Abstract
Maintaining wakefulness is associated with a progressive increase in the need for sleep. This phenomenon has been linked to changes in synaptic function. The synaptic adhesion molecule Neuroligin-1 (NLG1) controls the activity and synaptic localization of N-methyl-d-aspartate receptors, which activity is impaired by prolonged wakefulness. We here highlight that this pathway may underlie both the adverse effects of sleep loss on cognition and the subsequent changes in cortical synchrony. We found that the expression of specific Nlg1 transcript variants is changed by sleep deprivation in three mouse strains. These observations were associated with strain-specific changes in synaptic NLG1 protein content. Importantly, we showed that Nlg1 knockout mice are not able to sustain wakefulness and spend more time in nonrapid eye movement sleep than wild-type mice. These changes occurred with modifications in waking quality as exemplified by low theta/alpha activity during wakefulness and poor preference for social novelty, as well as altered delta synchrony during sleep. Finally, we identified a transcriptional pathway that could underlie the sleep/wake-dependent changes in Nlg1 expression and that involves clock transcription factors. We thus suggest that NLG1 is an element that contributes to the coupling of neuronal activity to sleep/wake regulation.
Keywords: ChIP, EEG, gene expression, sleep homeostasis, synaptic plasticity
Sleep is crucial for learning, memory, and other functions essential for proper functioning of the brain and body (1, 2). These functions have been associated with the sleep recovery process, which defines a level of pressure for sleep that increases with wakefulness and dissipates during sleep and that is reflected by changes in sleep intensity (3, 4). Sleep intensity is indexed by electroencephalographic (EEG) markers of neuronal synchrony in delta frequencies (1–4 Hz) measured during nonrapid eye movement (NREM) sleep (5). During wakefulness, mechanisms favoring desynchrony in the delta range predominate, and the brain can maintain cognition, whereas during sleep, events promoting network synchrony mostly take place with high delta activity thought to be permissive of recovery (3, 6). The sleep recovery process has been hypothesized to originate and contribute to the maintenance of both synaptic and network equilibrium (6–8). This notion is supported by the observation that specific plasticity-related genes may be directly involved in regulating sleep need (9). Certain clock genes may also directly contribute, in a circadian-independent manner, to the sleep recovery process (5, 9). However, the mechanisms underlying the capacity and requirement of the brain to switch from an alert desynchronized state to an unconscious synchronized state remain elusive.
Glutamate, the main excitatory neurotransmitter of the brain, can induce long-term modifications of synaptic transmission and, thus, changes in network connectivity. This is achieved mainly via glutamate’s action on two types of receptors: N-methyl-d-aspartate (NMDAR) and α-amino-3-hydroxy-5-methylisoazol-4-propionate (AMPAR) (10, 11). The balance between the activity of these receptors was proposed to determine neuronal firing synchrony (12, 13), with high NMDAR activity associated with desynchrony reminiscent of active wakefulness and low NMDAR linked to synchrony, similar to intense NREM sleep (14). Also, prolonged wakefulness achieved by sleep deprivation (SD), which triggers intense sleep, decreases NMDAR function and NMDAR-dependent forms of plasticity (15–17). It can thus be hypothesized that high glutamate transmission during wakefulness and rapid eye movement (REM) sleep (18) gradually desensitizes NMDAR and changes the balance in favor of AMPAR transmission, which would lead to high delta synchrony during NREM sleep. During NREM sleep, lowered glutamate transmission (18) may help recover NMDAR function, which would gradually dissipate high neuronal synchrony/sleep intensity. In this scenario, elements regulating NMDAR functioning are of unique relevance to the sleep recovery process.
Neuroligin-1 (NLG1) is a postsynaptic adhesion molecule localized at glutamatergic and GABAergic synapses with a prominent preference for the former (19, 20). At glutamatergic synapses, NLG1 determines the number of functional NMDAR (21–23) and is required for normal expression of NMDAR-dependent plasticity (23, 24). Also, the synaptic level of NLG1 is down-regulated by neuronal activity (25), its absence impairs spatial and associative fear memory (24, 26) in a way similar to SD, and SD seems to decrease Nlg1 mRNA expression (27). Therefore, through its response to neuronal activity and role in regulating glutamate transmission, NLG1 is a potent candidate to modulate both the deleterious consequences of sleep loss and the sleep/wake-dependent changes in sleep intensity. We indeed found that elevated sleep pressure changes not only the expression of specific Nlg1 transcript variants, but also the synaptic NLG1 level, and we identified a transcriptional pathway involving core clock transcription factors that likely contributes to these changes. Importantly, we revealed that these changes can directly regulate the duration and quality of wakefulness and sleep using EEG recording in mice lacking NLG1.
Results
Nlg1 Expression Is Changed by Sleep Deprivation.
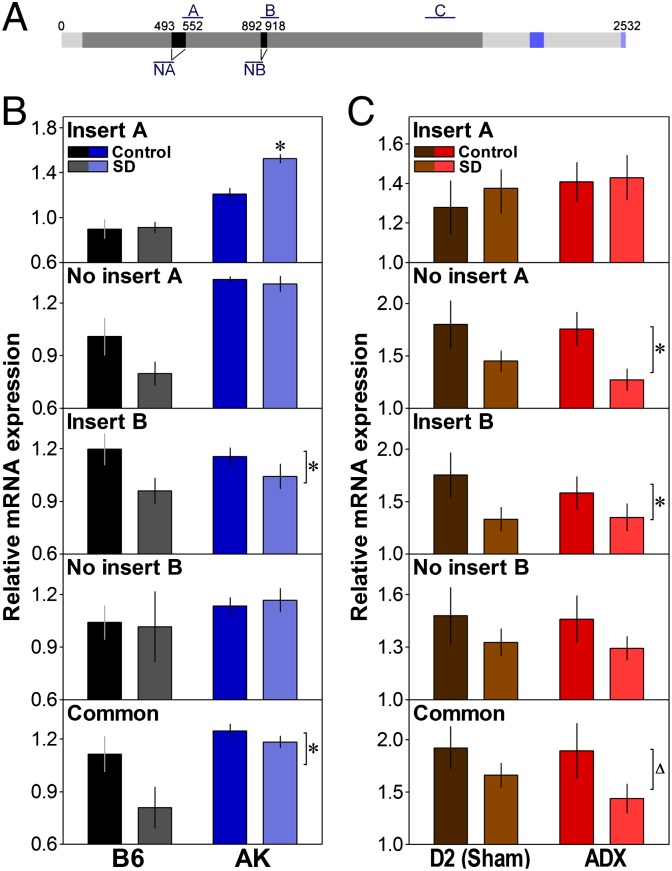
A role for NLG1 in the sleep recovery process may imply that its expression would track wakefulness duration. We thus measured the effect of SD on the expression of Nlg1 in two inbred mouse strains, C57BL/6J (B6) and AKR/J (AK), showing different degrees of delta power rebound following SD (28). This was quantified for different Nlg1 transcript variants because those were shown to change NLG1 properties (29). We used five probes targeting the following: insert in splice site A, absence of insert in splice site A, insert in splice site B, absence of insert in splice site B, and a sequence common to all variants (Fig. 1A). A 6-h SD starting at light onset significantly decreased the forebrain expression of Nlg1 with insert B and of the common probe in both strains (Fig. 1B). This decrease was already observed after SD of shorter durations (i.e., 1 and 3 h) and seemed to restore after 2 h of recovery sleep (Fig. S1). In AK mice only, the expression of Nlg1 with insert A was significantly increased by SD (Fig. 1B).
