Abstract
The branched-chain amino acids (BCAAs) leucine, isoleucine, and valine are elevated in maple syrup urine disease, heart failure, obesity, and type 2 diabetes. BCAA homeostasis is controlled by the mitochondrial branched-chain α-ketoacid dehydrogenase complex (BCKDC), which is negatively regulated by the specific BCKD kinase (BDK). Here, we used structure-based design to develop a BDK inhibitor, (S)-α-chloro-phenylpropionic acid [(S)-CPP]. Crystal structures of the BDK-(S)-CPP complex show that (S)-CPP binds to a unique allosteric site in the N-terminal domain, triggering helix movements in BDK. These conformational changes are communicated to the lipoyl-binding pocket, which nullifies BDK activity by blocking its binding to the BCKDC core. Administration of (S)-CPP to mice leads to the full activation and dephosphorylation of BCKDC with significant reduction in plasma BCAA concentrations. The results buttress the concept of targeting mitochondrial BDK as a pharmacological approach to mitigate BCAA accumulation in metabolic diseases and heart failure.
Keywords: branched-chain α-ketoacid dehydrogenase kinase inhibitor, structure-based inhibitor design, allosteric mechanisms, kinase-inhibitor complex structures, in vivo kinase inhibitor studies
The branched-chain amino acids (BCAA) leucine, isoleucine, and valine comprise 40% of essential amino acids in daily dietary intake (1). The catabolic pathways of BCAA begin with transamination by branched-chain aminotransferases giving rise to corresponding branch-chain α-ketoacids (BCKA). The second common step, the irreversible oxidative decarboxylation of BCKA, is catalyzed by the single mitochondrial branch-chain α-ketoacid dehydrogenase complex (BCKDC). The homeostasis of BCAA and BCKA in vivo is critical for health. The accumulation of BCAA and BCKA secondary to inherited BCKDC deficiency produces maple syrup urine disease (MSUD), which can lead to fatal acidosis, neurological derangement, and mental retardation (2, 3). In large-scale high-throughput metabolic profiling studies, high blood BCAA concentrations were found to be highly associated with the development of insulin resistance (4, 5) and can serve as useful metabolic markers in type 2 diabetes risk assessment (6, 7). Pathologic stresses produced by the accumulated BCKA are linked to congenital heart diseases and heart failure (8). The above findings underscore the pivotal role of aberrant BCAA metabolism in the pathogenesis of metabolic, cardiac, and neurological diseases.
The 4.5-MDa human BCKDC consists of three catalytic components: a heterotetrameric (α2β2) branched-chain α-ketoacid decarboxylase (E1), a homo-24 meric dihydrolipoyltransacylase (E2), and a homodimeric dihydrolipoamide dehydrogenase (E3). In addition, human BCKDC contains two regulatory enzymes: BCKD kinase (BDK) and BCKD phosphatase (BDP), the latter also called PP2Cm phosphatase; these enzymes tightly regulate activity of BCKDC through the phosphorylation (inactivation)/dephosphorylation (activation) of the E1α subunits (9, 10). BCKDC is organized around the cubic E2 core, to which 12 copies of E1, and undetermined numbers of E3, BDK, and BDP subunits are attached through noncovalent interactions. The docking of BDK and BDP to the E2 core of BCKDC via the lipoyl-bearing domain (LBD) of each E2 subunit markedly augments kinase and phosphatase activities, respectively (11–13).
BCKDC catalyzes the rate-limiting step in the disposal of BCAA (1, 14); the inactivation of BCKDC through phosphorylation by BDK results in increased BCAA concentrations in animal tissues. Therefore, modulation of BDK activity constitutes a major mechanism for BCAA homeostasis in vivo (15); BDK offers a therapeutic target for ameliorating the accumulation of BCAA and BCKA in disease conditions. BDK is inhibited by α-ketoisocaproate (KIC) from leucine, resulting in the activation of BCKDC in perfused rat hearts (16). Thus, leucine serves as a feedforward nutritional signal that promotes BCAA disposal through the inhibition of BDK activity. The inhibition of BDK by small molecules such as KIC prompted the development and identification of a series of KIC analogs that function as BDK inhibitors (16, 17). These BDK inhibitors include KIC analogs α-chloroisocaproate (CIC) (18), phenylpyruvate (17), clofibric acid (19), and recently phenylbutyrate (PB) (20). However, these BDK inhibitors are less than robust as BDK inhibitors, with reported I40 (concentration for 40% inhibition) in the submillimolar range (e.g., CIC, phenylpyruvate, and clofibric acid). To date, the structural basis for the inhibition of BDK has been unknown.
In the present study, we characterized the known BDK inhibitors α-ketoisocaproate, α-chloroisocaproate, and phenylbutyrate and determined crystal structures of BDK in complex with these compounds. We identified a unique allosteric site and used structure-based design to produce BDK inhibitors with improved IC50 and binding affinities for BDK. We show that one of these newly identified BDK inhibitors robustly augments BCKDC activity and reduces BCAA concentrations in WT mice. These “designer” BDK inhibitors have clinical ramifications for treating hereditary metabolic disorders that are associated with the accumulation of BCAA and BCKA.
Results
Modest Inhibition of BDK by α-Ketoisocaproate, α-Chloroisocaproate, and Phenylbutyrate.
KIC (Fig. 1), a physiological substrate of BCKDC, is a weak BDK inhibitor with a modest IC50 of 74.9 µM (Table 1; Fig. S1A). This value is comparable to the I40 of 65 µM for BDK inhibition, determined previously using the purified BCKDC from rabbit liver (16). The binding of KIC to BDK was measured by isothermal titration calorimetry (ITC), which shows a dissociation constant (Kd) of 8.5 µM for KIC (Table 1; Fig. S2A). CIC is an analog of KIC, in which the α-carbonyl group in KIC is replaced by a chlorine atom (Fig. 1). The (R)-enantiomer of CIC was reported to be a more potent BDK inhibitor than the (S)-enantiomer (21). This observation is confirmed by our in vitro kinase assays, with IC50 values of 12.0 and 53.4 µM for (R)-CIC and (S)-CIC, respectively (Table 1; Fig. S1B). ITC measurements of BDK binding showed similar binding affinities for these two enantiomers, with Kd values of 7.0 and 9.2 µM for (R)-CIC and (S)-CIC, respectively (Table 1; Fig. S2 B and C). The presence of nucleotides significantly enhances binding affinities of BDK for (S)-CIC; for example, the Kd of 9.2 µM in the absence of nucleotide is reduced to 2.4, 3.2, and 3.2 µM, respectively, when the nucleotides ATP, ATPγS, and ADP are present (Table S1). We reported previously that PB is also a BDK inhibitor (20). Here, PB shows an IC50 of 53.1 µM and a Kd of 5.7 µM (Table 1; Figs. S1C and S2D).
