Abstract
Eukaryotic translation initiation factor 3 (eIF3) plays a central role in translation initiation and consists of five core (conserved) subunits present in both budding yeast and higher eukaryotes. Higher eukaryotic eIF3 contains additional (noncore or nonconserved) subunits of poorly defined function, including sub-unit h (eIF3h), which in zebrafish is encoded by two distinct genes (eif3ha and eif3hb). Previously we showed that eif3ha encodes the predominant isoform during zebrafish embryogenesis and that depletion of this factor causes defects in the development of the brain and eyes. To investigate the molecular mechanism governing this regulation, we developed a genome-wide polysome-profiling strategy using stage-matched WT and eif3ha morphant zebrafish embryos. This strategy identified a large set of predominantly neural-associated translationally regulated mRNAs. A striking finding was a cohort of lens-associated crystallin isoform mRNAs lost from the eif3ha morphant polysomes, revealing a mechanism by which lens development is translationally controlled. We show that both UTR sequences of a targeted crystallin transcript are necessary but not sufficient for translational regulation by eif3ha. Therefore, our study reveals the role of a noncore eIF3 subunit in modulating a specific developmental program by regulating translation of defined transcripts and highlights the potential of the zebrafish system to identify translational regulatory mechanisms controlling vertebrate development.
Keywords: RNA sequencing, Crygm2d
Translation initiation in eukaryotic cells is defined as the process by which a 40S ribosomal subunit containing bound initiator methionyl-tRNA (Met-tRNAi) interacts with an mRNA and is positioned at the start AUG codon to form the 48S initiation complex. Subsequently, a 60S ribosomal subunit joins the 48S initiation complex to form the 80S ribosomal initiation complex (mRNA.80S.Met-tRNAi) that is competent to undergo peptide bond formation. The overall process requires the participation of a dozen protein factors, collectively called the “eukaryotic translation initiation factors” (eIFs) and involves the formation of multiple noncovalent intermediate biochemical complexes in a series of distinct steps that are highly conserved between the unicellular budding yeast Saccharomyces cerevisiae and higher eukaryotes, including mammals. Most initiation factors are conserved, and there are clear structural homologies between the budding yeast and mammalian factors (1–3). The only exception is the multiheteromeric initiation factor eIF3 that plays a central role in recruiting both the mRNA and the translation initiation machinery to achieve selection of the start codon.
The eIF3 from budding yeast contains only five subunits, whereas the multicellular higher eukaryotic eIF3 contains, in addition to the homologs of these five subunits, an additional five to eight subunits (4). Because the five evolutionarily conserved subunits (eIF3a, eIF3b, eIF3c, eIF3g, and eIF3i) are necessary and sufficient for global translation initiation of all mRNAs in yeast, they originally were designated as “core” subunits (4). It has been postulated that the additional subunits that are absent in the budding yeast might serve either as regulators of translation initiation or be required for other biological processes in higher eukaryotes. These—eIF3d, eIF3e, eIF3f, eIF3h, eIF3j, eIF3k, eIF3l, and eIF3m—were designated “non-core” subunits (4).
In contrast to the budding yeast, the genome of the fission yeast Schizosaccharomyces pombe contains structural homologs of at least five noncore (nonconserved) eIF3 subunits—eIF3d, eIF3e, eIF3f, eIF3h, and eIF3m. The gene encoding eIF3f is essential for growth, whereas eIF3d, eIF3e, and eIF3h are dispensable for growth and viability (5–11). However, deleted strains show specific phenotypes including defects in meiosis/sporulation (6, 9, 11). Genetic studies in Arabidopsis showed that inactivation by insertional mutagenesis of the eif3h+ gene leads to pleiotropic developmental defects. Translation efficiency of specific mRNAs containing multiple short ORFs in the 5′ UTR is reduced in the eif3h mutant (12–14). A recent study also revealed a positive role of the nonconserved subunit eIF3e in cellular proliferation and invasion by regulating the translation of distinct sets of functionally relevant mRNAs in a breast cancer cell line (15).
Because translational control plays an important role during early development (16–18), and because the role of the nonconserved eIF3 subunits is essentially untested in higher animals, including vertebrates, we investigated the role of one such eIF3 subunit, eIF3h, as a regulator of translation in the zebrafish model system. Zebrafish eIF3h (Eif3h) is encoded by two distinct genes, eif3ha and eif3hb; the two predicted proteins are 87% identical, showing 84% identity with human eIF3h. We reported previously that although both eif3h genes are expressed during early embryogenesis and display overlapping but distinct and highly dynamic spatial expression patterns, eif3ha is by far the predominant isoform during the early stages of embryogenesis, at least up to 2 d postfertilization (dpf). Although the loss of eif3ha causes brain and eye defects around 1 dpf, depletion of eif3hb does not cause any observable phenotype until later stages (19). For these reasons, we focused our studies on eif3ha and investigated whether this eIF3 subunit influences mRNA-specific translation.
Results
Development of a Protocol for the Isolation of Polysomes and Polysome-Associated mRNAs from Zebrafish Embryos.
