Abstract
Familial exudative vitreoretinopathy (FEVR) is a genetically heterogeneous disorder characterized by abnormal vascularization of the peripheral retina, which can result in retinal detachment and severe visual impairment. In a large Dutch FEVR family, we performed linkage analysis, exome sequencing, and segregation analysis of DNA variants. We identified putative disease-causing DNA variants in proline-alanine-rich ste20-related kinase (c.791dup; p.Ser265ValfsX64) and zinc finger protein 408 (ZNF408) (c.1363C>T; p.His455Tyr), the latter of which was also present in an additional Dutch FEVR family that subsequently appeared to share a common ancestor with the original family. Sequence analysis of ZNF408 in 132 additional individuals with FEVR revealed another potentially pathogenic missense variant, p.Ser126Asn, in a Japanese family. Immunolocalization studies in COS-1 cells transfected with constructs encoding the WT and mutant ZNF408 proteins, revealed that the WT and the p.Ser126Asn mutant protein show complete nuclear localization, whereas the p.His455Tyr mutant protein was localized almost exclusively in the cytoplasm. Moreover, in a cotransfection assay, the p.His455Tyr mutant protein retains the WT ZNF408 protein in the cytoplasm, suggesting that this mutation acts in a dominant-negative fashion. Finally, morpholino-induced knockdown of znf408 in zebrafish revealed defects in developing retinal and trunk vasculature, that could be rescued by coinjection of RNA encoding human WT ZNF408 but not p.His455Tyr mutant ZNF408. Together, our data strongly suggest that mutant ZNF408 results in abnormal retinal vasculogenesis in humans and is associated with FEVR.
Familial exudative vitreoretinopathy (FEVR) is a well-characterized developmental anomaly of the retinal vasculature that was first described by Criswick and Schepens in 1969 (1). The most prominent characteristics of the disease result from the incomplete and aberrant vascularization of the peripheral retina and/or retinal blood vessel differentiation (2). The latter can lead to various complications, such as retinal neovascularization and exudates, vitreous hemorrhage, vitreoretinal traction, ectopia of the macula, and retinal folds and detachments (3). The clinical signs in affected individuals can be diverse, ranging from hardly detectable vascular anomalies in the peripheral retina in asymptomatic individuals to bilateral retinal detachments leading to blindness.
FEVR displays all Mendelian forms of inheritance (4–8). To date, mutations in genes encoding frizzled 4 (FZD4), low-density lipoprotein receptor-related protein 5 (LRP5), tetraspanin-12 (TSPAN12), and Norrin [encoded by Norrie Disease Pseudoglioma (NDP)] have been shown to cause FEVR (4, 9–12). The proteins encoded by these genes participate in the norrin/β-catenin pathway, in which LRP5 and FZD4 act as coreceptors, NDP as a ligand for FZD4 (13) and TSPAN12 as an aid for FZD4 multimerization (14). Mutations in these genes explain up to ∼50% of all reported FEVR cases (3, 10, 11, 15).
We previously excluded mutations in FZD4, LRP5, and TSPAN12 in 10 Dutch probands with adFEVR (3, 15). In the largest genetically unsolved adFEVR family, linkage analysis was combined with an unbiased exome sequencing approach of two distantly related relatives, leading to identification of a missense mutation in zinc finger protein 408 (ZNF408) as a novel cause of autosomal dominant (ad) FEVR. Cellular transfection studies suggest a dominant-negative disease mechanism, and morpholino (MO)-induced knockdown of znf408 in zebrafish show its involvement in development of the retinal vasculature.
Results
Linkage Analysis.
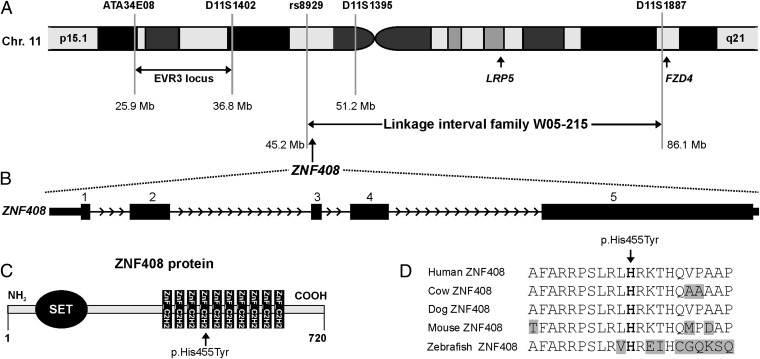
Family W05-215 consists of 11 persons with FEVR. The clinical characteristics of this family have been described previously (8) and are presented in SI Materials and Methods for individuals III-5 and V:2. The causative gene underlying FEVR was identified through linkage analysis combined with exome sequencing. Because nonpenetrance is frequently observed in adFEVR families, only affected individuals were analyzed. In eight affected individuals (Fig. S1A), SNP genotyping and linkage analysis revealed a maximum multipoint logarithm of the odds (LOD) score of 2.7 on chromosome 2q (13.6 Mb, between rs1403970 and rs1995496), and chromosome 11 (41.7 Mb, between rs8929 and rs1986778) (Fig. S1B). Intriguingly, the region on chromosome 11 contains two of the three known adFEVR genes, LRP5 and FZD4 (Fig. 1A). Mutations in the coding regions and flanking intronic sequences of these two genes had been excluded in this family through Sanger sequencing (3). In addition, fine-mapping with microsatellite markers had already identified D11S1887 as the flanking distal marker in individual V:6, thereby excluding FZD4 as the causative gene in this family (Fig. 1A).