Fig. 1.
Effect of SD on the expression of Nlg1 transcript variants. (A) Scheme of the Nlg1 mRNA showing position of splice sites and qPCR amplicons (A and NA or B and NB, with and without insert A or B, respectively; C, common probe). Dark gray, cholinesterase domain (94–1,878 bp); blue, transmembrane domain (2,086–2,142 bp); light blue, PSD95-binding domain (2,512–2,529 bp). (B) Relative expression of Nlg1 transcripts in the forebrain of C57BL/6J (B6) and AKR/J (AK) mice at ZT6 (6 h after light onset) under the control condition (n = 4 for B6 and AK) or after a 6-h SD (n = 3 for B6, 4 for AK). SD significantly decreased the expression of Nlg1 with insert B and common Nlg1 (condition effects: F1,12 ≥ 5.1, *P < 0.05) and increased the expression of Nlg1 with A only in AK mice (interaction: F1,12 = 6.8, *P < 0.05: compared with Control). AK mice also expressed more common Nlg1 and Nlg1 without A and without B than B6 mice (strain effects: F1,12 ≥ 9.8, P < 0.01). (C) Expression of Nlg1 transcripts in the forebrain of DBA/2J (D2) mice at ZT6 under the control condition or after a 6-h SD in mice submitted to either a sham surgery (n = 7/group) or an ADX (n = 6/group). SD decreased the expression of Nlg1 without A and with B (condition effects: F1,22 ≥ 4.2, *P ≤ 0.05), and a similar tendency was found for common Nlg1 (F1,22 = 3.7, ∆P < 0.07).
To verify that the SD-dependent changes in Nlg1 expression were not due to the associated surge in corticosterone, which importantly contributes to the SD-induced changes in brain transcriptome (9), expression was also measured in the mouse strain showing the larger surge in corticosterone with SD, DBA/2J (D2) mice, submitted to adrenalectomy (ADX) or sham lesion (9). In both ADX and sham mice, and similar to B6 and AK mice, SD significantly decreased the expression of Nlg1 with insert B and of the common probe (Fig. 1C). Moreover, a decrease in Nlg1 without insert A was found for ADX and sham D2 mice. These results indicate that elevated sleep pressure decreases the expression of Nlg1 containing an insert B (and likely no insert A) in various mouse strains and that this effect is independent of the corticosterone surge associated with SD.
Synaptic NLG1 Level Is Modified by Sleep Deprivation.
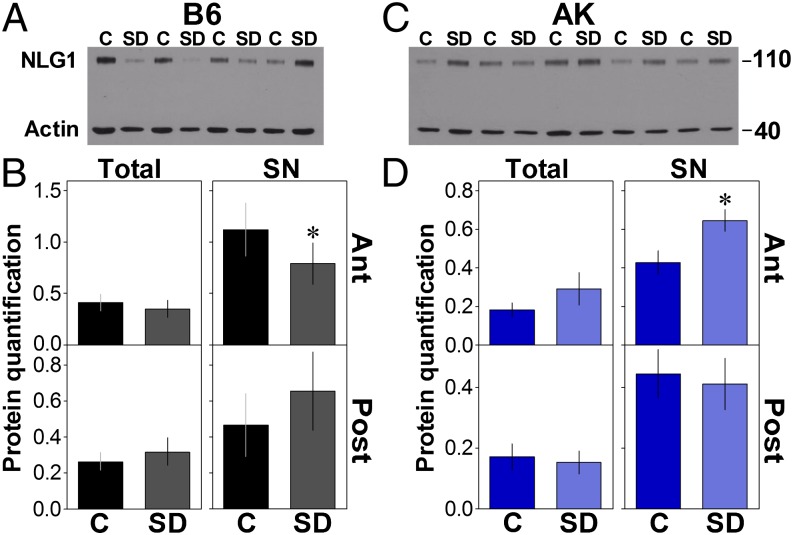
As changes in mRNA expression do not necessarily translate into changes in protein levels (30), we assessed the effect of a 6-h SD on the forebrain expression of total and synaptoneurosomal (SN) NLG1 protein content in B6 and AK mice. This was quantified for the anterior and the posterior forebrain separately, with the hypothesis that changes would predominate in the anterior area, which is largely activated by extended wakefulness (31). In both strains, SD did not affect the total NLG1 level in forebrain areas anterior or posterior to the bregma or the SN level in the posterior forebrain (Fig. 2 B and D). However, in the anterior forebrain, SD significantly decreased SN NLG1 in B6 mice (Fig. 2 A and B), whereas, in contrast, it increased SN NLG1 in AK mice (Fig. 2 C and D). The antibody used recognizes the N-terminal part of NLG1 that precedes both splice sites and, therefore, does not discriminate between NLG1 isoforms. However, the SD-dependent decrease in anterior SN NLG1 in B6 is reminiscent of the effect of SD on the expression of Nlg1 with insert B (Fig. 1B), whereas the SN NLG1 increase following SD in AK could be linked to the increase of Nlg1 with insert A, which was specific to this strain (Fig. 1B). These data suggest that elevated sleep pressure modulates the synaptic NLG1 level in a brain area- and strain-specific fashion.