We sought to determine whether the phenotypes caused by blocking eif3ha gene function were correlated with altered translation of a subset of mRNAs. Our strategy was to isolate the polysome-bound translationally active mRNAs by size fractionation using velocity-gradient centrifugation in sucrose gradients (20–22) and subsequently to identify changes in the translating mRNA profiles caused by loss of eif3ha using genome-wide RNA sequencing (RNA-seq) analysis (Fig. S1 A and B). As a first step, we optimized a reproducible protocol for generating polysome profiles which was amenable to a subsequent deep-sequencing strategy (described in Materials and Methods). As schematized in Fig. S1C, embryo-derived cell-free extracts were prepared and subjected to a 10–50% sucrose gradient centrifugation. A typical polysome profile pattern was obtained that is similar to that obtained from yeast using classical protocols for yeast cell-free extracts (Fig. S1D; see also ref. 23 for comparison with yeast cell-free extracts). Under the conditions of our gradient centrifugation, free 40S subunits are not well separated from the nonribosomal components at the top of the gradient. Also, the ratio of disomes (mRNAs containing two ribosomes engaged simultaneously in protein synthesis) to heavier polysomes (mRNAs containing more than two ribosomes) is higher in lysates isolated from zebrafish embryos at 24 h postfertilization (hpf) than typically observed in the yeast system. We further characterized the positions of 40S, 60S, 80S ribosomes and polysomes by identifying the constituent ribosomal RNAs using agarose gel electrophoresis (Fig. S1D, Lower). Identical profiles were obtained starting with deyolked embryos that were frozen in liquid nitrogen for 2 wk before use (Fig. S1E). Therefore, batches of frozen embryos can be stored and subsequently pooled if needed.
Comparison of the Polysome Profile Patterns Obtained from Cell-Free Extracts of WT and eif3ha Morphant Embryos.
We previously validated morpholinos (MOs) that specifically target eif3ha and cause reproducible brain-degeneration and somite-associated phenotypes (19). The phenotypes were validated by several criteria to be specific and p53-independent, correlating well with the spatiotemporal expression pattern of eif3ha during zebrafish embryogenesis (19).
We carried out polysome profile analyses with cell-free extracts of eif3ha morphant embryos at 1 dpf and compared them with profiles obtained from stage-matched WT control embryos (Fig. 1). We chose this time point because the initial phenotype for eif3ha morphants (brain defects) appeared around or after 1 dpf (19). The loss of eif3ha does not change the overall polysome profile significantly compared with that obtained from WT embryos (compare Fig. 1 A and B). Additionally, the yield of total RNA isolated from the polysomal fraction of the eif3ha morphants was similar to that obtained from WT embryos (30–50 μg), indicating that the total polysomal content did not change significantly upon depletion of Eif3ha.
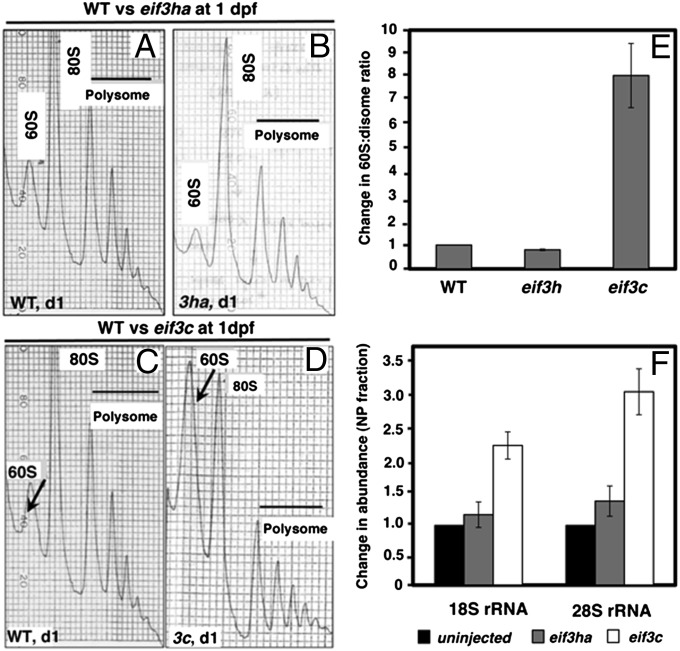
Fig. 1.
Comparison of the polysome profiles obtained from cell-free extracts of eif3ha or eif3c morphant embryos with the corresponding WT embryos. (A–D) Representative profiles derived from cell-free extracts of WT or morphant embryos, as indicated. The arrows in C and D indicate changes in the amounts of 60S subunits observed in the stage-matched WT and eif3c morphants at 1 dpf. (E) Quantitation showing the change of the 60S:disome ratios for individual eif3 morphants with respect to the corresponding WT embryos. (F) qPCR data measuring the relative abundance of 18S rRNA and 28S rRNA, as surrogate measures for the changes in abundance of 40S and 60S subunits, respectively.
For comparison, we also analyzed the polysome profiles generated from eif3c morphants at 1 dpf, targeting a bona fide core subunit of eIF3 that is presumed to be required for global mRNA translation. In contrast to the WT and eif3ha morphant profiles, this profile showed a striking increase in the relative ratio of free 60S subunits to polysomes (compare arrows in Fig. 1 C and D and quantification in Fig. 1E). To show that this increase is caused by general defective translation initiation, it was important to demonstrate that the level of 40S subunits also increased in the eif3c morphants. Because the 40S ribosomal subunits did not separate from the top of the gradient, we carried out quantitative RT-PCR (qPCR) to measure the relative levels of 18S and 28S ribosomal RNAs (rRNAs) in the nonpolysomal fractions. Indeed both rRNAs accumulate in the nonpolysomal fractions of the eif3c morphants, compared with the eif3ha morphants and stage-matched WT controls (Fig. 1F). Thus, a significant fraction of free ribosomal subunits is no longer engaged in mRNA translation in eif3c morphants, whereas in eif3ha morphants the abundance of ribosomes engaged in protein synthesis is similar to that observed for WT embryos. These results suggest general inhibition for translation of most of the mRNAs in eif3c morphants and are in agreement with our previous observation (19) that the eif3c morphant embryos are grossly disrupted for embryogenesis compared with the eif3ha morphants. It should be noted that polysome peaks are not eliminated entirely in the eif3c morphant profiles, perhaps as the result of an incomplete loss of zebrafish eIF3c protein (Eif3c) because of maternal contribution or incomplete blocking of the message.