Fig. 1.
ZNF408 chromosomal location, genomic structure, and ZNF408 protein characteristics. (A) Part of chromosome 11 showing the W05-215 linkage interval. The locations of the FEVR genes FZD4 and LRP5, as well as the boundaries of the EVR3 locus (16), are depicted. (B) Exon-intron structure of the ZNF408 gene. (C) Schematic overview of ZNF408, a protein of 720 amino acids that is predicted to contain one putative SET and 10 C2H2-type zinc-finger domains. The position of the histidine residue that is mutated to a tyrosine is indicated. (D) Sequence comparison of ZNF408 proteins from several vertebrate species. The alignment contains the histidine residue that is mutated (in bold type), along with 10 flanking amino acids on each side. Identical residues are black on a white background, whereas nonidentical residues are black on a gray background.
Exome Sequencing.
Exome sequencing was performed in two distantly related affected individuals from family W05-215 (III:5 and V:2; Fig. S1A). For each individual, ∼3.1 Gb of sequence data were obtained, and 76% and 70% of bases originated from the targeted exome, resulting in a mean coverage of 60- and 52-fold, respectively. More than 92% of the targeted exons were covered more than 10 times. After variant filtering (17) and removal of low-confident variants, ∼200 candidate variants remained for each affected individual (Table S1). After combining the exome data and assuming an autosomal dominant mode of inheritance, only 19 variants were present in both III:5 and V:2 (Tables S1 and S2), only 6 of which were located in the genomic regions with the highest LOD score, that is, on chromosomes 2 and 11 (Fig. S1B and Table S2). These six variants included a 1-bp duplication in proline–alanine-rich ste20-related kinase (PASK), predicted to result in the premature termination of protein synthesis, and five missense variants, in proline-rich protein 21 (PRR21), ZNF408, low density lipoprotein receptor-related protein 4 (LRP4), olfactory receptor, family 4, subfamily C, member 46 (OR4C46), and glycine-N-acyltransferase (GLYAT). A thorough bioinformatic analysis revealed that only the c.1363C>T change in ZNF408 had a high phyloP score and was predicted to be pathogenic by all three prediction programs used (Table S3). Thus, the frameshift variant in PASK (c.791dup; p.Ser265ValfsX64) and the missense variant in ZNF408 (c.1363C>T; p.His455Tyr) were considered the two prime candidates underlying FEVR in family W05-215.
Segregation Analysis of PASK and ZNF408 Variants.
Segregation analysis of the variants in PASK and ZNF408 in family W05-215 revealed that both were present in all affected individuals as well as in two nonaffected family members (PASK in IV:7 and V:4 and ZNF408 in V:1 and V:4; Fig. S1A). Nonpenetrance is observed in up to 25% of individuals carrying mutations in FZD4, LRP5, NDP, and TSPAN12 (3, 10). Neither of the two variants was detected in 220 ethnically matched control alleles. The c.1363C>T ZNF408 variant was not found in 1,154 in-house ethnically matched exomes unrelated to FEVR. The c.791dup PASK variant was found once in this database (Table S2), suggesting that this change might represent a rare polymorphism.
To determine whether one of the two variants was also present in other Dutch individuals with FEVR, restriction fragment length polymorphism analysis was performed on the probands of nine small FEVR families. Intriguingly, the proband of family W05-220 (IV:3) also carried the c.1363C>T change in ZNF408 heterozygously, as did his affected grand nephew V:2 (Fig. S1A). Clinical details of these two affected individuals are presented in SI Materials and Methods. The c.791dup variant in PASK was not detected in any of the other FEVR probands. Because the probands of families W05-215 and W05-220 originated from the same geographic region, microsatellite markers and SNPs surrounding ZNF408 were genotyped in both families, which revealed that the two families shared a disease haplotype of at least 9.9 Mb (Fig. S2). Finally, genealogical analysis revealed that families W05-215 and W05-220 shared a common ancestor five generations back.
To rule out the presence of a different PASK variant in the affected individuals of family W05-220, all 18 exons were sequenced in IV:3 and V:2. No sequence variants were observed. Thus, the missense mutation in ZNF408, and not the frameshift mutation in PASK, was considered the pathogenic mutation underlying FEVR in families W05-215 and W05-220.
Genome-wide SNP analysis of eight affected individuals of family W05-215 yielded a maximum LOD score of 2.7 for the locus on chromosome 11 harboring ZNF408. To determine the significance of linkage of this locus in the combined W05-215 and W05-220 pedigrees, two-point LOD scores were calculated using the microsatellite marker data from 10 affected individuals, individual IV:2 from family W05-220, and 8 unaffected relatives from both families. For affected individuals IV:10, IV:11, and V:7, all of whom carry the p.His455Tyr mutation, hypothetical marker alleles were assigned. A significant two-point LOD score of 3.05 was calculated for marker D11S1395.
The five exons of ZNF408 (Fig. 1B) encode a transcription factor of 720 amino acids that belongs to the class of C2H2 zinc finger proteins. ZNF408 is predicted to harbor a SET (su(var)3–9, enhancer-of-zeste, trithorax) domain, which is thought to be involved in protein–protein interactions in the regulation of chromatin-mediated gene expression (18–20). Ten C2H2-type zinc finger binding domain signatures are predicted, each consisting of two cysteine and two histidine residues thought to be crucial for DNA binding (Fig. 1C). The histidine residue at position 455, one of these two core histidines in the fourth zinc finger C2H2 domain, is highly conserved throughout vertebrate evolution (Fig. 1 C and D). Taken together, these data, along with the absence of the variant in 220 ethnically matched control alleles and 1,154 in-house exomes, and the bioinformatic prediction of pathogenicity indicate that the substitution of a tyrosine for a histidine at position 455 in ZNF408 is likely to affect the physiological function of this transcription factor.
