Significance
Microbes in contaminated environments often evolve new metabolic pathways for detoxification or degradation of pollutants. In some cases, intermediates in newly evolving pathways are more toxic than the initial compound. The initial step in the degradation of pentachlorophenol by Sphingobium chlorophenolicum generates a particularly toxic intermediate, tetrachlorobenzoquinone (TCBQ). This paper describes how the bacterium is protected from the toxic effects of TCBQ. In the presence of tetrachlorobenzoquinone reductase, TCBQ produced by pentachlorophenol hydroxylase is sequestered until it is reduced to the less toxic tetrachlorohydroquinone.
Keywords: biodegradation, molecular evolution, channeling, quinone reductase
Abstract
Microbes in contaminated environments often evolve new metabolic pathways for detoxification or degradation of pollutants. In some cases, intermediates in newly evolved pathways are more toxic than the initial compound. The initial step in the degradation of pentachlorophenol by Sphingobium chlorophenolicum generates a particularly reactive intermediate; tetrachlorobenzoquinone (TCBQ) is a potent alkylating agent that reacts with cellular thiols at a diffusion-controlled rate. TCBQ reductase (PcpD), an FMN- and NADH-dependent reductase, catalyzes the reduction of TCBQ to tetrachlorohydroquinone. In the presence of PcpD, TCBQ formed by pentachlorophenol hydroxylase (PcpB) is sequestered until it is reduced to the less toxic tetrachlorohydroquinone, protecting the bacterium from the toxic effects of TCBQ and maintaining flux through the pathway. The toxicity of TCBQ may have exerted selective pressure to maintain slow turnover of PcpB (0.02 s−1) so that a transient interaction between PcpB and PcpD can occur before TCBQ is released from the active site of PcpB.
Anthropogenic compounds used as pesticides, solvents, and explosives often persist in the environment and can cause toxicity to humans and wildlife. In some cases, microbes have evolved the ability to partially or completely degrade such compounds. Complete degradation of anthropogenic compounds requires the evolution of new metabolic pathways that convert these compounds into metabolites that can be used by existing metabolic networks. The sequence of steps in new degradative pathways is dictated by the repertoire of enzymes in exposed microbes and/or microbial communities that can catalyze newly needed reactions. The serendipitous assembly of new degradation pathways can result in the production of intermediates that are toxic due to their intrinsic chemical reactivity or adventitious interactions between these intermediates and cellular macromolecules. The toxicity of such intermediates can pose a significant challenge to the evolution of efficient pathways for the degradation of anthropogenic compounds (1).
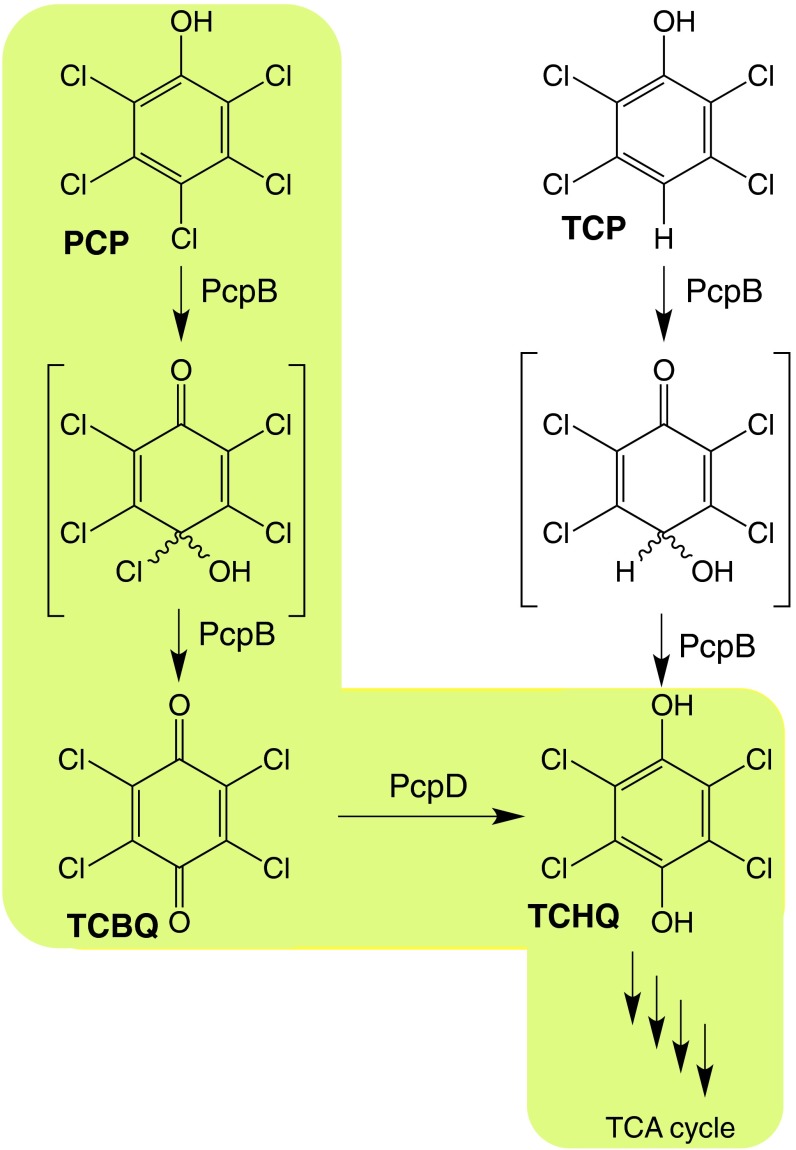
The pathway for degradation of pentachlorophenol (PCP) by Sphingobium chlorophenolicum (Fig. 1) (2, 3) includes the remarkably toxic intermediate tetrachlorobenzoquinone (TCBQ). The LD50 for Escherichia coli protoplasts is <1 µM (4). TCBQ is the most toxic of a set of 14 quinones (5), with an LD50 of 18 µM for primary rat hepatocytes. (For comparison, the LD50 for benzoquinone is 57 µM and for 2,3,4,6-tetramethylbenzoquinone is 800 µM.) TCBQ is a potent alkylating agent, capable of forming covalent adducts with proteins (6) and with the nucleobases of DNA (7). Thiols such as glutathione react with TCBQ particularly rapidly (3, 6). Depletion of glutathione eliminates a critical cellular mechanism for protection against alkylating agents and oxidative damage. Further, TCBQ can generate hydroxyl radicals by its interaction with hydrogen peroxide (8). Thus, TCBQ is a devastating toxin due to a plethora of molecular mechanisms.
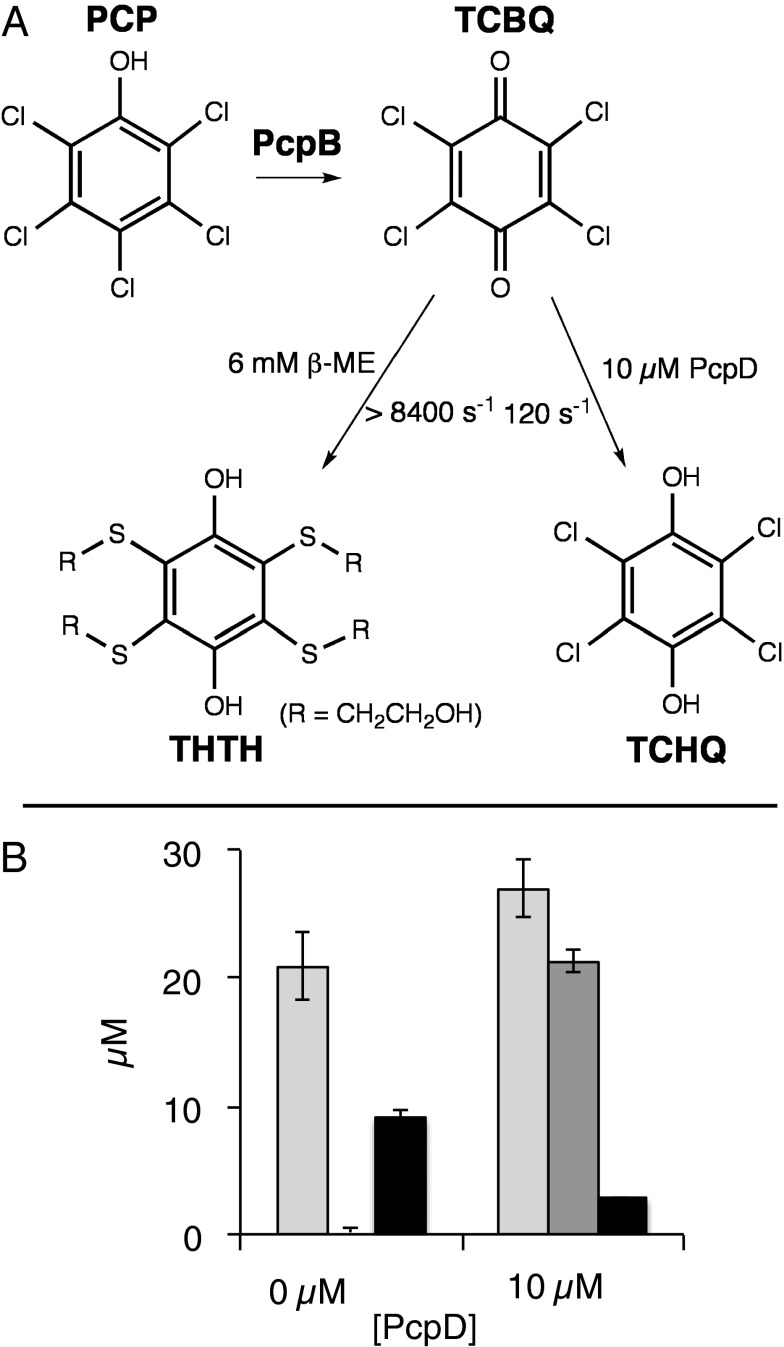
Fig. 1.
The initial steps in the pathway for degradation of PCP in S. chlorophenolicum via the intermediates TCBQ and TCHQ are highlighted in green. Hydroxylation of TCP by PcpB forms TCHQ directly.