Specific mRNAs Are Depleted from Polysomes in the eif3ha Morphants.
We used deep RNA-seq to compare the relative abundance of polysome-associated translating mRNAs between stage-matched WT and eif3ha morphant embryos. This comparison allowed us to identify at the genome-wide level cohorts of mRNAs that are regulated by Eif3ha for translation. It was important to distinguish transcripts lost from polysomes and those depleted by a transcriptional effect, which would be reflected by an overall loss in the total RNA pool. For this purpose, we defined “the change in translation state” as the ratio (Xmo/Xwt)poly, where Xmo and Xwt represent the abundance, in reads per kilobase per million reads (RPKM) units, of each specific transcript in the polysomal fraction in the morphant (mo) and WT embryos, respectively (see Materials and Methods for details). Likewise, we defined the change in total RNA pool as the ratio (Xmo/Xwt)total, where Xmo and Xwt represent the abundance, in RPKM units, of the same transcript in the total RNA pool in the morphants and WT embryos, respectively. Thus we could calculate quantitatively the change in translational state (∆TS) of a candidate mRNA relative to the total RNA pool, which is defined as the ratio of (Xmo/Xwt)poly to (Xmo/Xwt)total, i.e., [(Xmo/Xwt)poly/(Xmo/Xwt)total], because of Eif3ha loss of function. Thus, the term ∆TS should identify mRNAs primarily regulated by eif3ha at the level of polysome association (translation) by filtering out any mRNAs that are decreased significantly in the total RNA pool (through loss of mRNA transcription, mRNA decay, or cell/tissue degeneration). The characterization of all of the RNA samples used is shown in Table S1. The quality control and alignment results for the read-sequences after RNA-seq analysis are provided in SI Materials and Methods, and Fig. S2.
Transcripts with Decreased Translational State Are Identified in eif3ha Morphants at 1 dpf.
To identify translationally regulated genes, we applied the following criteria: (i) a change in translational state, [(Xmo/Xwt)poly/(Xmo/Xwt)total], ≤ 0.25 (these genes have a fourfold relative loss of transcripts from polysomes in the morphants, normalized to total mRNA); (ii) The abundance of an mRNA in the WT polysomal fraction, (Xwt)poly, ≥1 RPKM unit (this criterion limits analysis to genes that have a significant abundance in the WT polysomes); (iii) the change of an mRNA in the translating polysomal fraction, [(Xmo/Xwt)poly], ≤0.5 (this criterion excludes genes that show a significant increase in the total RNA pool but are not changed in the polysome-associated RNAs).
Using these criteria, we identified ∼300 genes significantly decreased in the translational state (Dataset S1), indicating that their transcripts are markedly depleted from polysomes because of the loss of eif3ha, but their total RNA abundance remained relatively unchanged. We generated three independent biological isolates of WT and morphant polysome fractions to validate the RNA-seq data using independent qPCR experiments for 30 randomly selected genes for which primers could be reliably tested (25 that were decreased in polysomes and five that were not altered significantly, according to the RNA-seq data). As shown in Fig. 2 and Table S2, 100% (all 25) of the polysome-depleted transcripts were validated qualitatively with significant reductions in translation state in all three independent isolates of the eif3ha morphant relative to WT. Likewise, the five randomly chosen genes that were not changed in polysomes of the eif3ha morphant according to RNA-seq data also were fully validated as unchanged by qPCR. Interestingly, when the full gene set depleted from polysomes in the eif3ha morphants was analyzed for gene ontology using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (24, 25), there was a strong correlation with the major expression domains for eif3ha during embryogenesis and the organs/tissues affected in eif3ha morphants, as described in our previous work (Fig. S3 and ref. 19). For example, the majority of transcripts are expressed in the nervous system, including the eye, hindbrain, central ganglion, diencephalon, telencephalon, neural tube, and basal plate midbrain. A significant number of transcripts also are expressed in the somites, where we previously observed a distinct and dynamic expression pattern for eif3ha transcripts (19).
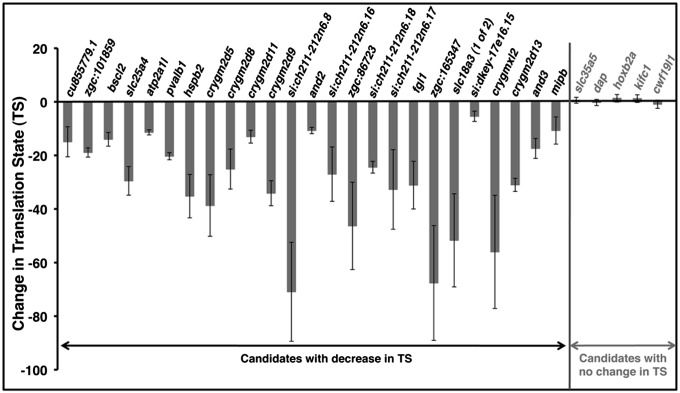
Fig. 2.
Validation of the change in translation state for genes identified through RNA-seq. Shown are qPCR results from three independent biological isolates of both WT and eif3ha morphants for 25 genes that had been found to be decreased in translational state (TS) and five randomly chosen genes that had shown no change in TS. The bars represent the mean and the error bars the SEM. The change in TS has been calculated using the formula [(WT/MO)poly/[(WT/MO)total], where (WT/MO)poly and (WT/MO)total represent the changes of individual mRNAs in polysomal and total RNA, respectively. A negative sign is added arbitrarily to the calculated values to indicate that the translation efficiency is reduced in the morphants relative to WT. Actual data for each independent set, compared with the RNA-seq data, are presented in Table S2.
