Abstract
When macroautophagy, a catabolic process that rids the cells of unwanted proteins, is initiated, 30–60 nm Atg9 vesicles move from the Golgi to the preautophagosomal structure (PAS) to initiate autophagosome formation. The Rab GTPase Ypt1 and its mammalian homolog Rab1 regulate macroautophagy and two other trafficking events: endoplasmic reticulum-Golgi and intra-Golgi traffic. How a Rab, which localizes to three distinct cellular locations, achieves specificity is unknown. Here we show that transport protein particle III (TRAPPIII), a conserved autophagy-specific guanine nucleotide exchange factor for Ypt1/Rab1, is recruited to the PAS by Atg17. We also show that activated Ypt1 recruits the putative membrane curvature sensor Atg1 to the PAS, bringing it into proximity to its binding partner Atg17. Since Atg17 resides at the PAS, these events ensure that Atg1 will specifically localize to the PAS and not to the other compartments where Ypt1 resides. We propose that Ypt1 regulates Atg9 vesicle tethering by modulating the delivery of Atg1 to the PAS. These events appear to be conserved in higher cells.
Keywords: GEF, membrane tethering
Defects in macroautophagy have been linked to a variety of human diseases including neurodegenerative diseases such as Parkinson disease (1). When macroautophagy is induced, 30–60 nm motile vesicles, which contain the transmembrane protein Atg9, are sorted from the Golgi and translocated to an assembly of ATG (autophagy-related) gene products at a perivacuolar structure called the preautophagosomal structure (PAS). At the PAS, approximately three Atg9 vesicles tether and fuse to become part of the phagophore or isolation membrane (2). The phagophore, which also expands from other compartments such as the endoplasmic reticulum (ER) and mitochondria (3), matures into an autophagosome. The autophagosome is a double-membrane structure that seals its contents from the cytosol and delivers them to the lysosome or vacuole for degradation (4).
The mechanism by which Atg9 vesicles tether and fuse to become part of the phagophore is an important unanswered question in the autophagy field. The recruitment of Atg9 to the PAS is dependent on Atg17 (5), the scaffold protein that organizes the other Atg proteins at the phagosome assembly site (6). Atg17 forms a stable complex with Atg29 and Atg31 (7, 8). When autophagy is induced, Atg1 and Atg13 associate with Atg17, Atg29, and Atg31 to form the Atg1 complex, which is required for phagophore assembly (9). Atg1 is a serine/threonine kinase that is fully active when it interacts with Atg17 and Atg13 (10). Recent studies have shown that Atg1, which is critical for phagophore initiation, is a putative membrane curvature sensor that tethers liposomes in vitro (8). Two pools of Atg1 have been described in the literature. One pool is associated with the Atg17 complex, while the larger pool is not (7).
Rab GTPases are molecular switches that regulate membrane traffic. They are active in their GTP-bound form and inactive when bound to GDP (11). The GTPase Ypt1, and its mammalian homolog Rab1, regulate membrane-tethering events on three different pathways: autophagy, ER-Golgi, and intra-Golgi traffic (12). In the yeast Saccharomyces cerevisiae, many of the Atg proteins needed for macroautophagy are shared with the biosynthetic cytoplasm to vacuole targeting (Cvt) pathway that transports certain hydrolases into the vacuole (13). Both pathways require Ypt1, however, only the macroautophagy pathway is conserved in higher eukaryotes (14, 15). Here we focus on the role of Ypt1 on the conserved macroautophagy pathway and show that it mediates the recruitment of Atg1 to the PAS. Our findings suggest that Ypt1 regulates macroautophagy by recruiting its effector, Atg1, to the PAS to tether Atg9 vesicles to each other or to other membranes.
Results
Trs85 Is a Component of a Transport Protein Particle III Complex in Mammalian Cells.
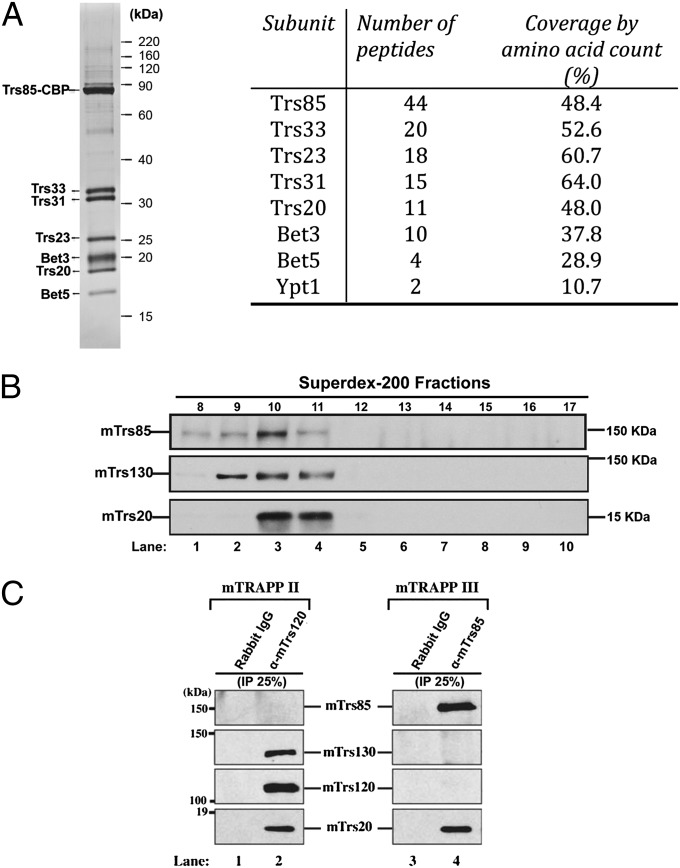
Previous studies demonstrated that the Ypt1 guanine nucleotide exchange factor (GEF), transport protein particle III (TRAPPIII), specifically activates Ypt1 on the autophagy pathway (14). TRAPPIII shares six subunits with two other Ypt1 GEFs, TRAPPI and TRAPPII, and contains one unique subunit, Trs85 (TRAPP subunit 85). Earlier studies did not definitively address if the TRAPPIII complex contains additional subunits (14). To address this question, we TAP (tandem affinity purification)-tagged Trs85 from yeast and analyzed the purified TRAPPIII complex by SDS gel electrophoresis and mass spectrometry. This analysis identified Trs85, Trs33, Trs31, Trs23, Bet3 (blocked early in transport 3), Trs20, and Bet5 as the only major subunits in the complex (Fig. 1A, Left). Mass spectrometry also identified Ypt1, which is known to bind TRAPPIII (Fig. 1A, Right).
