Abstract
Cytokinins are plant hormones that play critical roles in growth and development. In Arabidopsis, the transcriptional response to cytokinin is regulated by action of type-B Arabidopsis response regulators (ARRs). Although central elements in the cytokinin signal transduction pathway have been identified, mechanisms controlling output remain to be elucidated. Here we demonstrate that a family of F-box proteins, called the KISS ME DEADLY (KMD) family, targets type-B ARR proteins for degradation. KMD proteins form an S-PHASE KINASE-ASSOCIATED PROTEIN1 (SKP1)/Cullin/F-box protein (SCF) E3 ubiquitin ligase complex and directly interact with type-B ARR proteins. Loss-of-function KMD mutants stabilize type-B ARRs and exhibit an enhanced cytokinin response. In contrast, plants with elevated KMD expression destabilize type-B ARR proteins leading to cytokinin insensitivity. Our results support a model in which an SCFKMD complex negatively regulates cytokinin responses by controlling levels of a key family of transcription factors.
Keywords: cell signaling, root development, proteasome, two-component system
Cytokinins are mitogenic plant hormones that control multiple aspects of growth and development. Cytokinins were initially discovered and named based on their ability to promote cell division of plant cells grown in culture (1). Subsequent research defined a critical role for cytokinins in regulating cell division at stem cell niches throughout the life cycle of the plant (2, 3). Cytokinins also stimulate chloroplast development, modulate shoot and root development, delay leaf senescence, and regulate abiotic and biotic stress responses (4–6).
Cytokinin signals are transmitted through a multistep histidine-to-aspartate phosphorelay system, evolutionarily related to the two-component signaling systems of prokaryotes (4, 7). In Arabidopsis, cytokinins are perceived by the three receptors ARABIDOPSIS HISTIDINE KINASE 2 (AHK2), AHK3, and AHK4, which upon perception of the cytokinin signal autophosphorylate on a conserved His residue (8, 9). The regulatory phosphoryl group is passed from receptor to a histidine-containing phosphotransfer (AHP) protein, and from there to a type-B Arabidopsis response regulator (ARR). Phosphorylation of the Arabidopsis type-B ARR proteins modulates their ability to control gene expression as the key transcription factors in the primary response pathway (10, 11). Among the transcriptional targets of type-B ARRs are a second class of response regulators, type-A ARRs, which act as negative regulators of the signal transduction pathway (12). This model of cytokinin signal transduction has been established largely based on studies in Arabidopsis, but similar two-component signaling elements have been identified in other plant species such as monocot rice and the moss Physcomitrella patens, supporting a common pathway for the transmission of the cytokinin signal in land plants (13, 14). Although central elements in the cytokinin signal transduction pathway have been identified, mechanisms controlling output remain to be elucidated.
Control of protein stability through ubiquitin-mediated proteolysis has emerged as a central theme in plant growth and development over the past decade (15). E3 ubiquitin ligases have been identified that target key signaling elements for degradation in many plant hormone signaling pathways, including those for auxin, ethylene, gibberellin, and jasmonic acid (16). Identification of such a regulatory component for cytokinin signal transduction has proved elusive, although several studies suggest that the ubiquitin-proteasome system plays a role in the regulation of cytokinin signaling (17–20). Most significantly, the stability of the type-B response regulator ARR2 decreases in the presence of cytokinin, and a more stable mutant version of ARR2 enhances cytokinin sensitivity in various developmental processes (17). In this study, we identify a family of F-box proteins, designated the KISS ME DEADLY (KMD) family, that physically interact with and target type-B ARR proteins for degradation. Genetic analysis demonstrates that the KMD family members function as negative regulators of the cytokinin-signaling pathway. Thus, cytokinin joins the other classical plant hormones in having a key transcriptional regulator controlled by S-PHASE KINASE-ASSOCIATED PROTEIN1 (SKP1)/Cullin/F-box (SCF)/proteasome-mediated degradation.
Results
Cytokinin-Regulated KMD Genes Encode F-Box Proteins as Part of an SCF Complex.
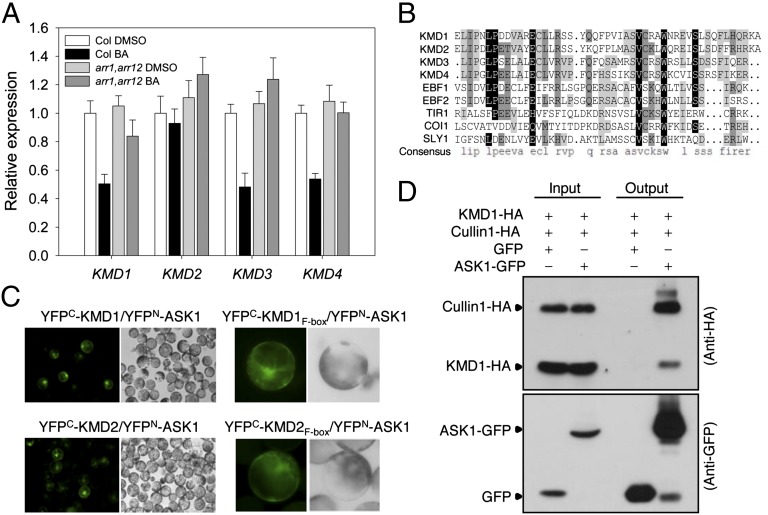
F-box proteins, which function as the specificity components within SCF-E3 ligases, are key regulators for several plant hormone-signaling pathways (15, 16). By examining microarray data (21, 22), we identified a cytokinin-regulated gene (accession no. At1g80440) encoding an F-box protein of unknown function. The protein encoded by At1g80440 contains an amino-terminal F-box motif followed by a Kelch-repeat domain that could mediate protein–protein interactions (23). We designated At1g80440 as KMD1 and its homologs At1g15670, At2g44130, and At3g59940 as KMD2, KMD3, and KMD4, respectively (Fig. S1 A and B). Expression of the three family members KMD1, -3, and -4 exhibited a twofold reduction in transcript levels within 1 h of cytokinin treatment (Fig. 1A). The effect of cytokinin on KMD gene expression was reduced in type-B ARR double mutant arr1, arr12 (24) (Fig. 1A), indicating the necessity of these type-B ARRs for mediating the transcriptional effect of cytokinin on KMD expression. Based on normalized microarray data from the Arabidopsis electronic fluorescent pictograph (eFP) browser (25, 26), KMD family members are broadly expressed, with expression at the shoot apical meristem (especially KMD1 and KMD2) and at the root tip (especially KMD2 and KMD3), tissues where cytokinin regulates cell division (2–4).