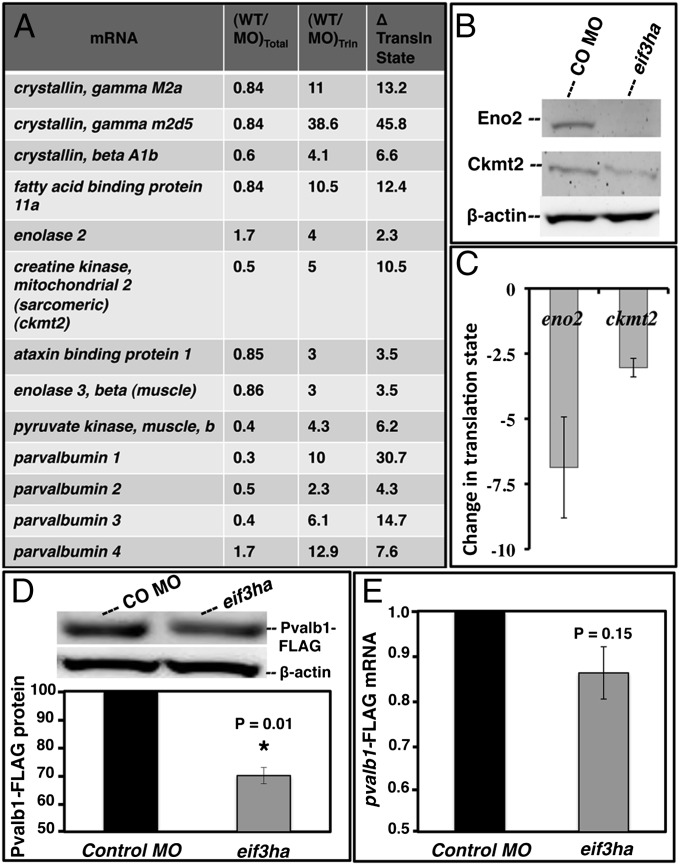
A representative list of transcripts that are most depleted from polysomes in the eif3ha morphant is shown in Fig. 3A. Immunological reagents to quantify zebrafish proteins are limited, but we could identify cross-reacting antibodies for two candidates, enolase 2 (Enos2) and creatine kinase mitochondrial 2 (Ckmt2). Western blotting experiments confirmed depletion of Eno2 and Ckmt2 proteins in the eif3ha morphant embryos (Fig. 3B). Again, independent qPCR experiments confirmed depletion of the corresponding mRNAs from polysomes (Fig. 3C). To test a candidate for which we lacked antibodies, we modified the cDNA encoding Parvalbumin 1 (Pvalb1) to incorporate a FLAG tag to provide an indirect read-out of translational capacity. When this RNA was injected into fertilized eggs, there was a significant reduction of translation by 24 hpf in eif3ha morphants compared with control-injected embryos (P < 0.01) (Fig. 3D). In contrast, the level of pvalb1-FLAG mRNA recovered from these embryos was unchanged (Fig. 3E). In sum, these results provide formal proof that our strategy successfully identified translationally regulated gene sets controlled by eif3ha.
Fig. 3.
Representative candidate mRNAs are specifically deregulated for translation in eif3ha morphant embryos. (A) Representative transcripts that are depleted in the polysome fractions obtained from eif3ha morphants. The complete list is presented in Dataset S1. The respective changes in total RNA [(WT/MO)total], polysomal RNA [(WT/MO)poly], and change in translation state relative to total RNA [(WT/MO)poly/(WT/MO)total] are indicated. (B) Eno2 and Ckmt2 are translationally regulated by eif3ha as shown by Western blotting of cell-free extracts prepared from embryos injected with control morpholino (CO MO) and eif3ha morphants. (C) qPCR analysis confirms a significant change in translational state for eno2 and ckmt2. (D) Representative Western blot and subsequent quantification from three independent experiments following coinjection of pvalb1-FLAG RNA into the control or eif3ha morphant embryos, as indicated. Lysates were isolated at 24 hpf. (E) Corresponding qPCR results of pvalb1-FLAG RNA confirming that total transcript levels are not significantly changed in the morphant.
Given the eventual brain degeneration phenotype of eif3ha morphants, one concern is whether tissue degeneration affects the loss of transcripts in the morphant embryos. However, ckmt2, pvalb1, and eno2 are expressed in tissues that do not show any apparent cell death/tissue degeneration phenotype in the eif3ha morphants at 24 hpf (ckmt2 and pvalb1 are expressed solely in the embryonic somites, and eno2 has overlapping expression in brain and notochord; www.zfin.org). Furthermore, only a subset of brain-associated transcripts shows a significant change in translational state. For example, homeobox B2a (hoxb2a), which serves as a control unchanged gene, is expressed exclusively in the embryonic brain (www.zfin.org). Thus, tissue degeneration is unlikely to have biased our identification of translationally regulated genes controlled by eif3ha.
Transcripts Encoding of Crystallin Gamma 2d, a Cohort of Crystallin Family Isoforms, Are Decreased Significantly in eif3ha Morphant Polysomes.