Fig. 2.
Effect of SD on NLG1 protein level. (A) Western blot showing NLG1 in the SN fraction of the anterior (Ant) forebrain in four control (C) and four SD C57BL/6J (B6) mice. Endogenous control Actin is also shown. (B) Quantification of Western blots for NLG1 in total protein fraction (Tot) and SN of the anterior and posterior (Post) parts of B6 mice forebrain (n = 10 or 11/group). SD significantly decreased the level of NLG1 in Ant SN (t = 2.4, *P < 0.05). (C) Western blot showing NLG1 (and Actin) in the SN of the Ant brain in five C and five SD AKR/J (AK) mice. (D) Quantification of NLG1 Western blots in Tot and SN of Ant and Post parts of AK mice forebrain (n = 5–7/group). SD significantly increases the level of NLG1 in Ant SN (t = −3.5, *P < 0.05).
Absence of NLG1 Decreases Wakefulness Duration.
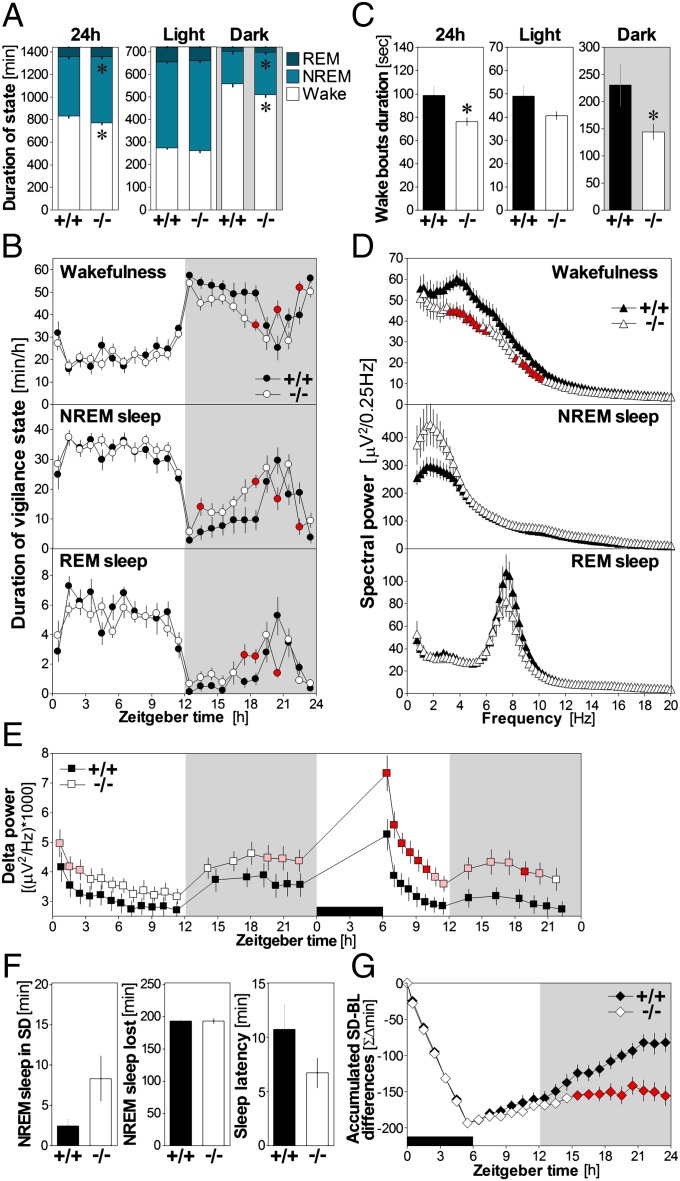
To assess if the observed SD-dependent changes in Nlg1 expression and protein really contribute to sleep/wake regulation, we verified if the absence of NLG1 affects wakefulness and sleep architecture. Hence, the 24-h time course of vigilance states during undisturbed condition was measured in Nlg1 knockout (KO) mice. NLG1 protein (all isoforms) is undetectable in these mice (Fig. S2A) generated by replacing coding exons 1 and 2 by a neo cassette (32). Lack of NLG1 decreased wakefulness and increased NREM sleep by more than an hour when analyzed over 24 h (Fig. 3A). These differences were restricted to the 12-h dark period (Fig. 3A). Heterozygous mice presented an intermediate phenotype (Fig. S2B). Furthermore, the time course of hourly sleep/wake values revealed that Nlg1 KO mice spent more time in NREM and REM sleep during the first two-thirds of the dark period, when WT mice are usually awake, and less time asleep in the last third of the dark period, when WT mice are usually “napping” (Fig. 3B). Because KO mice seemed to show impairment in maintaining wakefulness during the active phase, we computed the duration of individual wake bouts (Fig. 3C). Wake-bout duration was 37.3% shorter in Nlg1 KO than in WT mice during the 12-h dark period (intermediate duration in heterozygous; Fig. S2C). These data indicate that mice lacking NLG1 cannot sustain long bouts of wakefulness, which is accompanied by an overall decrease in wakefulness duration and an increase in NREM sleep duration.
Fig. 3.
Sleep structure and cortical synchrony in Nlg1 KO mice. (A) Vigilance state duration in WT (+/+, n = 10) and Nlg1 KO (−/−, n = 12) mice for 24 h, 12-h light, and 12-h dark periods. For the 24-h and 12-h dark periods, Nlg1 KO mice spent less time in wakefulness and more in NREM sleep (t ≥ 2.5 and t = −2.6, respectively; *P < 0.05). (B) State duration per hour in WT and KO mice. Genotype-by-time interactions were found for wake, NREM, and REM sleep (F23,460 > 1.9, P < 0.01). Differences between KO and WT are indicated by red symbols (P < 0.05; same in D, E, and G). (C) Mean duration of wake bouts in WT and KO mice for 24 h, 12-h light, and 12-h dark periods. For the 24 h and the 12-h dark period, the duration of wake bouts was lower in KO mice than in WT (t ≥ 2.2, *P < 0.05). (D) Spectral power for the three states in WT and KO mice. (E) Forty-eight-hour time course of delta power in WT and KO mice. Genotype-by-time interactions were found for the 24-h baseline (F17,340 = 2.3, P < 0.05) and for the 18-h recording after a 6-h SD (F13,247 = 3.7, P < 0.05). Pink symbols indicate a trend for difference between WT and KO (P < 0.1). (F) NREM sleep measured during the 6-h SD did not differ significantly between WT and KO (t = 1.8, P = 0.08). The same was observed for loss of time spent in NREM sleep and latency to sleep after SD (t = 1.5, P = 0.15). (G) Time course of accumulated differences in NREM sleep between the 6-h SD followed by 18 h of recovery and baseline conditions. Significant genotype-by-time interaction was found (F23,460 = 9.1, P < 0.001). Gray backgrounds indicate dark periods and black rectangles indicate the 6-h SD.