Fig. 1.
Cytokinin-regulated KMD genes encode F-box proteins interacting with components of the SCF complex. (A) Fourteen-day-old wild-type (Col) and arr1,arr12 mutant seedlings were treated with 10 µM benzyladenine (BA) or a DMSO vehicle control for 1 h, and expression of KMD1, KMD2, KMD3, and KMD4 analyzed by qRT-PCR in shoots excised from the treated seedlings. Error bars are ±SD. (B) Alignment of F-box motifs from Arabidopsis F-box proteins. Identities and similarities are highlighted by black and gray, respectively. (C) BiFC analysis of KMD1-ASK1 and KMD2-ASK1 interaction. The F-box domain (KMD1F-box or KMD2F-box) is sufficient for interaction with ASK1 in mesophyll protoplast cells. (D) Coimmunoprecipitation of KMD1-HA and Cullin1-HA with ASK1-GFP based on anti-GFP immunoprecipitation from transfected protoplasts. The immunoblot was probed with anti-HA and anti-GFP antibodies. GFP served as a negative control for the immunoprecipitation.
KMD proteins contain a conserved amino-terminal F-box motif (Fig. 1B and Fig. S1B). To determine if KMDs function as canonical F-box proteins, we examined their interactions with known components of the Arabidopsis SCF complex. All four of the full-length KMD proteins interacted with ASK1, an Arabidopsis Skp1 protein of the SCF complex, in a bimolecular fluorescence complementation (BiFC) assay in protoplast cells (Fig. 1C and Fig. S1 C and D). The F-box domains of KMD1 and KMD2 were sufficient for interaction with ASK1 (Fig. 1C). The BiFC fluorescent signal was detected in both the nucleus and cytoplasm (Fig. 1C and Fig. S1D), consistent with the subcellular localization of KMD proteins (Fig. S1E), implying that KMD proteins may function as F-box proteins in the cytoplasm as well as in the nucleus. We performed coimmunoprecipitation analysis to confirm interactions of KMD1 within an SCF complex (Fig. 1D). Both hemagglutinin (HA)-tagged KMD1 and Cullin1 coimmunoprecipitated with green-fluorescence-protein (GFP)-tagged ASK1, consistent with the KMD proteins functioning within a SCF complex in plant cells.
KMD Family Members Function as Negative Regulators of Cytokinin Responses.
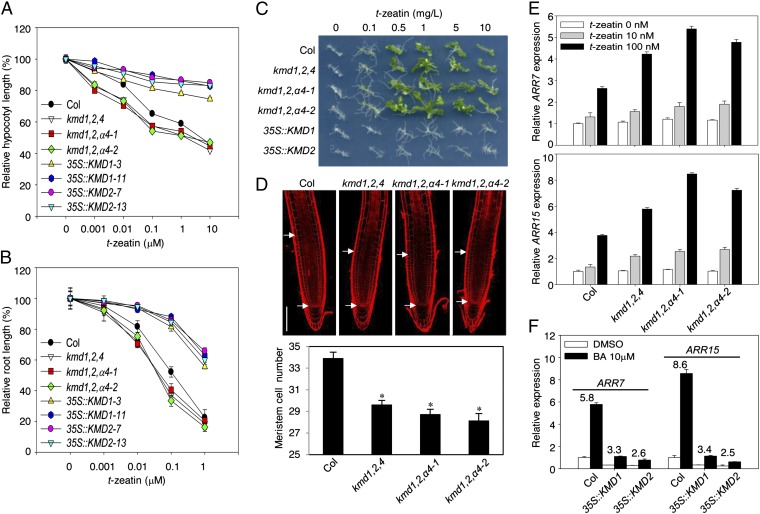
To investigate the functional role of KMDs in cytokinin signaling, we isolated transfer DNA (T-DNA) insertion mutations in the KMD1, KMD2, and KMD4 genes, and constructed single, double, and triple mutants. No full-length transcripts were detected for the mutant alleles based on RT-PCR analysis, suggesting, based on the position of the T-DNA inserts, that these represent null alleles (Fig. S2 A and B). We also generated independent antisense transgenic lines using a KMD4 antisense construct in the kmd1,2 background (kmd1,2,α4), which resulted in reduced transcript levels of both KMD3 and KMD4 (Fig. S2B). To examine the contributions of KMD genes to cytokinin responsiveness, the kmd mutants were examined for their inhibition of hypocotyl elongation in response to exogenous cytokinin. Single and double kmd mutants were comparable to the wild type, whereas the kmd1-1; kmd2-1; kmd4-1 (kmd1,2,4) and kmd1,2,α4 mutants showed enhanced sensitivity to cytokinin, indicating genetic redundancy among these genes (Fig. 2A and Fig. S2C). The higher order kmd mutants also exhibited increased sensitivity to cytokinin in a root growth response assay as well as in the ability of cytokinin to induce greening and shoot formation in tissue-cultured explants, the effects in tissue culture being most pronounced in the kmd1,2,α4–2 line (Fig. 2 A–C and Fig. S2C). The effect of cytokinin on root development is in part due to a negative effect on root meristem size (4, 27). Consistent with this, both kmd1,2,4 and kmd1,2,α4 lines had fewer cells in their root meristems (Fig. 2D), suggesting that the absence of endogenous KMD proteins mimics the effect of cytokinin by reducing the size of the root meristem.
Fig. 2.