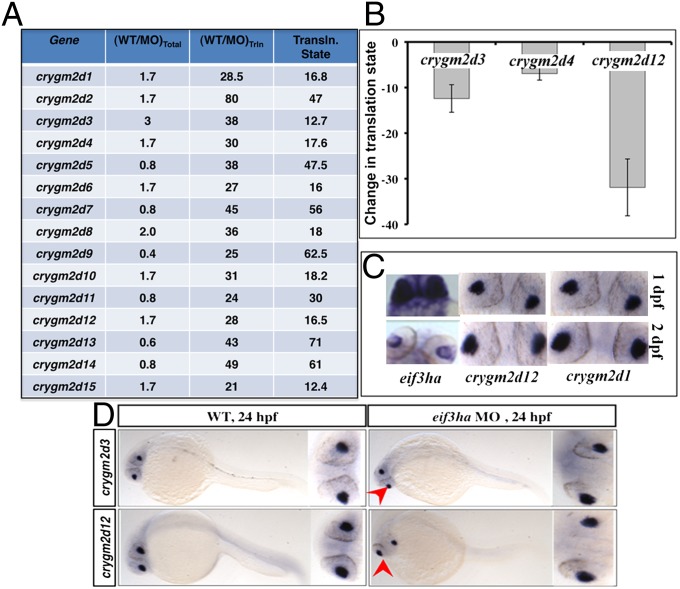
Interestingly, we identified a group of mRNAs encoding crystallin that decreased dramatically in the polysome-associated RNA pool of eif3ha morphants. In this group of transcripts encoding the specific subfamily of crystallin gamma 2d isoforms (crygm2d), there was a 10- to 60-fold decrease in the translational state, according to RNA-seq (Fig. 4A) and as validated in three independent morphant isolates (Fig. 4B). These results indicate that Eif3ha is required for efficient translation of this cohort of lens-specific mRNAs. We documented that eif3ha and at least some of these crystallins are coexpressed in the eye, including at 24 hpf, supporting a common domain for their functional interaction (Fig. 4C). Additionally, we compared the expression patterns of crygm2d3 and crygm2d12 mRNAs in the WT and eif3ha morphants at 24 hpf using in situ hybridization. The results show that these transcript patterns and levels are unaltered in the morphants as compared with the WT control embryos (Fig. 4D), further documenting that the identification of crygm2d transcripts in our polysome-based screen is not caused by a tissue-degeneration artifact of the lens. Taken together, the results obtained from RNA-seq, qPCR, and in situ hybridization show that these candidate mRNAs are specifically depleted from the translating mRNA population but are relatively unaltered in the total RNA pool, thus demonstrating that they are translationally regulated by Eif3ha. We note that a small-eye phenotype is one of the major features of eif3ha morphant embryos (19).
Fig. 4.
Crystallins of the crygm2d family are a cohort of genes that are translationally regulated by eif3ha. (A) Transcripts encoding crygm2d isoforms are shown according to their respective changes in the translation state relative to total RNA. (B) The change in translational state was validated in independent experiments by qPCR for crygm2d3, crygm2d4, and crygm2d12. (C) Representative in situ hybridization experiment showing that crygm2d1, crygm2d12, and eif3ha transcripts are colocalized in the developing lens at 1–2 dpf. (D) Representative in situ hybridization experiment showing that transcript patterns for crygm2d3 and crygm2d12 are unaltered in eif3ha morphants at 24 hpf compared with stage-matched WT controls.
UTR Sequences of crygm2d7 mRNA Mediate Regulation of Translation by eif3ha.
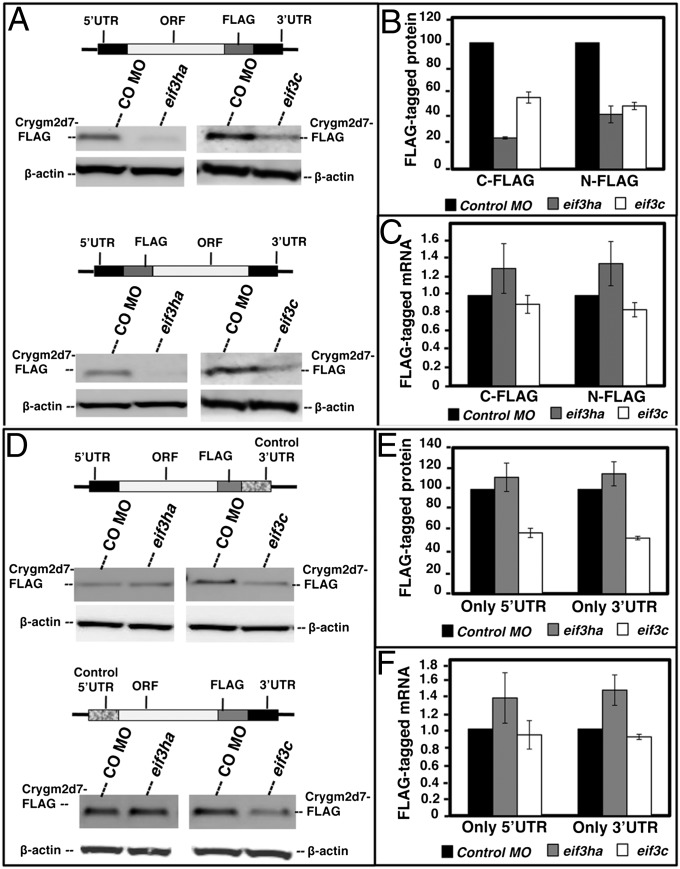
Because specific antibodies for the zebrafish Crygmd2 proteins are unavailable, we again used a tagging strategy to confirm indirectly that translation is regulated by Eif3ha. The sequence of all of the crystallin isoforms is highly conserved, so we chose one candidate mRNA, encoding Crygm2d7, that was associated with a significant decrease in the translational state (Fig. 4A). We generated constructs containing a FLAG tag in frame with the ORF of crygm2d7. The FLAG-crygm2d7 construct was flanked by the 5′ and 3′ UTRs of the crygm2d7 mRNA (Fig. S4A). RNA encoding FLAG-crygm2d7 was generated in vitro and injected into one-cell-stage embryos either with a control MO or with the eif3ha MO to compare the relative synthesis of the protein by subsequent Western blotting, as a measure of translation (Fig. S4B). We designed two constructs with the FLAG tag introduced either C terminally (crygm2d7-FLAG) or N terminally (FLAG-crygm2d7), as shown schematically in Fig. 5A.
Fig. 5.