The pathway for degradation of PCP in S. chlorophenolicum is believed to be still in the process of evolving because PCP was introduced as a pesticide less than 100 y ago (9, 10) and some of the enzymes in the pathway function quite poorly (3, 11). The intermediacy of a dangerous compound such as TCBQ in a newly evolving pathway brings up a number of interesting questions. First, why has the pathway evolved via a route that involves TCBQ? Second, how does the bacterium protect itself from the toxic effects of TCBQ? And third, does the intermediacy of TCBQ explain the inefficiency of PCP degradation by S. chlorophenolicum?
The answer to the first question emerges from a consideration of the enzymatic repertoire that exists to initiate degradation of naturally occurring phenols. Phenols are common natural products, produced by a range of organisms from microbes to insects and by degradation of lignin. Cleavage of phenols under aerobic conditions requires the introduction of a second hydroxyl group. This reaction can be carried out by cytochrome P450 enzymes or flavin monooxygenases. PCP hydroxylase (PcpB), the first enzyme in the pathway for degradation of PCP, is a flavin monooxygenase (3) and likely originated from an enzyme that hydroxylated a naturally occurring phenol. Hydroxylation of phenols at a position carrying a hydrogen results in formation of a catechol or hydroquinone rather than a benzoquinone (Fig. 1). However, when a good leaving group such as chloride is present at the position of hydroxylation, the unstable intermediate eliminates HCl to form a benzoquinone.
The answer to the second question is suggested by the observation that microbes that generate benzoquinones from phenols typically have a reductase that converts a benzoquinone to a hydroquinone. The gene encoding the reductase is located immediately downstream of the gene encoding the hydroxylase (12). Examples are found in the pathways for degradation of 2,4,6-trichlorophenol (TriCP), p-nitrophenol and o-nitrophenol in Cupriavidus necator JMP134 (13), Pseudomonas sp. Strain WBC-3 (14), and Alcaligenes sp. strain NyZ215 (15), respectively. Like these microbes, S. chlorophenolicum contains a gene encoding a reductase (PcpD) immediately downstream of the gene encoding the hydroxylase (PcpB). We have previously shown that pcpD is essential for survival of S. chlorophenolicum at high concentrations of PCP but is not essential for survival at high concentrations of 2,3,5,6-tetrachlorophenol (TCP) (2). In this work, we have addressed the mechanism of that protection. As shown in Fig. 1, hydroxylation of TCP produces tetrachlorohydroquinone (TCHQ) whereas hydroxylation of PCP produces the more toxic TCBQ. PcpD is an efficient TCBQ reductase. However, its ability to prevent the escape of TCBQ to the solvent before it is reduced to TCHQ is an even more critical aspect of its function.
Our results provide insights into the answer to the third question. Because PcpB and PcpD do not form a stable complex, the rate of their encounter is limited by diffusion. The slow rate of turnover by PcpB [at 0.02 s−1 (3), it is the rate-limiting step of the pathway (4)] may be a result of selective pressure to prevent production of TCBQ at a rate faster than it can be intercepted by PcpD. Thus, the efficiency of the interaction between PcpB and PcpD may limit the efficiency of the degradation of PCP and prevent selection of a more catalytically robust PcpB.
Results
TCBQ Reacts Extremely Rapidly with Thiols.
We attempted to measure the rate constants for reaction of TCBQ (25 µM) with a sampling of thiol compounds using stopped-flow spectroscopy. The rate of reaction with cysteine (250 µM; Fig. S1) was too fast to measure, as the reaction was complete within the dead-time of the instrument (2 ms). Given that at least 10 half-lives occurred during this interval, we can calculate that the second-order rate constant for reaction with cysteine is >1.3 × 107 M−1⋅s−1. Reaction of TCBQ with glutathione (250 µM) was slightly slower (Fig. S1), but the reaction was still over within a few milliseconds; we estimate that the second-order rate constant is >4 × 106 M−1⋅s−1. Similarly, we estimate that the second-order rate constant for reaction of TCBQ with β-mercaptoethanol (β-ME) is >1.4 × 106 M−1⋅s−1. In contrast to the extremely rapid reaction with thiols, reactions of TCBQ with nucleophilic amino acids (250 µM) such as lysine, serine, and histidine were quite slow, with half-lives >5 min. TCBQ did not react with ATP or GTP (as models for nucleobases in DNA) at a measurable rate over a period of 5 min.
TCBQ Reacts with PcpB.
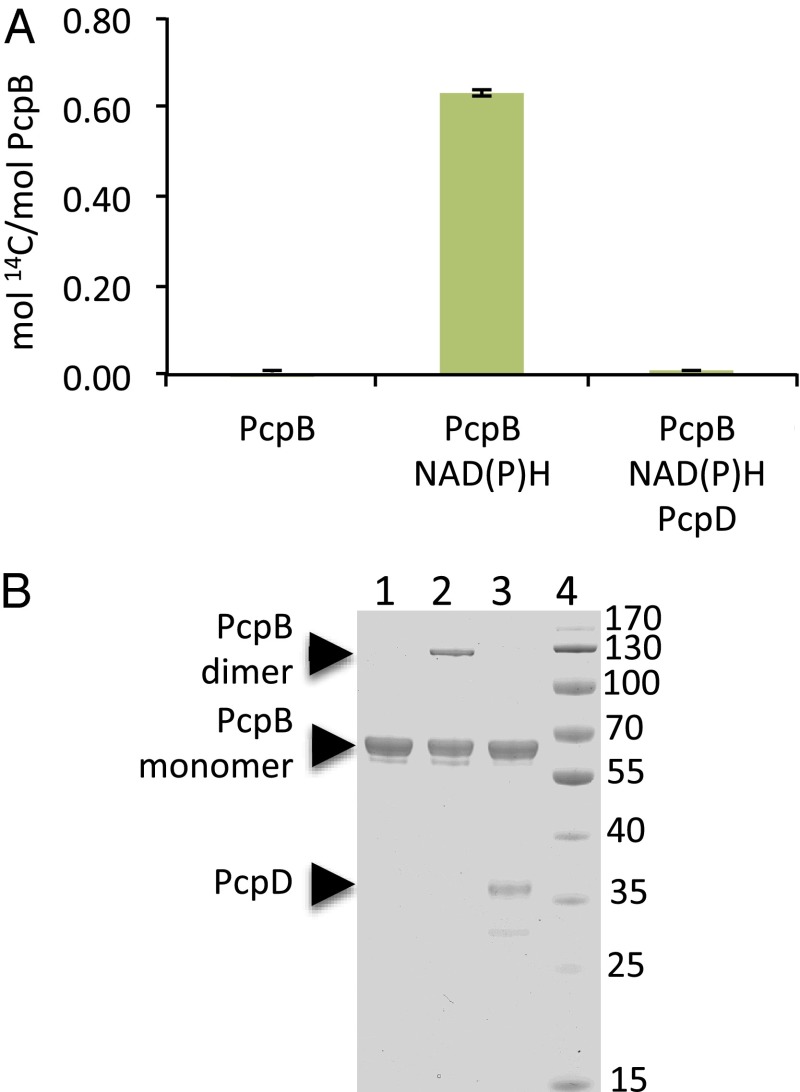
We examined the propensity of TCBQ to react with nucleophiles, most likely cysteine residues, on proteins by incubating PcpB with U-14C-PCP in the absence and presence of its cosubstrate, NADPH. As shown in Fig. 2A, 14C was incorporated into the protein only under turnover conditions, i.e., in the presence of NADPH. These data demonstrate that TCBQ reacts with the protein, but, as expected, PCP does not. Further evidence that TCBQ reacts with protein residues is provided by the observation that covalent dimers of PcpB are formed during turnover of PCP, but not during incubation with PCP in the absence of NADPH (Fig. 2B). In addition, we have previously reported that incubation of PcpB with TCBQ causes loss of catalytic activity (2).
Fig. 2.
TCBQ formed by PcpB forms covalent adducts with PcpB. (A) 14C incorporated into PcpB after incubation with U-14C-PCP and cofactors and PcpD as indicated for 10 min at room temperature. (B) SDS/PAGE analysis of reaction mixtures in which PcpB was incubated with PCP (100 µM) in the absence (lane 1) or presence (lane 2) of NADPH and NADH and in the presence of NADPH, NADH, and PcpD (10 μM) (lane 3) for 10 min at room temperature in 50 mM potassium phosphate, pH 7. Lane 4, molecular mass markers.
Deletion of pcpD from S. chlorophenolicum L1 Results in Increased Sensitivity to Phenols That Are Degraded via a Benzoquinone Intermediate.