Members of the KMD family negatively regulate cytokinin signaling. (A and B) Effect of cytokinin on hypocotyl (A) and root (B) growth of Col, kmd1,2,4, kmd1,2,α4 antisense lines, and KMD-overexpression lines (35S::KMD1-3, 35S::KMD1-11, 35S::KMD2-7, and 35S::KMD2-13). In A, etiolated seedlings were grown for 4 d on the indicated t-zeatin concentrations. Error bars are ±SE (not shown if smaller than symbol; n ≥ 20). In B, seedlings were grown on vertical plates supplemented with t-zeatin under constant light. Increase in root length from day 4 through day 7 was measured. Error bars are ±SE (not shown if smaller than symbol; n ≥ 18). (C) Effect of a cytokinin on callus formation and shoot initiation. Calli were prepared from the hypocotyl tissues of the indicated lines, then incubated on shoot induction medium containing indole-3-butyric acid (0.2 mg/L) and the indicated concentrations of t-zeatin. (D) Reduced expression of the KMD family results in a decrease in root meristem size. Roots of 7-d-old seedlings of Col, kmd1,2,4, and kmd1,2,α4 antisense lines were stained with propidium iodide and visualized by confocal fluorescence microscopy, and cortex meristematic cell numbers measured. White arrows indicate the transition zone where cells leave the meristem and enter the elongation-differentiation zone. (Scale bar, 100 μm.) Asterisks indicate significant differences (P < 0.0001). Error bars are ±SE (n ≥ 20). (E and F) RNA levels of ARR7 and ARR15 were analyzed by qRT-PCR in Col, kmd1,2,4, and kmd1,2,α4 antisense lines treated with 0, 10, or 100 nM t-zeatin for 1 h (E); or in Col, 35S::KMD1-11, and 35S::KMD2-13 lines treated with 10 μM BA or a DMSO vehicle control for 1 h, the fold change in response being indicated for the lines (F).
We confirmed that the KMD family functions as a negative regulator for cytokinin signaling by examining transgenic lines overexpressing KMD1 and KMD2 under the constitutive cauliflower mosaic virus (CaMV) 35S promoter (Fig. S2D). These overexpression lines exhibited a substantially reduced cytokinin response in several physiological assays, including hypocotyl and root growth in response to cytokinin as well as in shoot initiation assays (Fig. 2 A–C). Consistent with their reduced sensitivity to cytokinin in the root growth assay (4, 27), the overexpression lines exhibited larger root meristems (Fig. S2E). Significantly, the 35S::KMD1 and -2 lines responded normally to the ethylene precursor 1-aminocyclopropane-1-carboxylic acid (ACC) and the synthetic auxin naphthalene-1-acetic acid (NAA) (Fig. S2 F and G), indicating a specificity toward cytokinin for their effect on signaling. We also expressed KMD1 and KMD2 using the cassava vein mosaic virus (CsVMV) promoter, which yielded higher expression levels than that mediated by the CaMV 35S promoter (Fig. S2D). Seedlings of the CsVMV::KMD lines were stunted and exhibited premature termination of primary root growth (Fig. S3A). The shoot apical meristem (SAM) of the CsVMV::KMD lines exhibited a wild-type structure and organization, but was substantially smaller than that found in wild-type seedlings (Fig. S3B). In addition, the meristems of the prematurely aborted primary roots were substantially smaller in the CsVMV::KMD lines compared with wild type (Fig. S3 C and D). These meristematic changes resulting from CsVMV promoter-driven overexpression of KMD1 and KMD2 are similar to those previously reported for severe mutations in the primary cytokinin signaling pathway (11, 28, 29).
To determine if the increased cytokinin sensitivity of higher order kmd mutants is associated with enhanced cytokinin signal transduction, we examined expression of the cytokinin primary response genes ARR7 and ARR15 by qRT-PCR (Fig. 2E). In wild-type plants, ARR7 and ARR15 were induced two to fourfold by 1-h treatment with 100 nM t-zeatin. Both the kmd1,2,4 and kmd1,2,α4 lines exhibited a greater amplitude in cytokinin-induced ARR7 and ARR15 expression compared with wild type (1.5- to twofold stronger induction; Fig. 2E), a similar change in responsiveness to what has been found with mutations in other negative regulators of the pathway (12). The molecular response to cytokinin was also altered in the 35S::KMD1 and -2 lines. The basal RNA levels of ARR7 and ARR15 were decreased compared with that in the wild type (Fig. 2F), indicating a reduced response to endogenous cytokinin in the 35S::KMD lines. Moreover, the degree of induction of the ARR genes by exogenous cytokinin was also reduced by KMD overexpression (Fig. 2F). This molecular phenotype is similar to what has been observed with mutations in other positive regulators of the cytokinin signaling pathway (11, 28, 29). Overall, these physiological and molecular results reveal that the KMD family members function as negative regulators of the cytokinin signaling pathway.
KMD Proteins Physically Interact with Type-B ARR Proteins.