Injected RNA encoding FLAG-tagged Crygm2d7 is regulated at the translational level by Eif3ha, dependent on UTR sequences. (A) Schematics of C-terminal and N-terminal FLAG-tagged proteins along with the Western blots using anti-FLAG antibodies showing protein levels generated in the control, eif3ha, or eif3c morphant embryos. The endogenous level of β-actin in each lane served as control. (B) Quantitation of the respective Western blots from A. (C) Total RNA was isolated from each set of embryos and subjected to qPCR analysis, as indicated. (D) Schematics of the constructs containing either the 5′ UTR or the 3′ UTR of crygm2d7 along with the respective control β-actin UTR sequences along with Western blots showing the levels of corresponding FLAG-tagged proteins generated in control, eif3ha, or eif3c MO-injected embryos. The endogenous level of β-actin in each lane, served as control. (E) Quantification of the respective Western blots from D. (F) Total RNA was isolated from each set of embryos and subjected to qPCR analysis, as indicated.
After injection of fertilized eggs with MOs and in vitro-generated RNA, the embryos were allowed to develop until ∼24 hpf. Cell-free extracts were prepared, and FLAG-tagged protein was measured by Western blotting experiments using an anti-FLAG antibody. The synthesis of crygm2d7-FLAG and FLAG-crygm2d7 decreased significantly in eif3ha morphants as compared with the control MO-injected embryos (Fig. 5 A and B). As a control, we confirmed that the injected FLAG-mRNA construct is dispersed throughout the developing embryo (Fig. S5A). Thus, the result is not a consequence of selective stability of crystallin transcripts in lens, nor is it associated with potential tissue degeneration in eif3ha morphants. Furthermore, the relative abundance of ectopically injected FLAG-tagged mRNAs did not decrease in the morphants relative to the embryos injected with control MO, as determined by qPCR analysis of total RNA isolated from the respective embryos (Fig. 5C). These results show that decreased abundance of FLAG-tagged protein is not caused by loss of mRNA; rather it occurred because of inhibited synthesis of the tagged proteins in eif3ha morphants.
To test the role of UTR sequences, we replaced either the 3′ UTR or the 5′ UTR with the corresponding UTR sequences derived from the β-actin transcript (Fig. 5D). These elements were selected as controls because the RNA-seq and qPCR data indicate that β-actin is not regulated by eif3ha. The mRNA derived from each of these constructs was microinjected into fertilized eggs with control or eif3ha MOs, and translation was evaluated by anti-FLAG Western blotting experiments. For these constructs, the translation of FLAG-tagged protein was not reduced in the morphant embryos (Fig. 5 D and E). The relative abundance of the corresponding transcripts was equivalent in eif3ha and control morphant embryos (Fig. 5F). When injected into eif3c morphants, all the transcripts, regardless of UTR sequences, showed marked decrease of protein synthesis (Fig. 5), consistent with a general inhibition of translation in the eif3c morphant embryos, as suggested by the polysome profile analysis (Fig. 1). We did not observe a decrease in endogenous β-actin protein levels even in the eif3c morphant embryos, presumably because of a major maternal contribution.
Finally, the crygm2d7 5′ and 3′ UTR sequences were used to flank the coding sequence of the luciferase protein. When injected into fertilized eggs, no significant difference in luciferase activity was measured in control or eif3ha morphants (Fig. S5B), indicating that the UTR sequences were not sufficient to transfer eif3ha-dependent translation efficiency to a donor ORF. Therefore, eif3ha-mediated regulation of Crygm2d7 translation requires the participation of both native 5′- and 3′-flanking UTR sequences and also may require some internal sequences for enhanced recruitment into or detainment with the translationally active polysome complex. There already are examples of ORF sequences contributing to translational regulation of mRNAs in addition to the UTR [e.g., Nanos mRNA regulation during early development in Xenopus (26)]. It should be emphasized that, in addition to similar 5′ and 3′ UTR sequences, the mRNAs encoding crygm2d7 isoforms have markedly similar coding regions, and all these candidate mRNAs showed a significant decrease in the translation state in eif3ha morphants.
Discussion
In mammalian cells, the rates of initiation of translation from different sets of mRNAs are distinct, and the structure and/or the sequence features of the 5′ UTR are critical determinants of the relative efficiency of scanning (2, 27). The mRNA-specific translation rates by eIF3 might be governed by the differential efficiency with which eIF3-bound preinitiation complexes are able to scan through the 5′ UTR of a particular set of mRNAs, governed by the association of specific nonconserved subunits with the eIF3 conserved core protein complex. This notion predicts that regulatory codes are present in the 5′ UTRs and/or in the 3′ UTRs, because the translating mRNA is thought to be functionally circularized (28) and thus the 3′ UTR also interacts with the translation initiation machinery. Comparisons of the polysome profile patterns obtained from WT, eif3ha, and eif3c morphant embryos are consistent with a predicted regulatory role for eIF3h and a general or core role for eIF3c.