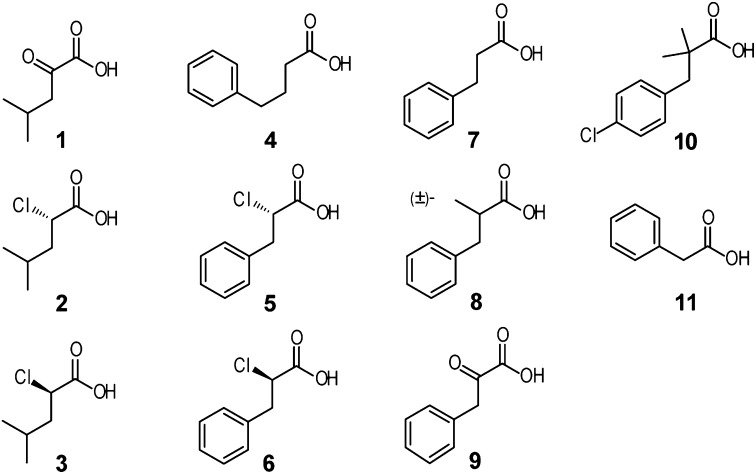
Fig. 1.
Structure-activity relationship of known and novel BDK inhibitors. Chemical structures of BDK inhibitors: α-ketoisocaproate (KIC) (compound 1); S-enantiomer of α-chloroisocaproate [(S)-CIC] (2); R-enantiomer of α-chloroisocaproate [(R)-CIC] (3); phenylbutyrate (PB) (4); S-enantiomer of α-chlorophenylpropionate [(S)-CPP] (5); R-enantiomer of α-chlorophenylpropionate [(R)-CPP] (6); hydrocinnamic acid (7); methyl-hydrocinnamic acid (8); phenylpyruvic acid (9); clofibric acid (10); phenylacetate (PA) (11).
Table 1.
IC50 and Kd values (in µM) of BDK inhibitors (compounds 1–11)
| Compound | Name or acronym | IC50 (μM) | Kd (μM) |
| 1 | KIC | 74.9 | 8.8 ± 0.5 |
| 2 | (S)-CIC | 53.4 | 9.2 ± 1.4 |
| 3 | (R)-CIC | 12.0 | 7.0 ± 1.5 |
| 4 | PB | 53.1 | 5.7 ± 0.7 |
| 5 | (S)-CPP | 6.3 | 2.4 ± 0.4 |
| 6 | (R)-CPP | 21.4 | 7.5 ± 2.2 |
| 7 | Hydrocinnamic acid | 43.1 | 9.1 ± 1.0 |
| 8 | Methyl-hydrocinnamic acid | 33.8 | 8.6 ± 2.0 |
| 9 | Phenylpyruvic acid | 346 | NM |
| 10 | Clofibric acid | 812 | NM |
| 11 | Phenylacetate | 1,270 | NM |
(S)-CPP shows the best IC50 and Kd as a novel BDK inhibitor. NM, immeasurable: enthalpy changes (ΔH) were too small for the binding analysis.
Crystal Structures Reveal Modes of Inhibitor Binding to BDK.
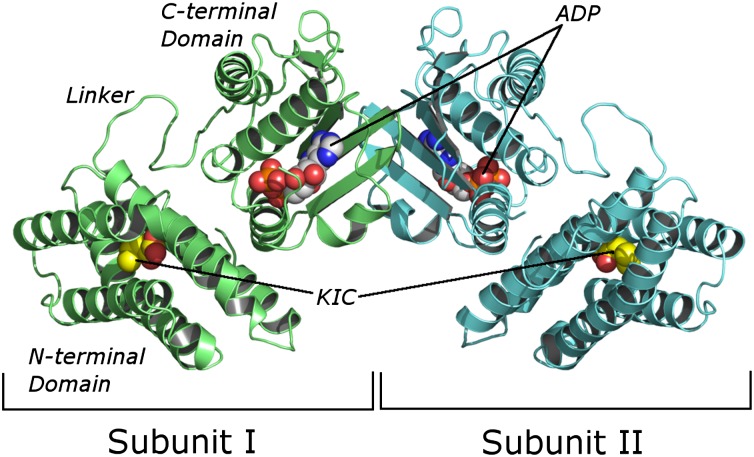
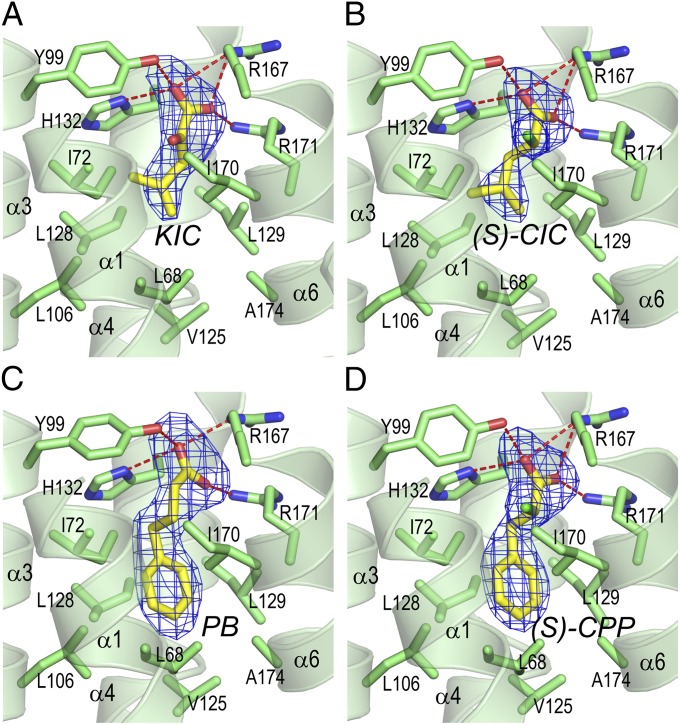
BDK-inhibitor complexes were produced by soaking BDK crystals (22) in 2 mM inhibitor solutions. Table S2 summarize X-ray diffraction statistics for crystals of BDK-inhibitor complexes with or without ADP. The resolution ranges from 2.05 to 2.85 Å. Fig. 2 shows that KIC binds to an allosteric site in the center of a four-helix bundle in the N-terminal domain of each subunit in the BDK dimer. The allosteric site is ∼10 Å away from the bound ADP located in the C-terminal domains of the BDK subunit. At the allosteric site, the bound KIC assumes two different conformations with the orientation of the carbonyl oxygen as the major discriminator. In conformation 1 (Fig. 3A, shown as the stick model in yellow), both carboxylate-oxygen atoms are in good distance (2.4–3.1A) to form hydrogen bonds with side-chains of His132, Tyr99, Arg167, and Arg171. In conformation 2 (Fig. S4), the two carboxylate-oxygen atoms make three hydrogen bonds with the side-chains of Tyr99, Arg167, and Arg171. An additional hydrogen bond is formed between the carbonyl oxygen and the side-chain of Arg171. It is unclear whether conformation 1 or 2 is more prevalent in the BDK-KIC-ADP structure. Interestingly, in the BDK-(S)-CIC-ADP structure (Fig. 3B), the ligand (S)-CIC incurs a single conformation similar to conformation 1 of KIC. Both carboxylate oxygen atoms maintain multiple hydrogen bonds with the above-mentioned residues in the pocket. The replacement of the carbonyl oxygen in KIC with a chlorine atom in (S)-CIC converts the planar sp2-carbon in the former ligand to the tetrahedral sp3-carbon in the latter. The α-chlorine atom of (S)-CIC protrudes into a space formed by side-chains of Ile170, Ile72, and Try99 and interacts with side-chains of these residues through hydrophobic interactions. PB binds to the BDK allosteric site (Fig. 3C), similar to (S)-CIC except that both carboxylate oxygen atoms shift, relative to their counterparts in the BDK-(S)-CIC-ADP structure (Fig. 3B). Thus, fewer and less favorable hydrogen bonds are formed between PB and side-chains of the interacting residues in the BDK-PB-ADP structure. PB compensates for the loss of hydrogen bonds by extending its phenyl group into a hydrophobic pocket formed by Ile72, Leu128, Leu106, Leu68, Val125, Ala174, Ile129, and Ile170. By comparison, the isobutyryl end of (S)-CIC is not extended fully into this hydrophobic pocket, resulting in only limited hydrophobic interactions with residues Ile72, Leu128, Ile129, and Ile170.
