Abstract
Subjective well-being (SWB) is a major topic of research across the social sciences. Twin and family studies have found that genetic factors may account for as much as 30–40% of the variance in SWB. Here, we study genetic contributions to SWB in a pooled sample of ≈11,500 unrelated, comprehensively-genotyped Swedish and Dutch individuals. We apply a recently developed method to estimate “common narrow heritability”: the fraction of variance in SWB that can be explained by the cumulative additive effects of genetic polymorphisms that are common in the population. Our estimates are 5–10% for single-question survey measures of SWB, and 12–18% after correction for measurement error in the SWB measures. Our results suggest guarded optimism about the prospects of using genetic data in SWB research because, although the common narrow heritability is not large, the polymorphisms that contribute to it could feasibly be discovered with a sufficiently large sample of individuals.
Keywords: happiness, GREML, GCTA
Subjective well-being (SWB)—most commonly measured by survey questions about a respondent’s happiness or life satisfaction—is a major topic of research across the social sciences (1, 2). SWB is conceptualized to include a continuous spectrum of positive feelings and subjective life assessments (3–5). In contrast to standard economic indicators, which focus on consumption of material goods, responses to SWB survey questions additionally convey information regarding a broad range of other determinants of well-being, including physical and mental health, social relationships, leisure, and subjective states, such as emotions and mental engagement (6, 7). Because SWB measures may represent a relatively comprehensive assessment of an individual’s feelings of well-being, much research aims to understand individual differences in SWB (8). Most of the literature examines social, economic, and psychological influences on SWB (4, 9), but there has also been recent interest in understanding how genetic factors influence SWB.
To date, most of these papers on the genetics of SWB are twin or family studies (10–17). These studies draw indirect inferences about the contribution of genes to SWB by contrasting the resemblance of relatives with different degrees of environmental and genetic similarity. The literature concludes that a moderate share, typically 30–40%, of the cross-sectional variation in SWB is accounted for by variation in genes.
Recently it has become possible to directly and inexpensively assay human genetic polymorphisms, segments of DNA that differ across individuals. For medical geneticists studying health outcomes, the availability of such data has ushered in a new era, as researchers are discovering an ever-increasing number of polymorphisms related to diseases and physical traits (18).
So far, however, the few attempts to find genetic polymorphisms associated with SWB have been unsuccessful (see ref. 19 for the earliest effort we know of). One study reported an association (20), but follow-up work on an augmented sample from the same data did not replicate the finding (21). This lack of success is not surprising, given the lessons that have emerged from genetics research across a range of medical and social-science traits. Among the central challenges for complex traits, such as height and probably even more so SWB, is that the heritability of these traits appears to be comprised of a huge number of tiny genetic effects. Consequently, large samples of individuals—several orders of magnitude larger than those used to date in gene-discovery work in the social sciences—are needed for adequate statistical power to identify specific genetic polymorphisms (22, 23).
Nevertheless, anticipating that polymorphisms related to SWB will soon be discovered, SWB researchers have expressed excitement about the transformative potential of genetic data for social-science research (21), which complements what can be learned from twin and family studies (22–24). Most directly, knowing the functions of the relevant genes could shed light on the biological pathways that matter for SWB. If a set of polymorphisms were found to be sufficiently predictive, then they could be used in social-science research as control variables. More speculatively, such polymorphisms could be used as instrumental variables (25, 26), in effect treating the Mendelian randomization that occurs at conception as a natural experiment to learn about the causal effects of SWB [which may be especially credible when used in family samples (27); for a critical perspective, see ref. 28]. Finally, the discovery of polymorphisms associated with SWB could catalyze the study of how genetic sources of individual differences are amplified or dampened by environmental factors and, conversely, how environmental effects are modulated by genetic pathways.
For evaluating the extent to which these promises of genetic data can be realized, a critical question is: How much of the variation in SWB will eventually be predictable using molecular genetic data? In this article, we provide empirical evidence on a quantity—the “common narrow heritability,” explained below—that may help calibrate reasonable expectations about the answer to this question. We also discuss the inferences that can and cannot legitimately be drawn from this estimate as well as from heritability estimates in general. For example, we scrutinize the logical coherence of invoking estimates of a trait’s heritability to draw conclusions about its responsiveness to environmental interventions.
The estimates of 30–40% mentioned above likely overstate the amount of predictive power that can be obtained from molecular genetic data for two distinct reasons. First, the numbers refer to what is known as “broad heritability,” but “narrow heritability” is more germane and is necessarily smaller. Narrow heritability is the fraction of variance that can be accounted for in aggregate by the cumulative additive effects of all genetic polymorphisms. Narrow heritability can be understood as the R2 from a population regression of SWB on its best linear genetic predictor; that is, a predictor in which each polymorphism enters additively, and the effect of each polymorphism is constrained to be linear in the number of reference alleles. Broad heritability, which is necessarily larger, is the fraction of variance in SWB that can be explained in aggregate by all genetic factors. Broad heritability can be understood as the R2 from a population regression of SWB on its best genetic predictor, allowing not only for linear and additive effects but also for interactions among different polymorphisms (“epistasis”) and nonlinear effects of specific polymorphisms (“dominance”). In a seminal report drawing together evidence from various twin and family comparisons, Lykken (29) proposed that for SWB (along with several other traits including personality), most—if not all—of the genetic influences stem from higher-order epistatic interactions among genetic polymorphisms. Lykken called this phenomenon the “emergenesis hypothesis” (for a recent and related discussion, see ref. 30). If true, then the narrow heritability of SWB is much smaller than its broad heritability. Several recent, large-scale, twin-family studies, including both twin and sibling pairs, have indeed documented evidence for the importance of both additive and nonadditive genetic effects in explaining individual differences in SWB (14, 16).