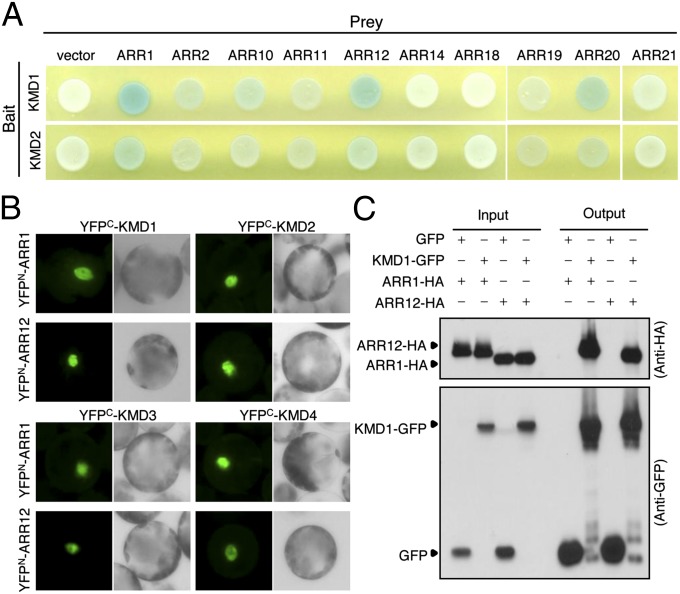
The similarity of KMD mutant phenotypes (Fig. 2) to those involving type-B ARR mutations (Fig. S3) (11, 24, 30, 31), suggested that these F-box proteins might target type-B ARR proteins for degradation. To test for interaction between KMD and type-B ARR proteins, yeast two-hybrid assays were performed (Fig. 3A and Fig. S4 A and B). The KMD proteins interacted with ARR1 and ARR12, and to a lesser extent ARR2 and ARR10, of the subfamily-1 type-B ARRs. The KMD proteins also interacted with ARR20 of type-B ARR subfamily-3. Significantly, the KMD proteins did not interact with representative members of type-A ARRs (ARR4 and ARR7), indicating that interaction with the KMD proteins is not a general characteristic of the plant response regulators. We focused our subsequent interaction analysis on ARR1 and ARR12 because genetic studies have demonstrated that these two type-B ARRs are major contributors to multiple cytokinin responses (11, 24, 30). All four KMD proteins interacted with both ARR1 and ARR12 in a BiFC assay (Fig. 3B). Strong fluorescence was detected in the nuclei of cells, consistent with the subcellular localization of type-B ARRs (32, 33). In addition, HA-tagged ARR1 or ARR12 proteins coimmunoprecipitated with a GFP-tagged KMD1 protein from extracts of transfected protoplasts (Fig. 3C). ARR2 also interacted in vivo with KMD1 based on BiFC analysis (Fig. S4C), even though this interaction was weaker based on the yeast-two hybrid analysis (Fig. 3A and Fig. S4A), supporting the significance of the detectable yeast-two hybrid interactions. We did not find any effect of cytokinin on the interaction of KMDs with ARRs based on yeast-two hybrid and BiFC analyses. These results indicate that multiple type-B ARRs serve as direct targets for the KMD F-box proteins. In a BiFC assay, we found that KMD1 interacted more strongly with the C-terminal domains of ARR1 and ARR12 [ARR1 (153–669) and ARR12 (133–596)] than with their receiver domains [ARR1 (1–154) and ARR12 (1–134)] (Fig. S4D).
Fig. 3.
KMD proteins physically interact with type-B ARR proteins. (A) Yeast two-hybrid assay between KMD proteins (KMD1 or KMD2) and type-B ARR proteins. Pairs of indicated bait and prey vectors were transformed into yeast cells. The growth of a blue yeast colony on selective medium containing X-gal indicates a positive interaction. (B) Interaction between KMD proteins and ARR1 or ARR12 in BiFC assay. The indicated constructs were cotransfected into the Arabidopsis mesophyll protoplasts. (C) Coimmunoprecipitation of type-B ARR proteins (ARR1 or ARR12) and KMD1. Protoplast cells were cotransfected with ARR1-HA or ARR12-HA along with either KMD1-GFP or a GFP control. The KMD1-GFP or GFP proteins were immunoprecipitated with an anti-GFP antibody, and an immunoblot was probed with anti-HA and anti-GFP antibodies.
KMD1 and KMD2 Target Type-B ARR Proteins for Degradation.
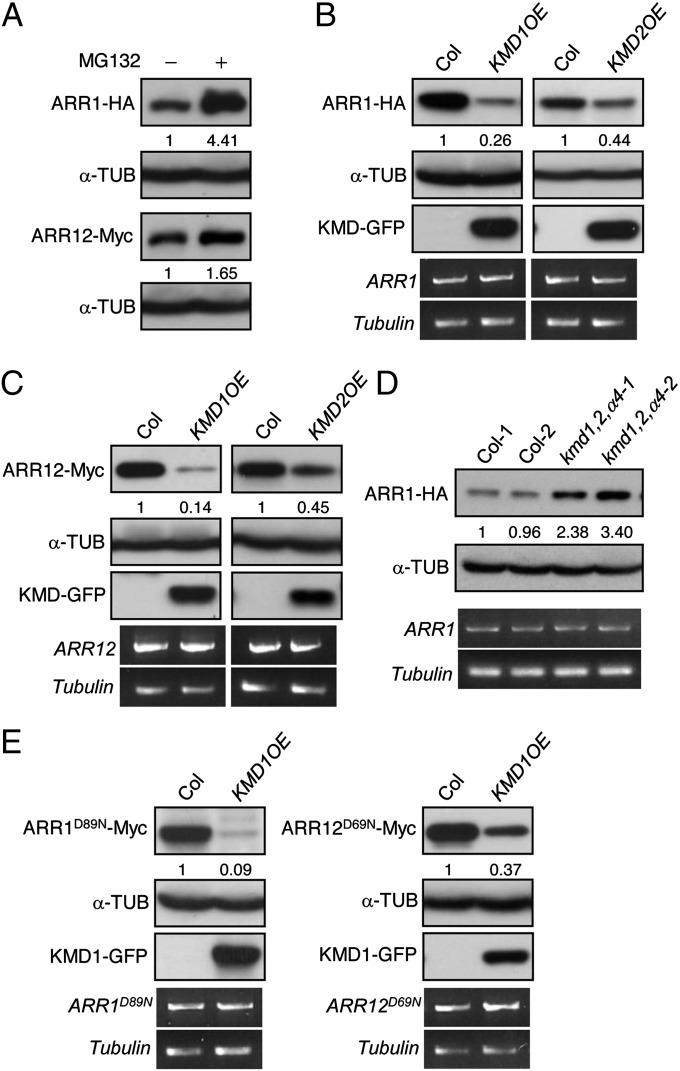
Protein levels of ARR1 and ARR12 increase in the presence of the proteasome inhibitor MG132 (Fig. 4A and Fig. S5A), consistent with their degradation being controlled by the ubiquitin/proteasome system. In addition, treatment with the protein biosynthesis inhibitor cycloheximide revealed that both ARR1 and ARR12 are unstable proteins. Interestingly, their turnover was not affected by exogenous cytokinin (Fig. S5A), suggesting that the lifespan of type-B ARR may contribute to its ability to propagate the cytokinin signal. To determine if type-B ARRs are targeted by KMDs for degradation in planta, we crossed transgenic lines expressing 35S::ARR1-HA, 35S::ARR2-HA, or 35S::ARR12-Myc with wild-type or 35S::KMD-GFP plants, and analyzed the abundance of type-B ARR proteins in the F1 seedlings. Elevated expression of KMD1 and KMD2 resulted in a decrease in ARR1, ARR2, and ARR12 protein levels (Fig. 4 B and C and Fig. S5B). Furthermore, analysis of the turnover kinetics for ARR1 indicates that elevated levels of KMD1 result in an enhanced turnover rate for ARR1 (Fig. S5C). MG132 retarded degradation of ARR1 in the KMD1 overexpression line (Fig. S5D), consistent with degradation being mediated by the ubiquitin/proteasome pathway. Based on the overexpression analysis, KMD1 was more effective than KMD2 in targeting type-B ARRs for degradation. Consistent with a role for KMD proteins in controlling type-B ARR levels, we also found that the ARR1 protein is more abundant in the kmd1,2,α4 lines (Fig. 4D and Fig. S5E), providing a mechanistic basis for why these lines display enhanced