Expression of ZNF408 in Fetal and Adult Tissues.
As a first step to unraveling the function of ZNF408, its expression pattern was assessed in a panel of various human adult and fetal tissues by quantitative RT-PCR according to previously published protocols (21). Although ZNF408 was expressed in all tissues or organs tested, retinal expression was the highest in the 11 human adult tissues or organs, almost ∼30 times higher than that in heart, placenta, or liver (Fig. S3). Interestingly, ZNF408 expression is also abundantly expressed in human fetal eye, supporting the idea that ZNF408 has an important role in eye development and homeostasis.
Sequence Analysis of ZNF408 in Additional Individuals with FEVR.
To determine the prevalence of ZNF408 mutations as a cause of FEVR, we studied another 77 European and 55 Japanese FEVR probands, in whom mutations in the known FEVR genes were previously excluded. One missense variant (p.Ser126Asn) was detected in an FEVR proband (F117; II:2; SI Materials and Methods) from Japan. This variant was also found in the father (I:1) of the proband, who exhibited not the typical features of FEVR, but rather an erosive vitreoretinopathy phenotype. The variant was not present in our in-house exome variant database or in 191 ethnically matched Japanese control individuals. The serine residue at position 126 was moderately conserved (Fig. S4) and predicted to have some pathogenic potential by bioinformatic analysis (Table S4).
Subcellular Localization of WT and Mutant ZNF408 Proteins.
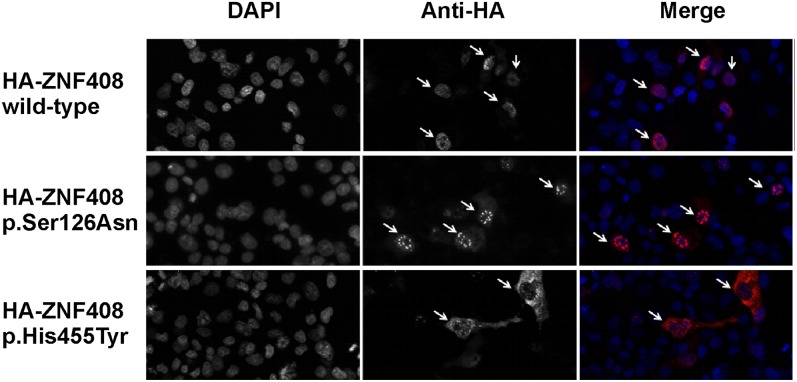
To determine the effect of the two missense changes on the predicted nuclear localization of ZNF408, constructs expressing N-terminally HA-tagged fusion proteins of WT ZNF408 or the ZNF408_Ser126Asn and ZNF408_His455Tyr mutants were generated. Transient transfection of COS-1 cells followed by immunocytochemical analysis revealed that localization of WT and mutant ZNF408_Ser126Asn proteins was restricted almost exclusively to the cell nucleus (Fig. 2). In contrast, the p.His455Tyr mutant ZNF408 protein was present predominantly in the cytoplasm. The latter observations are consistent with a possible role of ZNF408 as a transcriptional regulator and indicate that the p.His455Tyr mutation has an effect on the nuclear localization of ZNF408.
Fig. 2.
Immunocytochemical analysis of COS-1 cells transiently transfected with constructs encoding HA-tagged WT and mutant (p.Ser126Asn, p.His455Tyr) ZNF408 proteins. DAPI stains cell nuclei, whereas the anti-HA antibody stains the ZNF408 fusion proteins (indicated by arrows). (Right) Merged pictures (DAPI in blue, and HA-tagged ZNF408 proteins in red). Representative examples are shown for each transfection, indicating full nuclear localization for the WT and p.Ser126Asn ZNF408 proteins, but with the p.His455Tyr mutant ZNF408 confined mainly to the cytoplasm (each indicated by arrows).
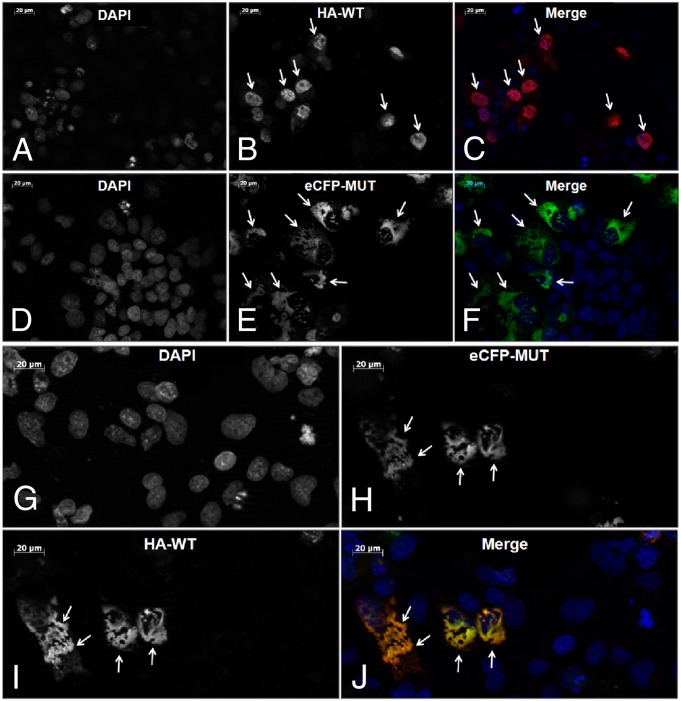
To test the hypothesis that the mutant p.His455Tyr ZNF408 protein interacts and possibly interferes with the routing of the WT counterpart, COS-1 cells were transiently transfected with both the HA-tagged WT ZNF408 construct and a mutant enhanced cyan fluorescent protein (eCFP)-tagged ZNF408_His455Tyr construct. Intriguingly, whereas on single transfection, WT ZNF408 is located almost exclusively in the nucleus, cotransfection of the WT and mutant constructs resulted in mislocalization of the WT ZNF408 in the cytoplasm (Fig. 3), suggesting oligomerization of ZNF408 WT and mutant proteins.