Because of the depletion of eif3ha, a dramatic change occurred for a group of mRNAs encoding the eye lens protein crystallin; specifically, ∼20 isoforms of the crygm2d family were depleted by at least 10- to 50-fold from the polysome-associated translating transcripts. Furthermore, the synthesis of tagged Crygm2d7 protein, a representative isoform of the family, is inhibited when the mRNA is injected into developing eif3ha morphants. Thus, this cohort of crystallins is positively regulated by eif3ha at the level of translation during eye development, as is consistent with our previous observation that failure in eye development is a major feature of eif3ha morphants (19). Additionally, these crystallin and eif3ha mRNAs are coexpressed in the developing lens, suggesting a common domain for their functional interaction. It appears that the crystallins are regulated as a cohort of mRNAs by a conserved translational mechanism through eif3ha. This level of control may be important for the precise integration of crystallin subunits into the lens matrix. Interestingly, a recent study (29) contradicted the long-standing notion that gene expression is regulated in the vertebrate lens predominantly at the level of transcription. Instead, a posttranscriptional mechanism mediated by cytoplasmic RNA granules was identified that could regulate subcellular localization and subsequent processing of lens mRNAs. Our study also supports posttranscriptional control in lens development mediated by translational regulation and provides mechanistic insight into how this regulation can be achieved through the translational initiation machinery. Our study suggests that this regulation can occur only when both the natural 5′ and 3′ UTRs are associated with the ORF of crygm2d7 mRNA. In this regard, it should be emphasized that both the 5′ and 3′ UTRs of the members of this family of crystallin mRNAs are highly conserved. It seems quite likely that such translational control mechanisms may occur in a wide variety of cell types during all stages of development, depending on the presence of eIF3h-specific regulatory codes in the UTRs of the mRNAs in those tissues. We note that many transcripts depleted from the polysomes show a concomitant increase in the total RNA pool. This increase might be caused by other biological processes that impact RNA abundance as the result of eif3ha loss. For example, developmentally important genes might activate compensatory mechanisms by a feedback mechanism. Given the complexities of such a dynamic system, altered transcript levels caused by a change in translation have been documented in previous studies (15, 30, 31).
Finally, our approach validates a useful general strategy for investigating the role of the other noncore (regulatory) eIF3 subunits or any putative translational regulatory factor in vertebrate development and for defining the capacity of these proteins for recruitment of specific mRNAs during translation initiation. Also, an attractive hypothesis is that distinct forms of eIF3 may exist, each containing all the conserved subunits but differing in their composition of the nonconserved subunits and thus giving rise to “eIF3-heterogeneity.” This heterogeneity, in turn, may depend on the type of nonconserved subunit being expressed in a particular type of tissue. In fact, the presence of distinct forms of eIF3 in fission yeast that differ in their composition of the nonconserved subunits has been reported (10, 11). It is possible that each distinct eIF3 protein complex containing a unique combination of nonconserved subunits might target specific as well as overlapping cohorts of transcripts to the polysomes for translation and that comparison of these transcripts, once identified, will enable the resolution of novel regulatory codes. In support of this hypothesis, a recent study (32) showed that RPL38, a protein component of the large ribosomal subunit 60S, influences translation of a specific subset of HOX mRNAs and highlighted the concept of ribosome heterogeneity through striking tissue-specific enrichment of different ribosomal proteins during vertebrate tissue patterning. Recent advances in ribosome footprinting (31, 33–35) and RNA-immunoprecipitation (36) have shown the potential for many levels of previously unappreciated control mechanisms. By applying classic biochemical assays coupled with next-generation sequencing technology to a well-established developmental system, it should be possible to probe deeply for the discovery of novel gene regulatory mechanisms.
Materials and Methods
Polysome Profiling.
Details on fish husbandry, isolation of polysomes, RNA sequencing, qPCR validation, and luciferase assays are provided in SI Materials and Methods. RNA-seq datasets are deposited in the Gene Expression Omnibus (accession GSE44584).
Gene-Expression Analysis.
The expression patterns of crystallin mRNAs were determined by in situ hybridization as described previously (37). The primer sequences used to PCR-amplify the probe sequences are given in SI Materials and Methods. For detection of endogenous protein levels of Eno2 and Ckmt2, cell-free extracts were prepared as described for polysome isolation. Rabbit antibodies specific for each of these zebrafish proteins (Aviva Systems Biology) were used at 1:1,000 dilution. Subsequently, an HRP-conjugated anti-rabbit secondary antibody was used to develop the signals.
Analysis of cis-Elements.
Details for construction and analysis of crygm2d7 constructs (Fig. S6) are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank F. Campagne for Gobyweb software to analyze raw RNA-seq data; T.-C. Liu for cDNA library construction; G. Rosenfeld for help with RNA-seq analysis; J. Warner, K. B. McIntosh, and A. Bhattacharya for help and suggestions during our use of their gradient fractionator; and F. Marlow for helpful suggestions. We also thank the Weill Cornell Medical College Genomics Facility for quality control and sequencing of the cDNA libraries. T.E. is supported by National Institutes of Health (NIH) Grants HL111400 and HL056182. U.M. is supported by NIH Grants GM15399 and P30CA13330.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE44584).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1302934110/-/DCSupplemental.