Fig. 2.
Dimeric BDK structure containing bound α-ketoisocaproate and ADP. The structure shows KIC (represented as space-filling model) binds to the N-terminal domain of each subunit. The bound ADP (also represented as space-filling model) is located in the C-terminal domain of each subunit.
Fig. 3.
Crystal structures of BDK inhibitors bound to the allosteric binding site in the N-terminal domain. (A) BDK-KIC-ADP. (B) BDK-(S)-CIC-ADP. (C) BDK-PB-ADP. (D) BDK-(S)-CPP-ADP. The inhibitors (with carbon atoms in yellow) are represented as the stick model, with the mesh (in blue) depicting the Fo-Fc omit map contoured at 3σ. Side-chains (with carbon atoms in green) of residues that interact with the inhibitors are also depicted in the stick model. Oxygen atoms are shown in red, nitrogen in blue, chlorine in light-green, and hydrogen bonds in red dotted lines. Portion of helixes α1 and α6 are omitted for clarity. The stereo views are shown in Fig. S3.
Mutational and Functional Studies of Allosteric Site Residues.
Residues at the allosteric site of BDK were altered by site-directed mutagenesis, and binding affinities of the BDK variants for (S)-CIC were analyzed by ITC. The Y99A and H132A mutant BDK proteins show slightly better binding affinities for (S)-CIC than WT BDK (Table S3). However, enthalpy changes (ΔH) exhibited by these BDK variants are significantly reduced compared with WT. The results suggest that hydrogen bonds formed between (S)-CIC, Tyr99, and His132 are not essential for high-affinity binding of (S)-CIC to BDK. Changing His132 to an alanine removes steric clashes of the histidine side-chain with one of the carboxylate-oxygen atoms in (S)-CIC, resulting in a slightly increased binding affinity. Arg167 and Arg171 make the essential salt bridges with both carboxylate-oxygen atoms of (S)-CIC, and conversion of either residue to an alanine completely abolishes (S)-CIC binding to the mutant BDK proteins (Table S3). The replacement of Ile170 with a bulky phenylalanine disrupts the apparently essential interaction between the α-chlorine atom of (S)-CIC and Ile170, also nullifying the binding of (S)-CIC to the I170F BDK variant. Unlike (S)-CIC, PB binding to BDK is adversely affected by mutations in Tyr99 and His132 (Table S3). These results suggest that the hydrogen bonding between the Tyr99 side-chain and one of the carboxylate-oxygen atoms is essential for PB binding to BDK. The mutagenesis study also highlights these subtle conformational differences between the carboxylate groups of the (S)-CIC and the longer PB in the respective BDK-inhibitor structures (Fig. 3 B and C). The salt bridges imparted by Arg167 and Arg171 side-chains are also indispensable for the binding of PB to BDK, as evidenced by the undetectable enthalpy changes in the R167A and R171A mutants. The phenylalanine side-chain poses a steric hindrance for PB binding, which is indicated by the immeasurable enthalpy changes in the I170F BDK mutant (Table S3).
Structure-Based Design of a Potent BDK Inhibitor, (S)-α-Chlorophenylpropionate.
Based on the above BDK-PB-ADP structure (Fig. 3C), the C-C-C bond angles of the BDK-bound PB at the α-, β-, and γ-carbons are 111.4°, 116.7°, and 99.3°, respectively, compared with the corresponding C-C-C bond angles of 114.2°, 113.1°, and 110.7° in an energy-minimized model (Fig. S5A). These results show that the bond angle at the γ-carbon (99.3°) of the BDK-bound PB is severely constrained due to the extended carbon chain of PB compared with the length of the ligand-binding pocket in BDK. In contrast, the C-C-C bond angles are similar between the BDK-bound (S)-CIC and the energy-minimized model of (S)-CIC (Fig. S5B), indicating the absence of conformational strains in the BDK-bound ligand. A comparison of PB and (S)-CIC structures, both in the BDK-bound form, shows that the carboxylate ends of both PB and (S)-CIC occupy the same location at the allosteric site (Fig. S5 C and D). However, (S)-CIC lacks the phenyl group from PB, which protrudes into the hydrophobic pocket opposite to the carboxylate end of the bound ligand. To remove the strained bond angle at the γ-carbon of PB and compensate for the absence of a phenyl ring in (S)-CIC, which is required for optimal hydrophobic interactions, a chimera molecule between PB and CIC, i.e., chiral α-chloro-phenylpropionate (CPP), was conceived (compounds 5 and 6 in Fig. 1). Enantiomerically pure (S)-CPP and (R)-CPP were synthesized from L- and D-phenylalanine, respectively. (S)-CPP shows a significantly better IC50 (6.3 µM) than (R)-CPP (21.4 µM) (Table 1; Fig. S1D). The Kd values for (S)-CPP and (R)-CPP are 2.4 and 7.5 µM, respectively (Table 1; Fig. S2 E and F). Thus, (S)-CPP proves to be a better BDK inhibitor than its parent compounds (S)-CIC, (R)-CIC, and PB (Table 1).