Fig. 3.
Cotransfection analysis of WT and p.His455Tyr mutant ZNF408. (A–C) Immunocytochemical analysis of transiently transfected HA-tagged WT ZNF408 proteins. Representative examples show WT ZNF408 expression inside the cell nuclei (arrows). Images are presented as DAPI (staining cell nuclei) (A), anti-HA (detecting HA-tagged ZNF408 proteins) (B), and merged pictures (DAPI in blue, HA-tagged ZNF408 in red) (C). (D–F) Fluorescence analysis of transiently transfected eCFP-p.His455Tyr mutant ZNF408 proteins. Mutant ZNF408 is located mainly outside the cell nuclei (indicated by arrows). Images are presented as DAPI (staining cell nuclei) (D), eCFP (detecting eCFP-ZNF408 mutant proteins) (E), and merged pictures (DAPI in blue, eCFP-tagged p.His455Tyr ZNF408 in green) (F). (G–J) Combined analysis of COS-1 cells cotransfected with HA-tagged WT ZNF408 as well as eCFP-p.His455Tyr mutant ZNF408 proteins. In cells that express both the mutant and the WT ZNF408, the WT ZNF408 is also retained in the cytoplasm (I and J). Images are presented as DAPI (staining cell nuclei) (G), eCFP (detecting eCFP-p.His455Tyr ZNF408 mutant proteins (H), anti-HA (detecting HA-tagged WT ZNF408 proteins) (I), and merged pictures (DAPI in blue, HA-tagged WT ZNF408 in red, eCFP-p.His455Tyr ZNF408 in green) (J).
Functional Characterization of znf408 in Zebrafish.
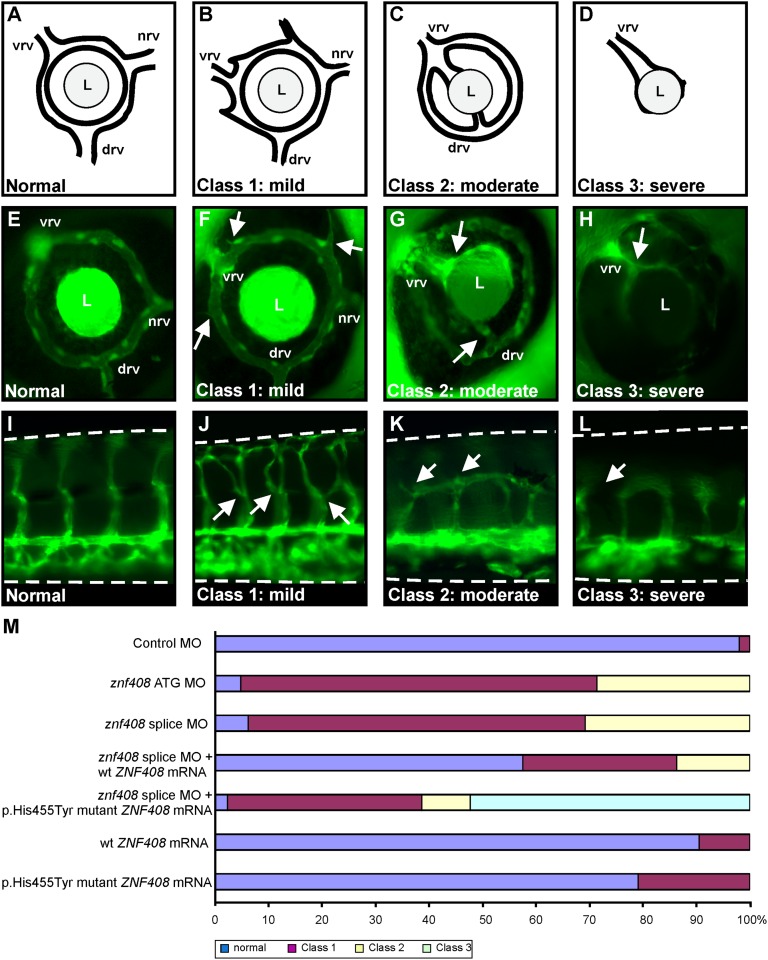
The role of ZNF408 in retinal vascularization was studied by examining the effects of znf408 knockdown in fli1:eGFP transgenic zebrafish. This zebrafish strain expresses eGFP under the control of the promoter of the early endothelial marker fli1 (22), allowing the monitoring of blood vessel growth during embryonic development. We designed a splice-blocking MO that was microinjected in different dosages in the yolk of embryos at the one- to two-cell stage. Control MO (10 ng) and znf408 MO-injected embryos were harvested at 2 d postfertilization (dpf) to evaluate the effect on splicing. A dose-dependent increase in aberrantly spliced znf408 was observed compared with control MO-injected embryos (Fig. S5). Morphological analysis of znf408 morphants identified 6 ng of splice-blocking MO as the optimal concentration for use in subsequent experiments. Injection of 10 ng znf408 MO resulted in embryos with curved tails, failure of ventral retinal fusion, and nearly complete absence of blood vessels, leading to necrosis and excessive embryonic death before 6 dpf (∼82% of injected embryos; n = 181). Embryos injected with 6 ng of MO appeared morphologically grossly normal. Approximately 92% (n = 297) of znf408 morphants showed defects in vessel sprouting at 3 dpf (Fig. 4M). Stage-matched uninjected controls and control MO-injected embryos from the same clutch developed a single intraocular ring vessel from which three radial vessels extend: the nasal (nrv), dorsal (drv), and ventral (vrv) radial vessels (Fig. 4 A and E).