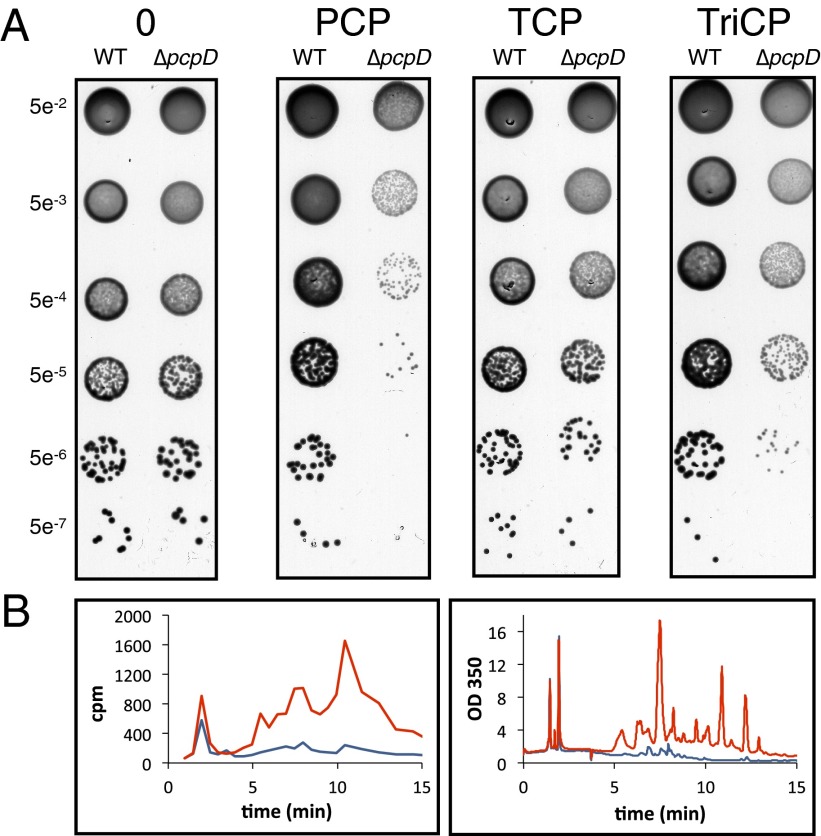
To investigate the toxicity of benzoquinones and hydroquinones generated by hydroxylation of chlorophenols, we deleted pcpD in S. chlorophenolicum L1. The phenotype of the ΔpcpD strain was characterized by growth on agar plates supplemented with PCP, TCP, or TriCP. PCP and TriCP, but not TCP, led to significant toxicity in the ΔpcpD strain compared with the wild type (Fig. 3A). Because both PCP and TriCP, but not TCP, generate benzoquinones, these results demonstrate the in vivo importance of PcpD in protecting cells from toxic benzoquinone intermediates. These results extend our earlier studies of the effects of disruption of pcpD in S. chlorophenolicum strain ATCC 39723 (2).
Fig. 3.
Phenotypic differences between wild-type and ΔpcpD S. chlorophenolicum. (A) Wild-type or ΔpcpD cells were spotted on ATCC 1687 agar plates containing glutamate as a carbon source. The plates were supplemented with 100 μM PCP, TCP, or TriCP and incubated in the dark for 5–7 d. (B) Products derived from U-14C-PCP are excreted into the medium by cells lacking PcpD (red), but not by wild-type cells (blue). Compounds eluting from an HPLC column were monitored by scintillation counting (Left) or by UV-Vis spectroscopy (Right).
We anticipated that TCBQ would not be converted to further intermediates in the PCP degradation pathway in the ΔpcpD strain but would react with various nucleophiles in the cytoplasm. To our surprise, we were unable to identify adducts between TCBQ and glutathione in cytoplasmic fractions. Rather, we found that a number of compounds derived from PCP accumulated in the medium during growth of the ΔpcpD strain, but not of the wild-type strain, in the presence of U-14C-labeled PCP. The presence of 14C in these products (Fig. 3B) suggests that they are derived from U-14C-labeled TCBQ because that is the last product expected to be generated by the ΔpcpD strain. Most of the products appear to contain an intact aromatic ring as they absorb at >320 nm (Fig. 3B). Analysis of some of these products by mass spectrometry indicates that they contain one, two, or three chlorines based upon their isotopic mass patterns. [Chlorine is present as a mixture of isotopes, including 35Cl (76%) and 37Cl (24%). The natural abundance of these isotopes causes a characteristic distribution of peaks separated by 2 atomic mass units.] The identity of these products is difficult to determine, as multiple isomeric species are possible for each, and pathways for decomposition of adducts of TCBQ are unknown.
Purification of Active PcpD.
To investigate the mechanism by which PcpD provides a protective effect against benzoquinones, we set out to characterize PcpD in vitro. PcpD from S. chlorophenolicum L1 is difficult to express and purify (2, 16). Consequently, we looked for other sources that might provide an enzyme more amenable to purification. A multiple sequence alignment of putative TCBQ reductases from other PCP-degrading Sphingomonads with three related phthalate dioxygenase reductases (Fig. S2) revealed that PcpD from S. chlorophenolicum L1 has a unique insertion of an arginine residue in the 2Fe-2S domain. Based on the structure of phthalate dioxygenase reductase from Burkholderia cepacia (17), this arginine should be located in a loop that may interact with the C terminus of the protein and stabilize the 2Fe-2S domain. We suspected that this insertion might interfere with expression and folding in a heterologous system. We therefore decided to produce the closely related protein from S. chlorophenolicum sp. RA2 (18), which differs from PcpD from S. chlorophenolicum sp. L1 in only two positions (position 241 is Val rather than Met, and there is no insertion of Arg at position 245). Overproduction of this PcpD, as well as a truncated version lacking the 2Fe-2S domain (PcpD-FeS), in E. coli resulted in high yields of soluble and active enzyme (50 and 100 mg/L respectively). Purified PcpD from S. chlorophenolicum RA2 is a golden-brown enzyme with a spectrum consistent with the presence of both a flavin and an iron-sulfur cluster (Fig. S3) (19). The truncated enzyme shows the spectrum typical of a flavoenzyme (Fig. S3). The native enzyme is trimeric based on gel filtration chromatography, eluting with an apparent molecular mass of 100 kDa. (The molecular mass of the monomer is 36 kDa.) In contrast, the truncated protein is a dimer rather than a trimer at moderate salt concentrations, eluting with an apparent molecular mass of 56 kDa. (The molecular mass of the monomer is 26 kDa.)
PcpD Has Broad Substrate Specificity.
Based on the homology between PcpD and enzymes such as phthalate dioxygenase reductase, PcpD most likely evolved from the reductase component of a two-component oxygenase system (2). Reductases in this family often have activity toward a range of substrates, both small molecules and proteins, and can donate either one or two electrons from either the flavin or the 2Fe-2S cluster (20). We therefore tested the ability of PcpD and PcpD-FeS to reduce the small molecule 2,6-dichloroindophenol (DCIP) and the protein cytochrome c using NADH as an electron donor (Table 1). DCIP is reduced efficiently by PcpD with a kcat/Km of 3.7 × 105 M−1⋅s−1 and a Km of 38 µM in the presence of 100 µM NADH. The protein substrate cytochrome c is also efficiently reduced by PcpD, with a kcat/KM of 5.1 × 105 M−1⋅s−1 and a Km of 18 µM at 100 µM NADH. The values of kcat/KM are about 10-fold lower than those of phthalate dioxygenase reductase for both substrates (kcat/KM is 1.8 × 106 M−1⋅s−1 for DCIP and 6.0 × 106 M−1⋅s−1 for cytochrome c) (20).
Table 1.
Kinetic parameters for reactions of PcpD with TCBQ, DCIP, and cytochrome c
| Substrate | Kinetic parameter | PcpD | PcpD-FeS |
| TCBQ (with 200 µM NADH) | kcat/Km (M−1⋅s−1) | (1.2 ± 0.17) x 107 | (2.51 ± 0.67) x 107 |
| Cytochrome c (with 100 µM NADH) | kcat (s−1) | 9.1 ± 0.49 | 0.46 ± 0.047 |
| Km (µM) | 17.6 ± 0.66 | 337.5 ± 44.6 | |
| kcat/Km (M−1⋅s−1) | (5.1 ± 0.4) x 105 | 1366 ± 42 | |
| DCIP (with 100 µM NADH) | kcat (s−1) | 14.1 ± 0.6 | 36.6 ± 1.2 |
| Km (µM) | 38.3 ± 2.5 | 208 ± 11 | |
| kcat/Km (M−1⋅s−1) | (3.68 ± 0.15) x 105 | (1.76 ± 0.05) x 105 | |
| NADH (with 90 µM cytochrome c) | kcat (s−1) | 20.1 ± 0.9 | |
| Km (µM) | 11.5 ± 0.7 | ||
| kcat/Km (M−1⋅s−1) | (1.74 ± 0.15) x 106 | ||
| NADPH (with 90 µM cytochrome c) | kcat (s−1) | 3.7 ± 0.3 | |
| Km (mM) | 1.21 ± 62 | ||
| kcat/Km (M−1⋅s−1) | (3.1 ± 0.1) x 103 |
PcpD Requires FMN and NADH.
The flavin cofactor in PcpD was identified by denaturation of the protein and HPLC analysis of the released flavin as described by Aliverti et al. (21). The cofactor was found to be flavin mononucleotide (FMN), which is consistent with the evolutionary relationship between PcpD and phthalate dioxygenase reductases, which also contain FMN.
We investigated the specificity of PcpD toward its nicotinamide cosubstrate using cytochrome c as a substrate. The reaction velocity in the presence of NADH showed saturation kinetics with a KM of 11.5 µM in the presence of 90 µM cytochrome c. In contrast, the KM for NADPH was 1.21 mM (Fig. S4). By comparing the values for kcat/KM, we conclude that PcpD is >550-fold more reactive with NADH than with NADPH (Table 1).
TCBQ Is Efficiently Reduced by PcpD.
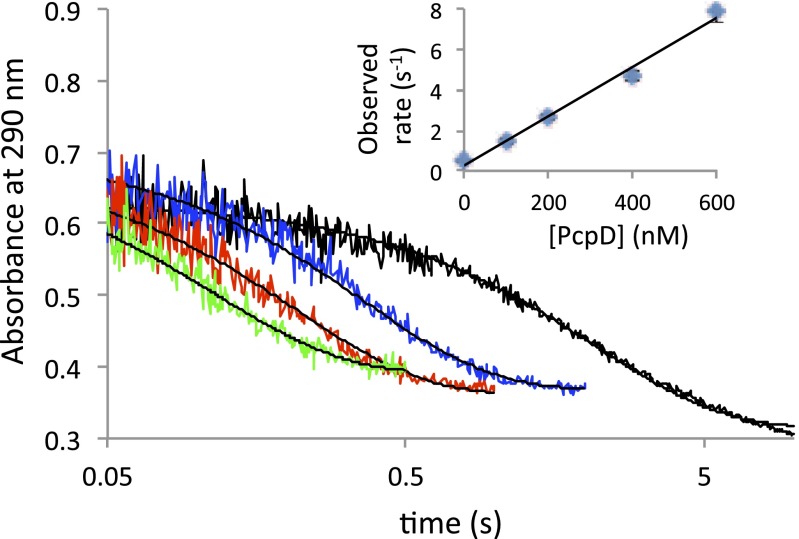
The high reactivity of TCBQ is a challenge when attempting to characterize the activity of PcpD in vitro because TCBQ is rapidly reduced nonenzymatically by NADH, the cosubstrate for PcpD, and also because TCBQ reacts rapidly with nucleophiles on enzymes, most especially cysteine (see above). It was necessary to use stopped-flow spectroscopy to determine the rate of the nonenzymatic reduction reaction. The second-order rate constant derived from the data is 2,400 M−1⋅s−1 (Fig. 4). Thus, the reduction of TCBQ to TCHQ in reactions containing 30 µM TCBQ and 200 µM NADH is essentially complete in 5 s.