Narrow heritability is more relevant than broad heritability for evaluating the predictive power that will be attainable using molecular genetic data because most interaction effects between polymorphisms are going to be extremely challenging to pinpoint. For genetically complex traits, we are not aware of a credible method for restricting the set of hypotheses about epistatic interactions that could be postulated. The number of possible combinations of polymorphisms that could be tested is therefore staggering, and this multiple hypothesis testing, in turn, necessitates imposing extremely stringent P-value thresholds. For a given sample and P-value threshold corrected for multiple testing, detecting even two polymorphisms whose interaction explains a given fraction of variance would require a sample size several orders of magnitude larger than the sample required to detect a single polymorphism that accounts for the same fraction of variance. As the order of the interaction increases, the requisite sample size quickly outstrips the number of people on the planet.
Second, even narrow heritability is likely to overstate the fraction of variance that discoverable polymorphisms are likely to capture. Estimates of narrow heritability from twin-family studies include additive variance attributable to any polymorphism, regardless of whether the polymorphism is common or rare among individuals. However, individual polymorphisms related to SWB that are rare in the population—which may collectively contribute much of the narrow heritability—will be much more difficult to reliably detect than polymorphisms that are common in the population.
In our empirical analysis, we estimate a parameter that cannot be estimated from twin or family data and that is necessarily smaller than narrow heritability, namely common narrow heritability: the fraction of variance that can be accounted for in aggregate by the cumulative additive effects of genetic polymorphisms that are common in the population (typically defined as minor allele frequency >1%). To do so, we use a recently developed method (31, 32) called genomic-relatedness-matrix restricted maximum likelihood, or GREML. We apply GREML to SWB, pooling data from two large datasets, TwinGene [TG; the genotyped subsample of the Swedish Twin Registry (33)] and the Rotterdam Study (RS) (34), both datasets in which dense SNP genetic data have been collected. GREML has previously been used to estimate the common narrow heritability of height (31), intelligence (35, 36), personality traits (37), several common diseases (38), schizophrenia (39), economic and political preferences (22), as well as smoking, glucose levels, and depression (40). GREML has not previously been applied to SWB.
GREML estimates a heritability parameter by examining how, across pairs of individuals, phenotypic similarity relates to genetic similarity, after controlling for observables (in our case: age, sex, 20 principal components of the variance-covariance matrix of the genotypic data, and an indicator for dataset) (Materials and Methods). However, unlike in twin-family studies, where expected genetic similarity (inferred from the family pedigree) is used, GREML proceeds by first estimating the realized genetic similarity between pairs of unrelated individuals using the dense SNP data. To be more precise, realized genetic similarity is estimated using the sample covariance matrix of the individuals’ genotypes. Because the genotypic data contains over half a million SNPs, this matrix is estimated very precisely. Because the covariance is a linear operator, the GREML method picks up the fraction of variance explained by the linear, additive action of the SNPs (i.e., the part of narrow heritability that is a result of the measured polymorphisms). Hence, GREML does not require the assumptions—about the degree of environmental and genetic resemblance between relatives and about the specific form of genetic effects (e.g., additive or dominance)—that tend to incite controversy when twin or family data are used to estimate narrow heritability. Because the current genotyping platforms from which our dense SNP data are obtained do not measure polymorphisms that are rare but do tag most of the genetic variation that is common in the population (41), GREML, as applied to these genotypic data, yields an estimate of common narrow heritability.
The key identifying assumption in GREML is that genetic similarity is uncorrelated with similarity in uncontrolled-for environmental factors that are exogenous to genotype (as defined by ref. 42). This assumption might be violated if the sample includes members of a shared extended family, such as siblings or close cousins. Therefore, it is standard to include in the sample only one individual from each family (in our case, one twin from a pair) and to drop individuals whose estimated genetic relatedness lies outside a small interval around zero. Because there is more random variation in the realized degree of genome sharing relative to the expected degree as the expected relatedness declines (43), uncontrolled-for environmental confounding factors are less likely to drive estimates that are based on realized relatedness among individuals whose expected relatedness is negligible.