Absence of NLG1 Modifies EEG Synchrony.
To demonstrate that NLG1 is implicated in shaping neuronal synchrony during sleep/wake states, EEG power spectra were calculated for the three states (Fig. 3D). During wakefulness, Nlg1 KO mice showed a pronounced reduction in high delta/low theta and alpha activity (3.25–5.75 and 8.25–10.5 Hz; see Fig. S2D for heterozygous). Conversely, during NREM sleep, KO mice showed a trend (P < 0.1) for higher spectral power in the delta and sigma frequency ranges (1.75–3 and 13.75–16 Hz), both previously linked to the sleep recovery process (33, 34). The analysis of the time course of NREM sleep delta activity (1–4 Hz) confirmed this (Fig. 3E); Nlg1 KO mice showed generally higher levels of delta, and this difference was most pronounced at the beginning of the light period and at the end of the dark period, when sleep pressure is usually highest (28). EEG activity during REM sleep was unaffected by the Nlg1 mutation. These data show that NLG1 is required to maintain normal synchrony of cerebral cortex activity during wakefulness and NREM sleep.
Absence of NLG1 Impacts Recovery Sleep.
To establish a role for NLG1 in the sleep recovery process, the response of Nlg1 KO mice to a 6-h SD was measured. Although KO mice were exceptionally difficult to keep awake, NREM sleep occurring during SD and latency to sleep after SD did not significantly differ from WT (Fig. 3F). However, even if KO mice did not lose more sleep than WT mice during SD (Fig. 3F), they could not catch up on their sleep loss to the same extent as WT during the first recovery day (Fig. 3G). Heterozygous mice showed an intermediate impairment (Fig. S2E). Furthermore, delta power measured during recovery NREM sleep was higher in KO compared with WT and remained higher for most of the recovery period (Fig. 3E and Fig. S2F). Those observations reveal that the absence of NLG1 amplified the cortical synchrony response to elevated sleep need.
Markers of Sleep Need and of Plasticity in Nlg1 KO Mice.
To investigate if Nlg1 KO mice express alterations in molecular markers of prolonged wakefulness, we measured, in the forebrain after SD, the expression of genes that have been associated with the sleep recovery process (9, 35–38). The expression of NMDAR subunits was also measured to evaluate the impact of the Nlg1 mutation on their transcriptional regulation during SD. The SD-dependent increase in expression in WT was equally observed in Nlg1 KO for the targets Homer homolog 1a (Homer1a), brain derived neurotrophic factor (Bdnf), and Period 2 (Per2) (Fig. S2G), as well as for NMDAR subunit 2a (Nr2a) (Fig. S2H), indicating that transcriptional pathways involving these markers are not affected by the mutation.
To understand the functional impact of sleep/wake modifications in Nlg1 KO, we assessed other quality indicators of the alert state. Although neurological function was preserved in KO (Fig. S3A), we found that KO mice failed to show preference for social novelty (Fig. S3B), similar to what was previously reported (26). In addition, we aimed to determine if sleep/wake modifications in Nlg1 KO could be associated with changes in the cerebrovascular response to neuronal stimulation, which is known to depend on NMDAR transmission (39). The somatosensory cortex of KO mice showed a 10% reduction in the number of responses to forepaw stimulation (Fig. S4A) and, compared with WT, reduced oxyhemoglobin change 4 s after stimulation (Fig. S4B). Overall, these findings indicate that NLG1 is likely a key modulator of the relationship between waking duration, waking quality, and the response to neuronal activity.
Nlg1 Expression Is Regulated by Clock Transcription Factors.
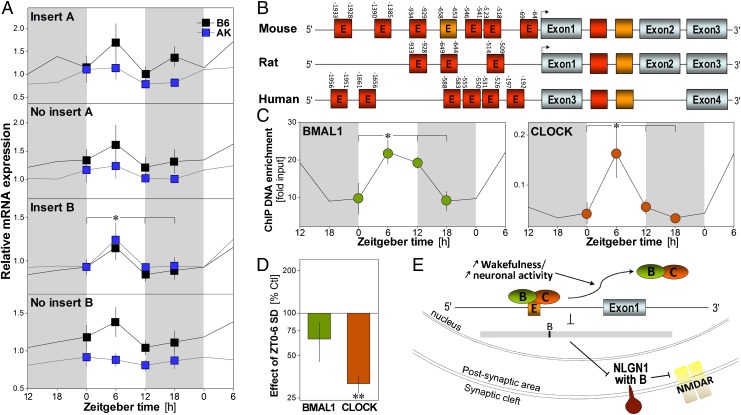
If Nlg1 has a role in regulating sleep and wakefulness, its expression should, in addition to being modulated by SD, vary with time of day as a function of the sleep/wake distribution. We indeed found that, in B6 and AK mouse forebrain, the expression of Nlg1 with insert B was significantly higher at Zeitgeber time 6 (ZT6, i.e., 6 h after light onset) than at other times of the day (Fig. 4A). The same was observed for the Nlg1 common probe (Fig. S5). In addition to sleep/wake-dependent changes, time-of-day–dependent expression of Nlg1 could also result from a circadian control because clock genes have been involved in shaping the dynamics of neuronal synchrony during sleep (5). We thus examined the Nlg1 gene and noted the presence of several E-boxes (Fig. 4B), which are cis-regulatory elements serving transcriptional control by the core clock transcription factors circadian locomotor output cycles kaput (CLOCK) and aryl hydrocarbon receptor nuclear translocator-like (Arntl) transcript variant 1 (BMAL1) (40). As some of these sequences are conserved across species, this suggests a role for transcription factors from the clock in regulating Nlg1 expression.
