Abstract
ADP-ribosylation is a posttranslational modification that modulates the functions of many target proteins. We previously showed that the fungal toxin brefeldin A (BFA) induces the ADP-ribosylation of C-terminal–binding protein-1 short-form/BFA–ADP-ribosylation substrate (CtBP1-S/BARS), a bifunctional protein with roles in the nucleus as a transcription factor and in the cytosol as a regulator of membrane fission during intracellular trafficking and mitotic partitioning of the Golgi complex. Here, we report that ADP-ribosylation of CtBP1-S/BARS by BFA occurs via a nonconventional mechanism that comprises two steps: (i) synthesis of a BFA–ADP-ribose conjugate by the ADP-ribosyl cyclase CD38 and (ii) covalent binding of the BFA–ADP-ribose conjugate into the CtBP1-S/BARS NAD+-binding pocket. This results in the locking of CtBP1-S/BARS in a dimeric conformation, which prevents its binding to interactors known to be involved in membrane fission and, hence, in the inhibition of the fission machinery involved in mitotic Golgi partitioning. As this inhibition may lead to arrest of the cell cycle in G2, these findings provide a strategy for the design of pharmacological blockers of cell cycle in tumor cells that express high levels of CD38.
Keywords: cell signaling, Golgi fragmentation, mitosis, anticancer molecules
The modification of proteins by mono-ADP-ribosylation involves the transfer of a single ADP-ribose (ADPR) from NAD+ to specific amino acids in target proteins by mono-ADP-ribosyltransferases (1–3). The mono-ADP-ribosylation reaction was characterized first in bacteria, in which ADP-ribosyltransferases have roles as toxins [e.g., cholera and pertussis toxins (1)]. More recently, this reaction was also characterized in eukaryotic cells, in which a large group of mono-ADP-ribosyltransferases has been identified and proposed to be involved in the regulation of numerous physiological functions (4). We previously reported that the fungal toxin brefeldin A (BFA), a macrocyclic lactone used widely in studies of membrane trafficking (see below), induces the ADP-ribosylation of the C-terminal–binding protein-1 short-form/BFA–ADP-ribosylation substrate (CtBP1-S/BARS; for brevity, BARS) with high affinity and selectivity, and of the glycolytic enzyme GAPDH with much lower efficiency (5, 6). Here, we explore the molecular mechanisms and possible function of the BFA-dependent ADP-ribosylation of BARS.
BARS is structurally related to the D2-hydroxy acid dehydrogenase family and is a member of the CtBP family (7). It is involved in two processes, one in the cell cytosol and the other in the nucleus (7). In the cytosol, BARS controls the membrane-fission machinery that drives the formation of post-Golgi carriers (8, 9), endocytic fluid-phase carriers (8, 10), and coatomer protein I (COP1)-coated vesicles (11), and the partitioning of Golgi during mitosis (12, 13). In the nucleus, members of the CtBP protein family act as transcription corepressors, thus regulating numerous cellular functions, including epithelial differentiation, tumorigenesis, and apoptosis (14, 15). It remains unclear whether the nuclear and cytoplasmic functions of BARS are related.
BFA is a toxin produced by several fungi (e.g., Eupenicillium brefeldianum, Alternaria carthami), and its role in nature is not well understood. It has been shown to induce necrosis of the leaf tissue in safflower (leaf spot diseases), probably to facilitate colonization by the fungi (16). As a research tool, BFA has been characterized extensively and used to analyze the mechanisms of membrane transport. Its best-known effect is the induction of the formation of numerous long tubules from the Golgi complex, which then fuse to the endoplasmic reticulum (ER), thereby mediating the redistribution of resident Golgi proteins into the ER (17) and hence causing a rapid and reversible block of secretion (17). At the molecular level, this effect of BFA is mediated by the inhibition of the GTPase exchange factor acting on the small Ras-like GTPase ARF, and by the release of ARF from the Golgi complex along with a set of proteins regulated by ARF (18). Whether the effects of BFA on Golgi tubulation and disassembly are linked to those of the ADP-ribosylation of BARS remains unclear (7). It has been proposed that ADP-ribosylation of BARS contributes to the disassembly of the Golgi complex, at least under certain conditions (19), by inhibiting the fission of Golgi tubules (7, 20). However, the molecular mechanisms and the functional effects of BARS ADP-ribosylation remain to be characterized.
In the present study, we show that BFA-induced ADP-ribosylation is a noncanonical two-step process that involves the enzymatic synthesis of a previously uncharacterized BFA-ADPR conjugate (BAC) by the ADP-ribosyl cyclase CD38. The formation of BAC then is followed by covalent binding of BAC to the NAD+-binding pocket of BARS. This ADP-ribosylation induces a change in the conformation of BARS that precludes its binding to key interactors and inhibits the BARS fission activity in mitotic Golgi partitioning. Because partitioning of the Golgi complex is required for completion of the cell cycle, these findings carry pharmacological implications for cancer treatment.
Results
Transfer of ADP-Ribose to BARS Is Mediated by Formation of a BAC.