We measure SWB in the TG and RS samples using responses to the two items from the Center for Epidemiologic Studies Depression Scale positive affect subscale that are available in both studies, namely responses regarding how frequently “During the past week, I was happy” and “During the past week, I enjoyed life.” We refer to these questions, respectively, as Happy and Enjoy. Responses are elicited using a four-point Likert scale ranging from “Rarely or none of the time (less than 1 day)” to “Most or all of the time (5–7 days).” Combined is a composite measure of the two variables. Because a substantial majority of responses to Happy and Enjoy are either in the highest-frequency category or the second-highest category (as is common with SWB survey measures), and because the software GCTA (genome-wide complex trait analysis) (32) that we use for the GREML analysis cannot presently handle multinomial variables, we converted the responses to binary variables (for details, see Materials and Methods).
Results
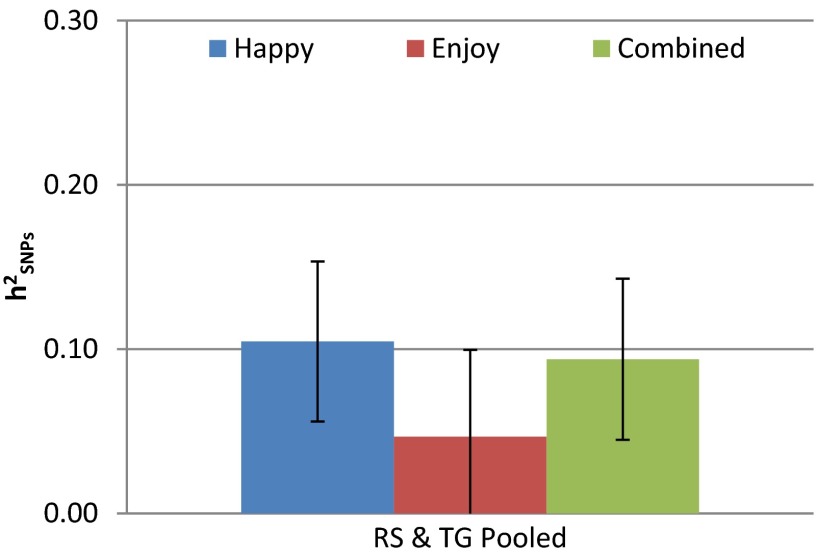
Table 1 reports the GREML estimates. For each sample, TG and RS, we report the fraction of variance explained by the measured SNPs for each of the two questions and for the combined SWB measure. We also report estimates with the TG and RS results pooled. In the TG sample, the GREML estimate for Happy is 0.10 (SE 0.10); for Enjoy, 0.06 (SE 0.10); and for Combined, 0.08 (SE 0.10). The corresponding figures in RS are 0.06 (SE 0.10), 0.04 (SE 0.10), and 0.08 (SE 0.10). Fig. 1 shows the estimates and SEs for the pooled sample. These estimates are of course more precise. For two of the three SWB measures, Happy (h2SNP = 0.10; SE 0.05) and Combined (h2SNP = 0.09; SE 0.05), the estimates are statistically distinguishable from zero at the 5% level.
Table 1.
GREML (common narrow heritability) estimates for SWB
| Sample | No. SNPs | Happy (“...I was happy”) | Enjoy (“…I enjoyed life”) | Combined | |||||||||
| n | h2SNP | SE | P value | n | h2SNP | SE | P value | n | h2SNP | SE | P value | ||
| TG | 627,011 | 5,682 | 0.10 | 0.10 | 0.16 | 5,742 | 0.06 | 0.10 | 0.28 | 5,670 | 0.08 | 0.10 | 0.22 |
| RS | 533,323 | 5,904 | 0.06 | 0.10 | 0.28 | 5,919 | 0.04 | 0.10 | 0.35 | 5,893 | 0.08 | 0.10 | 0.20 |
| Pooled | 852,597 | 11,484 | 0.10 | 0.05 | 0.01 | 11,558 | 0.05 | 0.05 | 0.19 | 11,461 | 0.09 | 0.05 | 0.03 |
| SALT retest | 105 | 0.55 | 0.12 | 105 | 0.41 | 0.15 | 105 | 0.61 | 0.12 | ||||
| RS retest | 7,845 | 0.41 | 0.03 | 7,916 | 0.42 | 0.03 | 7,795 | 0.41 | 0.03 | ||||
This table reports GREML estimates for Happy, Enjoy, and Combined. RS is the three Rotterdam cohorts pooled together; Pooled combines the TG and RS samples. We estimated the matrix of genetic relatedness after omitting one twin per pair in the Swedish data and then restricted the analyses to individuals whose relatedness did not exceed 0.025. n is the number of individuals used in the analyses after the relatedness threshold has been applied. In all analyses we control for sex, age, age-squared, and the first 20 principal components of the variance-covariance matrix of the genotypic data. The P value is from a likelihood ratio test of the null hypothesis that the fraction of variance explained is equal to zero. SALT retest is the sample of respondents in the SALT study who answered the survey twice (with 1 wk between survey occasions). RS retest is the retest correlation estimated using RS participants who answered the relevant questions in at least two different survey waves. These answers are at least 2 y apart in time.
Fig. 1.