cytokinin sensitivity (31). Because no changes in ARR1 and ARR12 transcript levels were observed in these lines (Fig. 4 B–D), our results are consistent with the KMDs mediating the posttranslational degradation of type-B ARRs. To investigate whether canonical phosphoregulation of type-B ARRs is necessary for their SCFKMD-mediated degradation in planta, we generated ARR1Asp89Asn (ARR1D89N) and ARR12D69N, in which the phosphotransfer to ARR1 and ARR12 is predicted to be abolished due to mutation of the conserved Asp in the receiver domain (34, 35). The levels of ARR1D89N and ARR12D69N decreased in the presence of overexpressed KMD1 (Fig. 4E), suggesting that a cytokinin-induced phosphorelay to this conserved residue is not required for the SCFKMD-mediated degradation of ARR1 and ARR12.
Fig. 4.
KMD1 and KMD2 target type-B ARR proteins for degradation. (A) Proteasome-dependent degradation of ARR1 and ARR12. Mesophyll protoplasts from 35S::ARR1-HA or 35S::ARR12-Myc lines were incubated for 3 h in the presence or absence of the proteasome inhibitor MG132 and protein extracts analyzed by immunoblotting. α-Tubulin served as a loading control. (B and C) Overexpression of KMD1 and KMD2 results in reduced protein levels of ARR1 (B) and ARR12 (C). Protein levels of the tagged proteins were determined by immunoblot analysis using anti-HA and anti-Myc antibodies, with α-tubulin as the loading control. Transcript levels for transgenes were detected using HA- or Myc-tag–specific primers by RT-PCR, with β-tubulin as the loading control. (D) Decreased expression of the KMD family results in increased protein levels of ARR1. Immunoblot detection of ARR1-HA in 10-d-old wild-type and kmd1,2,α4 seedlings, with α-tubulin as the loading control is shown. Each lane represents a protein sample from independent seedlings. (E) Degradation of ARR1 and ARR12 is not dependent on the conserved phosphorylation target Asp. The predicted phosphorylation target residues on ARR1 and ARR12 were mutated to Asn (ARR1D89N; ARR12D69N), and stability was examined in the presence or absence of KMD1 overexpression.
Discussion
We demonstrate a role for F-box proteins in regulating the cytokinin signal transduction pathway, identifying type-B ARR transcription factors as targets of the “orphan” KMD F-box proteins. A model integrating SCFKMD into the cytokinin signaling pathway is shown in Fig. S6A. SCFKMD interacts with and targets multiple type-B ARRs for degradation, notably the subfamily-1 members that mediate the majority of the cytokinin transcriptional response in Arabidopsis (11, 30, 35), changes in type-B ARR protein levels serving to regulate the cytokinin responsiveness of the plant. Consistent with this model are our data demonstrating that (i) loss-of-function mutations in the KMD family result in an enhanced sensitivity of plants to cytokinin, (ii) overexpression of KMD family members results in a decreased sensitivity of plants to cytokinin, (iii) KMDs physically interact with type-B ARRs, and (iv) the loss- and gain-of-function kmd mutant phenotypes correlate with changes in type-B ARR protein levels. Type-B ARRs are predicted to be direct targets of the KMD family based on the specificity of F-box proteins for their ubiquitination targets, our interaction data, and the proteasome-dependent turnover of type-B ARRs. The cytokinin signaling pathway is conserved in plants (13), and homologs to KMDs exist in the monocot rice (Fig. S1A) as well as the mosses P. patens (XP_001756872) and Selagenella moellendorffii (XP_002983138), supporting the broad conservation of this regulatory mechanism for controlling the cytokinin transcriptional output among the land plants.
Our results, coupled with prior analyses of type-A and type-B ARR stability, point to a central role of the ubiquitin-proteasome pathway in controlling the protein levels of two families of plant response regulators. A subset of type-A response regulators, which act as negative regulators of the cytokinin signaling pathway, are stabilized in response to cytokinin (20). In addition, proteasome-dependent degradation of ARR2, a type-B ARR, is enhanced by cytokinin-induced phosphorylation of the conserved Asp residue in the receiver domain (17). The two-component signaling system is evolutionarily ancient (36), and our study points to how plants, like bacteria, use degradation of response regulators as a pivotal mechanism for transcriptional regulation (37, 38). Plants, unlike bacteria, accomplish this by using the eukaryotic-specific ubiquitin-proteasome system.