Fig. 1.
TRAPPIII contains seven subunits and is conserved from yeast to man. (A, Left) Trs85–TAP was purified from yeast as described before (30). Purified TAP-tagged Trs85 yields a complex that contains seven major subunits. (Right) Mass spectrometry analysis of purified TAP-tagged Trs85 from yeast. (B) NIH 3T3 lysate was fractionated on a Superdex-200 column as described in Materials and Methods and immunoblotted for mTrs85, mTrs130, and mTrs20. (C) mTrs120 (Left) or mTrs85 (Right) were immunoprecipitated from lysates, and the immunoprecipitates were blotted for the presence of mTrs85, mTrs130, mTrs120, and mTrs20. Similar results were obtained when HeLa or COS-7 cells were used for the precipitations.
It was recently reported that depletion of the mammalian homolog of Trs85 (mTrs85) leads to a reduction in autophagosome formation (16), however, to date, only one TRAPP complex has been described in mammals (17, 18). Since the roles of Ypt1 and its homolog Rab1 appear to be highly conserved from yeast to man (12), we asked if mTrs85 is a component of a TRAPP complex that is analogous to yeast TRAPPIII. When we fractionated lysate prepared from NIH 3T3 cells on a Superdex-200 column, mTrs85 peaked in fraction 10, whereas mTrs130 was in a broader peak (fractions 9, 10, and 11) (Fig. 1B), suggesting that mTrs130 and mTrs85 might be in separate complexes. mTrs130 is a component of the TRAPPII complex, a Rab1 GEF that regulates Golgi traffic (19). TRAPPII shares six subunits with TRAPPIII and contains three specific subunits (Trs130, Trs120, and Trs65) that target it to the Golgi (19–21). When the TRAPPII complex was precipitated with anti-mTrs120 antibody, all TRAPP subunits coprecipitated with mTrs120 (Fig. 1C, Left), except for mTrs85. In contrast, anti-mTrs85 antibody precipitated mTrs20, but not mTrs130 and mTrs120 (Fig. 1C, Right). Together these findings show that mTrs85 is in a distinct TRAPP complex that includes mTrs20, but not TRAPPII-specific subunits. Thus, as was observed for yeast, mammalian cells appear to have a distinct TRAPPIII complex that activates Rab1 on the autophagy pathway.
Recruitment of Trs85 to the PAS Is Dependent on Atg17.
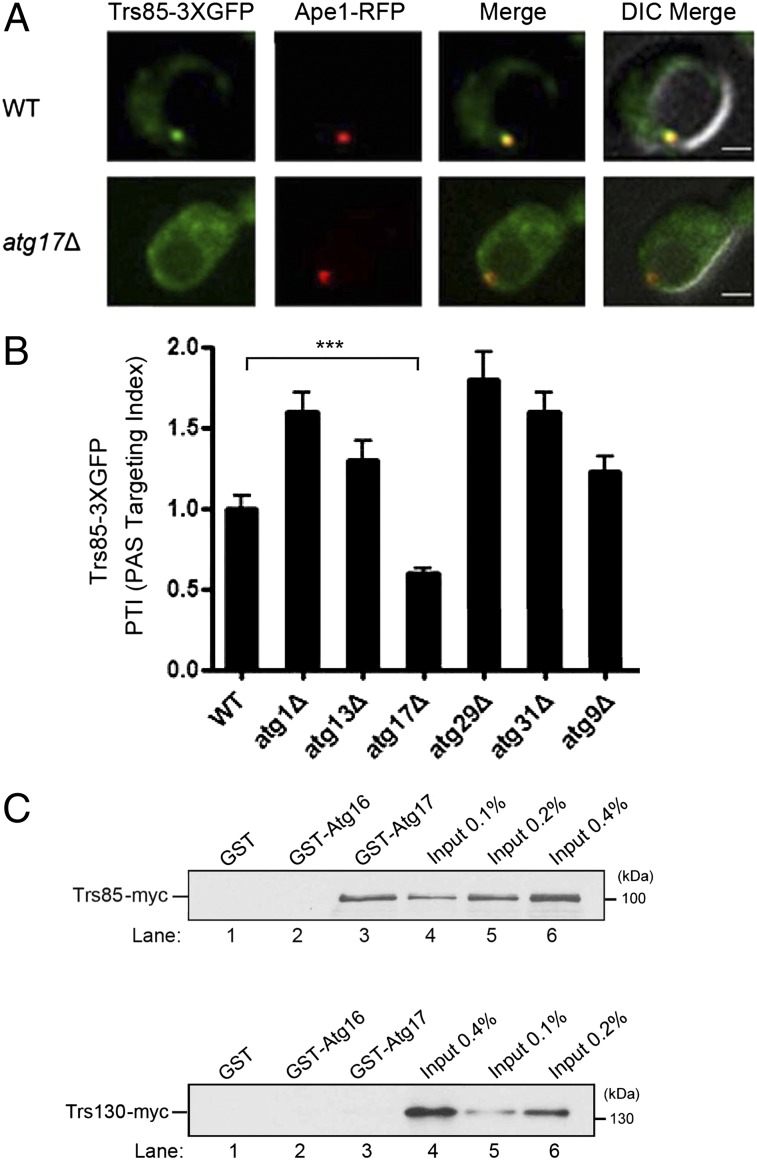
To begin to address where TRAPPIII and Ypt1 act on the conserved macroautophagy pathway (herein referred to as autophagy), we screened deletions of the known ATG genes that act on this pathway for defects in the recruitment of TRAPPIII to the PAS. We induced autophagy in the 22 atg mutants known to block this process, using a previously characterized construct (14), and examined the recruitment of the endogenous copy of Trs85 (Trs85–3XGFP) to the PAS. For this screen, the PAS was marked by the cargo protein amino peptidase I (Ape1) fused to red fluorescent protein (RFP) (Fig. S1A). A quantitative analysis revealed that Trs85–3XGFP was only mislocalized in the atg17Δ mutant, but not the other atg mutants (Fig. S1A). This result was confirmed in our strain background (Fig. 2 A and B). Like the Atg proteins, Trs85 accumulated at the PAS when autophagosome formation was blocked (6), however in the atg17Δ mutant, a defect in the recruitment of Trs85 was observed (Fig. 2 A and B). Consistent with the notion that Atg17 acts upstream of TRAPPIII, the recruitment of Atg17–GFP to the PAS was not decreased in the trs85Δ mutant (Fig. S1B). Although it has been suggested that Atg9 recruits TRAPPIII to the PAS (22), we could not detect a defect in the recruitment of Trs85–3XGFP to the PAS in the atg9Δ mutant. This analysis was done in our strain background (Fig. 2B) as well as two other strain backgrounds (Fig. S1A) (14). To test if Trs85 recruits TRAPPIII to the PAS via an interaction with Atg17, in vitro binding experiments were performed. As shown in Fig. 2C, Top, Trs85–myc bound specifically to GST–Atg17, but not GST or GST–Atg16. Additionally, no binding of Trs130–myc (Fig. 2C, Bottom) was seen. Together, these findings imply that TRAPPIII acts in the induction of the pathway and suggest that TRAPPIII binds directly or indirectly to Atg17.