This figure shows the GREML for Happy, Enjoy, and Combined. The error bars represent the point estimate ± 1 SD. The sample pools the three Rotterdam cohorts (RS) and the Swedish Twin Registry TwinGene sample (TG).
These estimates are attenuated by measurement error in SWB. Moreover, because our measures of SWB are based on only two questions, the attenuation is probably more severe compared with what is typically observed for lengthier personality batteries. We estimated reliability using data on 105 Screening Across the Lifespan Twin study (SALT) respondents who answered the SALT survey twice, with 2 wk between the measurement occasions. Our estimates are 0.55 (SE 0.12), 0.41 (SE 0.15), and 0.61 (SE 0.12) for Happy, Enjoy, and Combined, respectively. Although there are also individuals in the RS study who participated in multiple waves and for whom two or more responses are available, these responses are at least 2 y apart in time, and thus more of the change in measured SWB is likely to reflect true changes in SWB; nonetheless, for completeness, we also report the RS estimates in Table 1.
A simple adjustment for attenuation is to divide the heritability by the retest reliability. This adjustment assumes that any change in measured SWB between one survey occasion and the next is because of classic measurement error (which is uncorrelated with genotype) and not true change. Using this adjustment and the SALT retest reliabilities, we estimate that 18%, 12%, and 15% of the variance in Happy, Enjoy, and Combined would be accounted for by common SNPs if the SWB variables were measured without error.
In the SI Text, we report results of additional analyses that examine how sensitive our baseline results are to the choice of relatedness thresholds and to whether relatedness is estimated only using SNPs available on both platforms (Table S1). Reassuringly, these estimates are very close to those in our preferred specification. We also attempted to compare the GREML estimates to conventional twin-based estimates, when both are derived from the Swedish twin sample. The twin estimates, shown in Table S2, are in the same range as the GREML estimates, but unfortunately we cannot draw strong conclusions because the twin estimates are very imprecise.
Discussion
In this article we provide evidence on the common narrow heritability of SWB. We find that 5–10% of the variance in responses to single-question survey measures of SWB is accounted for by the additive effects of the SNPs measured on presently used genotyping platforms. A correction for measurement error in the SWB measures raises the point estimates to the range 12–18%.
We interpret our findings as indicating that the common narrow heritability is smaller than the typical estimates of narrow heritability from twin-family studies [although one recent, large-scale twin-family study estimated narrow heritability in the 10–20% range, as small as our estimates of narrow heritability (16)]. A caveat to this conclusion is that the twin-based heritability point estimates in our Swedish sample are actually lower than the GREML estimates, which raises the alternative possibility that our low GREML estimates are because of anomalously low “true” heritabilities in the data we happened to study, perhaps because the SWB measures that were available in our data tap into recently experienced SWB to a greater extent than do multi-item dispositional measures of SWB.
There are three reasons why we believe that our interpretation is more compelling than this alternative. First, the twin-based estimates that we report, which are only available from the Swedish sample, are sufficiently imprecise that we have little confidence in the point estimates. Moreover, with a retest reliability of ≈0.5 in the SWB measure, the measurement-error–adjusted 95% confidence intervals would overlap comfortably with the consensus estimates from the literature on twin-family studies. Second, we have relatively more confidence in our GREML estimates, which are similar across our Swedish and Dutch samples. Third and relatedly, our interpretation of the data also fits with the evidence regarding personality, another complex behavioral trait for which epistatic interactions have been hypothesized to be important (29). Twin-based analyses tend to produce heritability estimates for personality around 30–50% (44, 45), but a recent study finds evidence that a substantial share of the heritability of Neuroticism, Openness, and Agreeableness is due to nonadditive factors (46). Two studies applying GREML to personality traits have been published to date, with results remarkably close to those reported here. One study reports estimates of 9% and 12% for neuroticism and extraversion (37), respectively, and another report estimates in the range 4–10% for traits assessed by the Cloninger personality inventory (47).
For SWB, the gap between the common narrow heritability we estimate and the larger estimates of narrow heritability from twin-family studies may imply that some of the narrow heritability is due to rare polymorphisms. For most traits, it is not well understood to what extent rare polymorphisms with substantial effects account for the heritable variation (48). Until very recently, rare polymorphisms were not measured on standard genotyping platforms, and therefore most hypotheses regarding their role are based on indirect inferences such as those we make here (49).
Common narrow heritability is the quantity of most direct interest for assessing the potential contributions of genetic data to SWB research. Nonetheless, it may also be of interest to calibrate what our GREML results imply about narrow heritability, given that our GREML estimates do not rely on the same assumptions as the twin- and family-based estimates of narrow heritability. Two well-measured and widely studied complex traits for which reasonably reliable heritability estimates are available are height and cognitive ability. The twin-based estimates tend to fall in the range 50–80% for adult intelligence (50) and 80–90% for height (51). These estimates provide an upper bound on the narrow heritabilities. Other family-member comparisons—of full biological siblings, half-siblings, and parents and their children—suggest that the narrow heritabilities are unlikely to fall below 50% for adult intelligence (52) and 60% for height (53). By comparison, the one published GREML estimate for height is 45% (31), and GREML estimates for cognitive ability have also been around 45% (35, 36). This evidence suggests that the common narrow heritability that we estimate should be adjusted upward by a factor of roughly 1.5 to recover a ballpark estimate of narrow heritability.