Several additional studies implicate the ubiquitin-proteasome in the control of cytokinin signaling. Mutants of REGULATORY PARTICLE NON-ATPASE12 (RPN12), which is a subunit of proteasome regulatory structures, exhibit reduced cytokinin sensitivity, indicating that the 26S proteasome is required to degrade one or more repressors of cytokinin action in plants (19). Candidates for such a repressor include type-A ARRs as well as KMD proteins identified here. More recently, AUXIN UP-REGULATED F-BOX PROTEIN 1 (AUF1) was identified in the search for components of the ubiquitin-proteasome pathway that affect cytokinin signaling (18). No effects of AUF1 mutants were found on type-B ARR protein levels, suggesting that the role of AUF1 in cytokinin signaling is indirect, potentially via cross-talk through the auxin signaling pathway. Significantly, we found that the effects of KMD loss- and gain-of-function mutations are specific for cytokinin, mutant seedlings responding normally to auxin, based on several physiological assays, demonstrating the specificity of SCFKMD toward the cytokinin signaling pathway.
Activity of response regulators such as type-B ARRs is typically regulated by phosphorylation of a conserved Asp residue within the receiver domain, phosphorylation thought to induce conformational changes that activate type-B ARRs (31, 39, 40). However, based on our analysis, SCFKMD appears to regulate stability of type-B ARRs in a phospho-Asp-independent manner. This is consistent with known characteristics for type-B ARR turnover which, with the exception of ARR2 (17), is not affected by cytokinin treatment. An ability of SCFKMD to mediate degradation of both inactive and activated forms of type-B ARRs could serve two purposes based on well-established models for signal transduction (Fig. S6B) (41). First, SCFKMD could regulate the abundance of type-B ARRs, thereby determining the threshold level for a cytokinin response, a mechanism that may facilitate cross-talk with other signaling pathways; for example, expression of KMD1, -3, and -4 exhibits circadian dependence in response to light and temperature (25, 26, 42), and could thus regulate the interaction between cytokinin and the circadian clock (43). Second, SCFKMD could remove activated type-B ARRs, thereby preventing continued transcriptional activation by cytokinin. In its simplest form, this suggests a “timer” model for transcriptional regulation, the stability and lifespan of the active type-B ARR modulating its signal output. Differences in the efficacy of the KMD family members in targeting type-B ARRs for degradation may allow for fine tuning of cytokinin responses; differences in efficacy have also been observed in the EIN3 BINDING F-BOX1 (EBF1) F-box family that target the ETHYLENE-INSENSITIVE3 (EIN3) transcription factor family to control the ethylene response, EBF2 being more effective than EBF1 based on overexpression analysis (44, 45). Although we do not observe an effect of cytokinin on KMD-mediated degradation of type-B ARRs, KMD interaction with the C-terminal portion of type-B ARRs suggests the DNA-binding/activation domain of the transcription factor may play a role in its degradation. In animals and bacteria, transcriptional activation domains often overlap with the degrons that target the transcription factor for proteolysis, such that activation is closely linked to degradation (41).
With the identification of a role for SCFKMD, cytokinin joins the other classical phytohormones auxin, jasmonate, ethylene, and gibberellin in having key transcriptional regulators controlled by SCF/proteasome-mediated degradation (16, 46, 47). The role of degradation in the cytokinin signaling pathway, however, differs from that in these other hormonal signaling pathways. Auxin, jasmonate, and gibberellin all target transcriptional repressors for degradation. Ethylene, like cytokinin, employs a transcriptional activator (EIN3), but EIN3 is ubiquitinated and degraded in the absence of ethylene, stabilization of EIN3 in the presence of ethylene leading to activation of ethylene-response genes. In contrast, type-B ARRs functioning in cytokinin signaling are present in the absence of cytokinin but undergo continuous proteolysis. Although the mechanisms used for transcriptional control vary among these five phytohormones, constants are a role for SCF/proteasome-mediated degradation and a targeting of key elements involved in transcription as a means of modulating signal output.
Materials and Methods
Plant Materials and Growth Conditions.
Arabidopsis thaliana Columbia (Col-0) ecotype was the parent strain for mutants and transgenic lines. The kmd1-1 (SALK_008497) and kmd4-1 (SALK_080249) T-DNA insertion lines were obtained from the Salk T-DNA insertion collections (48), and kmd2-1 (GABI-KAT 079A01) plants were obtained from the GABI-KAT T-DNA insertion collection (http://www.gabi-kat.de). The genotype of each line was confirmed by PCR-based methodology (Table S1). For detailed description on the growth conditions, see SI Materials and Methods.
Cytokinin Growth and Molecular Response Analyses.
Hypocotyl and root growth analysis was performed as described previously (11) and hypocotyl and root lengths were measured using ImageJ software (version 1.32; National Institutes of Health). Root meristem size was determined as described (49). The apical meristem was visualized by bright field microscopy, with tissue sectioning, fixation, and staining performed as described (50). Total RNA isolation, cDNA production, and quantitative RT-PCR (qRT-PCR) were performed as described (11). Primers used for qRT-PCR are listed in Table S1. Transcript abundances were calculated using the comparative CT method, with β-TUBULIN3 (β-TUB3) (At5g62700) as the normalized control. See SI Materials and Methods for detailed procedures.
Tissue Regeneration Assay.
Hypocotyls from 4-d-old dark-grown seedlings were excised and transferred to 1× Murashige and Skoog (MS) plate medium containing 0.5 mg/L 2,4-dichlorophenoxyacetic acid (2,4-D) and 0.05 mg/L kinetin. After 4 d, hypocotyl segments were transferred to medium containing 0.2 mg/L indole-3-butyric acid (IBA) and t-zeatin ranging from 0 to 10 mg/L (24), and callus was examined after 3 wk.
Transient Expression in Arabidopsis Protoplasts and in Vivo Coimmunoprecipitation Assay.
Arabidopsis mesophyll protoplasts were isolated from mature leaves of the wild-type plants and transfected with various constructs as described (51). For BiFC assay, cDNA fragments were fused to plant expression vector containing either amino- or carboxyl-terminal fragments of the YFP (YFPN and YFPC) (52). Transfected protoplasts were examined with an Axioplan2 fluorescent microscope (Carl Zeiss). Coimmunoprecipitation (Co-IP) was performed using agarose-conjugated anti-GFP antibody (GFP-Trap; Chromotek) following the manufacturer’s protocol with slight modifications. To examine the role of the proteasome in ARR stability, protoplasts from ARR1-HA and ARR12-Myc overexpression lines were prepared and incubated for 3 h in the presence of 10 μM MG132 or 0.1% DMSO as a vehicle control, and protein levels were determined by immunoblot analysis. SI Materials and Methods has detailed information on BiFC and co-IP experiments.