At the structure-activity relationship level, the elimination of the α-chloro atom of (S)-CPP or (R)-CPP results in hydrocinnamic acid, which has significantly higher IC50 (43.1 µM) and Kd (9.1 µM) than (S)-CPP (Table 1). Similar results are obtained when the α-chlorine atom of (S)-CPP or (R)-CPP is replaced by a methyl group (as in racemic methyl-hydrocinnamic acid). When the α-chlorine atom of (S)-CPP or (R)-CPP is substituted for a keto group, the resultant compound phenylpyruvic acid displays a markedly increased IC50 (346 µM) and immeasurable Kd. The above results support the critical role of the α-chlorine atom for the high-affinity binding of (S)-CPP to BDK. Clofibric acid is a known BDK inhibitor (17), in which the α-position in (S)-CPP is occupied by two methyl groups with additional alterations in the β-carbon and phenyl ring. These structural modifications cause clofibric acid (IC50 = 812 µM and immeasurable Kd) to become a much weaker BDK inhibitor than (S)-CPP (Table 1). Surprisingly, phenylacetate (PA), which is a direct metabolite from PB through β-oxidation, shows an IC50 of 1,270 µM and immeasurable Kd (Table 1). The results provide evidence that PB itself, and not the metabolite PA, is the BDK inhibitor in vivo.
Structural Basis for High-Affinity Binding of (S)-CPP to BDK.
Crystal structures of the BDK-(S)-CPP complex reveal that the carboxylate and the α-chlorine atoms in (S)-CPP (Fig. 3D) assume the same configuration as those in (S)-CIC (Fig. 3B). The phenyl ring of (S)-CPP (Fig. 3D) occupies the hydrophobic pocket, similar to PB (Fig. 3C). However, the carbon chain of (S)-CPP is less constrained than that of PB. We posit that the latter property combined with the presence of the phenyl ring account for the markedly higher binding affinity and lower IC50 of (S)-CPP compared with (S)-CIC or PB (Table 1). The structure of the BDK-(R)-CPP complex was also determined, which showed a similar binding mode to (S)-CPP at the BDK allosteric site (Fig. S6B). A superimposition of the bound (S)-CPP (in yellow) with (R)-CPP (in magenta) in the BDK structures shows that the vertical phenyl ring in (S)-CPP intercalates in approximately equal distance between Leu106 (4.0 Å) and Leu129 (4.2 Å) (Fig. S6B). This configuration allows for optimal interactions of the phenyl ring with both hydrophobic residues. In contrast, the same phenyl moiety in (R)-CPP shifts toward Leu106, disrupting optimal interactions with Leu106 (3.8 Å) and Leu129 (4.9 Å). These results explain the significantly higher IC50 and Kd of (R)-CPP compared with its enantiomer (S)-CPP (Table 1). Interestingly, (S)-CPP does not inhibit pyruvate dehydrogenase kinase isoform 2 (PDK2) (Fig. S7A), despite high conservation at the allosteric sites between BDK and PDK isoforms.
(S)-CPP Blocks BDK Binding to the E2 Core.
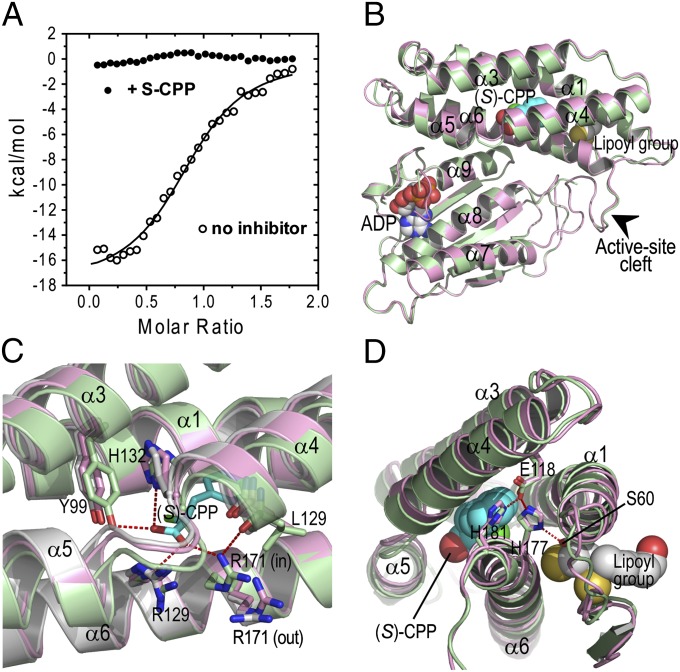
Effects of bound (S)-CPP on the binding of BDK to the E2 core were investigated. To facilitate ITC measurements, the E2 di-domain (DD) consisting of the LBD, which binds BDK, and the subunit-binding domain (SBD) was fused at the C terminus to GST. When GST-DD was titrated into the BDK solution, significant enthalpy changes (ΔH = −17.8 kcal/mol) were observed, with Kd = 2.1 µM (Fig. 4A). The results indicate a moderate binding affinity of BDK for GST-DD, which mimics interactions between BDK and the E2 core of BCKDC. In contrast, when GST-DD was titrated into BDK in the presence of 0.2 mM (S)-CPP, enthalpy changes reflecting the binding reaction were not detected (Fig. 4A). The results indicate that (S)-CPP mitigates interactions between BDK and LBD, interfering with the docking of BDK at the E2 core.
Fig. 4.
Structural mechanisms for inactivation of BDK by (S)-CPP. (A) Binding of GST-DD (LBD-SBD di-domain) to BDK measured by ITC. A Kd of 2.1 µM is derived from curve fittings of the binding isotherms without the inhibitor (open circle). Binding was not detected in the presence of 0.2 mM (S)-CPP (solid circle). (B) The C-terminal domain of the BDK-ADP-(S)-CPP monomer in green superimposed with the corresponding domain of the BDK-ADP monomer in pink. The binding of (S)-CPP to the allosteric site in the N-terminal domain causes the movements of the long helical rod comprising helices α4 and α5, resulting in narrowing the active-site cleft (arrow) between the N-terminal and C-terminal domain. (C) The allosteric site occupied by (S)-CPP depicted as the stick model (in cyan). The side-chain of Arg171 is in the out conformation in the apo-BDK structure (in gray). The Arg171 side-chain in the BDK-ADP structure (in pink) alternates between the in and the out conformations. In the BDK-ADP-(S)-CPP structure (in green), the Arg171 side-chain is locked in the in conformation through contacts with a carboxylate group of (S)-CPP and the main-chain oxygen of Leu129. (D) The hydrogen-bonding network connecting helix α4 to the putative lipoyl-binding pocket. The hydrogen-bonding network is formed between side-chains of Glu118, His177, His-181, and Ser60. The BDK-ADP-(S)-CPP structure (in green) is superimposed with the BDK-ADP structure (in pink).
