Measuring researchers’ scientific impact
Using publication data to measure researchers’ scientific impact has become increasingly complicated partly due to the rise in interdisciplinary collaboration. Direct measures of publication quantity and citations by other researchers may not accurately reflect individual contributions on multiauthor papers. Jonathan Stallings et al. (pp. 9680–9685) developed and analyzed new metrics for calculating the scientific impact of interdisciplinary researchers. The authors used an axiomatic approach to assign relative credits to the coauthors of a given paper, creating a metric called the A-index. The authors used the A-index to weight each peer-reviewed publication, and summed the credit shares to quantify the collaboration or C-index. The authors also used the A-index to weight corresponding journal impact factors, creating the productivity or P-index. Using publication data of 186 randomly selected biomedical engineering faculty members from top universities, the authors tested these indices against popular metrics, including a researcher’s total number of publications, and the commonly used H-index. The authors found that, by accounting for researchers’ collaborative tendencies, the new metrics better captured scientific caliber than did traditional measures. These easily computed new metrics might provide fair and precise evaluation of researchers with diverse collaborations, according to the authors. — B.A.
Linking clinical findings with genomics
Screenshot of the home page for the Clinical Genomic Database (available at http://research.nhgri.nih.gov/CGD/). Image courtesy of the National Human Genome Research Institute.
Advances in high-throughput DNA sequencing have enabled the generation of vast amounts of human genomic data, but the use of these data to inform medical care has lagged behind, partly due to the lack of a pertinent database of interventions for genetic disorders. To address this issue, Benjamin Solomon et al. (pp. 9851–9855) have constructed a database linking genetic diseases to available medical interventions. As of its inception in April 2013, the Clinical Genomic Database (CGD) includes 2,616 genes involved in human genetic disorders. For roughly half the genes listed in the database, specific medical interventions are available that would be warranted for disease-causing mutations. Each CGD entry includes the gene, condition(s), allelic condition(s), clinical categorization, mode of inheritance, intervention(s), age category for interventions, linked PubMed references, and gene-specific links to complementary databases. Contents of the database are freely downloadable from the CGD Web site or via a link from the article in PNAS online. According to the authors, broad dissemination of the CGD is intended to solicit feedback and input from experts. Such input will be used to continually revise the database through both automated and manual curation. Ultimately, the authors suggest that the CGD may serve as a tool for annotation of individual human genomes, generating a clinically relevant analysis of genomic data. — C.B.
Intestinal microbes, inflammation, and colorectal cancer
Colorectal cancer is the second deadliest malignancy in the United States, yet the underlying pathogenesis of the disease remains unclear. Researchers have long known that inflammation promotes cancers of the colon and rectum, and people with a long history of inflammatory bowel disease (IBD) are highly susceptible to colorectal cancer. Bo Hu et al. (pp. 9862–9867) explore another known risk factor for colorectal cancer—an abnormal intestinal microflora—and reveal an interplay between intestinal microbes and inflammatory stimuli that enhances the pathogenesis of colorectal cancer. Using a mouse model of colorectal cancer, the authors found that mice deficient in the NLRP6 inflammasome—a cytoplasmic complex of proteins that stimulate immune responses to microbes and other triggers—develop an altered intestinal microflora that enhances their susceptibility to colorectal cancer. Interestingly, colorectal cancer was also prevalent among wild-type mice that were housed with either NLRP6-deficient mice or mice that were unable to secrete the inflammatory protein IL-18. The results suggest that the transfer of an abnormal microbiome between individuals can exacerbate intestinal inflammation, which can promote the abnormal proliferation of epithelial cells and lead to tumor formation. The findings suggest that intestinal microbes may be capable of transferring cancer susceptibility between individuals. — A.G.