Fig. 4.
Reduction of TCBQ by NADH in the presence and absence of PcpD. TCBQ was mixed with NADH and varying concentrations of PcpD in a stopped-flow spectrophotometer to yield final concentrations of 30 µM TCBQ, 200 µM NADH, and 0 nM (black), 200 nM (blue), 400 nM (red), or 600 nM (green) PcpD. Depletion of TCBQ was monitored at 290 nm and was fit to a single exponential (shown in black for each curve). The Inset uses a compilation of additional experimental conditions and replicates to demonstrate the dependence of the observed rates on enzyme concentration.
Although the nonenzymatic reduction of TCBQ to TCHQ by 200 µM NADH is quite fast, we were able to determine the rate acceleration provided by PcpD using stopped-flow spectroscopy. In the presence of 200 µM NADH and 30 µM TCBQ, the disappearance of TCBQ followed first-order kinetics at all concentrations of PcpD (Fig. 4). A plot of the observed rate vs. PcpD concentration gives a rate constant of 1.2 × 107 M−1⋅s−1 (Fig. 4, Table 1). Thus, PcpD accelerates the rate of reduction of TCBQ over the nonenzymatic rate by 5,000-fold. These results differ from those reported by Chen and Yang (16), who measured a rate acceleration of only 2.9-fold. However, these workers added FAD and NADPH to assays using enzyme that had been reconstituted from inclusion bodies. Because the flavin cofactor is actually FMN and the nicotinamide cofactor is actually NADH, the low activity of the enzyme was likely due to the presence of the wrong cofactors.
TCBQ Generated from PCP by PcpB Is Efficiently Converted to TCHQ by PcpD.
To establish that PcpD is a physiologically relevant TCBQ reductase, we assayed PcpD with TCBQ generated in situ by PcpB. We incubated PcpB, PCP, NADPH (the cosubstrate for PcpB), and NADH (the cosubstrate for PcpD) under ambient O2 to generate ∼10 µM TCBQ in 1 min (10% turnover). In the absence of PcpD, some TCHQ was formed by nonenzymatic reduction of TCBQ by the NAD(P)H cosubstrates (Table 2). Notably, the amount of PCP consumed does not equal the amount of TCHQ formed; the “missing” material (∼3 µM) can be attributed to nonspecific covalent modification of PcpB (Fig. 2) by TCBQ. In the presence of PcpD, we observed a significant increase in the yield of TCHQ, the magnitude of which is dependent on the concentration of PcpD. Further, we observed a consistent PcpD-dependent increase in PcpB activity; in the presence of 12 µM PcpD, twice as much PCP is consumed during the reaction. The formation of TCHQ by PcpD depends on the presence of NADH, the cosubstrate specific for PcpD. When only NADPH is supplied, the amount of TCHQ formed is similar to that of the control reaction lacking PcpD. Thus, it must be the activity of PcpD, not merely its presence, that is important for reduction of TCBQ to TCHQ. The effect of PcpD might be due simply to a high rate of catalysis by PcpD, which sweeps TCBQ toward TCHQ [as suggested by Belchik et al. for a similar two-enzyme system involved in degradation of TriCP 2,4,6-trichlorophenol (13)], or to a physical interaction between PcpB and PcpD that prevents release of TCBQ to the solvent.
Table 2.
Products formed from PCP by PcpB
| PcpB, µM | PcpD, µM | PCP consumed, µM | TCHQ formed, µM |
| 2.8 | 0 | 9.0 ± 1.8 | 6.0 ± 0.5 |
| 2.8 | 3 | 13 ± 2.3 | 10 ± 0.9 |
| 2.8 | 6 | 14 ± 0.9 | 17 ± 0.5 |
| 2.8 | 12 | 20 ± 4.7 | 24 ± 2.3 |
| 2.8 | 6 (no NADH) | 9.0 ± 2.3 | 5.0 ± 0.1 |
| 2.8 | 10 (PcpD–FeS)* | 17 ± 1.6 | 0 |
| 2.8 | 40 (PcpD-FeS)* | 14 ± 5.2 | 0 |
| 5.6 | 20 (+ 0.5 M potassium phosphate)* | 23 ± 5.7 | 0.4 ± 0.9 |
Reaction mixtures contained 100 µM PCP, 50 µM NADPH, and 50 µM NADH, except where indicated.
Reactions carried out in the presence of 6 mM β-ME.
TCBQ Is Protected from Solvent in the Coupled PcpB–PcpD Reaction.
Given the high reactivity of TCBQ, we wondered whether PcpB and PcpD might interact to prevent release of TCBQ into the cytoplasm where it would react with nucleophiles in small molecules and proteins. We took advantage of the high reactivity of TCBQ with thiols to determine whether TCBQ is released to solution before it is reduced by PcpD. TCBQ reacts rapidly with β-ME to form the tetra-β-mercaptan adduct 2,3,5,6-tetrakis[(2-hydroxyethyl)thio]-1,4-hydroquinone (THTH) (Fig. 5A) (3). The rate constant for the reaction of β-ME with TCBQ is >1.4 × 106 M−1⋅s−1 (Fig. S1). Thus, the rate of the initial step in formation of THTH (i.e., attack of the first molecule of β-ME) would be >8,400 s−1 in the presence of 6 mM β-ME. This is 80-fold faster than the rate of reduction of TCBQ in solution in the presence of 10 µM PcpD (120 s−1). Any TCBQ that is released from the active site of PcpB under these reaction conditions will partition toward THTH rather than TCHQ with a ratio of 80:1.
Fig. 5.
Products formed from PCP by PcpB in the presence and absence of PcpD. (A) The rates of reaction of TCBQ with β−ME and PcpD under the experimental conditions. (B) The amount of PCP consumed is shown in light gray bars, the amount of TCHQ formed is shown in dark gray bars, and the amount of THTH formed is shown in black bars. Reactions contained 2.8 µM PcpB, 100 µM PCP, 50 µM NADPH, 50 µM NADH, 6 mM β-ME in the presence or absence of 10 µM PcpD. The discrepancies between the amounts of PCP used and the amounts of TCHQ or THTH formed are due to (i) the rapid reaction of TCBQ with thiols, especially in the absence of PcpD; and (ii) the difficulty in monitoring relatively small amounts of PCP disappearance by HPLC, which leads to relatively large errors in quantitation.
In the absence of PcpD, no TCHQ was observed when 6 mM β-ME was included in the reaction mixture (Fig. 5B). Rather, the only detectable product was THTH. This result confirms the extreme rapidity of the reaction of β-ME with TCBQ, as no TCHQ was formed by nonenzymatic reduction with NADPH. The amount of THTH formed does not equal the amount of PCP consumed, presumably because of the reactivity of TCBQ with other nucleophiles such as cysteines on PcpB (Fig. 2). In contrast, the primary product in the presence of 10 µM PcpD was TCHQ. These results demonstrate unambiguously that TCBQ produced by PcpB is sequestered from the solvent in the presence of PcpD and are consistent with the hypothesis that a physical interaction between PcpB and PcpD prevents release of TCBQ until it is reduced to TCHQ. The addition of PcpD did not completely prevent THTH formation, however, suggesting that release of TCBQ to solvent competes with formation of a PcpB:PcpD complex under these conditions.
PcpD Lacking the FeS-Cluster Domain Is Active and Kinetically Competent for Reduction of TCBQ.
To address the mechanism of reduction of small molecules and protein substrates by PcpD, we constructed PcpD-FeS, which lacks the 2Fe-2S domain. The reactivity of PcpD-FeS with both TCBQ and DCIP is similar to that of PcpD itself (Table 1). These results suggest that electrons flow from NADH to the flavin and then to small molecule substrates, as has been observed for other quinone reductases (22). In contrast, PcpD-FeS is much less efficient at reducing cytochrome c than the full-length protein. The Km is increased and the kcat is decreased, leading to a >350-fold decrease in kcat/Km (Table 1), suggesting that the iron-sulfur domain participates significantly in the transfer of electrons from PcpD to cytochrome c, due either to a role in achieving the protein–protein interaction required for electron transfer, or because the 2Fe-2S cluster actually provides the electron transferred to the heme of cytochrome c. (If the latter were the case, then the source of electrons would differ for small molecule substrates and proteins, which seems unlikely.) This result is reminiscent of that seen for phthalate dioxygenase reductase, where removal of the iron-sulfur domain caused a similar change in kcat toward cytochrome c, although the Km remained unchanged (20).
Removal of the Fe-S Domain of PcpD Abrogates Its Ability to Protect TCBQ from the Solvent.
PcpD-FeS is capable of reducing TCBQ free in solution at a rate twice as fast as that of full-length PcpD. However, TCBQ formed by PcpB is not sequestered from the solvent in the presence of PcpD-FeS (Table 2). No TCHQ was formed when PcpD-FeS was present at either a 3.6-fold or a 14.4-fold excess over the concentration of PcpB. Because the FeS-domain is not required for reduction of TCBQ, we hypothesize that it is required for a protein–protein interaction between PcpB and PcpD.