Fig. 4.
Functional characterization of znf408 in zebrafish. (A–D) Schematic overview of zebrafish eye vasculature phenotypes observed on MO-induced knockdown, or expression of WT or mutant ZNF408. Embryos were divided into four subclasses: normal, mildly affected (class 1), moderately affected (class 2), and severely affected (class 3). In the normal situation, blood enters through the nasal vessel (nrv) and exits via the intraocular ring vessel through the dorsal (drv) and ventral (vrv) vessels. L, lens. (E–H) Representative images of GFP expression in the eyes of fli1:EGFP transgenic zebrafish larvae. Abnormal sprouting of the vessels in class 1 embryos is indicated by white arrows (F). In class 2 (G), the nrv is absent, the vrv is enlarged, and both vrv and drv are directly attached to the lens. In class 3 (H), the complete intraocular ring vessel is absent. (I–L) Lateral views of the trunk vasculature (dorsal side is up) in fli1:EGFP transgenic larvae. Intersegmental vessels project from the dorsal aorta (bottom horizontal vessel) toward the dorsal longitudinal vessel at the top. (J) In class 1 embryos, intersegmental vessel branching is disrupted (indicated by white arrows). (K and L) In class 2 and class 3 embryos, the intersegmental vessels failed to reach the dorsal longitudinal vessel. (M) Phenotypic classification of zebrafish larvae injected with control MO (10 ng), znf408-ATG MO (6 ng) or znf408-splice MO (6 ng), mRNA encoding human WT or p.His455Tyr mutant ZNF408, or combinations of splice MO and mRNAs. At least 50 larvae per condition were evaluated.
Znf408 morphants were divided into three distinct classes based on the severity of the observed phenotype: normal, class 1 (mildly affected), and class 2 (moderately affected). Nineteen out of the 297 morphants analyzed were classified as normal. Retinal radial vessel sprouting defects were observed in the class 1 morphants (182 of the 297 morphants). Along with the nrv, drv, and vrv, additional radial vessels were found to extend from the intraocular ring vessel surrounding the lens (Fig. 4 B and F). The moderately affected class 2 morphants (97 of the 297 morphants) exhibited only two radial vessels (vrv and drv) that projected toward and connected with the lens (Fig. 4 C and G).
In addition to the retinal vascularization defects, abnormal trunk vascularization was observed. In the control MO-injected zebrafish, intersegmental vessels (ISVs) projected from the dorsal aorta (bottom horizontal vessel) toward the dorsal longitudinal vessel at the top (Fig. 4I). In class 1 morphants, the ISVs extended from the dorsal aorta, branched at the midline, and eventually reached the dorsal longitudinal vessel (Fig. 4J). In class 2 morphants, the ISVs branched at the midline and failed to connect to the dorsal longitudinal vessel (Fig. 4K).
The specificity of the morphant phenotype was further corroborated using a second MO (ATG MO; 6 ng) that blocks translation of znf408. The results were highly comparable to those obtained using the splice-blocking MO (Fig. 4M). These data indicate that the developmental abnormalities are specifically caused by reduced znf408 levels, and suggest that znf408 function is essential for normal blood vessel development in zebrafish.
Rescue experiments were performed to further support the specificity of the znf408 MO-induced phenotype. Coinjection of RNA encoding the human WT ZNF408 together with the splice MO clearly rescued the vascular phenotype, with more than one-half of all embryos appearing normal (Fig. 4M). In contrast, coinjection of RNA encoding the human p.His455Tyr mutant ZNF408 with the splice MO did not rescue the phenotype, but made it even worse. More than one-half of the embryos displayed a complete loss of the intraocular ring vessel surrounding the lens (Fig. 4 D and H). Severe defects in the trunk vasculature were observed as well (Fig. 4L). Thus, these embryos were classified as severely affected class 3 embryos (Fig. 4M).
To determine whether human p.His455Tyr mutant ZNF408 could exert a dominant-negative effect on the zebrafish WT protein, RNA encoding human WT or p.His455Tyr mutant ZNF408 was injected into zebrafish embryos. Injection of RNA encoding WT ZNF408 resulted in ∼9% of all animals displaying a mild class 1 vascular phenotype. Injection of the same amount of RNA encoding p.His455Tyr mutant ZNF408 resulted in 21% of embryos with a vascular phenotype (Fig. 4M), suggesting that overexpression of the p.His455Tyr mutant protein is able to induce a mild vascular phenotype. Taken together, these data, along with the inability of p.His455Tyr mutant ZNF408 to rescue the MO-induced vascular phenotype, further prove the pathogenic nature of this variant.
