, G. T., J. L. Bruce, P. L. McDonough, J. Scarlett, K. J. Boor, and M. Wiedmann. 2001. Comparative genetic characterization of Listeria monocytogenes isolates from human and animal listeriosis cases. Microbiology 147:1095-1104. [DOI] [PubMed] [Google Scholar]
- 20.Kathariou, S. 2002. Listeria monocytogenes virulence and pathogenicity, a food safety perspective. J. Food Prot. 65:1811-1829. [DOI] [PubMed] [Google Scholar]
- 21.Kay, S., and G. Van den Eede. 2001. The limits of GMO detection. Nat. Biotechnol. 19:405. [DOI] [PubMed] [Google Scholar]
- 22.Klein, D. 2002. Quantification using real-time PCR technology: applications and limitations. Trends Mol. Med. 8:257-260. [DOI] [PubMed] [Google Scholar]
- 23.Kohler, S., M. Leimeister-Wachter, T. Chakraborty, F. Lottspeich, and W. Goebel. 1990. The gene coding for protein p60 of Listeria monocytogenes and its use as a specific probe for Listeria monocytogenes. Infect. Immun. 58:1943-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koo, K., and L. A. Jaykus. 2003. Detection of Listeria monocytogenes from a model food by fluorescence resonance energy transfer-based PCR with an asymmetric fluorogenic probe set. Appl. Environ. Microbiol. 69:1082-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lou, Y., and A. E. Yousef. 1999. Characteristics of Listeria monocytogenes important to food processors, p. 131-224. In E. T. Ryser and E. H. Marth (ed.), Listeria, listeriosis, and food safety, 2nd ed. Marcel Dekker, Inc., New York, N.Y.
- 26.Manzano, M., L. Cocolin, C. Pipan, E. Falasca, G. A. Botta, C. Cantoni, and G. Comi. 1997. Single-strand conformation polymorphism (SSCP) analysis of Listeria monocytogenes iap gene as tool to detect different serogroups. Mol. Cell. Probes 11:459-462. [DOI] [PubMed] [Google Scholar]
- 27.Mengaud, J., M. F. Vicente, J. Chenevert, J. M. Pereira, C. Geoffroy, B. Gicquel-Sanzey, F. Baquero, J. C. Perez-Diaz, and P. Cossart. 1988. Expression in Escherichia coli and sequence analysis of the listeriolysin O determinant of Listeria monocytogenes. Infect. Immun. 56:766-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nadon, C. A., D. L. Woodward, C. Young, F. G. Rodgers, and M. Wiedmann. 2001. Correlations between molecular subtyping and serotyping of Listeria monocytogenes. J. Clin. Microbiol. 39:2704-2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nogva, H. K., K. Rudi, K. Naterstad, A. Holck, and D. Lillehaug. 2000. Application of 5′-nuclease PCR for quantitative detection of Listeria monocytogenes in pure cultures, water, skim milk, and unpasteurized whole milk. Appl. Environ. Microbiol. 66:4266-4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oliver, A., R. Canton, P. Campo, F. Baquero, and J. Blazquez. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288:1251-1254. [DOI] [PubMed] [Google Scholar]
- 31.Phillips, C. 1998. Listeria monocytogenes, p. 63-68. In (ed.) Food, bacteria and health. A practical guide. Chandos Publishing Limited, Oxford, England.
- 32.Piffaretti, J. C., H. Kressebuch, M. Aeschbacher, J. Bille, E. Bannermann, J. M. Musser, R. K. Selander, and J. Rocourt. 1989. Genetic characterization of clones of the bacterium Listeria monocytogenes causing epidemic disease. Proc. Natl. Acad. Sci. USA 86:3818-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rasmussen, O. F., P. Skouboe, L. Dons, L. Rossen, and J. E. Olsen. 1995. Listeria monocytogenes exists in at least three evolutionary lines: evidence from flagellin, invasive associated protein and listeriolysin O genes. Microbiology 141:2053-2061. [DOI] [PubMed] [Google Scholar]
- 34.Ripio, M.-T., G. Dominquez-Bernal, M. Lara, M. Suárez, and J.-A. Vázquez-Boland. 1997. A Gly145Ser substitution in the transcriptional activator PrfA causes constitutive overexpression of virulence factors in Listeria monocytogenes. J. Bacteriol. 179:1533-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roberts, T. A., A. C. Baird-Parker, and R. B. Tompkin (ed.). 1996. Microorganisms in foods, vol. 5. Characteristics of microbial pathogens, p. 141-182. Blackie Academic & Professional, London, England.
- 36.Ryser, E. T. 1999. Foodborne listeriosis, p. 299-358. In E. T. Ryser and E. H. Marth (ed.), Listeria, listeriosis, and food safety, 2nd ed. Marcel Dekker, Inc., New York, N.Y.
- 37.Slutsker, L., and A. Schuchat. 1999. Listeriosis in humans, p. 75-95. In E. T. Ryser and E. H. Marth (ed.), Listeria, listeriosis, and food safety, 2nd ed. Marcel Dekker, Inc., New York, N.Y.
- 38.Tran, H. L., and S. Kathariou. 2002. Restriction fragment length polymorphisms detected with novel DNA probes differentiate among diverse lineages of serogroup 4 Listeria monocytogenes and identify four distinct lineages in serotype 4b. Appl. Environ. Microbiol. 68:59-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Unnerstad, H., I. Nilsson, H. Ericsson, M. Danielsson-Tham, J. Bille, E. Bannerman, and W. Tham. 1999. Division of Listeria monocytogenes serovar 1/2a strains into two groups by PCR and restriction enzyme analysis. Appl. Environ. Microbiol. 65:2054-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vazquez-Boland, J. A., G. Domínguez-Bernal, B. González-Zorn, J. Kreft, and W. Goebel. 2001. Pathogenicity islands and virulence evolution in Listeria. Microb. Infect. 3:571-584. [DOI] [PubMed] [Google Scholar]
- 41.Vázquez-Boland, J. A., M. Kuhn, P. Berche, T. Chakraborty, G. Domínguez-Bernal, W. Goebel, B. González-Zorn, J. Wehland, and J. Kreft. 2001. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14:584-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vines, A., M. W. Reeves, S. Hunter, and B. Swaminathan. 1992. Restriction fragment length polymorphism in four virulence-associated genes of Listeria monocytogenes. Res. Microbiol. 143:281-294. [DOI] [PubMed] [Google Scholar]
- 43.Vines, A., and B. Swaminathan. 1998. Identification and characterization of nucleotide sequence differences in three virulence-associated genes of Listeria monocytogenes strains representing clinically important serotypes. Curr. Microbiol. 36:309-318. [DOI] [PubMed] [Google Scholar]
- 44.Wiedmann, M., J. L. Bruce, C. Keating, A. E. Johnson, P. L. McDonough, and C. A. Batt. 1997. Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in pathogenic potential. Infect. Immun. 65:2707-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wiedmann, M. 2002. Molecular subtyping methods for Listeria monocytogenes. J. AOAC Int. 85:524-530. [PubMed] [Google Scholar]