Although our empirical contribution in this article focuses on estimating the common narrow heritability of SWB, we also believe it is important to highlight for SWB researchers that the conclusions that can be drawn from heritability estimates are more limited than is generally understood (for a related discussion, see ref. 54). Two misconceptions in particular appear to be widespread. First, some scholars erroneously conclude that higher heritability implies less variation left over to be explained by environmental factors. As the authors of the World Happiness Report (8) put it, twin studies are often misleadingly understood as “estimating the extent to which happiness depends on genetically based personality differences rather than differing circumstances.” However, as Jencks (42) explained, heritability comprises any genetic variation that ultimately contributes to phenotypic variation, regardless of the pathway, and many plausible pathways are in fact mediated by environmental factors. In the terminology of econometrics, the population regression of SWB on all genes—for which heritability is the R2—is a reduced-form regression, but the structural equations describing the true relationship between a gene and SWB may involve many intervening environmental variables. Although some genes may affect SWB via relatively direct physical pathways—for example, by affecting baseline serotonin levels or dopamine-receptor density—it also seems likely that many genes matter for SWB through their effects on preferences, personality, and abilities, which in turn influence individuals’ choices about friendships, marriage, fertility, and occupational choice. Consequently, some of the variance in SWB explained by genes is the same variance that is explained by these environmental factors. Because genetic effects may operate indirectly through environmental variables, the heritability of SWB does not put any bound on the proportion of variance that could be explained by the full set of relevant environmental variables.
Second, findings of higher heritability are sometimes misinterpreted as demonstrating that there is less scope for interventions to increase SWB. For example, in their seminal paper, Lykken and Tellegen (11) conclude that “trying to be happier [may be] as futile as trying to be taller.” Such a claim may or may not be true, but it is in no way implied by the finding that SWB is heritable. The conclusion is incorrect for two distinct reasons. Related to the point above, some genetic effects may be mediated by modifiable environmental variables. The very same genotype may cause a person to grow to 5 feet or 6 feet tall, depending on nutritional intake. Furthermore, as Goldberger (55) pointed out, even if heritability were 100% and the genetic effects operated entirely through mechanisms that are difficult to modify, there may still exist powerful environmental interventions that do not contribute to outcome variance in the current population. As Bang (54) emphasized in her discussion of genetics and SWB, a heritability estimate represents the fraction of variance explained by genes in a specific population at a specific point in time. Using econometric terminology, one set of explanatory variables (in this case, genes) having a high R2 does not rule out a large coefficient on another variable (an intervention), if the latter explanatory variable varies little across individuals in the population under study. In Goldberger’s (55) example, the introduction of eyeglasses dramatically improves vision even though eyesight is highly heritable.
To summarize what can be concluded from our findings, the magnitude of common narrow heritability provides useful information regarding the potential contributions of genetic data for research on SWB. Because our estimates are lower than typical heritability estimates for SWB, our results suggest that the scope for uses of genetic data that rely on substantial predictive power—such as using a set of polymorphisms as control variables, instrumental variables, or moderators in social-science research—may be more limited than has been assumed. At the same time, the fact that our estimates of common narrow heritability are nonnegligible suggests that—even if much of the broad heritability is due to epistatic interactions—some of the SNPs measured on existing platforms have main effects on SWB. Therefore, gene-discovery efforts with a large enough sample size are likely to be successful.
Materials and Methods
Our study combines data from the Swedish Twin Registry (STR) and the Rotterdam Study. STR is a large, population-based twin registry. Between 1998 and 2002, STR administered to twins born in 1958 or earlier a survey called the Screening Across the Lifespan Twin study (33). A subsample of SALT was recently genotyped using the Illumina HumanOmniExpress BeadChip technology as part of the TwinGene project (22). We refer to these ≈10,000 genotyped SALT respondents as the TG sample. TG participants are all born between 1911 and 1958.
RS is a large, population-based prospective cohort study of elderly people ongoing since 1990 in the city of Rotterdam in the Netherlands (34). Approximately 11,000 subjects in the RS have been genotyped using the Illumina 550 K and 610 K arrays. RS respondents are divided into three cohorts, which we refer to as RS-I, RS-II, and RS-III.
To minimize the expected relatedness of the individuals in our sample, we only included one twin per family in the TG sample. If only one twin from a pair had answered both survey questions, the individual with complete phenotypic data was included in the analysis. If both twins had complete phenotypic data, one of them was chosen at random. We then pooled the resulting sample with the RS sample and used the GCTA software to estimate the pairwise relatedness between all individuals in the pooled dataset. Following convention, we restricted the sample to individuals whose pairwise relatedness did not exceed 0.025 in absolute value.