References
- 1.Kapp LD, Lorsch JR. The molecular mechanics of eukaryotic translation. Annu Rev Biochem. 2004;73:657–704. doi: 10.1146/annurev.biochem.73.030403.080419. [DOI] [PubMed] [Google Scholar]
- 2.Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234(2):187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- 3.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11(2):113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hinnebusch AG. eIF3: A versatile scaffold for translation initiation complexes. Trends Biochem Sci. 2006;31(10):553–562. doi: 10.1016/j.tibs.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 5.Akiyoshi Y, et al. Fission yeast homolog of murine Int-6 protein, encoded by mouse mammary tumor virus integration site, is associated with the conserved core subunits of eukaryotic translation initiation factor 3. J Biol Chem. 2001;276(13):10056–10062. doi: 10.1074/jbc.M010188200. [DOI] [PubMed] [Google Scholar]
- 6.Bandyopadhyay A, Matsumoto T, Maitra U. Fission yeast Int6 is not essential for global translation initiation, but deletion of int6(+) causes hypersensitivity to caffeine and affects spore formation. Mol Biol Cell. 2000;11(11):4005–4018. doi: 10.1091/mbc.11.11.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yen HC, Chang EC. Yin6, a fission yeast Int6 homolog, complexes with Moe1 and plays a role in chromosome segregation. Proc Natl Acad Sci USA. 2000;97(26):14370–14375. doi: 10.1073/pnas.97.26.14370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crane R, et al. A fission yeast homolog of Int-6, the mammalian oncoprotein and eIF3 subunit, induces drug resistance when overexpressed. Mol Biol Cell. 2000;11(11):3993–4003. doi: 10.1091/mbc.11.11.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bandyopadhyay A, Lakshmanan V, Matsumoto T, Chang EC, Maitra U. Moe1 and spInt6, the fission yeast homologues of mammalian translation initiation factor 3 subunits p66 (eIF3d) and p48 (eIF3e), respectively, are required for stable association of eIF3 subunits. J Biol Chem. 2002;277(3):2360–2367. doi: 10.1074/jbc.M107790200. [DOI] [PubMed] [Google Scholar]
- 10.Zhou C, et al. PCI proteins eIF3e and eIF3m define distinct translation initiation factor 3 complexes. BMC Biol. 2005;3:14. doi: 10.1186/1741-7007-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ray A, Bandyopadhyay A, Matsumoto T, Deng H, Maitra U. Fission yeast translation initiation factor 3 subunit eIF3h is not essential for global translation initiation, but deletion of eif3h+ affects spore formation. Yeast. 2008;25(11):809–823. doi: 10.1002/yea.1635. [DOI] [PubMed] [Google Scholar]
- 12.Kim TH, Kim BH, Yahalom A, Chamovitz DA, von Arnim AG. Translational regulation via 5′ mRNA leader sequences revealed by mutational analysis of the Arabidopsis translation initiation factor subunit eIF3h. Plant Cell. 2004;16(12):3341–3356. doi: 10.1105/tpc.104.026880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim BH, Cai X, Vaughn JN, von Arnim AG. On the functions of the h subunit of eukaryotic initiation factor 3 in late stages of translation initiation. Genome Biol. 2007;8(4):R60. doi: 10.1186/gb-2007-8-4-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roy B, et al. The h subunit of eIF3 promotes reinitiation competence during translation of mRNAs harboring upstream open reading frames. RNA. 2010;16(4):748–761. doi: 10.1261/rna.2056010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grzmil M, et al. An oncogenic role of eIF3e/INT6 in human breast cancer. Oncogene. 2010;29(28):4080–4089. doi: 10.1038/onc.2010.152. [DOI] [PubMed] [Google Scholar]
- 16.Sonenberg N, Hinnebusch AG. New modes of translational control in development, behavior, and disease. Mol Cell. 2007;28(5):721–729. doi: 10.1016/j.molcel.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 17.Groisman I, Huang YS, Mendez R, Cao Q, Richter JD. Translational control of embryonic cell division by CPEB and maskin. Cold Spring Harb Symp Quant Biol. 2001;66:345–351. doi: 10.1101/sqb.2001.66.345. [DOI] [PubMed] [Google Scholar]
- 18.Curtis D, Lehmann R, Zamore PD. Translational regulation in development. Cell. 1995;81(2):171–178. doi: 10.1016/0092-8674(95)90325-9. [DOI] [PubMed] [Google Scholar]
- 19.Choudhuri A, Evans T, Maitra U. 2010. Non-core subunit eIF3h of translation initiation factor eIF3 regulates zebrafish embryonic development. Dev Dyn 239(6):1632–1644.
- 20.Maiti T, Maitra U. Characterization of translation initiation factor 5 (eIF5) from Saccharomyces cerevisiae. Functional homology with mammalian eIF5 and the effect of depletion of eIF5 on protein synthesis in vivo and in vitro. J Biol Chem. 1997;272(29):18333–18340. doi: 10.1074/jbc.272.29.18333. [DOI] [PubMed] [Google Scholar]
- 21.Warner JR, Knopf PM, Rich A. A multiple ribosomal structure in protein synthesis. Proc Natl Acad Sci USA. 1963;49:122–129. doi: 10.1073/pnas.49.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Si K, Maitra U. The Saccharomyces cerevisiae homologue of mammalian translation initiation factor 6 does not function as a translation initiation factor. Mol Cell Biol. 1999;19(2):1416–1426. doi: 10.1128/mcb.19.2.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ray P, et al. The Saccharomyces cerevisiae 60 S ribosome biogenesis factor Tif6p is regulated by Hrr25p-mediated phosphorylation. J Biol Chem. 2008;283(15):9681–9691. doi: 10.1074/jbc.M710294200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 26.Luo X, Nerlick S, An W, King ML. Xenopus germline nanos1 is translationally repressed by a novel structure-based mechanism. Development. 2011;138(3):589–598. doi: 10.1242/dev.056705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Child SJ, Miller MK, Geballe AP. Translational control by an upstream open reading frame in the HER-2/neu transcript. J Biol Chem. 1999;274(34):24335–24341. doi: 10.1074/jbc.274.34.24335. [DOI] [PubMed] [Google Scholar]
- 28.Wells SE, Hillner PE, Vale RD, Sachs AB. Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell. 1998;2(1):135–140. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- 29.Lachke SA, et al. Mutations in the RNA granule component TDRD7 cause cataract and glaucoma. Science. 2011;331(6024):1571–1576. doi: 10.1126/science.1195970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sampath P, et al. A hierarchical network controls protein translation during murine embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2008;2(5):448–460. doi: 10.1016/j.stem.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 31.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147(4):789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kondrashov N, et al. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell. 2011;145(3):383–397. doi: 10.1016/j.cell.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324(5924):218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brar GA, et al. High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science. 2012;335(6068):552–557. doi: 10.1126/science.1215110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bazzini AA, Lee MT, Giraldez AJ. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science. 2012;336(6078):233–237. doi: 10.1126/science.1215704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hafner M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141(1):129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alexander J, Stainier DY, Yelon D. Screening mosaic F1 females for mutations affecting zebrafish heart induction and patterning. Dev Genet. 1998;22(3):288–299. doi: 10.1002/(SICI)1520-6408(1998)22:3<288::AID-DVG10>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.