To investigate the possibility of a protein–protein interaction between PcpB and PcpD, we attempted to disrupt the interaction by adding detergent or high concentrations of salt, which can disrupt hydrophobic and electrostatic interactions, respectively. Addition of Triton X-100 (2% final concentration) or PEG 2000 (0.2% final concentration) did not affect the protection of TCBQ in the coupled assay in the presence of β-ME. In contrast, addition of 500 mM potassium phosphate resulted in loss of the protection of TCBQ (Table 2). [In these experiments the concentrations of both PcpB and PcpD were doubled to compensate for the modest decreases in activity seen for each of these enzymes in the presence of high salt concentrations (Fig. S5).] Taken together with the observation that the FeS-domain of PcpD is required for sequestration of TCBQ, these results are consistent with the hypothesis that an electrostatic interaction between PcpB and the FeS-domain of PcpD enables reduction of TCBQ before it is released to the solvent.
Attempts to Demonstrate a Physical Interaction Between PcpB and PcpD.
We were unable to demonstrate a stable complex between PcpB and PcpD by native gel electrophoresis, gel filtration, dynamic light scattering, or surface plasmon resonance. We also failed to detect cross-linking between PcpB and PcpD during substrate turnover using 1-ethyl-3-[3-dimethylaminopropyl]carbodiimide, dimethyl-suberimidate, glutaraldehyde, and di-succinimidyl-glutarate, as well as the photo-cross-linkers Tris(2-2′-bipyridyl)dichlororuthenium(II) and 2-azido-5-amino benzoic acid N-hydroxysuccinimide ester. However, the monomers of PcpB were cross-linked to dimers and the monomers of PcpD to dimers and trimers, under all of these conditions.
Discussion
Although previous studies have addressed the consequences of treating cells (5) or nucleosides (7) with TCBQ, the rates of physiologically relevant reactions between TCBQ and nucleophiles have not been examined. TCBQ reacts with thiols with rate constants >1.4 × 106–1.3 × 107 M−1⋅s−1 at pH 7. The apparent decrease in reactivity going from cysteine to glutathione to β-ME correlates with their pKas (8.2, 9.2, and 9.6, respectively) (Fig. S1) and is consistent with the reactive moiety being the thiolate. Given these pKa values, 6%, 0.6%, and 0.3% of the thiols will be deprotonated at pH 7 for cysteine, glutathione, and β-ME, respectively. Thus, the rate constant for reaction between TCBQ and thiolates is >108 M−1⋅s−1 , essentially the diffusion-controlled limit. It is important to note that each molecule of TCBQ can react with multiple thiols, leading to depletion of up to four molecules of glutathione per TCBQ (23). Further, reaction of TCBQ with multiple thiols can lead to cross-linking of proteins (Fig. 2B).
The rate constants for reaction of TCBQ with thiols are more than three orders of magnitude higher than those for reactions with hydroxyl and amino groups of amino acids. A similar reactivity profile has been observed for the benzoquinones of polychlorinated biphenyls (24). Reactions with nucleotides are even slower, consistent with the prolonged reaction times used to generate adducts from TCBQ and nucleosides or DNA (reactions were carried out for 2 h at 65 °C and for 72 h at 37 °C, respectively (7)) and also with a report that quinone adducts are formed at a frequency of only 8 per 107 nucleotides in rats that were chronically exposed to 1,000 ppm (3.8 mM) PCP for 27 wk (25). In agreement with these previous qualitative reports, our kinetic data suggest that, in cells, TCBQ will primarily react with glutathione and cysteine (both free and in proteins), and at a slower rate with hydroxyl and amino groups.
We have shown that S. chlorophenolicum lacking pcpD can grow in the presence of high concentrations of TCP but not in the presence of high concentrations of PCP [300 µM in liquid cultures (2) and 100 μM on agar plates; Fig. 3A]. Similar results are seen with TriCP, which also generates a benzoquinone after hydroxylation by PcpB. The results described here provide a molecular explanation for this observation. Our data confirm that PcpD is a TCBQ reductase, and further suggest that its most important role in PCP degradation is to enable reduction of TCBQ before it is released to the cytoplasm.
Sequestration of highly reactive intermediates is important both because escape of such intermediates to the solvent decreases flux through a pathway and because it can result in formation of useless by-products or damage to cellular molecules. Passage of an intermediate directly from one active site to another without equilibration with the solvent is called channeling (26–30). Examples of reactive intermediates that are channeled from one active site to another include carbamate (31), ammonia (32), indole (33), aspartyl phosphate (34), glutamyl phosphate (35), and phosphoribosylamine (26). TCBQ is even more reactive than these intermediates, undergoing reaction with thiols at physiologically relevant concentrations in milliseconds. The protection of TCBQ from thiols in the presence of PcpD suggests that an interaction between PcpB and PcpD prevents release of TCBQ to the solvent before it is reduced to TCHQ.
We were not able to demonstrate a physical interaction between PcpB and PcpD using various physical methods. However, we have presented strong kinetic evidence for such an interaction. We have shown that TCBQ is trapped by thiols in the solvent in the absence of PcpD, but is protected in the presence of PcpD. Trapping of a reaction intermediate that has been released to the solvent by a very fast reaction is one of the most rigorous tests for channeling. This approach has been used to measure the efficiency of channeling between the active sites of BphI and BphJ (36), dihydroxyacetone kinase and aldolase in a fused bifunctional enzyme (37), and orthologous and chimeric aldolase–dehydrogenase complexes (38). In the latter case, channeling occurs in orthologous complexes, but not in chimeric complexes, even though stable complexes are formed in both cases. Similarly, channeling occurs between the active sites of ATP sulfurylase and 3′adenosine-5′-phosphosulfate in a type III sulfate activating complex, but not in a type I complex, even though the enzymes are in the same polypeptide chain in both cases (39). Thus, the existence of a stable association between proteins does not ensure that channeling will occur; the kinetic evidence is considered definitive.
Kinetic evidence that demonstrates protection of a reactive intermediate from the solvent is taken as prima facie evidence that a protein–protein interaction takes place. The PcpB:PcpD system is not the first case in which kinetic evidence for a protein–protein interaction is compelling, but a physical interaction cannot be detected. Other examples include aspartyl kinase:aspartate semialdehyde dehydrogenase (34), glutamyl kinase:glutamate semialdehyde dehydrogenase (35), and glutamine phosphoribosylpyrophosphate amidotransferase:glycinamide ribonucleotide synthetase (26). The hypothesis that a protein–protein interaction between PcpD and PcpB is responsible for the sequestration of TCBQ is supported by experiments showing that TCBQ is released to the solvent when PcpD-FeS is present, even though this truncated enzyme is kinetically competent for reduction of TCBQ. Finally, addition of high salt concentrations but not detergent abrogates sequestration of TCBQ. PcpD is active under these conditions so these results cannot be attributed to lack of TCBQ reductase activity. It is difficult to imagine a mechanism for these effects that does not involve a protein–protein interaction.
The combination of kinetic evidence for a protein–protein interaction in the absence of confirmation of such an interaction by physical methods suggests the possibility that the interaction between PcpD and PcpB is transient. Such interactions are often not amenable to detection by methods such as native gel electrophoresis, gel filtration, chemical cross-linking, or surface plasmon resonance (26, 40, 41) although their importance is well recognized (42). A transient interaction between PcpB and PcpD might be expected given that PcpB is expected to undergo a number of conformational changes during its catalytic cycle. The catalytic cycle of related enzymes such as p-hydroxybenzoate hydroxylase (43) begins with a conformational change that opens a channel to the active site, allowing substrate to bind. The flavin then moves to the “out” conformation, where it can be reduced by NADPH. The reduced flavin reacts with O2 to form C4a-hydroperoxyflavin, which moves to the “in” conformation and transfers a hydroxyl group to the substrate. Finally, another conformational change is required to release the products. It is unlikely that PcpD could bind stably near the active site without interfering with some of these conformational changes. We suspect that PcpD binds to the enzyme after TCBQ is formed. At that point, TCBQ might slide through a channel to the active site of PcpD. Alternatively, by analogy with the ancestral function of donation of electrons to FeS clusters in proteins and its current ability to reduce cytochrome c, PcpD might simply transfer electrons to TCBQ still bound at the active site of PcpB. In this case, reduced PcpD could be considered a substrate for PcpB, and interaction between the two proteins would certainly be required for electron transfer to occur. Distinguishing between substrate channeling and electron transfer is not an easy problem given that both proteins contain flavins with overlapping spectroscopic properties. In either case, a physical interaction between the two proteins is required.
The profound reactivity and toxicity of TCBQ suggest that the PCP degradation pathway could only have emerged in a bacterium in which a reductase capable of reducing TCBQ was already present, and furthermore, capable of an interaction with the hydroxylase that produces TCBQ. Taken together with the tight association between the genes encoding PcpB and PcpD in the genome, we infer that this two-enzyme system originated as a module in a bacterium that can degrade a phenol with a leaving group such as chloride, bromide, or nitrite located para to the hydroxyl group. A large number of halogenated phenols are found in nature (44, 45), including 2,6-dichlorophenol (46) and 2,4-dichlorophenol (47). The marine acorn worm produces 10 different chlorinated and brominated phenols (48). Naturally occurring nitrophenols are less common, but include o-nitrophenol (46) and a variety of more complex nitrophenols with additional substituents synthesized by some bacteria and fungi (49). Degradation pathways for only a handful of substituted phenols that are degraded via benzoquinone intermediates have been reported. Examples include the previously mentioned pathways for degradation of TriCP, p-nitrophenol, and o-nitrophenol in TriCP C. necator JMP134 (13), Pseudomonas sp. Strain WBC-3 (14), and Alcaligenes sp. strain NyZ215 (15), respectively. However, despite the similarities in reaction sequences and genetic organization, the reductase genes in these microbes are not necessarily homologous to each other or to pcpD (12). The reductases in the degradation of TriCP (TcpB) and p-nitrophenol (PnpB) belong to the nitroreductase-like family 5 and NADPH-dependent FMN reductase superfamilies, respectively. PcpD and the reductase involved in o-nitrophenol degradation (OnpB) belong to the FNR-like superfamily, but they share only 39% identity. Thus, it is not possible at this point to identify a pair of genes that is homologous to pcpB and pcpD and has high enough sequence similarity to indicate the ancestral functions of PcpB and PcpD.