Discussion
In this study, we combined linkage analysis, next-generation sequencing (NGS) technology, and bioinformatic analysis of sequence variants to identify a missense mutation in ZNF408 (p.His455Tyr) underlying FEVR in two distantly related Dutch families. Sequence analysis of ZNF408 in 132 other FEVR probands revealed only one other missense variant of unknown pathogenicity, p.Ser126Asn, in an individual of Japanese origin.
ZNF408 belongs to the family of zinc finger transcription factors. Zinc fingers are versatile DNA-recognition elements found in numerous regulatory proteins that participate in a variety of cellular activities, including embryonic development and cellular differentiation. The zinc finger gene family can be divided into many subclasses based on the number and type of zinc fingers that they contain (23). Each finger constitutes a self-contained domain stabilized by a zinc ion ligated to a pair of cysteines and a pair of histidines, along with an inner structural hydrophobic core. Although differential use of the two residues gives rise to several types of zinc fingers (24), the C2H2 finger has emerged as the most prevalent protein motif, found in 2% of all human proteins (25). C2H2 zinc fingers also have been implicated in protein–protein interactions. In general, the greater the number of finger domains, the higher the versatility and the more fingers with specific affinity for different ligands in specific classes of proteins, for example, the class of multiple adjacent C2H2 (maC2H2) finger proteins (26, 27). ZNF408 is predicted to consist of 10 zinc finger motifs arranged in a tandem array, located at the C-terminal half of the protein (Fig. 1C). The large number of these motifs suggests that ZNF408 may be involved in a wide range of functions apart from high-affinity DNA binding.
To establish a clear link between ZNF408 and the development of retinal vasculature, we used the zebrafish model system to down-regulate znf408 expression by MO injections and determine the effects on retinal blood vessel formation. Using two different znf408 MOs, we found that more than 90% of the morphants displayed aberrant retinal vasculature. The most prominent abnormalities included sprouting defects of the retinal radial vessels, which clearly demonstrate a functional link between znf408 and the development of retinal vasculature. Along with retinal vessel abnormalities, zebrafish morphants also displayed aberrant development of the trunk vasculature, suggesting that the role of znf408, at least in zebrafish, might not be restricted to the eye. This vascular phenotype was convincingly rescued by coinjection of RNA encoding the human WT, but not the p.His455Tyr mutant ZNF408. Previous work has identified several proteins that are crucial for the development of (retinal) vasculature in zebrafish, including heparin sulfate proteoglycans, laminin alpha, and plexin D1 (28). Other studies have implicated vascular endothelial growth factor A, sphingosine-1 phosphate receptor 1, and the E3 ubiquitin ligase as important to the development of retinal vasculature (29, 30). The wide variety of signaling pathways in which these proteins act directly demonstrates the complexity of retinal vasculature development. The position of znf408 within this complex event remains elusive.
In our cohort of individuals with FEVR, two ZNF408 missense variants were detected, p.Ser126Asn and p.His455Tyr. Transient expression studies in COS-1 cells showed that the p.His455Tyr mutant is localized not in the nucleus, but rather in the cytoplasm. In addition, the mutant p.His455Tyr ZNF408 protein was also capable of retaining the WT ZNF408 protein in the cytoplasm, indicating that ZNF408 can oligomerize. Indeed, C2H2 zinc finger proteins containing a tandem repeat of zinc finger motifs have been shown to be able to dimerize or homopolymerize, for instance, in the Ikaros family members (31, 32). Taken together, these data suggest that the p.His455Tyr mutation acts in a dominant-negative fashion. In contrast, the p.Ser126Asn mutant ZNF408 protein was correctly routed to the nucleus and did not show a mislocalization in the cytoplasm on transient expression. Because the serine residue at position 126 does not reside within the DNA-binding domain but within a putative SET domain, the effect of this variant on the ZNF408 protein may be different compared with p.His455Tyr. The small size of the Japanese family in which the p.Ser126Asn variant is segregating, the variable phenotypes observed in this family, and the absence of misrouting of the mutant protein in cellular transfection studies shed doubt on the causality of this variant.
Screening of a large FEVR cohort failed to identify unequivocal heterozygous loss of function changes such as nonsense mutations, and also did not reveal other clearly pathogenic changes in ZNF408. Using ZNF408 WT/mutant cotransfection studies in COS-1 cells, and (co)injections of RNA encoding mutant ZNF408 in zebrafish, we obtained strong evidence that the p.His455Tyr variant acts in a dominant-negative manner. Taken together, these data suggest that only highly selective amino acid changes in ZNF408 give rise to adFEVR. ZNF408 may have several crucial and as-yet unknown cellular functions in other organs. As a result, ZNF408 haploinsufficiency might not have a phenotypic effect, might give rise to a completely different phenotype, or might not be compatible with life. This is supported in part by the data obtained from our zebrafish studies. Whereas the p.His455Tyr mutant found in persons with FEVR appears to act in a dominant-negative fashion, the MO injections result in reduced expression of znf408, mimicking loss of function. The RT-PCR analysis of splice-MO injected zebrafish showed that there is still a significant amount of znf408 that is correctly spliced, indicating that the MO injections knock down the expression of znf408 only partially. Nevertheless, morphant zebrafish exhibit extraocular vascular abnormalities, suggesting that other classes of ZNF408 mutations also may cause a broader phenotype in humans.