These restrictions brought the sample size to just below 6,000 individuals in each sample. Our analysis is restricted to individuals with SNP data that passed quality controls and who answered both questions. Results in TG are based on 627,011 SNPs; in RS, on 533,323 SNPs. Because of incomplete overlap in the two samples, the number of SNPs in the pooled sample is larger: 852,597.
To convert the Happy and Enjoy measures to binary variables, we coded responses as “high” if they were the highest-frequency category and “not high” otherwise. We also constructed a composite measure of the two variables, which we call Combined, the value of which is “high” if both Happy and Enjoy are high and “not high” otherwise. By generating the binary variables in this way, the fraction coded as high (or equivalently, not high) is made as close as possible to one half, thereby maximizing the variance and hence statistical power. The distributions of SWB measures before they were binarized are given in Tables S3 and S4. In the analyses we assume that each binary variable we observe results from the realization of an underlying, normally distributed random variable for liability falling above or below some threshold.
Table S5 reports age and sex, as well as the fraction of individuals coded as high for the three variables. Throughout, we control for sex, age, age-squared, dummies for each of the three RS cohorts (TG is the omitted category), and to guard against population stratification, the first 20 principal components of the genotype data.
Supplementary Material
Acknowledgments
We thank Pascal Arp, Mila Jhamai, Michael Moorhouse, Marijn Verkerk, and Sander Bervoets for their assistance in creating the genome-wide association study database; and Martha Widger and Cody Xuereb for research assistance. This work was supported in part by the Ragnar Söderberg Foundation (E9/11); the National Science Foundation (EArly Concept Grants for Exploratory Research: “Workshop for the Formation of a Social Science Genetic Association Consortium”, SES-1064089) as supplemented by the National Institutes of Health’s (NIH) Office of Behavioral and Social Sciences Research, and the National Institute on Aging/NIH through Grants P01-AG005842, P01-AG005842-20S2, P30-AG012810, and T32-AG000186-23 to the National Bureau of Economic Research. The Swedish Twin Registry is supported by the Swedish Department of Higher Education, the European Commission European Network for Genetic and Genomic Epidemiology [ENGAGE: 7th Framework Program (FP7/2007-2013)/Grant agreement HEALTH-F4-2007-201413; and GenomEUtwin: 5th Framework program “Quality of Life and Management of the Living Resources” Grant QLG2-CT-2002-01254]; the NIH (DK U01-066134); the Swedish Research Council (M-2005-1112 and 2009-2298); the Swedish Foundation for Strategic Research (ICA08-0047); the Jan Wallander and Tom Hedelius Foundation; and the Swedish Council for Working Life and Social Research. The Rotterdam Study is funded by the Erasmus Medical Center; Erasmus University, Rotterdam; the Netherlands Organization for the Health Research and Development (ZonMw); the Research Institute for Diseases in the Elderly; the Ministry of Education, Culture, and Science; the Ministry for Health, Welfare, and Sports; the European Commission (DG XII); and the Municipality of Rotterdam. The genome-wide association study database of the Rotterdam Study was funded by the Netherlands Organization for Scientific Research Investments (175.010.2005.011, 911-03-012); the Research Institute for Diseases in the Elderly (014-93-015), and the Netherlands Genomics Initiative/Netherlands Consortium for Healthy Aging Project 050-060-810. Statistical analyses were carried out on the Genetic Cluster Computer (www.geneticcluster.org), which is financially supported by the Netherlands Scientific Organization (NWO 480-05-003, PI: Posthuma) along with a supplement from the Dutch Brain Foundation and the Vrije Universiteit Amsterdam. D.J.B. received financial support through National Institute on Aging/NIH Grants R01-AG040787 to the University of Michigan and T32-AG00186 to the National Bureau of Economic Research. J.-E.D.N. acknowledges financial support from the UK Department for Work and Pensions and National Institute on Aging/NIH Grant RO1-AG040640.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1222171110/-/DCSupplemental.