Fig. 4.
Rhythmic expression of Nlg1 and binding of its gene by clock transcription factors. (A) Relative expression of Nlg1 transcripts measured at ZT0, -6, -12, and -18 in the forebrain of C57BL/6J (B6) and AKR/J (AK) mice (n = 5–8/group). The expression of Nlg1 containing insert B is highest at ZT6 (time effect: F3,49 = 11.7, P < 0.01). A significant strain effect was observed for Nlg1 with A and without B (F3,49 ≥ 4.4, P < 0.05). Gray backgrounds indicate dark periods. *P < 0.05 between indicated values (also in C). (B) Diagram of the mouse, rat, and human Nlg1 gene showing the position of E-boxes relative to the transcription start sites (arrows). Red E-boxes represent CANNTG and orange CACGTG. Absence of E in the box indicates the presence of multiple E-boxes. (C) Binding of BMAL1 and CLOCK to the CACGTG of Nlg1 in B6 mouse cerebral cortex measured at ZT0, -6, -12, and -18 (n = 3/point, except n = 2 for ZT12). BMAL1 and CLOCK bind to Nlg1 in a time-of-day–dependent manner (F3,7 ≥ 4.2, P = 0.05). (D) Binding measured at ZT6 after 6 h of SD and expressed relative to the binding in undisturbed control (n = 5/group). SD decreased CLOCK binding to Nlg1 (t = −15.1, **P < 0.001), whereas a tendency was observed for BMAL1 (t = −1.7, P = 0.17). (E) Model showing how elevated sleep pressure may change Nlg1 expression: prolonged wakefulness could lead to detachment of CLOCK/BMAL1 from the gene, which would decrease expression of Nlg1 with insert B and synaptic NLG1 and down-regulate NMDAR function.
Therefore, we assessed if core clock transcription factors bind to the Nlg1 gene as a function of time of day and sleep need in the mouse cerebral cortex. The binding of BMAL1 and CLOCK to one E-box located 653 bp before the Nlg1 transcription start site varied with time of day (Fig. 4C), showing a peak at ZT6 and a trough around dark–light transition (ZT18–ZT0). This phase is similar to what we recently observed for binding to several clock genes (41). Furthermore, and also similar to our previous data (41), we found that SD decreases the binding of those transcription factors to the Nlg1 E-box (Fig. 4D), although the effect reached statistical significance for CLOCK only. In sum, these data imply a role for the clock molecular machinery in determining the strength of glutamatergic synapses via regulation of Nlg1 expression.
Discussion
Our study highlights the involvement of Nlg1 in bridging waking duration and quality to neuronal synchrony. We first showed that sleep pressure determines the expression of specific Nlg1 transcript variants as well as synaptic NLG1 protein content in the forebrain. Importantly, we uncovered that the absence of NLG1 alters sleep/wake duration and EEG activity during wakefulness and NREM sleep. And finally, we found that core clock transcription factors are likely part of the mechanisms underlying the sleep/wake-dependent changes in Nlg1 expression.
Previous work showed that SD decreases the expression of several adhesion molecules (9, 37), including Nlg1 in the mouse cerebral cortex (27). The latter study (27) used an amplicon targeting all transcript variants although four different NLG1 isoforms exist. Indeed, NLG1 can contain (or not) inserts in splice sites A and B (29). In line with changes observed for total Nlg1 expression (27), we observed that enforced wakefulness consistently decreased the expression of Nlg1 with insert B in three different inbred mouse strains. In addition, in B6 mice, synaptic NLG1 protein level was also decreased by SD in the anterior forebrain, which could be more representative of NLG1 with B, given the absence of changes for the other variants. Indeed, recent data indicate that neuronal activity decreases NLG1 level at the synapse (25, 42), and our findings suggest that this decrease mainly concerns the isoform containing insert B. NLG1 with B, the expression of which in the brain prevails over isoforms lacking B (19), is mostly localized at glutamatergic synapses and is specifically implicated in the interaction with presynaptic Neurexins (NRX) (19, 29). Studies have shown that proper NMDAR function requires both the presence of NRX and NLG (21, 43, 44). Accordingly, decreased synaptic level of NLG1 with B following elevated sleep pressure may disrupt NRX-NLG1 complexes and impair glutamatergic transmission. Alternatively, decreased NLG1 after SD might also represent a neuroprotective pathway against glutamate toxicity triggered by high neuronal activity because it was recently shown to negatively feed back on presynaptic neurotransmitter release by means of NLG1 cleavage (25). This mechanism may explain the observation of reduced neuronal loss following NMDA-induced toxicity after chronic sleep restriction (45). We already proposed a role for sleep in neuroprotection as the expression of many transcripts involved in neuroprotection is reliably affected by sleep loss (9).
We also established that Nlg1 expression varied with time of day, again specifically for the insert B variant in both B6 and AK mice. Time-of-day– and sleep pressure–dependent expression likely involves regulation by clock genes, which independently underlie both circadian and sleep/wake driven effects and act as modulators of synaptic function (5). We indeed found that Nlg1 gene contains E-boxes; that CLOCK and BMAL1 can bind to one of these with maximum binding during the main sleep episode; and that binding of CLOCK and BMAL1 to the Nlg1 E-box is decreased by SD in the cerebral cortex. These last two observations parallel changes in the expression of Nlg1 with B, which provide support for a transcriptional control of Nlg1 with insert B by clock transcription factors. Thus, during enforced wakefulness, prolonged high neuronal activity could decrease CLOCK/BMAL1 binding to Nlg1 E-boxes as it does for other CLOCK/BMAL1 targets (41). This would lead to reduced expression of Nlg1 with B and NLG1 at the synapse and consequently to impaired NMDAR function (Fig. 4E) and high delta synchrony. In fact, glutamate transmission was shown to induce BMAL1 degradation (46). During sleep, transcriptional activation by core clock transcription factors may restore the NLG1 pool at the synapse and eventually NMDAR function. In this context, we propose that the time-of-day–dependent expression of Nlg1 with B mainly reflects the short-term sleep/wake history. However, implication of CLOCK/BMAL1 could also indicate a spontaneous self-sustained circadian oscillation, and transcriptional in vitro assays combined with mutagenesis are currently being performed to delineate their exact contribution to Nlg1 transcriptional regulation.