Fig. 2.
The localization of Trs85–3XGFP to the PAS is disrupted in the atg17Δ mutant. (A) Cells expressing Trs85–3XGFP and Ape1–RFP were grown to log phase in synthetic complete (SC)–Leu medium, pelleted, and resuspended in SD-N medium for 4 h. (Scale bar, 2 μm.) (B) Quantitation of the data in A, and several other atg mutants, in our laboratory strain background, which is derived from S288C. The PAS targeting index (PTI) was calculated in 50 cells by multiplying the percent of cells that contain colocalized Trs85–3XGFP and Ape1–RFP to the total Trs85–3XGFP signal that resides at the PAS. The PTI in wild-type was set at 1.00. Error bars represent SEM, n = 150 cells from three separate experiments. ***P < 0.001 Student t test. (C) Trs85 specifically binds to Atg17. Yeast lysate containing Trs85–myc (Top) or Trs130–myc (Bottom) was incubated with glutathione-Sepharose beads. Bound protein was eluted and analyzed.
Ypt1 Regulates the Recruitment of Atg1 to the PAS.
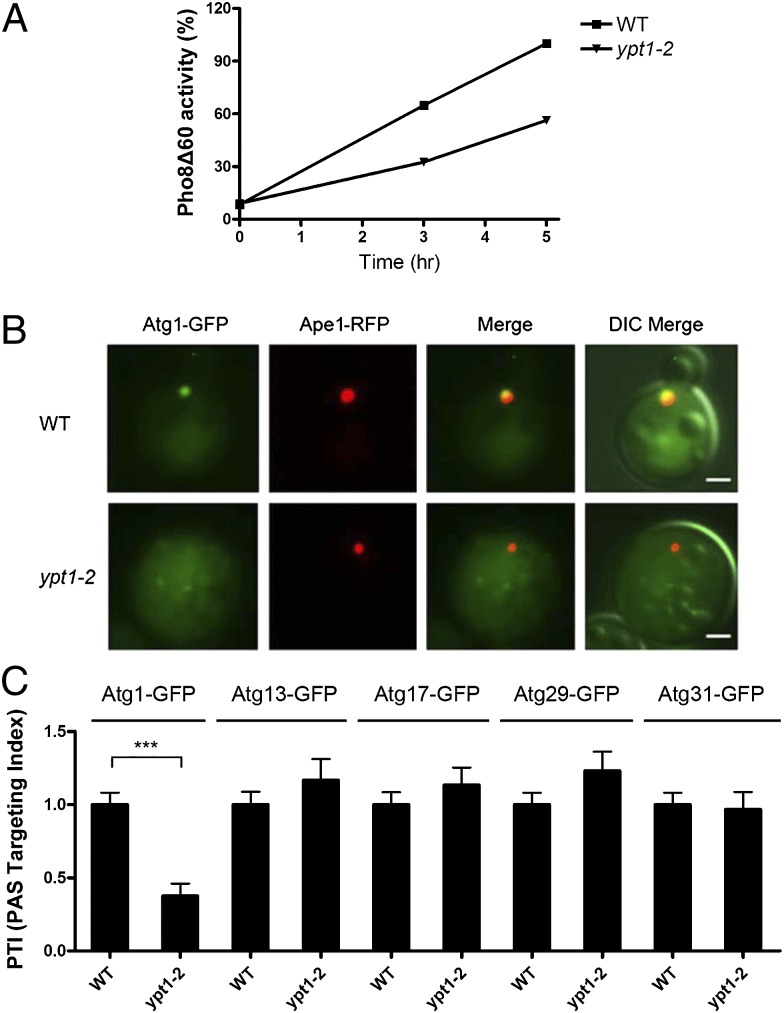
To address if Ypt1 regulates the recruitment of one or more of the five Atg proteins that initiate autophagy, we induced autophagy by nitrogen starvation and then examined the localization of chromosomally expressed Atg1–GFP, Atg13–GFP, Atg17–GFP, Atg29–GFP, and Atg31–GFP in the ypt1–2 mutant. We chose the ypt1–2 mutant for our analysis because it delays autophagy at 25 °C (Fig. 3A), but has no effect on secretion in vivo (23). Interestingly, only Atg1–GFP was mislocalized in this mutant (Fig. 3 B and C). Under nutrient-rich conditions, no significant defect in the localization of Atg1–GFP was observed (Fig. S1D), suggesting that in this mutant allele mislocalization is dependent on the induction of macroautophagy. Additionally, the defect in the recruitment of Atg1 appeared to be specific for Ypt1, as no defect was observed in the Rab mutant ypt51Δ (Fig. S1C).
Fig. 3.
The recruitment of Atg1 to the PAS is reduced in the ypt1–2 mutant. (A) Cells were grown at 25 °C and shifted to SD-N medium for 0, 3, and 5 h. Protein extracts were assayed for vacuolar alkaline phosphatase activity as described in Materials and Methods. (B) The localization of Atg1–GFP to the PAS is disrupted in the ypt1–2 mutant. Wild-type and ypt1–2 cells expressing Atg1–GFP and Ape1–RFP were grown to log phase and shifted to SD-N medium as in Fig. 2. (Scale bar, 2 μm.) (C) The PAS localization of Atg13–GFP, Atg17–GFP, Atg29–GFP, and Atg31–GFP is not disrupted in the ypt1–2 mutant. Cells were grown and analyzed as in B. The PTI in wild-type was set at 1.00. Error bars represent SEM, n = 150 cells from three separate experiments. ***P < 0.001 Student t test. The slight increase seen with Atg13–GFP, Atg17–GFP, and Atg29–GFP is not statistically significant.
The ATG genes have been mapped into a hierarchy of six groups of Atg proteins that are required for autophagosome formation (Fig. S2A) (4, 6). When we fused the Atgs in the other five groups to GFP and examined their localization in the ypt1–2 mutant, none were mislocalized (Fig. S2B). These findings suggest that the delay in autophagy in the ypt1–2 mutant is due to a decrease in the recruitment of Atg1 to the PAS.