Degradation of PCP by S. chlorophenolicum is slow, and the bacterium cannot tolerate high concentrations of PCP (50). We previously reported that PCP hydroxylase is an unusually inefficient enzyme, with a kcat of only 0.02 s−1 (3). Given that the transient interaction between PcpB and PcpD that results in reduction of TCBQ is a second-order process, its rate will be determined by the rate of diffusion in the cytoplasm as well as the local concentration of the two enzymes. If release of TCBQ from the active site of PcpB is faster than the rate of encounter of PcpB and PcpD, the fitness of the cell will be compromised by the damage caused by TCBQ. Thus, there may be a strictly enforced upper limit on the efficiency of PCP hydroxylase that limits the rate of degradation. If a more stable association between PcpB and PcpD were to evolve, this limit might increase, consequently increasing flux through the entire pathway.
Formation of highly reactive and/or toxic intermediates in metabolic pathways is uncommon in nature, but it does occur. It occurs more frequently when synthetic biologists engineer novel pathways for production of pharmaceuticals, biofuels, or commodity chemicals. The most elegant solution to handling such intermediates in nature is the construction of tunnels linking two active sites so that reactive or toxic intermediates are never released to the solvent (51). In the absence of a tunnel, a transient physical interaction that allows channeling of a product from the active site at which it is formed to the active site at which it is consumed is also an effective mechanism for sequestering problematic intermediates (52). Because these mechanisms are difficult to generate by protein engineering, a practical approach used by synthetic biologists is to overproduce the enzyme immediately downstream of the enzyme that produces the toxic intermediate. The snapshot we have obtained of PCP degradation at this stage in the evolution of the pathway suggests an additional strategy for controlling damage by a highly reactive intermediate—handicapping of the upstream enzyme to simply slow production of the intermediate so that it can be efficiently converted to a stable product in a transient complex with the downstream enzyme without being released to the solvent. Unfortunately, this strategy minimizes toxicity at the expense of flux through the pathway. Our data suggest that the best strategy for improving flux through the pathway would be to genetically engineer the bacterium so that higher levels of PcpD are produced, while improving the specificity of PcpD to prevent promiscuous reduction of other targets.
Methods
Deletion of pcpD in S. chlorophenolicum L1.
In our earlier work with PcpD, we generated a strain of S. chlorophenolicum strain ATCC 39723 in which pcpD had been disrupted by insertion of a kanamycin resistance cassette (2). Because the genome sequence of S. chlorophenolicum L1 is now available (12), we generated a strain completely devoid of pcpD by replacing pcpD with a kanamycin resistance cassette. Details are provided in SI Methods.
Assays of the Toxicity of Chlorophenols During Growth on Agar Plates.
Wild-type and ΔpcpD S. chlorophenolicum L1 were grown on 1/4 TSB agar plates supplemented with 50 μM PCP (wild-type) or 7.5 μg/mL kanamycin (ΔpcpD). A single colony was used to inoculate 4 mL of S. chlorophenolicum medium [per liter: 0.65 g of K2HPO4, 0.19 g of KH2PO4, 0.1 g of MgSO4 7H2O, 0.5 g of NaNO3, 4.0 g of sodium glutamate, 2 mL of 0.01 M FeSO4, 0.01 g of CaCl2, and trace elements (53)]. Cultures were grown to stationary phase and subcultured into 4 mL of the same medium until mid log phase. The cells were then washed three times with 5 mM potassium phosphate, pH 7.4, containing 5 mM NaNO3 and diluted to an OD600 of 0.05. Aliquots of these cells and further 10-fold dilutions were spotted on ATCC 1687 agar plates (the medium described above containing 1.5% agar) supplemented with varying concentrations of chlorophenols. The plates were incubated at room temperature for 5–7 d in the dark.
PcpD Expression and Purification.
The pcpD gene from Sphingomonas sp. RA2 was amplified by PCR and cloned into a pET28 vector (Novagen) downstream of and in-frame with a His10-tagged Smt3 protein [small ubiquitin-related modifier (SUMO)] from Saccharomyces cerevisiae (54). The sequence of pcpD was verified by DNA sequencing. The plasmid pET28-His10-Smt3-pcpDRA2 was introduced into E. coli BL21(DE3) cells, and the protein was expressed according to Jaganaman et al. (55).
The cell pellet from 0.5 L of induced cells was resuspended in 60 mL of buffer B (50 mM sodium phosphate, pH 7.6, containing 500 mM NaCl and 0.2 mM DTT) supplemented with 20 mM imidazole, 1 mM phenylmethanesulfonylfluoride, and protease inhibitor mixture (Sigma P8849; 1:500 dilution). The cells were lysed by two passages through a French Press. Nucleic acids were precipitated with 0.3% protamine sulfate, and the clarified lysate was loaded on an Ni2+-column (5 mL of HisTrap HP from GE Healthcare), and eluted using a linear gradient of 100–500 mM imidazole in buffer B. Fractions containing PcpD were pooled and treated with Ulp1 (SUMO protease 1) to remove the Smt3 tag using a protocol described in Malakhov et al. (54). Tag-free PcpD was concentrated and applied to a Superdex 200 16/60 gel filtration column (GE Healthcare) and eluted with buffer B. Fractions containing PcpD as determined by absorption spectrum and SDS/PAGE were pooled, concentrated, flash frozen, and stored at −80 °C. Additional details are provided in SI Methods.
Preparation of PcpD Lacking the 2Fe-2S Domain.
The gene segment encoding the N terminus of PcpD from Sphingomonas sp. RA2, which contains the FMN and NADH binding domains, was amplified by PCR so as to truncate the gene after the codon for Ala229. [This location was chosen based on the multiple sequence alignment shown in Fig. S2 and previous work that identified the domain boundary in phthalate dioxygenase reductase (56).] The digested PCR product was cloned into a modified pET20b vector (Novagen) in which pelB, which encodes an N-terminal signal sequence, had been replaced with a His6-tag. The plasmid pET20His6-[pcpD-FeS] was introduced into E. coli BL21(DE3) cells. Growth of cells and induction of PcpD-FeS was performed as for PcpD, omitting the supplementation with cysteine, iron, and sulfur. PcpD-FeS was purified by nickel affinity chromatography and gel filtration as described for PcpD in SI Methods.
Purification of PcpB.
PcpB from S. chlorophenolicum l-1 was purified as previously reported (3). Protein concentrations were determined by quantifying enzyme-bound flavin, as described therein.
Nonenzymatic Reactions of TCBQ.
TCBQ was dissolved in N′N′-dimethyl formamide and subsequently diluted to 10–60 µM in 50 mM potassium phosphate, pH 7.0 (ε290 = 15,100 M−1⋅cm−1). Glutathione, amino acids, ATP, GTP, NADH, or β-ME were diluted from stock solutions (1–140 mM) into the same buffer to yield final concentrations of 0.2–6 mM after mixing with TCBQ in a Hi-Tech SF-61DX stopped-flow instrument. The loss of signal at 290 nm was used to monitor loss of TCBQ.
PcpD Activity Measurements.
Enzymatic activity was measured at room temperature in 50 mM sodium phosphate, pH 7.6, containing 150 mM NaCl (PBS), or 50 mM potassium phosphate, pH 7.0, and 100 µM NADH, unless otherwise described. For experiments in which the enzyme was used at low concentrations, it was diluted to the appropriate concentration in PBS supplemented with 0.5 mg/mL BSA (NEB). Reactions were followed using a Varioskan plate reader (Thermo). The activity of PcpD with 2,6-dichloroindophenol (DCIP) was measured by following the disappearance of absorbance at 600 nm (ε600 = 15,400 M−1⋅cm−1) using 10 nM PcpD. The cytochrome c reductase activity of PcpD was measured by following the appearance of reduced cytochrome c at 550 nm (ε550 = 21,000 M−1⋅cm−1) using 10 nM PcpD or 500 nM PcpD-FeS. The cofactor dependence of PcpD was determined using 90 µM cytochrome c and 5 and 100 nM PcpD, while varying NADH (2–125 µM) or NADPH (18–2000 µM), respectively. Activity of PcpD with TCBQ was monitored by stopped-flow spectroscopy by mixing 30–60 µM TCBQ with variable concentrations of PcpD (10 nM–600 nM) in the presence of 200 µM NADH. The loss of signal at 290 nm was used to monitor depletion of TCBQ.
Identification of Products Formed from PCP by PcpB.
The depletion of PCP and accumulation of TCHQ and/or TCBQ catalyzed by PcpB in the presence or absence of PcpD was monitored by HPLC. Reaction mixtures in 200 µL contained 50 mM potassium phosphate, pH 7.0, 100 µM PCP, 50 µM NADPH, 50 µM NADH, 1–3 µM PcpB, and 1–40 µM PcpD. In some reactions, β-ME (6 mM) was included in the reaction mixture to trap TCBQ as 2,3,5,6-tetrakis[(2-hydroxyethyl)thio]-1,4-hydroquinone (THTH) (3). After 1 min, reactions were quenched by addition of 250 µL of ethyl acetate containing 50 µM p-nitrophenol (internal standard), followed by 15 s of vortexing. The quenched reactions were analyzed by HPLC as described previously (3). The p-nitrophenol internal standard was used to normalize each sample for work-up and or sample-loading discrepancies. All assays were performed in quadruplicate.
Gel Filtration Chromatography.
PcpB (10 µM) and PcpD (20 µM) in 50 mM sodium phosphate, pH 7.6, or the same buffer supplemented with 150 mM NaCl, were coinjected on a Superdex 200 10/30 column (GE Healthcare). Eluted fractions were analyzed by SDS/PAGE.
Chemical Cross-Linking.
Cross-linking reactions contained PcpB (12 µM) and PcpD (12–48 µM) in PBS or PBS supplemented with 10% glycerol. To initiate the cross-linking reaction, proteins were incubated with 5 mM 1-ethyl-3-[3-dimethylaminopropyl]carbodiimide and 20 mM N-hydroxysulfosuccinimide (Sigma), 0.1% glutaraldehyde (Sigma), or 1 mM di-succinimidyl-glutarate (Sigma) in the presence or absence of 100 mM PCP and/or 100 µM NAD(P)H for 1 h at room temperature. Products were analyzed using 4–20% gradient SDS/PAGE.