We cannot completely exclude the possibility that other variants segregating in families W05-215 and W05-220 are not covered by our exome NGS. However, the identification of a ZNF408 missense variant affecting a highly conserved amino acid and nucleotide position, in combination with the mislocalization of the mutant ZNF408 protein in a cellular transfection assay and the vasculogenesis defects observed in znf408 knockdown zebrafish embryos, strongly suggest that ZNF408 is implicated in FEVR.
Identification of the transcriptional targets of ZNF408 could pinpoint other genes important for retinal angiogenesis and thus reveal prime candidates for mutations in unsolved cases with FEVR. In conclusion, we have identified ZNF408 as a gene underlying autosomal dominant FEVR, and hypothesize that we have revealed a crucial transcription factor that plays an important role in retinal vasculogenesis.
Materials and Methods
Details on the human subjects; linkage analysis, exome sequencing methodology, and mutation analysis of PASK and ZNF408; generation of plasmids, transient transfection of COS-1 cells, and subcellular localization studies of mutant and WT ZNF408; analysis of the evolutionary conservation of ZNF408; and MO and zebrafish embryo manipulations are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank C. Beumer, D. T. Cremers, S. Feil, M. Kwint, D. Mans, K. Neveling, T. A. Peters, S. D. van der Velde-Visser, I. de Wijs, N. Wieskamp, and M. Wittmer for their excellent technical assistance. We also thank all of the participating individuals with FEVR and their relatives. This work was supported by the European Union Research Training Network (RETNET; Grant MRTN-CT-2003-5040032, to F.P.M.C.); Foundation Fighting Blindness Center (Grant C-CMM-0811-0547-RAD03, to H. Kremer and E.v.W.); Netherlands Organization for Health Research and Development (Grants 917-66-363 and 911-08-025, to J.A.V.); the European Union TECHGENE project (Grant Health-F5-2009-223143 to J.A.V.); the AnEUploidy project (Grant LSHG-CT-2006-37627, to A.H. and J.A.V.); funding from the Algemene Nederlandse Vereniging ter Voorkoming van Blindheid, F.P. Fischer Stichting, Gelderse Blinden Stichting, Landelijke Stichting Blinden en Slechtzienden, Rotterdamse Stichting Blindenbelangen, Stichting Blindenhulp, Stichting Blinden-Penning, Stichting Nederlands Oogheelkundig Onderzoek, Stichting OOG, Stichting voor Ooglijders, and Stichting tot Verbetering van het Lot der Blinden (to F.P.M.C., F.N.B., and C.E.v.N.); the Vereniging Bartiméus (to F.N.B.); the Stichting Blindenhulp, Stichting Nederlands Oogheelkundig Onderzoek, and Stichting Researchfonds oogheelkunde (to H. Kremer, E.v.W.); the Swiss National Science Foundation (Grant 31003A_122359, to W.B.); the Royal Society (to C.T.); and Grant-in-Aid 22591956 for Scientific Research (C) from the Japan Society for the Promotion of Science (to H. Kondo, T.T., and E.U.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1220864110/-/DCSupplemental.
References
- 1.Criswick VG, Schepens CL. Familial exudative vitreoretinopathy. Am J Ophthalmol. 1969;68(4):578–594. doi: 10.1016/0002-9394(69)91237-9. [DOI] [PubMed] [Google Scholar]
- 2.Canny CLB, Oliver GL. Fluorescein angiographic findings in familial exudative vitreoretinopathy. Arch Ophthalmol. 1976;94(7):1114–1120. doi: 10.1001/archopht.1976.03910040034006. [DOI] [PubMed] [Google Scholar]
- 3.Boonstra FN, et al. Clinical and molecular evaluation of probands and family members with familial exudative vitreoretinopathy. Invest Ophthalmol Vis Sci. 2009;50(9):4379–4385. doi: 10.1167/iovs.08-3320. [DOI] [PubMed] [Google Scholar]
- 4.Chen ZY, et al. A mutation in the Norrie disease gene (NDP) associated with X-linked familial exudative vitreoretinopathy. Nat Genet. 1993;5(2):180–183. doi: 10.1038/ng1093-180. [DOI] [PubMed] [Google Scholar]
- 5.Gow J, Oliver GL. Familial exudative vitreoretinopathy: An expanded view. Arch Ophthalmol. 1971;86(2):150–155. doi: 10.1001/archopht.1971.01000010152007. [DOI] [PubMed] [Google Scholar]
- 6.Jiao X, Ventruto V, Trese MT, Shastry BS, Hejtmancik JF. Autosomal recessive familial exudative vitreoretinopathy is associated with mutations in LRP5. Am J Hum Genet. 2004;75(5):878–884. doi: 10.1086/425080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Nouhuys CE. Dominant exudative vitreoretinopathy. Ophthalmic Paediatr Genet. 1985;5(1-2):31–38. doi: 10.3109/13816818509007853. [DOI] [PubMed] [Google Scholar]
- 8.van Nouhuys CE. Dominant exudative vitreoretinopathy and other vascular developmental disorders of the peripheral retina. Doc Ophthalmol. 1982;54(1-4):1–414. doi: 10.1007/BF00183127. [DOI] [PubMed] [Google Scholar]
- 9.Berger W, et al. Mutations in the candidate gene for Norrie disease. Hum Mol Genet. 1992;1(7):461–465. doi: 10.1093/hmg/1.7.461. [DOI] [PubMed] [Google Scholar]