Additional materials and methods are included in SI Materials and Methods.
Supplementary Material
Acknowledgments
The authors thank the other members of the Schaller and Kieber labs, particularly Kristine Hill and Ian Street, for sharing reagents and advice. This work was supported by grants from the US Department of Agriculture, Agriculture Food and Research Initiative/National Research Institute Grant 2007-35304-18323 (to G.E.S.), National Science Foundation Grant IOS-0618286 to J.J.K. and G.E.S.), and the Human Frontier Science Program Grant LT000757/2009-L to H.J.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1300403110/-/DCSupplemental.
References
- 1.Miller CO, Skoog F, Okomura FS, von Saltza MH, Strong FM. Isolation, structure and synthesis of kinetin, a substance promoting cell division. J Am Chem Soc. 1956;78(7):1375–1380. [Google Scholar]
- 2.Werner T, Motyka V, Strnad M, Schmülling T. Regulation of plant growth by cytokinin. Proc Natl Acad Sci USA. 2001;98(18):10487–10492. doi: 10.1073/pnas.171304098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gordon SP, Chickarmane VS, Ohno C, Meyerowitz EM. Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proc Natl Acad Sci USA. 2009;106(38):16529–16534. doi: 10.1073/pnas.0908122106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hwang I, Sheen J, Müller B. Cytokinin signaling networks. Annu Rev Plant Biol. 2012;63:353–380. doi: 10.1146/annurev-arplant-042811-105503. [DOI] [PubMed] [Google Scholar]
- 5.Werner T, Schmülling T. Cytokinin action in plant development. Curr Opin Plant Biol. 2009;12(5):527–538. doi: 10.1016/j.pbi.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 6.Argueso CT, Raines T, Kieber JJ. Cytokinin signaling and transcriptional networks. Curr Opin Plant Biol. 2010;13(5):533–539. doi: 10.1016/j.pbi.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 7.Schaller GE, Shiu SH, Armitage JP. Two-component systems and their co-option for eukaryotic signal transduction. Curr Biol. 2011;21(9):R320–R330. doi: 10.1016/j.cub.2011.02.045. [DOI] [PubMed] [Google Scholar]
- 8.Inoue T, et al. Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature. 2001;409(6823):1060–1063. doi: 10.1038/35059117. [DOI] [PubMed] [Google Scholar]
- 9.Ueguchi C, Sato S, Kato T, Tabata S. The AHK4 gene involved in the cytokinin-signaling pathway as a direct receptor molecule in Arabidopsis thaliana. Plant Cell Physiol. 2001;42(7):751–755. doi: 10.1093/pcp/pce094. [DOI] [PubMed] [Google Scholar]
- 10.Kim HJ, et al. Cytokinin-mediated control of leaf longevity by AHK3 through phosphorylation of ARR2 in Arabidopsis. Proc Natl Acad Sci USA. 2006;103(3):814–819. doi: 10.1073/pnas.0505150103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Argyros RD, et al. Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell. 2008;20(8):2102–2116. doi: 10.1105/tpc.108.059584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.To JP, et al. Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell. 2004;16(3):658–671. doi: 10.1105/tpc.018978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pils B, Heyl A. Unraveling the evolution of cytokinin signaling. Plant Physiol. 2009;151(2):782–791. doi: 10.1104/pp.109.139188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsai YC, et al. Characterization of genes involved in cytokinin signaling and metabolism from rice. Plant Physiol. 2012;158(4):1666–1684. doi: 10.1104/pp.111.192765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hua Z, Vierstra RD. The cullin-RING ubiquitin-protein ligases. Annu Rev Plant Biol. 2011;62:299–334. doi: 10.1146/annurev-arplant-042809-112256. [DOI] [PubMed] [Google Scholar]
- 16.Santner A, Estelle M. The ubiquitin-proteasome system regulates plant hormone signaling. Plant J. 2010;61(6):1029–1040. doi: 10.1111/j.1365-313X.2010.04112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim K, et al. Cytokinin-facilitated proteolysis of ARABIDOPSIS RESPONSE REGULATOR2 attenuates signaling output in two-component circuitry. Plant J. 2012;69(6):934–945. doi: 10.1111/j.1365-313X.2011.04843.x. [DOI] [PubMed] [Google Scholar]
- 18.Zheng X, et al. AUXIN UP-REGULATED F-BOX PROTEIN1 regulates the cross talk between auxin transport and cytokinin signaling during plant root growth. Plant Physiol. 2011;156(4):1878–1893. doi: 10.1104/pp.111.179812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smalle J, et al. Cytokinin growth responses in Arabidopsis involve the 26S proteasome subunit RPN12. Plant Cell. 2002;14(1):17–32. doi: 10.1105/tpc.010381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.To JP, et al. Cytokinin regulates type-A Arabidopsis Response Regulator activity and protein stability via two-component phosphorelay. Plant Cell. 2007;19(12):3901–3914. doi: 10.1105/tpc.107.052662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kiba T, et al. Combinatorial microarray analysis revealing arabidopsis genes implicated in cytokinin responses through the His->Asp Phosphorelay circuitry. Plant Cell Physiol. 2005;46(2):339–355. doi: 10.1093/pcp/pci033. [DOI] [PubMed] [Google Scholar]
- 22.Goda H, et al. The AtGenExpress hormone and chemical treatment data set: experimental design, data evaluation, model data analysis and data access. Plant J. 2008;55(3):526–542. doi: 10.1111/j.0960-7412.2008.03510.x. [DOI] [PubMed] [Google Scholar]
- 23.Andrade MA, González-Guzmán M, Serrano R, Rodríguez PL. A combination of the F-box motif and kelch repeats defines a large Arabidopsis family of F-box proteins. Plant Mol Biol. 2001;46(5):603–614. doi: 10.1023/a:1010650809272. [DOI] [PubMed] [Google Scholar]