Photocrosslinking reactions were carried out with N-5-azido-2-nitrobenzoyloxysuccinimide (ANB-NOS) (Sigma) and Tris(2,2’-bipyridyl)-ruthenium(II) chloride hexahydrate Ru(bpy). ANB-NOS (1 mM) was incubated with PcpB (45 µM) or PcpD (45 µM) in 50 mM phosphate, pH 7, for 1 h in the dark to derivatize primary amines. The reaction was quenched by addition of glycine to a final concentration of 2 mM. The derivatized proteins (15 µM final concentration) were mixed with the nonderivatized partner (derivatized PcpB with nonderivatized PcpD and vice versa) in the absence or presence of 100 µM PCP, NADPH, and NADH, and the cross-linking reaction was initiated by exposure to a 6W UV lamp at 365 nm for 10 min. Reaction mixtures were analyzed by 12% (wt/vol) SDS/PAGE. Products were visualized by Western blots with antibodies against the His-tagged PcpB.
Ru(bpy) (0.125 mM) and ammonium persulfate (1 mM) were incubated in the dark with PcpB (15 µM) and PcpD (30 µM) in 50 mM phosphate, pH 7, in the absence or presence of 100 μM PCP, NADPH, and NADH. The reaction was initiated by exposure to light from a mini Maglite from which the plastic filter was removed for 30 s. Reaction mixtures were analyzed by 12% SDS/PAGE. Products were visualized by Western blots with antibodies against the His-tagged PcpB.
Detection of Adducts Between TCBQ and PcpB.
PcpB (20 µM) was incubated with U-14C-PCP (100 µM) only and with U-14C-PCP (100 µM), NADPH (50 µM), and NADH (50 µM) in the absence or presence of PcpD (20 µM) for 10 min at room temperature in 50 mM potassium phosphate, pH 7. The reaction mixtures were loaded onto a 9 mL G-25 Sephadex column equilibrated in 50 mM Tris⋅HCl, pH 8.0, and the fractions containing protein were collected and subjected to scintillation counting.
Cross-Linking of PcpB by TCBQ.
PcpB (10 µM) was incubated with PCP (100 µM) in the absence or presence of NADPH and NADH and in the presence of NADPH, NADH, and PcpD (10 µM) for 10 min at room temperature in 50 mM potassium phosphate, pH 7. Reaction mixtures were analyzed by reducing 12% SDS/PAGE in 50 mM Tris⋅HCl, pH 8.8.
Detection of Metabolites of PCP in the Growth Medium.
Wild-type or ΔpcpD S. chlorophenolicum L1 were grown to midlog phase as described above. Cells (25–100 mL) were harvested by centrifugation at 3,000 × g and room temperature for 20 min and then resuspended at an OD600 of 100 in fresh medium containing 100–200 µM PCP or U-14C-PCP (American Radiolabeled Chemicals; 1.6 mCi/mmol). After 90 min at 30 °C, cells were harvested by centrifugation at 12,000 × g and 4 °C for 3 min. The supernatant was extracted with ethyl acetate to remove the small amount of residual PCP. The aqueous layer was acidified by addition of TFA to 0.05% and analyzed by HPLC as described below. Cell pellets were lysed by incubation in 100 µL of BugBuster (Novagen) for 30 min at room temperature, and insoluble material was removed by centrifugation at 12,000 × g for 10 min at 4 °C. Following acidification of the cell lysate by addition of TFA to 0.05%, the samples were analyzed by HPLC on a Zorbax SB-C18 (4.6 mm × 150 mm) HPLC column (Agilent Technologies) using a 0–60% gradient of acetonitrile containing 0.1% aqueous TFA over 20 min at a flow rate of 1 mL/min. Control experiments using TCBQ incubated with glutathione demonstrated the elution of various TCBQ-GSH adducts over the range of 7–12 min under these conditions. Molecular weights of degradation products were determined using negative-mode electrospray ionization mass spectrometry on a Synapt G2 high-definition instrument (Waters).
Supplementary Material
Acknowledgments
This work was funded by National Institutes of Health Grant GM078554 (to S.D.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences for the previously unsequenced PcpD proteins reported in this paper have been deposited in the GenBank database [accession nos. JX514946 (Sphingobium chlorophenolicum sp. RA2), JX514945 (Sphingobium chlorophenolicum ATCC 33790), and JX514944 (Novosphingobium lentum)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1214052110/-/DCSupplemental.
References
- 1.Copley SD. Evolution of efficient pathways for degradation of anthropogenic chemicals. Nat Chem Biol. 2009;5(8):559–566. doi: 10.1038/nchembio.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dai M, Rogers JB, Warner JR, Copley SD. A previously unrecognized step in pentachlorophenol degradation in Sphingobium chlorophenolicum is catalyzed by tetrachlorobenzoquinone reductase (PcpD) J Bacteriol. 2003;185(1):302–310. doi: 10.1128/JB.185.1.302-310.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hlouchova K, Rudolph J, Pietari JM, Behlen LS, Copley SD. Pentachlorophenol hydroxylase, a poorly functioning enzyme required for degradation of pentachlorophenol by Sphingobium chlorophenolicum. Biochemistry. 2012;51(18):3848–3860. doi: 10.1021/bi300261p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCarthy DL, Claude AA, Copley SD. In vivo levels of chlorinated hydroquinones in a pentachlorophenol-degrading bacterium. Appl Environ Microbiol. 1997;63(5):1883–1888. doi: 10.1128/aem.63.5.1883-1888.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siraki AG, Chan TS, O’Brien PJ. Application of quantitative structure-toxicity relationships for the comparison of the cytotoxicity of 14 p-benzoquinone congeners in primary cultured rat hepatocytes versus PC12 cells. Toxicol Sci. 2004;81(1):148–159. doi: 10.1093/toxsci/kfh182. [DOI] [PubMed] [Google Scholar]
- 6.van Ommen B, Adang A, Müller F, van Bladeren PJ. The microsomal metabolism of pentachlorophenol and its covalent binding to protein and DNA. Chem Biol Interact. 1986;60(1):1–11. doi: 10.1016/0009-2797(86)90013-x. [DOI] [PubMed] [Google Scholar]
- 7.Vaidyanathan VG, Villalta PW, Sturla SJ. Nucleobase-dependent reactivity of a quinone metabolite of pentachlorophenol. Chem Res Toxicol. 2007;20(6):913–919. doi: 10.1021/tx600359d. [DOI] [PubMed] [Google Scholar]
- 8.Zhu BZ, Kalyanaraman B, Jiang GB. Molecular mechanism for metal-independent production of hydroxyl radicals by hydrogen peroxide and halogenated quinones. Proc Natl Acad Sci USA. 2007;104(45):17575–17578. doi: 10.1073/pnas.0704030104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Copley SD. Evolution of a metabolic pathway for degradation of a toxic xenobiotic: The patchwork approach. Trends Biochem Sci. 2000;25(6):261–265. doi: 10.1016/s0968-0004(00)01562-0. [DOI] [PubMed] [Google Scholar]
- 10.Cline RE, Hill RH, Jr, Phillips DL, Needham LL. Pentachlorophenol measurements in body fluids of people in log homes and workplaces. Arch Environ Contam Toxicol. 1989;18(4):475–481. doi: 10.1007/BF01055012. [DOI] [PubMed] [Google Scholar]
- 11.Warner JR, Copley SD. Mechanism of the severe inhibition of tetrachlorohydroquinone dehalogenase by its aromatic substrates. Biochemistry. 2007;46(14):4438–4447. doi: 10.1021/bi0620104. [DOI] [PubMed] [Google Scholar]
- 12. Copley SD, et al. (2012) The whole genome sequence of Sphingobium chlorophenolicum L-1: insights into the evolution of the pentachlorophenol degradation pathway. Genome Biol Evol 4(2):184–198. [DOI] [PMC free article] [PubMed]
- 13.Belchik SM, Xun L. Functions of flavin reductase and quinone reductase in 2,4,6-trichlorophenol degradation by Cupriavidus necator JMP134. J Bacteriol. 2008;190(5):1615–1619. doi: 10.1128/JB.01697-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang JJ, Liu H, Xiao Y, Zhang XE, Zhou NY. Identification and characterization of catabolic para-nitrophenol 4-monooxygenase and para-benzoquinone reductase from Pseudomonas sp. strain WBC-3. J Bacteriol. 2009;191(8):2703–2710. doi: 10.1128/JB.01566-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiao Y, Zhang JJ, Liu H, Zhou NY. Molecular characterization of a novel ortho-nitrophenol catabolic gene cluster in Alcaligenes sp. strain NyZ215. J Bacteriol. 2007;189(18):6587–6593. doi: 10.1128/JB.00654-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen L, Yang J. Biochemical characterization of the tetrachlorobenzoquinone reductase involved in the biodegradation of pentachlorophenol. Int J Mol Sci. 2008;9(3):198–212. doi: 10.3390/ijms9030198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Correll CC, Batie CJ, Ballou DP, Ludwig ML. Phthalate dioxygenase reductase: A modular structure for electron transfer from pyridine nucleotides to [2Fe-2S] Science. 1992;258(5088):1604–1610. doi: 10.1126/science.1280857. [DOI] [PubMed] [Google Scholar]