- 10.Nikopoulos K, et al. Next-generation sequencing of a 40-Mb linkage interval reveals TSPAN12 mutations in patients with familial exudative vitreoretinopathy. Am J Hum Genet. 2010;86(2):240–247. doi: 10.1016/j.ajhg.2009.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poulter JA, et al. Mutations in TSPAN12 cause autosomal-dominant familial exudative vitreoretinopathy. Am J Hum Genet. 2010;86(2):248–253. doi: 10.1016/j.ajhg.2010.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toomes C, et al. Mutations in LRP5 or FZD4 underlie the common familial exudative vitreoretinopathy locus on chromosome 11q. Am J Hum Genet. 2004;74(4):721–730. doi: 10.1086/383202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu Q, et al. Vascular development in the retina and inner ear: Control by Norrin and Frizzled-4, a high-affinity ligand–receptor pair. Cell. 2004;116(6):883–895. doi: 10.1016/s0092-8674(04)00216-8. [DOI] [PubMed] [Google Scholar]
- 14.Junge HJ, et al. TSPAN12 regulates retinal vascular development by promoting Norrin- but not Wnt-induced FZD4/beta-catenin signaling. Cell. 2009;139(2):299–311. doi: 10.1016/j.cell.2009.07.048. [DOI] [PubMed] [Google Scholar]
- 15.Nikopoulos K, et al. Overview of the mutation spectrum in familial exudative vitreoretinopathy and Norrie disease with identification of 21 novel variants in FZD4, LRP5, and NDP. Hum Mutat. 2010;31(6):656–666. doi: 10.1002/humu.21250. [DOI] [PubMed] [Google Scholar]
- 16.Downey LM, et al. A new locus for autosomal dominant familial exudative vitreoretinopathy maps to chromosome 11p12-13. Am J Hum Genet. 2001;68(3):778–781. doi: 10.1086/318790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoischen A, et al. De novo mutations of SETBP1 cause Schinzel–Giedion syndrome. Nat Genet. 2010;42(6):483–485. doi: 10.1038/ng.581. [DOI] [PubMed] [Google Scholar]
- 18.Cui XM, et al. Association of SET domain and myotubularin-related proteins modulates growth control. Nat Genet. 1998;18(4):331–337. doi: 10.1038/ng0498-331. [DOI] [PubMed] [Google Scholar]
- 19.Huang S, Shao G, Liu LM. The PR domain of the Rb-binding zinc finger protein RIZ1 is a protein-binding interface and is related to the SET domain functioning in chromatin-mediated gene expression. J Biol Chem. 1998;273(26):15933–15939. doi: 10.1074/jbc.273.26.15933. [DOI] [PubMed] [Google Scholar]
- 20.Min JR, Zhang X, Cheng XD, Grewal SIS, Xu RM. Structure of the SET domain histone lysine methyltransferase Clr4. Nat Struct Biol. 2002;9(11):828–832. doi: 10.1038/nsb860. [DOI] [PubMed] [Google Scholar]
- 21.Ayyagari R, et al. Late-onset macular degeneration and long anterior lens zonules result from a CTRP5 gene mutation. Invest Ophthalmol Vis Sci. 2005;46(9):3363–3371. doi: 10.1167/iovs.05-0159. [DOI] [PubMed] [Google Scholar]
- 22.Ellertsdóttir E, et al. Vascular morphogenesis in the zebrafish embryo. Dev Biol. 2010;341(1):56–65. doi: 10.1016/j.ydbio.2009.10.035. [DOI] [PubMed] [Google Scholar]
- 23.Pavletich NP, Pabo CO. Crystal structure of a five-finger GLI–DNA complex: New perspectives on zinc fingers. Science. 1993;261(5129):1701–1707. doi: 10.1126/science.8378770. [DOI] [PubMed] [Google Scholar]
- 24.Mackay JP, Crossley M. Zinc fingers are sticking together. Trends Biochem Sci. 1998;23(1):1–4. doi: 10.1016/s0968-0004(97)01168-7. [DOI] [PubMed] [Google Scholar]
- 25.Lander ES, et al. International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 26.Morgan B, et al. Aiolos, a lymphoid restricted transcription factor that interacts with Ikaros to regulate lymphocyte differentiation. EMBO J. 1997;16(8):2004–2013. doi: 10.1093/emboj/16.8.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsai RY, Reed RR. Identification of DNA recognition sequences and protein interaction domains of the multiple-Zn-finger protein Roaz. Mol Cell Biol. 1998;18(11):6447–6456. doi: 10.1128/mcb.18.11.6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alvarez Y, et al. Genetic determinants of hyaloid and retinal vasculature in zebrafish. BMC Dev Biol. 2007;7:114. doi: 10.1186/1471-213X-7-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Izumi N, et al. Fbxw7 controls angiogenesis by regulating endothelial Notch activity. PLoS ONE. 2012;7(7):e41116. doi: 10.1371/journal.pone.0041116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ben Shoham A, et al. S1P1 inhibits sprouting angiogenesis during vascular development. Development. 2012;139(20):3859–3869. doi: 10.1242/dev.078550. [DOI] [PubMed] [Google Scholar]
- 31.McCarty AS, Kleiger G, Eisenberg D, Smale ST. Selective dimerization of a C2H2 zinc finger subfamily. Mol Cell. 2003;11(2):459–470. doi: 10.1016/s1097-2765(03)00043-1. [DOI] [PubMed] [Google Scholar]
- 32.Sun L, Liu A, Georgopoulos K. Zinc finger-mediated protein interactions modulate Ikaros activity, a molecular control of lymphocyte development. EMBO J. 1996;15(19):5358–5369. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.