- 24.Mason MG, et al. Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell. 2005;17(11):3007–3018. doi: 10.1105/tpc.105.035451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmid M, et al. A gene expression map of Arabidopsis thaliana development. Nat Genet. 2005;37(5):501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- 26.Winter D, et al. An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE. 2007;2(8):e718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dello Ioio R, et al. A genetic framework for the control of cell division and differentiation in the root meristem. Science. 2008;322(5906):1380–1384. doi: 10.1126/science.1164147. [DOI] [PubMed] [Google Scholar]
- 28.Nishimura C, et al. Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell. 2004;16(6):1365–1377. doi: 10.1105/tpc.021477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hutchison CE, et al. The Arabidopsis histidine phosphotransfer proteins are redundant positive regulators of cytokinin signaling. Plant Cell. 2006;18(11):3073–3087. doi: 10.1105/tpc.106.045674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ishida K, Yamashino T, Yokoyama A, Mizuno T. Three type-B response regulators, ARR1, ARR10 and ARR12, play essential but redundant roles in cytokinin signal transduction throughout the life cycle of Arabidopsis thaliana. Plant Cell Physiol. 2008;49(1):47–57. doi: 10.1093/pcp/pcm165. [DOI] [PubMed] [Google Scholar]
- 31.Sakai H, et al. ARR1, a transcription factor for genes immediately responsive to cytokinins. Science. 2001;294(5546):1519–1521. doi: 10.1126/science.1065201. [DOI] [PubMed] [Google Scholar]
- 32.Hwang I, Sheen J. Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature. 2001;413(6854):383–389. doi: 10.1038/35096500. [DOI] [PubMed] [Google Scholar]
- 33.Mason MG, Li J, Mathews DE, Kieber JJ, Schaller GE. Type-B response regulators display overlapping expression patterns in Arabidopsis. Plant Physiol. 2004;135(2):927–937. doi: 10.1104/pp.103.038109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Imamura A, Kiba T, Tajima Y, Yamashino T, Mizuno T. In vivo and in vitro characterization of the ARR11 response regulator implicated in the His-to-Asp phosphorelay signal transduction in Arabidopsis thaliana. Plant Cell Physiol. 2003;44(2):122–131. doi: 10.1093/pcp/pcg014. [DOI] [PubMed] [Google Scholar]
- 35.Imamura A, Yoshino Y, Mizuno T. Cellular localization of the signaling components of Arabidopsis His-to-Asp phosphorelay. Biosci Biotechnol Biochem. 2001;65(9):2113–2117. doi: 10.1271/bbb.65.2113. [DOI] [PubMed] [Google Scholar]
- 36.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 37.Ogura M, Tsukahara K. Autoregulation of the Bacillus subtilis response regulator gene degU is coupled with the proteolysis of DegU-P by ClpCP. Mol Microbiol. 2010;75(5):1244–1259. doi: 10.1111/j.1365-2958.2010.07047.x. [DOI] [PubMed] [Google Scholar]
- 38.Li L, Kehoe DM. Abundance changes of the response regulator RcaC require specific aspartate and histidine residues and are necessary for normal light color responsiveness. J Bacteriol. 2008;190(21):7241–7250. doi: 10.1128/JB.00762-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perraud AL, Weiss V, Gross R. Signalling pathways in two-component phosphorelay systems. Trends Microbiol. 1999;7(3):115–120. doi: 10.1016/s0966-842x(99)01458-4. [DOI] [PubMed] [Google Scholar]
- 40.Hass C, et al. The response regulator 2 mediates ethylene signalling and hormone signal integration in Arabidopsis. EMBO J. 2004;23(16):3290–3302. doi: 10.1038/sj.emboj.7600337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muratani M, Tansey WP. How the ubiquitin-proteasome system controls transcription. Nat Rev Mol Cell Biol. 2003;4(3):192–201. doi: 10.1038/nrm1049. [DOI] [PubMed] [Google Scholar]
- 42.Michael TP, et al. Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet. 2008;4(2):e14. doi: 10.1371/journal.pgen.0040014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hanano S, Domagalska MA, Nagy F, Davis SJ. Multiple phytohormones influence distinct parameters of the plant circadian clock. Genes Cells. 2006;11(12):1381–1392. doi: 10.1111/j.1365-2443.2006.01026.x. [DOI] [PubMed] [Google Scholar]
- 44.Gagne JM, et al. Arabidopsis EIN3-binding F-box 1 and 2 form ubiquitin-protein ligases that repress ethylene action and promote growth by directing EIN3 degradation. Proc Natl Acad Sci USA. 2004;101(17):6803–6808. doi: 10.1073/pnas.0401698101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo H, Ecker JR. Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell. 2003;115(6):667–677. doi: 10.1016/s0092-8674(03)00969-3. [DOI] [PubMed] [Google Scholar]
- 46.Yu H, Wu J, Xu N, Peng M. Roles of F-box proteins in plant hormone responses. Acta Biochim Biophys Sin (Shanghai) 2007;39(12):915–922. doi: 10.1111/j.1745-7270.2007.00358.x. [DOI] [PubMed] [Google Scholar]
- 47.Vierstra RD. The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol. 2009;10(6):385–397. doi: 10.1038/nrm2688. [DOI] [PubMed] [Google Scholar]
- 48.Alonso JM, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301(5633):653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 49.Perilli S, Sabatini S. Analysis of root meristem size development. Methods Mol Biol. 2010;655:177–187. doi: 10.1007/978-1-60761-765-5_12. [DOI] [PubMed] [Google Scholar]
- 50.Chiang YH, et al. Functional characterization of the GATA transcription factors GNC and CGA1 reveals their key role in chloroplast development, growth, and division in Arabidopsis. Plant Physiol. 2012;160(1):332–348. doi: 10.1104/pp.112.198705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kovtun Y, Chiu WL, Tena G, Sheen J. Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc Natl Acad Sci USA. 2000;97(6):2940–2945. doi: 10.1073/pnas.97.6.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walter M, et al. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 2004;40(3):428–438. doi: 10.1111/j.1365-313X.2004.02219.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.