Robust Enhancement of BCKDC Activity by (S)-CPP in Vivo.
The in vitro results clearly establish that (S)-CPP is the most effective BDK inhibitor among all inhibitors studied (Table 1). To assess the efficacy of (S)-CPP in vivo, the stability of the compound was examined in murine hepatocytes and S-9 fractions containing the cytosol and microsomes. (S)-CPP showed half-lives of 408 and 187 min in hepatocytes and S-9 fractions, respectively (Fig. S8A). Under the assay conditions used here, these data predict that this compound will have a low clearance in vivo based on a rearrangement of the well-stirred model (23). This prediction is supported by the in vivo pharmacokinetic data, in which relative clearance was calculated to be only 0.113 mL/min after i.p. administration to WT female CD-1 mice. These data also revealed that (S)-CPP has a low volume of distribution (Vz/F), which suggests it does not penetrate efficiently into tissues and is likely due to the polarity and water solubility of (S)-CPP (Fig. S8 B and C).
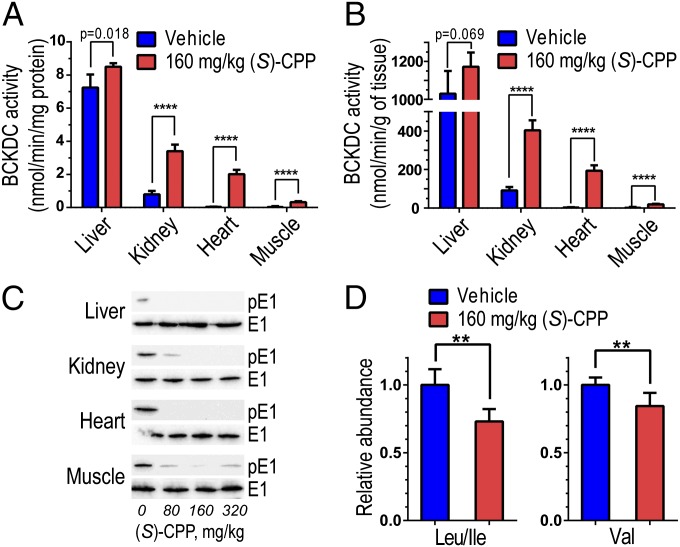
WT CD-1 male mice were dosed with vehicle or (S)-CPP at 160 mg/kg body weight by i.p. injections. At 60 min after the injection, mice were dissected, and the liver, kidney, heart, and hind-leg quadricep muscle were harvested and assayed for BCKDC activity, which is elevated significantly in tissues from (S)-CPP–treated mice vs. vehicle-treated mice. The increases in BCKDC activity (expressed as nmol/min per milligram protein) are 1.2-, 4.3-, 50.5-, and 11.0-fold (n = 6) in (S)-CPP–treated liver, kidney, heart, and muscle, respectively (Fig. 5A). When expressed in nmol/min per gram wet weight tissue, similar 1.1-, 4.4-, 55.4-, and 9.3-fold (n = 5) increases in BCKDC activity, respectively, were obtained (Fig. 5B). In (S)-CPP–treated mice, but not vehicle-treated mice, the phospho-E1α subunit was completely undetectable in liver, kidney, and heart but slightly visible in skeletal muscle (Fig. 5C). Therefore, the activity levels in tissues from mice treated with saturating (S)-CPP concentrations represent the total or full BCKDC activity. We conclude that short-term treatments with (S)-CPP systemically convert BCKDC from the partially active (and partially dephosphorylated) state to the fully active (and completely dephosphorylated) state by the intrinsic BDP through efficient inhibition of BDK in vivo. (S)-CPP shows no direct effect on BDP activity (Fig. S7B). The result rules out increased BCKDC activity, in part or in full, due to the activation of BDP by (S)-CPP. The drastic enhancement of BCKDC activity leads to significant reduction in plasma BCAA levels; leucine/isoleucine and valine concentrations in (S)-CPP–treated mice are 27% and 16% lower, respectively, than those in vehicle-treated mice (Fig. 5D).
Fig. 5.
Robust activation and complete dephosphorylation of BCKDC in vivo by (S)-CPP. WT CD-1 male mice were treated with vehicle or (S)-CPP (n = 6) by i.p. injections at a dosage of 160 mg/kg. At 60 min after the injection, mice were killed and tissues analyzed. (A) BCKDC activity, expressed in nmol/min/mg protein, in vehicle-treated vs. (S)-CPP–treated tissues. (B) BCKDC activity expressed in nmol/min/g wet weight. (C) Western blotting of tissues from vehicle-treated and (S)-CPP–treated mice (at 80, 160, and 320 mg/kg). Affinity-purified antibodies to phospho-serine 293 of the E1α subunit (p-E1α) and to the E1α subunit were used. Signals obtained with the latter antibody serve as loading controls. (D) Plasma BCAA concentrations determined by LC-MS/MS are significantly reduced in (S)-CPP–treated mice compared with vehicle-treated (n = 6). Relative abundance of each BCAA in (S)-CPP-treated mice was obtained by designating the corresponding peak in vehicle-treated mice as 1.0. The graph of Leu/Ile shows the sum of both amino acids as the LC-MS/MS method cannot distinguish between Leu and Ile. **P < 0.01; ****P < 0.0001.
Discussion
The present study supports the notion of using the physiological allosteric site to advance therapeutics for metabolic diseases. In variant forms of MSUD, the mutant BCKDC protein is present with residual BCKDC activity (2, 3). We propose that a systemic BDK inhibitor–mediated increase in residual BCKDC activity can reduce plasma BCAA/BCKA concentrations and ameliorate the MSUD phenotype. We reported earlier that the treatments of intermediate MSUD patients with PB result in significantly lower circulating concentrations of BCAA and BCKA (20). Clofibric acid, which is derived from the anti-lipidemic drug clofibrate and functions as a weak BDK inhibitor (17), has also been used to lower BCAA concentration in rats (19, 24–27). However, both PB and clofibric acid are significantly less potent BDK inhibitors (IC50 of 53.1 and 812 µM, respectively), than (S)-CPP (IC50 of 6.3 µM; Table 1). PB has multiple targets in cells (28) and is rapidly metabolized to PA in vivo (29); the latter metabolite is inefficient in abating BDK activity (IC50 = 1.27 mM). Smaller BDK inhibitors such as (R)- and (S)-CIC are also competitive inhibitors of BCKDC (18) and have not been studied in vivo. Thus, far, only the racemic mixture of CIC has been shown to inhibit BDK activity in perfused tissues (16, 30, 31). These properties render the known BDK inhibitors less ideal than (S)-CPP for use to reduce BCAA/BCKA levels in MSUD.