References
- 1.Easterlin R. Explaining happiness. Proc Natl Acad Sci USA. 2003;100(19):11176–11183. doi: 10.1073/pnas.1633144100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kahneman D, Deaton A. High income improves evaluation of life but not emotional well-being. Proc Natl Acad Sci USA. 2010;107(38):16489–16493. doi: 10.1073/pnas.1011492107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diener E. Subjective well-being. The science of happiness and a proposal for a national index. Am Psychol. 2000;55(1):34–43. [PubMed] [Google Scholar]
- 4.Seligman ME, Csikszentmihalyi M. Positive psychology. An introduction. Am Psychol. 2000;55(1):5–14. doi: 10.1037//0003-066x.55.1.5. [DOI] [PubMed] [Google Scholar]
- 5.Vaillant GE. Positive mental health: Is there a cross-cultural definition? World Psychiatry. 2012;11(2):93–99. doi: 10.1016/j.wpsyc.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diener E, Seligman MEP. Beyond money: Toward an economy of well-being. Psychol Sci Public Interest. 2004;5(1):1–31. doi: 10.1111/j.0963-7214.2004.00501001.x. [DOI] [PubMed] [Google Scholar]
- 7.Kahneman D, Krueger AB. Developments in the measurement of subjective well-being. J Econ Perspect. 2006;20(1):3–24. [Google Scholar]
- 8.Helliwell J, Layard R, Sachs J, editors. World Happiness Report. New York: The Earth Institute; 2012. [Google Scholar]
- 9.Headey B, Muffels R, Wagner GG. Long-running German panel survey shows that personal and economic choices, not just genes, matter for happiness. Proc Natl Acad Sci USA. 2010;107(42):17922–17926. doi: 10.1073/pnas.1008612107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harris JR, Pedersen NL, Stacey C, McClearn GE. Age differences in the etiology of the relationship between life satisfaction and self-rated health. J Aging Health. 1992;4(3):349–368. [Google Scholar]
- 11.Lykken DT, Tellegen A. Happiness is a stochastic phenomenon. Psych Sci. 1996;7(3):186–189. [Google Scholar]
- 12.Tellegen A, et al. Personality similarity in twins reared apart and together. J Pers Soc Psychol. 1988;54(6):1031–1039. doi: 10.1037//0022-3514.54.6.1031. [DOI] [PubMed] [Google Scholar]
- 13.Røysamb E, Harris JR, Magnus P, Vitterso J, Tambs K. Subjective well-being. Sex-specific effects of genetic and environmental factors. Pers Individ Dif. 2002;32(2):211–223. [Google Scholar]
- 14.Stubbe JH, Posthuma D, Boomsma DI, De Geus EJ. Heritability of life satisfaction in adults: A twin-family study. Psychol Med. 2005;35(11):1581–1588. doi: 10.1017/S0033291705005374. [DOI] [PubMed] [Google Scholar]
- 15.Nes RB, Røysamb E, Tambs K, Harris J-R, Reichborn-Kjennerud T. Subjective well-being: Genetic and environmental contributions to stability and change. Psychol Med. 2006;36(7):1033–1042. doi: 10.1017/S0033291706007409. [DOI] [PubMed] [Google Scholar]
- 16.Bartels M, Boomsma DI. Born to be happy? The etiology of subjective well-being. Behav Genet. 2009;39(6):605–615. doi: 10.1007/s10519-009-9294-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franz CE, et al. Genetic and environmental multidimensionality of well- and ill-being in middle aged twin men. Behav Genet. 2012;42(4):579–591. doi: 10.1007/s10519-012-9538-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. Am J Hum Genet. 2012;90(1):7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartels M, et al. Heritability and genome-wide linkage scan of subjective happiness. Twin Res Hum Genet. 2010;13(2):135–142. doi: 10.1375/twin.13.2.135. [DOI] [PubMed] [Google Scholar]
- 20.De Neve JE. Functional polymorphism (5-HTTLPR) in the serotonin transporter gene is associated with subjective well-being: Evidence from a US nationally representative sample. J Hum Genet. 2011;56(6):456–459. doi: 10.1038/jhg.2011.39. [DOI] [PubMed] [Google Scholar]
- 21.De Neve JE, Christakis NA, Fowler JH, Frey BS. Genes, economics, and happiness. J Neuroscience Psychology Econ. 2012;5(4):193–211. doi: 10.1037/a0030292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benjamin DJ, et al. The genetic architecture of economic and political preferences. Proc Natl Acad Sci USA. 2012;109(21):8026–8031. doi: 10.1073/pnas.1120666109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benjamin DJ, et al. The promises and pitfalls of genoeconomics. Ann Rev Econ. 2012;4(1):627–662. doi: 10.1146/annurev-economics-080511-110939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beauchamp JP, et al. Molecular genetics and economics. J Econ Perspect. 2011;25(4):57–82. doi: 10.1257/jep.25.4.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith GD, Ebrahim S. ‘Mendelian randomization’: Can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32(1):1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 26.Ding W, Lehrer SF, Rosenquist JN, Audrain-McGovern J. The impact of poor health on academic performance: New evidence using genetic markers. J Health Econ. 2009;28(3):578–597. doi: 10.1016/j.jhealeco.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 27.Fletcher JM, Lehrer SF. Genetic lotteries within families. J Health Econ. 2011;30(4):647–659. doi: 10.1016/j.jhealeco.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 28.Conley D. The promise and challenges of incorporating genetic data into longitudinal social science surveys and research. Biodemography Soc Biol. 2009;55(2):238–251. doi: 10.1080/19485560903415807. [DOI] [PubMed] [Google Scholar]