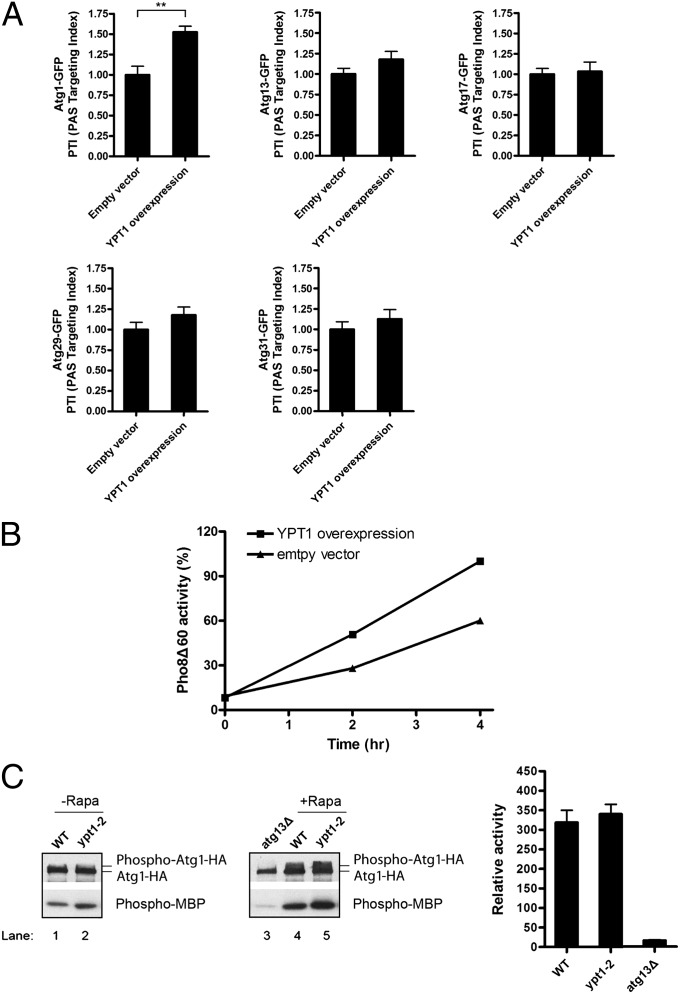
In contrast to what we observed for ypt1–2, the overexpression of Ypt1 increased the percentage of cells that contained Atg1–GFP at the PAS and augmented autophagy (Fig. 4 A and B). The increase in PAS recruitment was specific for Atg1, as no significant increase in the recruitment of Atg13, Atg17, Atg29, or Atg31 was observed when Ypt1 was overexpressed (Fig. 4A). Because the substrates of Atg1 are unknown, the effect of Ypt1 on kinase activity was assessed by analyzing the phosphorylation of myelin basic protein (MBP). As shown in Fig. 4C, Atg1 kinase activity was unaltered in the ypt1–2 mutant. Together these findings indicate that when autophagy is induced, Ypt1 recruits Atg1, but not its regulators (Atg13 and Atg17), to the PAS.
Fig. 4.
The overexpression of Ypt1 increases the recruitment of Atg1–GFP to the PAS. (A) Autophagy was induced as described in Fig. 2 in cells harboring Atg1–GFP and a 2 µm vector, without or with YPT1, and the recruitment of Atg1–GFP, Atg13–GFP, Atg17–GFP, Atg29–GFP, and Atg31–GFP to the PAS was measured. Error bars represent SEM, n = 150 cells from three separate experiments. **P < 0.01 Student t test. (B) YPT1 overexpression enhances autophagy. Cells were grown in SC–Ura medium at 25 °C and shifted to SD-N medium for 0, 2, and 4 h. Protein extracts were assayed for vacuolar alkaline phosphatase activity. (C) Atg1 kinase activity is not affected in the ypt1–2 mutant. Wild-type and ypt1–2 cells were treated for 30 min at 30 °C without (Left) or with rapamycin (Center) and assayed for activity using MBP as substrate. The atg13Δ mutant was used as a negative control. The data in the center panel are quantitated on the right. Error bars represent SD, n = 3 from three separate experiments.
Atg1 Interacts with Ypt1 in Yeast and Mammalian Cells.
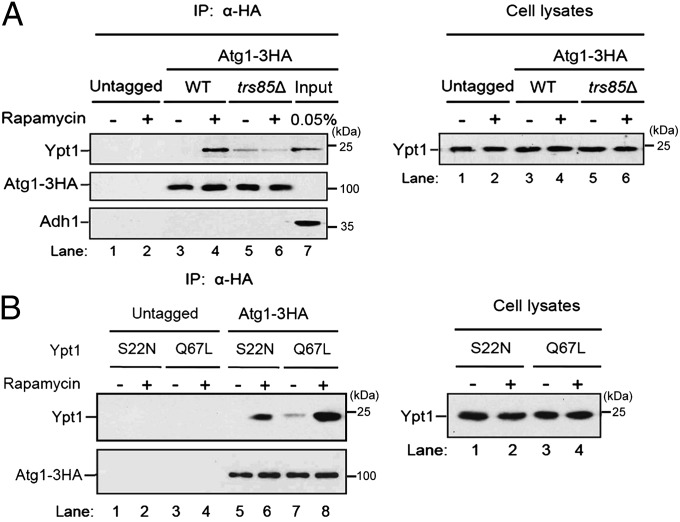
To begin to address if Atg1 interacts with activated Ypt1 when autophagy is induced, we removed the cell wall from wild-type and trs85Δ cells and induced autophagy in spheroplasts with rapamycin. Interestingly, Ypt1 coprecipitated with Atg1–HA from wild-type but not trs85Δ cells, suggesting that the coprecipitation of Atg1 with Ypt1 required activated Ypt1. In our strain background this interaction was enhanced in rapamycin-treated spheroplasts (Fig. 5A, Left, lanes 3–6). The absence of Ypt1 in the trs85Δ precipitate did not appear to be due to decreased levels of Ypt1 (Fig. 5A, Right). Additionally, the interaction between Atg1–HA and Ypt1 was specific since no Ypt1 was precipitated from lysates that lacked the HA tag (Fig. 5A, Left, lanes 1 and 2), and neither the cytosolic protein Adh1 (Fig. 5A, Lower) nor the Rab Ypt32 (Fig. S3A) was detected in the precipitate. To directly address if the coprecipitation of Atg1 and Ypt1 is dependent on activated Ypt1, we constructed strains in which Ypt1 was locked in its GTP-bound (Q67L) or GDP-bound (S22N) form and precipitated Atg1–HA from lysates prepared from these strains. Although the expression of GTP- and GDP-locked Ypt1 was the same in yeast lysates (Fig. 5B, Right), Atg1–HA preferentially coprecipitated with the activated form of Ypt1 (Fig. 5B, Left and Fig. S3B).