In parallel, we showed that the effects of SD on NLG1 are isoform- and strain-dependent. SD increased the expression of Nlg1 with insert A and synaptic NLG1 level only in AK mice. Our NLG1 antibody (20) targeted an epitope in close proximity to insert A with one overlapping amino acid, which makes it possible for this antibody to preferentially identify NLG1 with A. AK mice were shown to express the highest sleep amount and the largest increase in delta power in response to SD (47), and our results suggest that sleep pressure differentially affects synaptic function in this strain. The presence of insert A does not affect binding of NLG1 to its NRX partners (29), but rather seems to target NLG1 to GABAergic synapses in the absence of insert B (19). Consequently, and specifically in AK mice, increased NLG1 with A may change inhibitory GABAergic transmission with elevated sleep need. Alternative splicing of Nlg1 may thus represent a pathway relevant to interindividual differences in the susceptibility to sleep loss.
Importantly, we reveal that changes in NLG1 with sleep pressure are linked to a functional role in sleep/wake regulation because Nlg1 KO mice showed a decrease in the overall duration of wakefulness and in the mean duration of wake bouts. These findings point to a specific implication of NLG1 in promoting and maintaining wakefulness. Reduced wake maintenance in Nlg1 KO mice is reminiscent of the observation made in mice lacking Homer1a (48), which also regulates glutamatergic transmission. Although the SD-dependent increase in Homer1a expression was not affected by the Nlg1 mutation, these observations suggest that NLG1 and HOMER1A could target similar molecular pathways in the regulation of wakefulness.
The absence of NLG1 did not only affect wake duration but also impaired wake quality because theta and alpha EEG activity during wakefulness was blunted in KO mice, which also failed to respond to social novelty. Theta/low-alpha activity during wakefulness is increased during active wakefulness, for example, during exploration, compared with quiet waking in rodents (49). These alterations in waking quality occurred simultaneously with a higher delta synchrony during NREM sleep, which was more prominent when sleep need is at its highest, especially after SD. This overall phenotype (i.e., increased NREM sleep, shorter wake-bout duration, higher delta response to SD) is similar to what has been observed in AK mice in comparison with other strains (28, 47) and might thus indicate a steeper buildup of sleep pressure during wakefulness in Nlg1 KO mice as reported for AK mice (28). Even if we cannot exclude, at this point, a developmental defect in arousal systems in the full-KO studied here, these data strongly suggest that NLG1 is required to maintain wakefulness quality and normal synchrony of cerebral cortex activity during wakefulness and sleep. This is likely triggered by the ability of NLG1 to tune NMDAR function (21–24), which is also supported by our observation of reduced hemodynamic response to neuronal activation in Nlg1 KO, a response involving NMDAR (39).
In sum, we identify NLG1 as a part of a molecular pathway underlying the capacity of the brain to sustain the desynchronized alert state and to respond to its environment. Because autism spectrum disorder was repeatedly linked to mutation in elements of the NRX-NLG system (50), our findings also suggest that dysfunctions in NLG1 or in other NRX-NLG elements could directly mediate the severe sleep alterations observed in autistic patients (51).
Materials and Methods
Detailed methodology is provided in SI Materials and Methods.
Animals.
Male mice used in this study were from four genetic backgrounds: C57BL/6J (B6), AKR/J (AK), DBA/2J (D2), and B6;129-Nlgn1tm1Bros/J (B6;129). Mice were purchased from Jackson Laboratories and studied directly [quantitative PCR (qPCR), Western blot] or after being bred on site (EEG, qPCR, ChIP). Mice were maintained under standard housing conditions (free access to food and water, 12h-light/12h-dark cycle, 22–25 °C ambient temperature) for 2 wk, and then studied under the same conditions between 8 and 16 wk of age. Mice used for qPCR were the same as those used in refs. 52 (Fig. 1B), 9 (Fig. 1C), and 37 (Fig. 4A). Mice used for ChIP (Fig. 4 C and D) were from ref. 41. Experiments were approved by the Ethical Committee for Animal Experimentation of the Hôpital du Sacré-Coeur de Montréal and the Ethical Committee of the State of Vaud Veterinary Office.
qPCR and Western Blotting.
RNA extraction, reverse transcription, and qPCR experiments and analysis were performed as detailed before (9, 35, 52). Primer and probe sequences are provided in Table S1. Protein extractions were done similarly as previously described (53), and extracts were revealed on nitrocellulose membranes using the following antibodies: anti-NLG1 (Synaptic System), mouse anti-Actin (Sigma-Aldrich), and HRP-conjugated donkey anti-mouse (Santa Cruz Biotechnology).
EEG/Electromyography Recording and Analyses.
KO mice for the Nlg1 gene (mouse line B6;129-Nlgn1tm1Bros/J) and their WT littermates were implanted with EEG and electromyography (EMG) electrodes as detailed previously (9, 38). EEG/EMG was recorded continuously for 48 h starting at light onset during 24 h of undisturbed [baseline (BL)] conditions and during a 6-h SD and 18 h of recovery. Behavioral state duration was averaged for different intervals. The EEG of artifact-free epochs was subjected to spectral analysis to calculate power density between 0.5 and 20 Hz for the BL recording and delta power (1–4 Hz) during NREM sleep.
ChIP.
Chromatin extraction and immunoprecipitation were performed as previously described (41). Anti-BMAL1 used was from Abcam, and anti-CLOCK was from Abcam or Santa Cruz Biotechnology. The amplicon targeted the CACGTG sequence located at position −653:−658 of the Nlg1 gene.
Supplementary Material
Acknowledgments
We thank Mehdi Tafti for sharing some of the samples used here for qPCR (Fig. 4A); colleagues who helped with SD (B. Petit, S. Jimenez, C. Pfister, A. Vassalli, V. Hinard, J. Vienne, H. Hor, S. R. Netedu, C. Koumar, and H. Blais); Caroline Bouchard, Jean Paquet, Gaétan Poirier, and Gaétan Tremblay for technical help; Guy Doucet and Alexandre Lebel for help with the social interaction apparatus; and Hannes Richter and Johann Weber of the Genomic Technologies Facility (University of Lausanne) for expert help with ChIP experiments. This work was supported by the Canadian Institutes of Health Research (Operating Grant 231095-111021), the Fonds de la Recherche du Québec-Santé, the Université de Lausanne, the Swiss National Science Foundation, and the Research Center of the Hôpital du Sacré-Coeur de Montréal.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1221381110/-/DCSupplemental.