Despite the relative low BDP (PP2Cm) expression in skeletal muscle (13), the BCKDC E1α subunit was close to completely dephosphorylated within 1 h of (S)-CPP treatment (Fig. 5C). The nearly complete dephosphorylation hence the close to maximal activation of BCKDC in muscle is potentially significant. BCAA oxidation involves an extensive interplay between muscle and liver. In skeletal muscle, branched-chain aminotransferase (BCAT) activity is high (32, 33), but BCKDC activity is low due to inactivation by phosphorylation (19, 34). The reverse situation exists in the liver, with both cytosolic and mitochondrial BCAT activities undetectable in the rat liver (33). Therefore, the primary role of muscle is thought to be the transamination of BCAA, which provides the major source of circulating BCKA; the latter are transported to liver, kidney, and heart, where they are oxidized by BCKDC (34, 35). Because muscle makes up 40% of the body weight, the (S)-CPP–mediated robust activation of BCKDC in muscle could make this tissue an important site, in addition to the liver, for in situ BCAA oxidation. The relatively high dose of 160 mg/kg for (S)-CPP used in i.p. injection was designed to trigger the maximal acute dephosphorylation of BCKDC. Similar results could have been achieved at a lower dose, e.g., 80 mg/kg of the inhibitor (Fig. 5C). The robust augmentation of BCKDC activity in mouse tissues results in only moderate reduction in plasma BCAA concentrations (Fig. 5D). We attribute this to the short duration (1 h) of the treatment with (S)-CPP. It is expected that prolonged treatments with (S)-CPP could further reduce BCAA concentrations in vivo.
The molecular mechanism for the feedforward regulation of BCKDC by its physiological substrate KIC through the inactivation of BDK has, until now, never been elucidated. Comparisons of apo-BDK and BDK-(S)-CPP structures offer significant insights into the structural mechanisms for the inactivation of BDK by its allosteric inhibitors including KIC. As shown in Fig. 4B, each BDK monomer contains an allosteric site, a putative lipoyl-binding pocket, both located in the N-terminal domain, as well as a nucleotide-binding pocket situated inside the active-site cleft. Signals from ligand binding to any one of the three ligand-binding pockets/sites are mediated by a long helical rod, formed by helices α4 and α5, to the other two pockets/sites (Fig. 4B; Movie S1). The side-chain of Arg171 mostly assumes the “out” conformation in apo-BDK (Fig. 4C), exposing the allosteric site to the milieu (Fig. 4D). In contrast, in the BDK-ADP-(S)-CPP structure, the Arg171 side-chain is locked in the “in” conformation, which completely shields the ligand from the solvent. The Arg171 side-chain in the same (S)-CPP–bound structure is in contact with the main-chain oxygen of Leu129 with a concomitant movement of helix α4 (Fig. 4C; Movie S2). The side-chain of Glu118 forms a hydrogen-bonding network with side-chains of His177, His181, and Ser60 (Fig. 4D). Helices α1 and α6 harbor the conserved amino acid residues that contact the dihydrolipoamide group in the lipoyl-binding pocket, based on the related PDK3-inner lipoyl domain (L2) structure (36). Therefore, the (S)-CPP–induced displacement of helix α4 is likely communicated to the lipoyl-binding pocket through the hydrogen-bonding network, resulting in the perturbation of the lipoyl-binding pocket. This mechanism explains the undetectable binding of the GST-DD construct harboring LBD to BDK in the presence of (S)-CPP (Fig. 4A). Because BDK requires the E2 core for the bulk of kinase activity (11), abrogating BDK binding to LBD from the E2 core will deactivate the kinase. In support of this mechanism, the presence of KIC in HEK293T cells (13, 37) or the dietary supplementation of BCAA in type 2 diabetes rat models (38) promotes the dissociation of BDK from the E2 core, resulting in the dephosphorylation of BCKDC.
On the other hand, an (S)-CPP–promoted shift in helix α5 of the helical rod results in the narrowing the active-site cleft bordered between helices α5 and α8 (Fig. 4B; Movie S1), which is consistent with the inactive closed conformation observed in the related PDK3-L2 structure (36). A restricted active-site cleft will interfere with the access of the E1 substrate in the phosphoryl transfer reaction. Moreover, an ordered ATP lid in the BDK-ADP-(S)-CPP complex (Fig. 4B) traps ADP (22), thwarting the removal of reaction product ADP that competes with substrate ATP for binding to the nucleotide-binding pocket. The effects of (S)-CPP on the BDK active site provides a complementary mechanism for the inhibition of BDK by the allosteric ligands. The above structural insights, taken together, will be useful for further drug discovery and development targeting the unique allosteric site in BDK.
Materials and Methods
Crystallization of the BDK and BDK Inhibitor Complexes.
Purified C-terminal His6-tagged BDK protein (22) was concentrated to 36 mg/mL and stored at −80 °C in small aliquots. Crystals were obtained by the hanging-drop vapor-diffusion method (22). The recombinant BDK-His6 protein (5–10 μg/μL) in 2 μL buffer A was mixed with 2 μL well solution [12% (wt/vol) PEG-8000, 0.1 M Tris⋅HCl, pH 8.5, 1.3–1.5 M NaCl] and kept in a 20 °C incubator. Crystals developed within 24 h and matured to 250 × 450 μm in 10 d. Mature crystals were first transferred to the basic soaking solution (BSS) [0.1 M Tris, pH 8.5, 12% (wt/vol) PEG-8000, 1 M NaCl, 0.1 M KCl, 2 mM MgCl2, 5% (vol/vol) glycerol], then transferred to the BSS containing 10 mM peptide (NH2-G-H-H-S-T-S-D-COOH from the E1 phosphorylation loop) (10), 2 mM of various inhibitors, and/or nucleotides, and were incubated at 20 °C overnight. Soaked crystals were gradually transferred to a cryo-solution containing 25% (vol/vol) glycerol and were snap frozen in liquid nitrogen. The structure determination and refinements are described in SI Materials and Methods.
Assay for Inhibition of BDK Activity.