Fig. 5.
The interaction between Atg1 and activated Ypt1 is enhanced in rapamycin-treated cells. (A) Atg1–HA was integrated into wild-type or trs85Δ cells. (Left) Lysed spheroplasts were treated with or without rapamycin, immunoprecipitated with anti-HA antibody, and immunoblotted. (Right) Lysates were immunoblotted with anti-Ypt1 antibody. (B) Atg1–HA preferentially coprecipitates with the GTP form of Ypt1. (Left) Cells expressing the GTP (Q67L) or GDP (S22N) form of Ypt1 were converted to spheroplasts, treated with rapamycin, and processed as in A. (Right) Lysates were immunoblotted with anti-Ypt1 antibody.
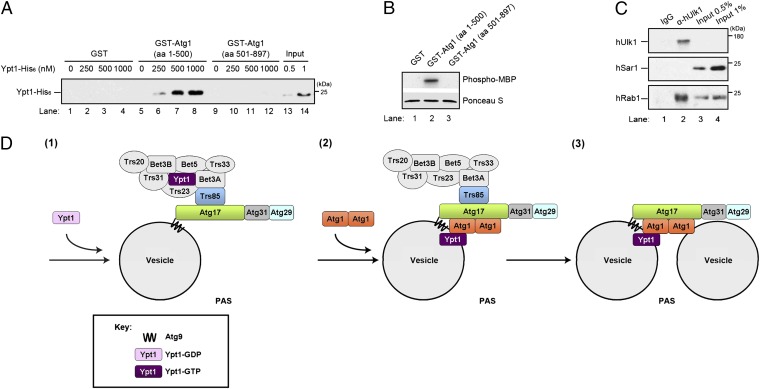
To address if Atg1 binds directly to Ypt1, we performed in vitro binding studies with the amino (amino acids 1–500) or carboxy (amino acids 501–897) terminus of Atg1 fused to GST. The N terminus of Atg1 contains the kinase domain, while the C terminus contains the Atg13 binding domain (24, 25). GST, GST–Atg1 (1–500), and GST–Atg1 (501–897), purified from bacteria and immobilized on glutathione beads, were incubated with increasing concentrations of Ypt1–His6 (Fig. 6A). Ypt1–His6 bound specifically to GST–Atg1 (1–500), which contains a functional kinase domain (Fig. 6B), but not to GST or GST–Atg1 (501–897). These findings indicate that Ypt1 binds directly to the N terminus of Atg1.
Fig. 6.
Ypt1 binds to the N terminus of Atg1. (A) Purified Ypt1–His6 was incubated for 3 h at 4 °C with equimolar amounts (0.1 µM) of immobilized GST, GST–Atg1 (aa 1–500), or GST–Atg1 (aa 501–897). The beads were pelleted, washed, and bound protein was eluted and analyzed by Western blotting. (B) GST–Atg1 (aa 1–500), but not GST or GST–Atg1 (aa 501–897), can phosphorylate MBP. (Upper) 32P-labeled MBP is shown in lane 2. (Lower) Ponceau S staining of MBP in each sample. Equal amounts of GST and GST fusion proteins were used in each sample. (C) Rab1 coprecipitates with Ulk1 from HeLa cell lysates. HeLa cell lysates were incubated for 2 h at 4 °C with anti-Ulk1 antibody or rabbit IgG, followed by the addition of Protein-A–conjugated agarose beads for an additional hour. The beads were pelleted, washed, and bound protein was eluted and analyzed by Western blotting. The induction of autophagy with rapamycin did not stimulate the coprecipitation of Rab1 with Ulk1. (D) (1) TRAPPIII binds to Atg17 and activates Ypt1 (2). Activated Ypt1 recruits Atg1 (3). Dimeric Atg1 tethers Atg9 vesicles to each other or to other membranes.
To determine if the interaction of Atg1 and Ypt1 is conserved in higher cells, we asked if the mammalian homolog of Atg1, Ulk1, interacts with Rab1 in HeLa cells. Using an antibody directed against Ulk1, we were able to coprecipitate ∼2% of the Rab1 in a cell lysate with Ulk1 (Fig. 6C). This interaction was specific, since Rab1 could not be precipitated with control IgG, and anti-Ulk1 antibody did not precipitate the GTPases Sar1 (Fig. 6C), Rab 5, or Rab 9 (Fig. S3C). Together these findings show that Ypt1/Rab1 interacts with Atg1/Ulk1 in yeast and mammalian cells.
Discussion
The GTPase Ypt1 and its mammalian homolog Rab1 regulate three different trafficking events: autophagy, ER-Golgi, and intra-Golgi traffic (12). During autophagy, Ypt1 is recruited to the phagophore by the TRAPPIII complex, one of three multimeric Ypt1/Rab1 GEFs that localize to distinct cellular locations (12). These complexes share several essential subunits that are required for GEF activity. The TRAPPIII-specific subunit Trs85 specifically directs TRAPPIII to the phagophore where it recruits and activates Ypt1 on the autophagy pathway (14).
To determine where on the autophagy pathway TRAPPIII and Ypt1 act, we screened all known autophagy-deficient atg mutants for defects in the recruitment of Trs85 to the PAS. This screen revealed that the recruitment of TRAPPIII to the PAS is dependent on Atg17 and suggested that Ypt1 and its GEF act in the induction step of the pathway. Five Atg proteins act in induction: Atg1, Atg13, Atg17, Atg29, and Atg31 (6, 9). Of these, only Atg1 is recruited to the PAS in a Ypt1-dependent manner. The loss of Ypt1 function decreases the recruitment of Atg1 to the PAS and autophagy, while the overexpression of Ypt1 has the opposite effect. Atg1 binds directly to Ypt1 and it preferentially interacts with the activated form.
The C terminus of Atg1 acts early in phagophore initiation, while the N-terminal kinase domain is required later in the autophagy pathway (24). The conserved C terminus of Atg1 (amino acids 562–831), which forms a stable dimer, targets Atg1 to membranes and is sufficient to tether liposomes in vitro (8, 26). The C-terminal domain also contains the Atg13 binding site (25), suggesting that it plays a key role in both targeting Atg1 to the PAS and assembling Atg1 into a complex with its binding partners.