References
- 1.Ohlmann KK, O’Sullivan MI. The costs of short sleep. AAOHN J. 2009;57(9):381–385, quiz 386–387. doi: 10.3928/08910162-20090817-02. [DOI] [PubMed] [Google Scholar]
- 2.Diekelmann S, Born J. The memory function of sleep. Nat Rev Neurosci. 2010;11(2):114–126. doi: 10.1038/nrn2762. [DOI] [PubMed] [Google Scholar]
- 3.Borbély AA. A two process model of sleep regulation. Hum Neurobiol. 1982;1(3):195–204. [PubMed] [Google Scholar]
- 4.Daan S, Beersma DG, Borbély AA. Timing of human sleep: Recovery process gated by a circadian pacemaker. Am J Physiol. 1984;246(2 Pt 2):R161–R183. doi: 10.1152/ajpregu.1984.246.2.R161. [DOI] [PubMed] [Google Scholar]
- 5. Mongrain V, Franken P. Genetic interaction between circadian and homeostatic regulation of sleep. The Genetic Basis of Sleep and Sleep Disorders, eds. Shaw P, Tafti M, Thorpy M (Cambridge Univ Press, Cambridge, UK), in press.
- 6.Tononi G, Cirelli C. Sleep function and synaptic homeostasis. Sleep Med Rev. 2006;10(1):49–62. doi: 10.1016/j.smrv.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 7.Steriade M, Timofeev I. Neuronal plasticity in thalamocortical networks during sleep and waking oscillations. Neuron. 2003;37(4):563–576. doi: 10.1016/s0896-6273(03)00065-5. [DOI] [PubMed] [Google Scholar]
- 8.Krueger JM, et al. Sleep as a fundamental property of neuronal assemblies. Nat Rev Neurosci. 2008;9(12):910–919. doi: 10.1038/nrn2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mongrain V, et al. Separating the contribution of glucocorticoids and wakefulness to the molecular and electrophysiological correlates of sleep homeostasis. Sleep. 2010;33(9):1147–1157. doi: 10.1093/sleep/33.9.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malinow R, Malenka RC. AMPA receptor trafficking and synaptic plasticity. Annu Rev Neurosci. 2002;25:103–126. doi: 10.1146/annurev.neuro.25.112701.142758. [DOI] [PubMed] [Google Scholar]
- 11.Rebola N, Srikumar BN, Mulle C. Activity-dependent synaptic plasticity of NMDA receptors. J Physiol. 2010;588(Pt 1):93–99. doi: 10.1113/jphysiol.2009.179382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Golomb D, Shedmi A, Curtu R, Ermentrout GB. Persistent synchronized bursting activity in cortical tissues with low magnesium concentration: A modeling study. J Neurophysiol. 2006;95(2):1049–1067. doi: 10.1152/jn.00932.2005. [DOI] [PubMed] [Google Scholar]
- 13.Tegnér J, Compte A, Wang XJ. The dynamical stability of reverberatory neural circuits. Biol Cybern. 2002;87(5–6):471–481. doi: 10.1007/s00422-002-0363-9. [DOI] [PubMed] [Google Scholar]
- 14.Vyazovskiy VV, et al. Cortical firing and sleep homeostasis. Neuron. 2009;63(6):865–878. doi: 10.1016/j.neuron.2009.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen C, Hardy M, Zhang J, LaHoste GJ, Bazan NG. Altered NMDA receptor trafficking contributes to sleep deprivation-induced hippocampal synaptic and cognitive impairments. Biochem Biophys Res Commun. 2006;340(2):435–440. doi: 10.1016/j.bbrc.2005.12.021. [DOI] [PubMed] [Google Scholar]
- 16.Kopp C, Longordo F, Nicholson JR, Lüthi A. Insufficient sleep reversibly alters bidirectional synaptic plasticity and NMDA receptor function. J Neurosci. 2006;26(48):12456–12465. doi: 10.1523/JNEUROSCI.2702-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aton SJ, et al. Mechanisms of sleep-dependent consolidation of cortical plasticity. Neuron. 2009;61(3):454–466. doi: 10.1016/j.neuron.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dash MB, Douglas CL, Vyazovskiy VV, Cirelli C, Tononi G. Long-term homeostasis of extracellular glutamate in the rat cerebral cortex across sleep and waking states. J Neurosci. 2009;29(3):620–629. doi: 10.1523/JNEUROSCI.5486-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chih B, Gollan L, Scheiffele P. Alternative splicing controls selective trans-synaptic interactions of the neuroligin-neurexin complex. Neuron. 2006;51(2):171–178. doi: 10.1016/j.neuron.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 20.Song JY, Ichtchenko K, Südhof TC, Brose N. Neuroligin 1 is a postsynaptic cell-adhesion molecule of excitatory synapses. Proc Natl Acad Sci USA. 1999;96(3):1100–1105. doi: 10.1073/pnas.96.3.1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barrow SL, et al. Neuroligin1: A cell adhesion molecule that recruits PSD-95 and NMDA receptors by distinct mechanisms during synaptogenesis. Neural Dev. 2009;4:17. doi: 10.1186/1749-8104-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wittenmayer N, et al. Postsynaptic Neuroligin1 regulates presynaptic maturation. Proc Natl Acad Sci USA. 2009;106(32):13564–13569. doi: 10.1073/pnas.0905819106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jung SY, et al. Input-specific synaptic plasticity in the amygdala is regulated by neuroligin-1 via postsynaptic NMDA receptors. Proc Natl Acad Sci USA. 2010;107(10):4710–4715. doi: 10.1073/pnas.1001084107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim J, et al. Neuroligin-1 is required for normal expression of LTP and associative fear memory in the amygdala of adult animals. Proc Natl Acad Sci USA. 2008;105(26):9087–9092. doi: 10.1073/pnas.0803448105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peixoto RT, et al. Transsynaptic signaling by activity-dependent cleavage of neuroligin-1. Neuron. 2012;76(2):396–409. doi: 10.1016/j.neuron.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blundell J, et al. Neuroligin-1 deletion results in impaired spatial memory and increased repetitive behavior. J Neurosci. 2010;30(6):2115–2129. doi: 10.1523/JNEUROSCI.4517-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang H, Liu Y, Briesemann M, Yan J. Computational analysis of gene regulation in animal sleep deprivation. Physiol Genomics. 2010;42(3):427–436. doi: 10.1152/physiolgenomics.00205.2009. [DOI] [PubMed] [Google Scholar]