To determine the IC50 for BDK inhibitors, a mixture containing 0.2 µM BDK, 6 µM E1, 0.5 µM of E2, and various amounts of inhibitor was incubated at 25 °C for 10 min in a buffer of 20 mM Tris⋅HCl (pH 7.5), 100 mM KCl, 5 mM MgCl2, 2 mM DTT, 0.02% (vol/vol) Tween-20, and 0.1 mg/mL BSA before the reaction. All inhibition titrations were performed at nine dose points ranging from 316 to 3.16 mM in a 3.162-fold dilution series, with each inhibitor concentration tested in duplicate. The remaining steps were described previously (39).
Assay for BCKDC Activity in Tissues.
Untreated and (S)-CPP–treated mice were killed by CO2 asphyxiation, and hearts, livers, kidneys, and skeletal muscle were removed and immediately frozen in liquid nitrogen. Tissue samples were thawed on ice. Tissues (100–400 mg) from kidneys, hearts, muscle, and liver were manually homogenized in an ice-cold glass homogenizers containing 1 mL of a homogenizing buffer containing 30 mM KPi, pH 7.5, 3 mM EDTA, 5 mM DTT, 1 mM KIC, 3% (vol/vol) fetal bovine serum (FBS), 5% (vol/vol) Triton X-100, and 1 μM leupeptin. Samples were spun at 4 °C in an ultracentrifuge at 25,000 × g for 10 min; supernatants were collected and diluted on ice (1:3 for muscle, 1:6 for kidneys and heart, and 1:20 for liver) with a dilution buffer containing 50 mM Hepes, pH 7.5, 0.5 mM DTT, 0.1% Triton X-100, 3% (vol/vol) FBS, and 1 μM leupeptin. Diluted samples (50 μL) were placed in 24-well assay plates containing 295 μL/well of the assay mixture, which comprised 30 mM KPi, pH 7.5, 0.4 mM CoA, 3 mM NAD+, 5% (vol/vol) FBS, 2 mM thiamine diphosphate, 2 mM MgCl2, and 65 μg human E3. The oxidative decarboxylation reaction was initiated by the addition of the α-keto[1-14C]isovalerate substrate (0.5 mM in 25 μL; specific radioactivity: 1,000 cpm/nmol). The radiochemical assay for BCKDC activity on 24-well plates was performed as described previously (40).
The statistical significance of differences between groups was determined by Student’s unpaired t test using Prism 6 software. Results are presented as means ± SD for the indicated number of independent samples tested.
Supplementary Material
Acknowledgments
We thank Dr. Christopher Lynch for the generous supply of the antibody to phospho-Ser293 of the E1α subunit for Western blot analysis and Dr. Chang-guang Wang for help in pharmacokinetics assays. This work was supported by National Institutes of Health Grants DK62306, DK26758, and DK92921 and Welch Foundation Grant I-1286. Crystal structures presented in this report are derived from work performed at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source, operated under Department of Energy Contract DE-AC02-06CH11357.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. R.A.H. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4H7Q, 3TZ5, 3TZ4, 4DZY, 3TZ2, 3TZ0, 4H85, and 4H81).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1303220110/-/DCSupplemental.
References
- 1.Harper AE, Miller RH, Block KP. Branched-chain amino acid metabolism. Annu Rev Nutr. 1984;4:409–454. doi: 10.1146/annurev.nu.04.070184.002205. [DOI] [PubMed] [Google Scholar]
- 2.Chuang DT, Shih VE. Maple syrup urine disease (Branched-chain ketoaciduria) In: Scriver CR, et al., editors. The Metabolic and Molecular Basis of Inherited Disease. 8th Ed. New York: McGraw-Hill; 2001. pp. 1971–2006. [Google Scholar]
- 3.Chuang DT, Wynn RM, Shih VE. Maple syrup urine disease (branched-chain ketoaciduria): An update. In: Scriver CR, et al., editors. The Online Metabolic and Molecular Basis of Inherited Disease. New York: McGraw Hill; 2008. pp. 1–42. [Google Scholar]
- 4.Newgard CB, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009;9(4):311–326. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Newgard CB. Interplay between lipids and branched-chain amino acids in development of insulin resistance. Cell Metab. 2012;15(5):606–614. doi: 10.1016/j.cmet.2012.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang TJ, et al. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17(4):448–453. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wurtz P, et al. Branched-chain and aromatic amino acids are predictors of insulin resistance in young adults. Diabetes Care. 2013;36(3):648–655. doi: 10.2337/dc12-0895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun H, Lu G, Ren S, Chen J, Wang Y. Catabolism of branched-chain amino acids in heart failure: Insights from genetic models. Pediatr Cardiol. 2011;32(3):305–310. doi: 10.1007/s00246-010-9856-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao Y, et al. Site-directed mutagenesis of phosphorylation sites of the branched chain alpha-ketoacid dehydrogenase complex. J Biol Chem. 1994;269(28):18583–18587. [PubMed] [Google Scholar]
- 10.Wynn RM, et al. Molecular mechanism for regulation of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase complex by phosphorylation. Structure. 2004;12(12):2185–2196. doi: 10.1016/j.str.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 11.Davie JR, et al. Expression and characterization of branched-chain alpha-ketoacid dehydrogenase kinase from the rat. Is it a histidine-protein kinase? J Biol Chem. 1995;270(34):19861–19867. doi: 10.1074/jbc.270.34.19861. [DOI] [PubMed] [Google Scholar]
- 12.Wynn RM, Li J, Brautigam CA, Chuang JL, Chuang DT. Structural and biochemical characterization of human mitochondrial branched-chain α-ketoacid dehydrogenase phosphatase. J Biol Chem. 2012;287(12):9178–9192. doi: 10.1074/jbc.M111.314963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou M, Lu G, Gao C, Wang Y, Sun H. Tissue-specific and nutrient regulation of the branched-chain α-keto acid dehydrogenase phosphatase, protein phosphatase 2Cm (PP2Cm) J Biol Chem. 2012;287(28):23397–23406. doi: 10.1074/jbc.M112.351031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harris RA, Popov KM, Zhao Y, Shimomura Y. Regulation of branched-chain amino acid catabolism. J Nutr. 1994;124(8, Suppl):1499S–1502S. doi: 10.1093/jn/124.suppl_8.1499S. [DOI] [PubMed] [Google Scholar]