When autophagy is induced, three Atg9 vesicles coalesce at the PAS to initiate phagophore formation (2). Other membranes derived from the ER and mitochondria are also likely to contribute to the expanding phagophore (3). Like Trs85, the targeting of Atg9 to the PAS is dependent on Atg17 (2, 5), a component of the scaffold complex that organizes this site (6). We propose that Ypt1 regulates autophagy by recruiting its effector Atg1 to Atg9 vesicles to tether them to each other or to other membranes at the PAS (Fig. 6D) (steps 2 and 3). This proposal is consistent with the observation that Ypt1 partially colocalizes with Atg9 and is present on immunoisolated Atg9 vesicles (14, 22). We speculate that TRAPPIII arrives at the PAS before or after the arrival of Atg9 vesicles (Fig. 6D) (step 1). Previous studies have shown that Ypt1 is recruited to the phagophore assembly site by the TRAPPIII complex (14). Here we show that when Ypt1 is activated, it recruits Atg1. Our findings also suggest that Atg1 kinase activity is not regulated by Ypt1, although a definitive answer awaits the identification of Atg1 substrates. One intriguing possibility is that Atg1 kinase activity is required later for membrane fusion. Consistent with this hypothesis is the observation that Atg9 vesicles cluster at the PAS when kinase activity is abrogated (2). Precedence for the role of a kinase in membrane fusion has recently been documented for ER to Golgi traffic (27). Although we did not see a defect in the recruitment of Atg1 to the PAS under nutrient-rich conditions in the ypt1–2 mutant, we cannot rule out the possibility that more severely impaired alleles of ypt1 would show a defect. In another study, Atg11 was suggested to be an effector of Ypt1 on the Cvt pathway (15).
The interactions we describe here explain how a Rab that localizes to three different cellular compartments achieves specificity on the autophagy pathway through a distinct GEF. In addition to specifically activating Ypt1 on the autophagy pathway, the TRAPPIII complex marks the site where the binding partners for Atg1 reside. Because the Atg1 binding partners (Atg13 and Atg17) are concentrated at the PAS, TRAPPIII ensures that Atg1 is specifically recruited to the PAS and not the ER-Golgi vesicles or the Golgi, the two other compartments where Ypt1 functions (12). In summary, our findings explain how a key regulator of phagophore formation, the Atg1 kinase, is targeted to the PAS. The role of Ypt1 in autophagy also appears to be conserved in higher cells.
Materials and Methods
Media and Growth Conditions.
Cells were grown at 25 °C in YPD [yeast extract (1%), peptone (2% mass/vol), dextrose (2% mass/vol)] or SMD [synthetic minimal media: yeast nitrogen base (0.67%), dextrose (2% mass/vol) and amino acids] media. To induce autophagy by nitrogen starvation, SD-N [synthetic minimal medium lacking nitrogen; yeast nitrogen base without amino acids (0.17%) and ammonium sulfate and dextrose (2% mass/vol)] medium was used. For solid media, agar was added to a final concentration of 2%.
Column Fractionation and Immunoprecipitation from Mammalian Cell Lysates.
NIH 3T3 cells from five confluent 15 cm plates were collected, resuspended in 0.5 mL of lysis buffer (20 mM Hepes, pH 7.2, 150 mM NaCl, 2 mM EDTA, 1 mM DTT, 1× Protease Inhibitor Mixture, 1× PMSF) and lysed by dounce homogenization. The lysates were then centrifuged at 100,000 × g for 1 h. The cleared lysates were collected and the protein concentration was estimated by the Bradford assay. A volume equivalent to 2.5 mg of total protein was loaded on a Superdex-200 column, and 1 mL fractions were collected. Each fraction was trichloroacetic acid (TCA) precipitated, resuspended in 1× sample buffer containing SDS, and heated to 100 °C. The samples were electrophoresed on a 6% (mTrs85 and mTrs130) and 15% (mTrs20) SDS polyacrylamide gel, transferred to nitrocellulose membranes, and probed with the appropriate antibody.
To precipitate mTrs120 and mTrs85, HeLa or COS-7 cells were grown at 37 °C in 10 cm dishes until they were subconfluent, washed with PBS, and lysed in lysis buffer (20 mM Tris pH 7.2, 150 mM NaCl, 1 mM EDTA, 100 µg/mL PMSF, 1 µg/mL aprotinin, 1 µg/mL leupeptin, 1 µg/mL pepstatin A) that contained 1% Nonidet P-40. After a 30-min incubation at 4 °C, the lysate was centrifuged at 16,100 × g for 15 min and the supernatant was collected. The cell lysate (3 mg) was incubated overnight at 4 °C with 10 µL of a protein G bead suspension (purchased from GE and washed 1× in the Nonidet P-40 wash buffer described below) and 10 µg of either rabbit IgG, affinity-purified anti-mTrs120, or anti-mTrs85 antibodies. The beads were then washed three times with Nonidet P-40 wash buffer (20 mM Tris, pH 7.2, 150 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40) and heated to ∼100 °C for 5 min with 1× sample buffer (25 mM Tris, pH 6.8, 5% glycerol, 1% SDS, 1% 2-mercaptoethanol, 0.05% bromophenol blue). The samples were analyzed by Western blot analysis using the ECL method. mTrs20 was analyzed on a 15% SDS polyacrylamide gel, while mTrs130, mTrs120, and mTrs85 were analyzed on 6% SDS polyacrylamide gels.
To precipitate Ulk1, HeLa cells were lysed in 1 mL of lysis buffer (25 mM Hepes pH 7.4, 150 mM NaCl, 1% Triton 100, 1 mM DTT with protease inhibitors), and the lysate was centrifuged at 13, 200 × g at 4 °C for 15 min. The clarified lysate was incubated for 2 h at 4 °C with anti-Ulk1 antibody (1 µg/mg of lysate; Santa Cruz Biotechnology) or rabbit IgG (1 µg/mg of lysate). Protein-A agarose beads prewashed in lysis buffer were then added for an additional 1 h before the beads were processed as described above and analyzed on a 13% polyacrylamide gel using the ECL method.
Fluorescence Microscopy.
Cells were cultured in SMD selective medium at 25 °C to log phase. For the nitrogen starvation experiments, cells were washed twice and shifted to SD-N medium for 4 h. Fluorescent cells were visualized on an Axio Imager Z1 fluorescence microscope using a 100× oil-immersion objective. Images were captured with an AxioCam MRm digital camera and AxioVision software, and deconvolved using OpenLab software.