- 18.Radehaus PM, Schmidt SK. Characterization of a novel Pseudomonas sp. that mineralizes high concentrations of pentachlorophenol. Appl Environ Microbiol. 1992;58(9):2879–2885. doi: 10.1128/aem.58.9.2879-2885.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamaguchi M, Fujisawa H. Reconstitution of iron-sulfur cluster of NADH-cytochrome c reductase, a component of benzoate 1,2-dioxygenase system from Pseudomonas arvilla C-1. J Biol Chem. 1981;256(13):6783–6787. [PubMed] [Google Scholar]
- 20.Gassner GT, Ludwig ML, Gatti DL, Correll CC, Ballou DP. Structure and mechanism of the iron-sulfur flavoprotein phthalate dioxygenase reductase. FASEB J. 1995;9(14):1411–1418. doi: 10.1096/fasebj.9.14.7589982. [DOI] [PubMed] [Google Scholar]
- 21.Aliverti A, Curti B, Vanoni MA. Identifying and quantitating FAD and FMN in simple and in iron-sulfur-containing flavoproteins. Methods Mol Biol. 1999;131:9–23. doi: 10.1385/1-59259-266-X:9. [DOI] [PubMed] [Google Scholar]
- 22.Colucci MA, Moody CJ, Couch GD. Natural and synthetic quinones and their reduction by the quinone reductase enzyme NQO1: From synthetic organic chemistry to compounds with anticancer potential. Org Biomol Chem. 2008;6(4):637–656. doi: 10.1039/b715270a. [DOI] [PubMed] [Google Scholar]
- 23.van Ommen B, et al. Active site-directed irreversible inhibition of glutathione S-transferases by the glutathione conjugate of tetrachloro-1,4-benzoquinone. J Biol Chem. 1988;263(26):12939–12942. [PubMed] [Google Scholar]
- 24.Amaro AR, Oakley GG, Bauer U, Spielmann HP, Robertson LW. Metabolic activation of PCBs to quinones: Reactivity toward nitrogen and sulfur nucleophiles and influence of superoxide dismutase. Chem Res Toxicol. 1996;9(3):623–629. doi: 10.1021/tx950117e. [DOI] [PubMed] [Google Scholar]
- 25.Lin PH, La DK, Upton PB, Swenberg JA. Analysis of DNA adducts in rats exposed to pentachlorophenol. Carcinogenesis. 2002;23(2):365–369. doi: 10.1093/carcin/23.2.365. [DOI] [PubMed] [Google Scholar]
- 26.Rudolph J, Stubbe J. Investigation of the mechanism of phosphoribosylamine transfer from glutamine phosphoribosylpyrophosphate amidotransferase to glycinamide ribonucleotide synthetase. Biochemistry. 1995;34(7):2241–2250. doi: 10.1021/bi00007a019. [DOI] [PubMed] [Google Scholar]
- 27.Huang X, Holden HM, Raushel FM. Channeling of substrates and intermediates in enzyme-catalyzed reactions. Annu Rev Biochem. 2001;70:149–180. doi: 10.1146/annurev.biochem.70.1.149. [DOI] [PubMed] [Google Scholar]
- 28.Spivey HO, Ovadi J. Substrate channeling. Methods. 1999;19(2):306–321. doi: 10.1006/meth.1999.0858. [DOI] [PubMed] [Google Scholar]
- 29.Geck MK, Kirsch JF. A novel, definitive test for substrate channeling illustrated with the aspartate aminotransferase/malate dehydrogenase system. Biochemistry. 1999;38(25):8032–8037. doi: 10.1021/bi983029c. [DOI] [PubMed] [Google Scholar]
- 30.Rakus D, Pasek M, Krotkiewski H, Dzugaj A. Interaction between muscle aldolase and muscle fructose 1,6-bisphosphatase results in the substrate channeling. Biochemistry. 2004;43(47):14948–14957. doi: 10.1021/bi048886x. [DOI] [PubMed] [Google Scholar]
- 31.Thoden JB, Holden HM, Wesenberg G, Raushel FM, Rayment I. Structure of carbamoyl phosphate synthetase: A journey of 96 Å from substrate to product. Biochemistry. 1997;36(21):6305–6316. doi: 10.1021/bi970503q. [DOI] [PubMed] [Google Scholar]
- 32.Fan Y, Lund L, Shao Q, Gao YQ, Raushel FM. A combined theoretical and experimental study of the ammonia tunnel in carbamoyl phosphate synthetase. J Am Chem Soc. 2009;131(29):10211–10219. doi: 10.1021/ja902557r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hyde CC, Ahmed SA, Padlan EA, Miles EW, Davies DR. Three-dimensional structure of the tryptophan synthase alpha 2 beta 2 multienzyme complex from Salmonella typhimurium. J Biol Chem. 1988;263(33):17857–17871. [PubMed] [Google Scholar]
- 34.James CL, Viola RE. Production and characterization of bifunctional enzymes. Substrate channeling in the aspartate pathway. Biochemistry. 2002;41(11):3726–3731. doi: 10.1021/bi0159074. [DOI] [PubMed] [Google Scholar]
- 35.Arentson BW, Sanyal N, Becker DF. Substrate channeling in proline metabolism. Front Biosci. 2012;17(1):375–388. doi: 10.2741/3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carere J, Baker P, Seah SY. Investigating the molecular determinants for substrate channeling in BphI-BphJ, an aldolase-dehydrogenase complex from the polychlorinated biphenyls degradation pathway. Biochemistry. 2011;50(39):8407–8416. doi: 10.1021/bi200960j. [DOI] [PubMed] [Google Scholar]
- 37.Iturrate L, Sanchez-Moreno I, Doyaguez EG, Garcia-Junceda E. Substrate channelling in an engineered bifunctional aldolase/kinase enzyme confers catalytic advantage for C-C bond formation. Chem Commun (Camb) 2009;(13):1721–1723. doi: 10.1039/b822345a. [DOI] [PubMed] [Google Scholar]
- 38.Baker P, Hillis C, Carere J, Seah SY. Protein-protein interactions and substrate channeling in orthologous and chimeric aldolase-dehydrogenase complexes. Biochemistry. 2012;51(9):1942–1952. doi: 10.1021/bi201832a. [DOI] [PubMed] [Google Scholar]
- 39.Sun M, Leyh TS. Channeling in sulfate activating complexes. Biochemistry. 2006;45(38):11304–11311. doi: 10.1021/bi060421e. [DOI] [PubMed] [Google Scholar]
- 40.Roux B, Walsh CT. p-aminobenzoate synthesis in Escherichia coli: Kinetic and mechanistic characterization of the amidotransferase PabA. Biochemistry. 1992;31(30):6904–6910. doi: 10.1021/bi00145a006. [DOI] [PubMed] [Google Scholar]
- 41.Mathews CK, Sjoberg BM, Reichard P. Ribonucleotide reductase of Escherichia coli. Cross-linking agents as probes of quaternary and quinary structure. Eur J Biochem. 1987;166(2):279–285. doi: 10.1111/j.1432-1033.1987.tb13512.x. [DOI] [PubMed] [Google Scholar]
- 42.Perkins JR, Diboun , Dessailly BH, Lees JG, Orengo C. Transient protein-protein interactions: Structural, functional, and network properties. Structure. 2010;18(10):1233–1243. doi: 10.1016/j.str.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 43.Entsch B, Cole LJ, Ballou DP. Protein dynamics and electrostatics in the function of p-hydroxybenzoate hydroxylase. Arch Biochem Biophys. 2005;433(1):297–311. doi: 10.1016/j.abb.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 44.Siuda JF, DeBernardis JF. Naturally occurring halogenated organic compounds. Lloydia. 1973;36(2):107–143. [PubMed] [Google Scholar]
- 45.Gribble GW. The natural production of chlorinated compounds. Environ Sci Technol. 1994;28(7):311A–319A. doi: 10.1021/es00056a712. [DOI] [PubMed] [Google Scholar]
- 46.Sonenshine DE. Tick pheromones and their use in tick control. Annu Rev Entomol. 2006;51:557–580. doi: 10.1146/annurev.ento.51.110104.151150. [DOI] [PubMed] [Google Scholar]
- 47.Ando K, Kato A, Suzuki S. Isolation of 2.4-dichlorophenol from a soil fungus and its biological significance. Biochem Biophys Res Commun. 1970;39(6):1104–1107. doi: 10.1016/0006-291x(70)90672-8. [DOI] [PubMed] [Google Scholar]
- 48.Corgiat JM, Dobbs FC, Burger MW, Scheuer PJ. Organohalogen constituents of the acorn worm Ptychodera bahamensis. Comp Biochem Physiol. 1993;106B(1):83–86. [Google Scholar]
- 49.Parry R, Nishino S, Spain J. Naturally-occurring nitro compounds. Nat Prod Rep. 2011;28(1):152–167. doi: 10.1039/c0np00024h. [DOI] [PubMed] [Google Scholar]
- 50.Dai M, Copley SD. Genome shuffling improves degradation of the anthropogenic pesticide pentachlorophenol by Sphingobium chlorophenolicum ATCC 39723. Appl Environ Microbiol. 2004;70(4):2391–2397. doi: 10.1128/AEM.70.4.2391-2397.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weeks A, Lund L, Raushel FM. Tunneling of intermediates in enzyme-catalyzed reactions. Curr Opin Chem Biol. 2006;10(5):465–472. doi: 10.1016/j.cbpa.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 52.Miles EW, Rhee S, Davies DR. The molecular basis of substrate channeling. J Biol Chem. 1999;274(18):12193–12196. doi: 10.1074/jbc.274.18.12193. [DOI] [PubMed] [Google Scholar]
- 53.Pfennig N. Photosynthetic bacteria. Annu Rev Microbiol. 1967;21:285–324. doi: 10.1146/annurev.mi.21.100167.001441. [DOI] [PubMed] [Google Scholar]
- 54.Malakhov MP, et al. SUMO fusions and SUMO-specific protease for efficient expression and purification of proteins. J Struct Funct Genomics. 2004;5(1–2):75–86. doi: 10.1023/B:JSFG.0000029237.70316.52. [DOI] [PubMed] [Google Scholar]
- 55.Jaganaman S, Pinto A, Tarasev M, Ballou DP. High levels of expression of the iron-sulfur proteins phthalate dioxygenase and phthalate dioxygenase reductase in Escherichia coli. Protein Expr Purif. 2007;52(2):273–279. doi: 10.1016/j.pep.2006.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gassner GT, Ballou DP. Preparation and characterization of a truncated form of phthalate dioxygenase reductase that lacks an iron-sulfur domain. Biochemistry. 1995;34(41):13460–13471. doi: 10.1021/bi00041a025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.