- 29.Lykken DT. Presidential address, 1981. Research with twins: The concept of emergenesis. Psychophysiology. 1982;19(4):361–373. doi: 10.1111/j.1469-8986.1982.tb02489.x. [DOI] [PubMed] [Google Scholar]
- 30.Zuk O, Hechter E, Sunyaev SR, Lander ES. The mystery of missing heritability: Genetic interactions create phantom heritability. Proc Natl Acad Sci USA. 2012;109(4):1193–1198. doi: 10.1073/pnas.1119675109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang J, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42(7):565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: A tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88(1):76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lichtenstein P, et al. The Swedish Twin Registry in the third millennium: An update. Twin Res Hum Genet. 2006;9(6):875–882. doi: 10.1375/183242706779462444. [DOI] [PubMed] [Google Scholar]
- 34.Hofman A, et al. The Rotterdam Study: 2012 objectives and design update. Eur J Epidemiol. 2011;26(8):657–686. doi: 10.1007/s10654-011-9610-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davies G, et al. Genome-wide association studies establish that human intelligence is highly heritable and polygenic. Mol Psychiatry. 2011;16(10):996–1005. doi: 10.1038/mp.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chabris CF, et al. Most reported genetic associations with general intelligence are probably false positives. Psychol Sci. 2012;23(11):1314–1323. doi: 10.1177/0956797611435528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vinkhuyzen AAE, et al. Common SNPs explain some of the variation in the personality dimensions of neuroticism and extraversion. Transl Psychiatry. 2012;2(4):e102. doi: 10.1038/tp.2012.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee SH, Wray NR, Goddard ME, Visscher PM. Estimating missing heritability for disease from genome-wide association studies. Am J Hum Genet. 2011;88(3):294–305. doi: 10.1016/j.ajhg.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee SH, et al. Schizophrenia Psychiatric Genome-Wide Association Study Consortium (PGC-SCZ) International Schizophrenia Consortium (ISC) Molecular Genetics of Schizophrenia Collaboration (MGS) Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nat Genet. 2012;44(3):247–250. doi: 10.1038/ng.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lubke GH, et al. Estimating the genetic variance of major depressive disorder due to all single nucleotide polymorphisms. Biol Psychiatry. 2012;72(8):707–709. doi: 10.1016/j.biopsych.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barrett JC, Cardon LR. Evaluating coverage of genome-wide association studies. Nat Genet. 2006;38(6):659–662. doi: 10.1038/ng1801. [DOI] [PubMed] [Google Scholar]
- 42.Jencks C. Heredity, environment, and public policy reconsidered. Am Sociol Rev. 1980;45(5):723–736. [PubMed] [Google Scholar]
- 43.Hill WG, Weir BS. Variation in actual relationship as a consequence of Mendelian sampling and linkage. Genet Res (Camb) 2011;93(1):47–64. doi: 10.1017/S0016672310000480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jang KL, Livesley WJ, Vernon PA. Heritability of the big five personality dimensions and their facets: A twin study. J Pers. 1996;64(3):577–591. doi: 10.1111/j.1467-6494.1996.tb00522.x. [DOI] [PubMed] [Google Scholar]
- 45.Johnson AM, Vernon PA, Feiler RA. Behavioral genetic studies of personality: An introduction and review of the results of 50+ years of research. In: Boyle G, Saklofske D-H, Matthews G, editors. Handbook of Personality Theory and Assessment. London: Sage; 2008. pp. 145–173. [Google Scholar]
- 46.Hahn E, et al. The complexity of personality: advantages of a genetically sensitive multi-group design. Behav Genet. 2012;42(2):221–233. doi: 10.1007/s10519-011-9493-y. [DOI] [PubMed] [Google Scholar]
- 47.Verweij KJ, et al. Maintenance of genetic variation in human personality: testing evolutionary models by estimating heritability due to common causal variants and investigating the effect of distant inbreeding. Evolution. 2012;66(10):3238–3251. doi: 10.1111/j.1558-5646.2012.01679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gibson G. Rare and common variants: Twenty arguments. Nat Rev Genet. 2011;13(2):135–145. doi: 10.1038/nrg3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Visscher PM, Goddard ME, Derks EM, Wray NR. Evidence-based psychiatric genetics, AKA the false dichotomy between common and rare variant hypotheses. Mol Psychiatry. 2012;17(5):474–485. doi: 10.1038/mp.2011.65. [DOI] [PubMed] [Google Scholar]
- 50.Bouchard TJ, Jr, McGue M. Genetic and environmental influences on human psychological differences. J Neurobiol. 2003;54(1):4–45. doi: 10.1002/neu.10160. [DOI] [PubMed] [Google Scholar]
- 51.Silventoinen K, et al. Heritability of adult body height: A comparative study of twin cohorts in eight countries. Twin Res. 2003;6(5):399–408. doi: 10.1375/136905203770326402. [DOI] [PubMed] [Google Scholar]
- 52.Bouchard TJ, Jr, McGue M. Familial studies of intelligence: A review. Science. 1981;212(4498):1055–1059. doi: 10.1126/science.7195071. [DOI] [PubMed] [Google Scholar]
- 53.Silventoinen K. Determinants of variation in adult body height. J Biosoc Sci. 2003;35(2):263–285. doi: 10.1017/s0021932003002633. [DOI] [PubMed] [Google Scholar]
- 54.Bang RH. Happiness in behaviour genetics: Findings and implications. J Happiness Stud. 2010;11(3):369–381. [Google Scholar]
- 55.Goldberger A. Heritability. Economica. 1979;46(184):327–347. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.