Circadian patterns of gene expression in the brain disrupted in depression
Circadian rhythms in physiology and behavior are frequently disrupted in individuals with major depressive disorder, but the molecular basis for circadian clock disruption in the human brain remains unclear. Jun Li et al. (pp. 9950–9955) characterized the genome-wide gene expression patterns in postmortem brains from 55 individuals with no history of psychiatric or neurological illness and 34 patients with clinical depression. The authors analyzed multiple RNA samples isolated from six regions of each brain, and arranged the gene expression data around a 24-hour cycle based on time of death. The authors report that several hundred genes in each brain region displayed rhythmic patterns of expression over the 24-hour cycle, including several genes essential to the body’s circadian machinery, and more than 100 genes had consistent cyclic patterns across all six brain regions. However, the cyclic patterns in individuals with clinical depression were disrupted. The disruption may be involved in the abnormal circadian rhythms in hormonal, body temperature, sleep, and behavioral patterns frequently associated with depressive disorders, the authors suggest. The findings reveal hundreds of genes in the human brain that may be involved in rhythmic daily processes, and may aid the identification of molecular targets for the treatment of mood disorders, according to the authors. — N.Z.
A single vaccine component against multiple pathogens
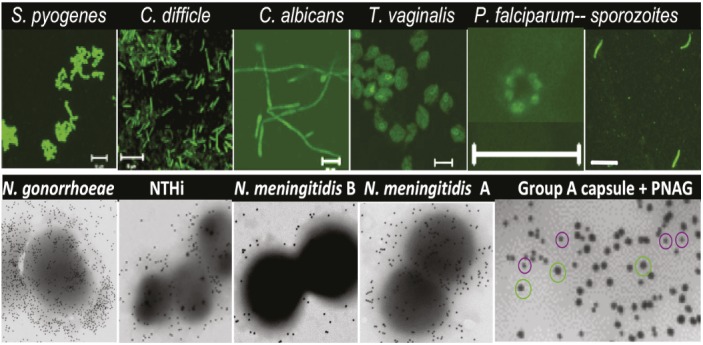
PNAG production by various microbial pathogens.
The development of vaccines is fraught with challenges, notably the difficulty of finding a single component that can protect against diverse microbial pathogens. One potential component is a β-(1→6)–linked polymeric-N-acetyl glucosamine sugar known as PNAG, found on the surface of some bacterial species. Colette Cywes-Bentley et al. (pp. E2209–E2218) report that PNAG is produced by more microbes than previously thought, including the causative agents of trichomoniasis and malaria as well as a diverse group of gram-positive, gram-negative, fungal, and protozoan pathogens. Noting that natural antibodies produced by humans and animals do not protect against pathogens that express PNAG, the authors raised animal antibodies to a synthetic form of PNAG, in which the sugar residues were altered. The authors also used a human antibody capable of binding to both the natural and modified forms of PNAG. Vaccination with both antibodies protected mice against local and systemic infections caused by a number of pathogens, including Streptococcus pyogenes, Streptococcus pneumoniae, Listeria monocytogenes, Neisseria meningitidis serogroup B, Candida albicans, and the mouse malarial parasite Plasmodium berghei ANKA.
The findings suggest that vaccines targeting PNAG could potentially protect against many deadly prokaryotic and eukaryotic pathogens, according to the authors. — A.G.
Charting the evolution of Western music styles

Charting music styles through mathematical patterns. Image courtesy of Daniel Migliorelli.
The evolution of stylistic changes in music is driven by intrinsic dynamics, despite the influence of culture and technology. To characterize the evolution of music styles over the last two centuries, Pablo Rodriguez Zivic et al. (pp. 10034–10038) analyzed a large Western music data set called the Peachnote Corpus, which includes information about the number of individual melodic interval patterns used in compositions each year between 1730 and 1930, a period spanning the Baroque, Classical, Romantic, and post-Romantic music styles. Using an approach based on machine learning and cognitive theories of music expectation, the authors report that changes in the probability distributions of the melodic intervals, defined as the distance between adjacent notes and measured in semitones, reflected the evolution of music styles in the historical record. Further, the authors uncovered a set of quantitative factors that could identify transitions between the style epochs and point to cognitively relevant structures in different music styles. Because the human brain perceives music by making predictions based on temporal statistics, the authors suggest that the data-driven principles uncovered by the study might help better understand melodic expectation in the perception of music. — P.N.