- 28.Franken P, Chollet D, Tafti M. The homeostatic regulation of sleep need is under genetic control. J Neurosci. 2001;21(8):2610–2621. doi: 10.1523/JNEUROSCI.21-08-02610.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boucard AA, Chubykin AA, Comoletti D, Taylor P, Südhof TC. A splice code for trans-synaptic cell adhesion mediated by binding of neuroligin 1 to alpha- and beta-neurexins. Neuron. 2005;48(2):229–236. doi: 10.1016/j.neuron.2005.08.026. [DOI] [PubMed] [Google Scholar]
- 30.Reddy AB, et al. Circadian orchestration of the hepatic proteome. Curr Biol. 2006;16(11):1107–1115. doi: 10.1016/j.cub.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 31.Thompson CL, et al. Molecular and anatomical signatures of sleep deprivation in the mouse brain. Front Neurosci. 2010;4:165. doi: 10.3389/fnins.2010.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Varoqueaux F, et al. Neuroligins determine synapse maturation and function. Neuron. 2006;51(6):741–754. doi: 10.1016/j.neuron.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 33.Franken P, et al. NPAS2 as a transcriptional regulator of non-rapid eye movement sleep: Genotype and sex interactions. Proc Natl Acad Sci USA. 2006;103(18):7118–7123. doi: 10.1073/pnas.0602006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Landolt HP, et al. Caffeine attenuates waking and sleep electroencephalographic markers of sleep homeostasis in humans. Neuropsychopharmacology. 2004;29(10):1933–1939. doi: 10.1038/sj.npp.1300526. [DOI] [PubMed] [Google Scholar]
- 35.Cirelli C, Tononi G. The search for the molecular correlates of sleep and wakefulness. Sleep Med Rev. 2001;5(5):397–408. doi: 10.1053/smrv.2001.0160. [DOI] [PubMed] [Google Scholar]
- 36.Huber R, Tononi G, Cirelli C. Exploratory behavior, cortical BDNF expression, and sleep homeostasis. Sleep. 2007;30(2):129–139. doi: 10.1093/sleep/30.2.129. [DOI] [PubMed] [Google Scholar]
- 37.Maret S, et al. Homer1a is a core brain molecular correlate of sleep loss. Proc Natl Acad Sci USA. 2007;104(50):20090–20095. doi: 10.1073/pnas.0710131104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Curie T, et al. Homeostatic and circadian contribution to EEG and molecular state variables of sleep regulation. Sleep. 2013;36(3):311–323. doi: 10.5665/sleep.2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gsell W, et al. Differential effects of NMDA and AMPA glutamate receptors on functional magnetic resonance imaging signals and evoked neuronal activity during forepaw stimulation of the rat. J Neurosci. 2006;26(33):8409–8416. doi: 10.1523/JNEUROSCI.4615-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ko CH, Takahashi JS. 2006. Molecular components of the mammalian circadian clock. Hum Mol Genet 15(Spec 2): R271–R277.
- 41.Mongrain V, La Spada F, Curie T, Franken P. Sleep loss reduces the DNA-binding of BMAL1, CLOCK, and NPAS2 to specific clock genes in the mouse cerebral cortex. PLoS ONE. 2011;6(10):e26622. doi: 10.1371/journal.pone.0026622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suzuki K, et al. Activity-dependent proteolytic cleavage of neuroligin-1. Neuron. 2012;76(2):410–422. doi: 10.1016/j.neuron.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 43.Kattenstroth G, Tantalaki E, Südhof TC, Gottmann K, Missler M. Postsynaptic N-methyl-D-aspartate receptor function requires alpha-neurexins. Proc Natl Acad Sci USA. 2004;101(8):2607–2612. doi: 10.1073/pnas.0308626100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chubykin AA, et al. Activity-dependent validation of excitatory versus inhibitory synapses by neuroligin-1 versus neuroligin-2. Neuron. 2007;54(6):919–931. doi: 10.1016/j.neuron.2007.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Novati A, Hulshof HJ, Granic I, Meerlo P. Chronic partial sleep deprivation reduces brain sensitivity to glutamate N-methyl-D-aspartate receptor-mediated neurotoxicity. J Sleep Res. 2012;21(1):3–9. doi: 10.1111/j.1365-2869.2011.00932.x. [DOI] [PubMed] [Google Scholar]
- 46.Tamaru T, et al. Light and glutamate-induced degradation of the circadian oscillating protein BMAL1 during the mammalian clock resetting. J Neurosci. 2000;20(20):7525–7530. doi: 10.1523/JNEUROSCI.20-20-07525.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Franken P, Malafosse A, Tafti M. Genetic determinants of sleep regulation in inbred mice. Sleep. 1999;22(2):155–169. [PubMed] [Google Scholar]
- 48.Naidoo N, et al. Role of Homer proteins in the maintenance of sleep-wake states. PLoS ONE. 2012;7(4):e35174. doi: 10.1371/journal.pone.0035174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leemburg S, et al. Sleep homeostasis in the rat is preserved during chronic sleep restriction. Proc Natl Acad Sci USA. 2010;107(36):15939–15944. doi: 10.1073/pnas.1002570107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Südhof TC. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 2008;455(7215):903–911. doi: 10.1038/nature07456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kotagal S, Broomall E. Sleep in children with autism spectrum disorder. Pediatr Neurol. 2012;47(4):242–251. doi: 10.1016/j.pediatrneurol.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 52.Franken P, Thomason R, Heller HC, O’Hara BF. A non-circadian role for clock-genes in sleep homeostasis: A strain comparison. BMC Neurosci. 2007;8:87. doi: 10.1186/1471-2202-8-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seibt J, et al. Protein synthesis during sleep consolidates cortical plasticity in vivo. Curr Biol. 2012;22(8):676–682. doi: 10.1016/j.cub.2012.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.