- 15.Harris RA, Joshi M, Jeoung NH. Mechanisms responsible for regulation of branched-chain amino acid catabolism. Biochem Biophys Res Commun. 2004;313(2):391–396. doi: 10.1016/j.bbrc.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 16.Paxton R, Harris RA. Regulation of branched-chain alpha-ketoacid dehydrogenase kinase. Arch Biochem Biophys. 1984;231(1):48–57. doi: 10.1016/0003-9861(84)90361-8. [DOI] [PubMed] [Google Scholar]
- 17.Paxton R, Harris RA. Clofibric acid, phenylpyruvate, and dichloroacetate inhibition of branched-chain alpha-ketoacid dehydrogenase kinase in vitro and in perfused rat heart. Arch Biochem Biophys. 1984;231(1):58–66. doi: 10.1016/0003-9861(84)90362-x. [DOI] [PubMed] [Google Scholar]
- 18.Harris RA, Paxton R, DePaoli-Roach AA. Inhibition of branched chain alpha-ketoacid dehydrogenase kinase activity by alpha-chloroisocaproate. J Biol Chem. 1982;257(23):13915–13918. [PubMed] [Google Scholar]
- 19.Kobayashi R, et al. Clofibric acid stimulates branched-chain amino acid catabolism by three mechanisms. Arch Biochem Biophys. 2002;407(2):231–240. doi: 10.1016/s0003-9861(02)00472-1. [DOI] [PubMed] [Google Scholar]
- 20.Brunetti-Pierri N, et al. Phenylbutyrate therapy for maple syrup urine disease. Hum Mol Genet. 2011;20(4):631–640. doi: 10.1093/hmg/ddq507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harris RA, Kuntz MJ, Simpson R. Inhibition of branched-chain alpha-keto acid dehydrogenase kinase by alpha-chloroisocaproate. Methods Enzymol. 1988;166:114–123. doi: 10.1016/s0076-6879(88)66017-4. [DOI] [PubMed] [Google Scholar]
- 22.Machius M, Chuang JL, Wynn RM, Tomchick DR, Chuang DT. Structure of rat BCKD kinase: Nucleotide-induced domain communication in a mitochondrial protein kinase. Proc Natl Acad Sci USA. 2001;98(20):11218–11223. doi: 10.1073/pnas.201220098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Houston JB. Utility of in vitro drug metabolism data in predicting in vivo metabolic clearance. Biochem Pharmacol. 1994;47(9):1469–1479. doi: 10.1016/0006-2952(94)90520-7. [DOI] [PubMed] [Google Scholar]
- 24.Paul HS, Liu WQ, Adibi SA. Alteration in gene expression of branched-chain keto acid dehydrogenase kinase but not in gene expression of its substrate in the liver of clofibrate-treated rats. Biochem J. 1996;317(Pt 2):411–417. doi: 10.1042/bj3170411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao Y, Jaskiewicz J, Harris RA. Effects of clofibric acid on the activity and activity state of the hepatic branched-chain 2-oxo acid dehydrogenase complex. Biochem J. 1992;285(Pt 1):167–172. doi: 10.1042/bj2850167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ono K, et al. Regulation by induction of branched-chain 2-oxo acid dehydrogenase complex in clofibrate-fed rat liver. Biochem Biophys Res Commun. 1990;172(1):243–248. doi: 10.1016/s0006-291x(05)80200-1. [DOI] [PubMed] [Google Scholar]
- 27.Kadota Y, Kazama S, Bajotto G, Kitaura Y, Shimomura Y. Clofibrate-induced reduction of plasma branched-chain amino acid concentrations impairs glucose tolerance in rats. JPEN J Parenter Enteral Nutr. 2012;36(3):337–343. doi: 10.1177/0148607111414578. [DOI] [PubMed] [Google Scholar]
- 28.Iannitti T, Palmieri B. Clinical and experimental applications of sodium phenylbutyrate. Drugs R D. 2011;11(3):227–249. doi: 10.2165/11591280-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mokhtarani M, et al. Urinary phenylacetylglutamine as dosing biomarker for patients with urea cycle disorders. Mol Genet Metab. 2012;107(3):308–314. doi: 10.1016/j.ymgme.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hood DA, Terjung RL. Effect of alpha-ketoacid dehydrogenase phosphorylation on branched-chain amino acid metabolism in muscle. Am J Physiol. 1991;261(5 Pt 1):E628–E634. doi: 10.1152/ajpendo.1991.261.5.E628. [DOI] [PubMed] [Google Scholar]
- 31.Su Y, et al. Hypothalamic leucine metabolism regulates liver glucose production. Diabetes. 2012;61(1):85–93. doi: 10.2337/db11-0857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ichihara A, Koyama E. Transaminase of branched chain amino acids. I. Branched chain amino acids-alpha-ketoglutarate transaminase. J Biochem. 1966;59(2):160–169. doi: 10.1093/oxfordjournals.jbchem.a128277. [DOI] [PubMed] [Google Scholar]
- 33.Hutson SM. Subcellular distribution of branched-chain aminotransferase activity in rat tissues. J Nutr. 1988;118(12):1475–1481. doi: 10.1093/jn/118.12.1475. [DOI] [PubMed] [Google Scholar]
- 34.Shinnick FL, Harper AE. Branched-chain amino acid oxidation by isolated rat tissue preparations. Biochim Biophys Acta. 1976;437(2):477–486. doi: 10.1016/0304-4165(76)90016-7. [DOI] [PubMed] [Google Scholar]
- 35.Livesey G, Lund P. Enzymic determination of branched-chain amino acids and 2-oxoacids in rat tissues. Transfer of 2-oxoacids from skeletal muscle to liver in vivo. Biochem J. 1980;188(3):705–713. doi: 10.1042/bj1880705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kato M, Chuang JL, Tso SC, Wynn RM, Chuang DT. Crystal structure of pyruvate dehydrogenase kinase 3 bound to lipoyl domain 2 of human pyruvate dehydrogenase complex. EMBO J. 2005;24(10):1763–1774. doi: 10.1038/sj.emboj.7600663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu G, et al. Protein phosphatase 2Cm is a critical regulator of branched-chain amino acid catabolism in mice and cultured cells. J Clin Invest. 2009;119(6):1678–1687. doi: 10.1172/JCI38151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuzuya T, et al. Regulation of branched-chain amino acid catabolism in rat models for spontaneous type 2 diabetes mellitus. Biochem Biophys Res Commun. 2008;373(1):94–98. doi: 10.1016/j.bbrc.2008.05.167. [DOI] [PubMed] [Google Scholar]
- 39.Wynn RM, et al. Pyruvate dehydrogenase kinase-4 structures reveal a metastable open conformation fostering robust core-free basal activity. J Biol Chem. 2008;283(37):25305–25315. doi: 10.1074/jbc.M802249200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chuang JL, Chuang DT. Diagnosis and mutational analysis of maple syrup urine disease using cell cultures. Methods Enzymol. 2000;324:413–423. doi: 10.1016/s0076-6879(00)24250-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.