In Vitro Binding Assays.
Yeast lysates were prepared by converting 3, 000 OD600 units of cells to spheroplasts and lysing the spheroplasts in 15 mL of lysis buffer [25 mM Hepes, pH 7.4, 150 mM KCl, 2 mM EDTA, 1 mM DTT, and 1× protease inhibitor cocktail (PIC)]. The lysate was centrifuged at 16,000 × g for 15 min, and the supernatant (5–10 mg) was incubated overnight at 4 °C with equimolar amounts (0.5 µM) of purified GST, GST–Atg16, or GST–Atg17 immobilized on beads. Because purified GST–Atg17 tended to adhere to proteins in vitro, all binding experiments were performed with yeast lysate to minimize nonspecific binding. After the incubation, the beads were washed and eluted in SDS/PAGE sample buffer. The eluate was then fractionated on a 13% SDS polyacrylamide gel, and immunoblot analysis was performed using the ECL method. Anti-Myc antibody was used at 1:1,000 dilution.
In vitro binding studies with purified Ypt1–His6, GST, GST–Atg1 (aa 1–500), and GST–Atg1 (aa 501–897) were performed with recombinant proteins expressed in bacteria. Varying concentrations of Ypt1–His6 were incubated for 3 h at 4 °C with equimolar amounts (0.1 µM) of immobilized GST or GST fusion proteins in binding buffer (50 mM Hepes, pH 7.2, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.5 mM MgCl2, 2% Triton X-100, and 1× PIC). The beads were washed, eluted in SDS/PAGE sample buffer, and the eluate was fractionated on a 13% SDS polyacrylamide gel. Immunoblot analysis was performed with the ECL method using anti-His6 antibody at 1:1,000 dilution.
Pho8Δ60 Assay.
Alkaline phosphatase assays were performed using a cytosolic form of Pho8 (vacuolar alkaline phosphatase) as described previously (28). To induce autophagy, exponentially grown yeast cells were washed twice and shifted to SD-N medium. At each time point, an aliquot of cells was resuspended in ice-cold lysis buffer (20 mM Pipes, pH 7.2, 0.5% Triton X-100, 50 mM KCl, 100 mM potassium acetate, 10 mM MgSO4, 10 μM ZnSO4, and 1mM PMSF), vortexed with glass beads, and centrifuged. The supernatant was transferred to assay buffer (1.25 mM p-nitrophenyl phosphate, 250 mM Tris⋅HCl pH 8.5, 0.4% Triton X-100, 10 mM MgSO4, and 10 μM ZnSO4) and assayed at 37 °C. The reaction was stopped with 500 µL of stop buffer (1 M glycine/KOH, pH 11.0) and the OD400 value was measured using an Ultrospec 3100 pro UV/Visible spectrophotometer. Protein concentration was measured using the Bradford method.
Atg1 Kinase Assay.
Cells grown exponentially in YPD medium were treated for 30 min at 30 °C with or without rapamycin (200 nM). Cells were collected by centrifugation at 500 × g in tubes filled with ice and washed once with Tris⋅HCl buffer (50 mM Tris⋅Cl pH 7.4, 100 mM NaCl). The washed cells (5 × 108) were lysed in 0.5 mL of lysis buffer containing 50 mM Tris⋅HCl pH 7.4, 100 mM NaCl, 5 mM EDTA, 1 mM PMSF, and 1× protease inhibitor mixture (Roche) by vortexing with glass beads. The lysates were then transferred to fresh tubes, and the remaining glass beads were washed twice with 0.5 mL of lysis buffer. Upon combining the washes with lysates, Triton X-100 was added to the samples to a final concentration of 0.75%. The samples were then incubated at 4 °C on a nutator for 15 min and centrifuged at 10,000 × g for 10 min.
Clarified lysates (2 mg) were incubated with 2 µg of anti-HA antibody (12CA5) for 2 h at 4 °C. Protein-A–conjugated agarose beads (20 µL) were then added to the lysates and incubated for an additional 1.5 h. The beads were washed twice with lysis buffer containing 0.75% of Triton X-100 and twice with kinase buffer containing 50 mM Hepes pH 7.4, 5 mM MgCl2, 0.2% Nonidet P-40, and 1 mM DTT. The kinase activity of immunopurified Atg1–3HA was assayed using MBP as substrate (29). The purified beads (20 µL) were incubated at 30 °C for 30 min with 2 µg of MBP, 100 µM ATP, and 5 µCi of 32P γATP in 50 µL of kinase buffer. The reaction was terminated by adding 20 µL of 4× SDS sample buffer and incubated at 95 °C for 5 min. The reaction mixture was then subjected to SDS/PAGE. The incorporation of radiolabeled 32P into MBP was determined by radioautography and the amount of immunopurified Atg1–3HA was determined by immunoblotting.
For assays with bacterially expressed GST–Atg1 fragments, 3 µg of purified GST or GST fusion protein was used for each reaction.
Immunoprecipitation Using Yeast Lysates.
One hundred OD600 units of exponentially grown yeast cells were converted to spheroplasts with zymolyase 100T in Z buffer (50 mM Tris⋅HCl pH 7.5, 1 M sorbitol, 1% yeast extract, 2% polypeptone, and 1% glucose). The resulting spheroplasts were divided into two aliquots, treated with or without rapamycin (0.2 μg/mL) for 30 min, and lysed with ice-cold lysis buffer (PBS, 200 mM sorbitol, 1 mM MgCl2, 0.1% Tween 20) supplemented with 1 mM PMSF and 1× protease inhibitor mixture (5). The extracts were centrifuged for 15 min at 16,000 × g, and the supernatants were incubated with 15 µL of HA affinity matrix at 4 °C for 4 h. The immunoprecipitates were analyzed by SDS/PAGE and immunoblot analysis.
Note added in proof.
While this manuscript was in revision, a study [Bassik MC, et al. (2013) Cell 152(4):909–922] measuring genetic interactions in high throughput in mammalian cells reported two distinct complexes that are equivalent to the mTRAPPII and mTRAPPIII complexes we describe here.
Supplementary Material
Acknowledgments
We thank Dan Klionsky for plasmids, Chris Fromme for advice on the in vitro bindings studies with Ypt1, and Suzanne Pfeffer for anti-Rab9 antibody. We also thank W. Zhou for technical assistance. Salary support for J.W., S.M., A.Y., H.-T.C., T.W., and S.F.-N. was provided by the Howard Hughes Medical Institute. Work at the University of Pittsburgh was supported by National Institutes of Health (NIH) Grant R01 CA129821 (to Y.J.). Work at Harvard Medical School was also supported by an NIH grant (P01 GM0622580 to Stephen C. Harrison).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1302337110/-/DCSupplemental.
References
- 1.Friedman LG, et al. Disrupted autophagy leads to dopaminergic axon and dendrite degeneration and promotes presynaptic accumulation of α-synuclein and LRRK2 in the brain. J Neurosci. 2012;32(22):7585–7593. doi: 10.1523/JNEUROSCI.5809-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yamamoto H, et al. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol. 2012;198(2):219–233. doi: 10.1083/jcb.201202061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tooze SA, Yoshimori T. The origin of the autophagosomal membrane. Nat Cell Biol. 2010;12(9):831–835. doi: 10.1038/ncb0910-831. [DOI] [PubMed] [Google Scholar]
- 4.Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10(7):458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- 5.Sekito T, Kawamata T, Ichikawa R, Suzuki K, Ohsumi Y. Atg17 recruits Atg9 to organize the pre-autophagosomal structure. Genes Cells. 2009;14(5):525–538. doi: 10.1111/j.1365-2443.2009.01299.x. [DOI] [PubMed] [Google Scholar]
- 6.Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12(2):209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- 7.Kabeya Y, et al. Characterization of the Atg17-Atg29-Atg31 complex specifically required for starvation-induced autophagy in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2009;389(4):612–615. doi: 10.1016/j.bbrc.2009.09.034. [DOI] [PubMed] [Google Scholar]
- 8.Ragusa MJ, Stanley RE, Hurley JH. Architecture of the Atg17 complex as a scaffold for autophagosome biogenesis. Cell. 2012;151(7):1501–1512. doi: 10.1016/j.cell.2012.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawamata T, Kamada Y, Kabeya Y, Sekito T, Ohsumi Y. Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol Biol Cell. 2008;19(5):2039–2050. doi: 10.1091/mbc.E07-10-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kamada Y, et al. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150(6):1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mizuno-Yamasaki E, Rivera-Molina F, Novick P. GTPase networks in membrane traffic. Annu Rev Biochem. 2012;81:637–659. doi: 10.1146/annurev-biochem-052810-093700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barrowman J, Bhandari D, Reinisch K, Ferro-Novick S. TRAPP complexes in membrane traffic: Convergence through a common Rab. Nat Rev Mol Cell Biol. 2010;11(11):759–763. doi: 10.1038/nrm2999. [DOI] [PubMed] [Google Scholar]
- 13.Harding TM, Hefner-Gravink A, Thumm M, Klionsky DJ. Genetic and phenotypic overlap between autophagy and the cytoplasm to vacuole protein targeting pathway. J Biol Chem. 1996;271(30):17621–17624. doi: 10.1074/jbc.271.30.17621. [DOI] [PubMed] [Google Scholar]
- 14.Lynch-Day MA, et al. Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc Natl Acad Sci USA. 2010;107(17):7811–7816. doi: 10.1073/pnas.1000063107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lipatova Z, et al. Regulation of selective autophagy onset by a Ypt/Rab GTPase module. Proc Natl Acad Sci USA. 2012;109(18):6981–6986. doi: 10.1073/pnas.1121299109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466(7302):68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scrivens PJ, et al. C4orf41 and TTC-15 are mammalian TRAPP components with a role at an early stage in ER-to-Golgi trafficking. Mol Biol Cell. 2011;22(12):2083–2093. doi: 10.1091/mbc.E10-11-0873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi C, et al. Organization and assembly of the TRAPPII complex. Traffic. 2011;12(6):715–725. doi: 10.1111/j.1600-0854.2011.01181.x. [DOI] [PubMed] [Google Scholar]
- 19.Yamasaki A, et al. mTrs130 is a component of a mammalian TRAPPII complex, a Rab1 GEF that binds to COPI-coated vesicles. Mol Biol Cell. 2009;20(19):4205–4215. doi: 10.1091/mbc.E09-05-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cai H, Zhang Y, Pypaert M, Walker L, Ferro-Novick S. Mutants in trs120 disrupt traffic from the early endosome to the late Golgi. J Cell Biol. 2005;171(5):823–833. doi: 10.1083/jcb.200505145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen S, et al. Trs65p, a subunit of the Ypt1p GEF TRAPPII, interacts with the Arf1p exchange factor Gea2p to facilitate COPI-mediated vesicle traffic. Mol Biol Cell. 2011;22(19):3634–3644. doi: 10.1091/mbc.E11-03-0197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kakuta S, et al. Atg9 vesicles recruit vesicle-tethering proteins Trs85 and Ypt1 to the autophagosome formation site. J Biol Chem. 2012;287(53):44261–44269. doi: 10.1074/jbc.M112.411454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bacon RA, Salminen A, Ruohola H, Novick P, Ferro-Novick S. The GTP-binding protein Ypt1 is required for transport in vitro: The Golgi apparatus is defective in ypt1 mutants. J Cell Biol. 1989;109(3):1015–1022. doi: 10.1083/jcb.109.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheong H, Nair U, Geng J, Klionsky DJ. The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2008;19(2):668–681. doi: 10.1091/mbc.E07-08-0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yeh YY, Shah KH, Herman PK. An Atg13 protein-mediated self-association of the Atg1 protein kinase is important for the induction of autophagy. J Biol Chem. 2011;286(33):28931–28939. doi: 10.1074/jbc.M111.250324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan EY, Longatti A, McKnight NC, Tooze SA. Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism. Mol Cell Biol. 2009;29(1):157–171. doi: 10.1128/MCB.01082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lord C, et al. Sequential interactions with Sec23 control the direction of vesicle traffic. Nature. 2011;473(7346):181–186. doi: 10.1038/nature09969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klionsky DJ. Monitoring autophagy in yeast: The Pho8Delta60 assay. Methods Mol Biol. 2007;390:363–371. doi: 10.1007/978-1-59745-466-7_24. [DOI] [PubMed] [Google Scholar]
- 29.Kijanska M, et al. Activation of Atg1 kinase in autophagy by regulated phosphorylation. Autophagy. 2010;6(8):1168–1178. doi: 10.4161/auto.6.8.13849. [DOI] [PubMed] [Google Scholar]
- 30.Yip CK, Berscheminski J, Walz T. Molecular architecture of the TRAPPII complex and implications for vesicle tethering. Nat Struct Mol Biol. 2010;17(11):1298–1304. doi: 10.1038/nsmb.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.