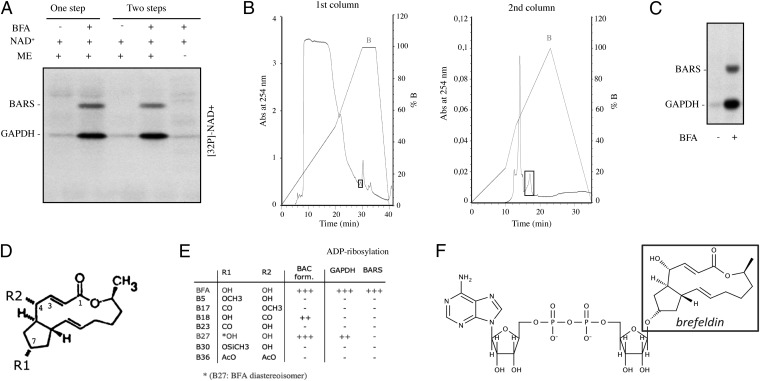
To investigate the molecular mechanisms of the BFA-induced ADP-ribosylation reaction, we incubated a mixture of rat brain membranes and cytosol with BFA and [32P]-NAD+ for 1 h at 37 °C (Fig. 1A, one step). As expected, this complete mixture gave rise to ADP-ribosylation of the BARS in the cytosol (5, 6). Next, we carried out the reaction in two steps, using different mixtures. For the first step, we incubated the rat brain membranes (as a source of the enzyme that catalyzes the ADP-ribosylation reaction) with NAD+ without or with BFA, and NAD+ and BFA without the membranes. The samples then were centrifuged to remove the membranes and ultrafiltered (with a 3,000-Da cutoff). In the second step, we mixed this ultrafiltrate with cytosol. Here, the ultrafiltrate that had been generated in the presence of the membranes, NAD+, and BFA (but not the ultrafiltrate produced in the absence of the membranes or of BFA) resulted in ADP-ribosylation of the cytosolic BARS, as shown by denaturing SDS/PAGE analysis (Fig. 1A, two steps). This indicates that in the presence of BFA and NAD+, a membrane-bound enzyme catalyzes the synthesis of a soluble BFA/NAD+ derivative that then leads to the modification of BARS.
Fig. 1.
ADP-ribosylation of BARS is mediated by formation of a conjugate between BFA and ADPR. (A) One-step reaction: total rat brain membrane fractions (ME) and rat cytosol were incubated for 1 h at 37 °C with 30 µM total NAD+ (spiked with 5 µCi [32P]-NAD+), in the absence and presence of BFA (80 µg/mL). Two-step reaction: total rat brain membrane fractions, 30 µM total NAD+ (spiked with 5 µCi [32P]-NAD+), and BFA (80 µg/mL) were combined as indicated and incubated for 1 h at 37 °C; the membranes then were removed by centrifugation. The resulting supernatant was ultrafiltered (Microcon YM-3) and incubated with rat cytosol for 1 h at 37 °C. The samples were analyzed by SDS/PAGE and autoradiography. (B) Rat brain membranes, [32P]-NAD+, and BFA were incubated as indicated in SI Materials and Methods. Membranes were removed by centrifugation, and the supernatants were ultrafiltered. The resulting filtrate was loaded onto an HPLC C18 reverse-phase (RP) column and eluted with a nonlinear gradient of phosphate buffer containing tetrabutylammonium chloride (first column). The fractions that induced ADP-ribosylation were eluted at 100% nonlinear gradient of buffer B and recovered, lyophilized, and loaded onto an HPLC C18 RP column for the second purification step in the absence of tetrabutylammonium chloride. The metabolite of interest eluted at 50% buffer B after 17 min of the gradient (second column). (C) The purified metabolite was incubated with rat brain cytosol, separated by SDS/PAGE, and analyzed by autoradiography. (D) Molecular structure of BFA. (E) Effect of BFA analogs on ADP-ribosylation of GADPH and BARS. (F) Proposed structure of BAC.
We thus set out to isolate this active derivative by HPLC using ADP-ribosylation of cytosolic BARS for its detection (Fig. 1 B and C) (5, 6). The derivative showed an HPLC elution profile that did not match the elution times of NAD+ and its known analogs, i.e., ADPR, cyclic ADPR (cADPR), nicotinamide adenine dinucleotide phosphate, and nicotinic acid adenine dinucleotide phosphate. Next, to characterize the active derivative further, we sought to synthesize it using other ADPR-containing molecules as substrates, instead of NAD+. Strikingly, the active derivative also was synthesized in the presence of cADPR (albeit with lower efficiency; SI Materials and Methods) but not ADPR. This finding offered a clue toward determining the molecular mechanisms of the generation of this BFA/NAD+ derivative. Both NAD+ and cADPR are substrates of a class of enzymes known as ADP-ribosyl cyclases. These cyclases catalyze the conversion of NAD+ to cADPR through cleavage of the nicotinamide–ribose glycosidic bond and the formation of an enzyme-stabilized ADP-ribosyl–oxocarbenium ion intermediate (Fig. S1) (21, 22). This intermediate is a good electrophile and can react with water to form ADPR, or intramolecularly, with the N1 atom of the purinic ring of the adenine moiety of NAD+, to form cADPR. In addition, the same ADP-ribosyl cyclases catalyze the hydrolysis of cADPR to ADPR via generation of the same ADP-ribosyl–oxocarbenium ion (Fig. S1, 3) (23). As BFA has two hydroxyl groups (positions 4 and 7; Fig. 1 D and E), we hypothesized that these might react with the oxocarbenium ion intermediate. This hypothesis was tested by producing the active derivative through the incubation of [3H]-BFA with [32P]-NAD+, with rat brain membranes as the source of enzyme, and analyzing the product by HPLC. The results showed that the NAD+ derivative contained both [3H] and [32P] at a 1:1 ratio (Fig. S2), indicating that the active derivative is a BAC. The expected mass of a product composed of BFA and ADPR is 822 Da (see below).
To determine the BFA structural features required to induce BAC formation, we tested several BFA analogs that were modified at one or both of the BFA hydroxyl groups (positions 4 and 7; Fig. 1 D and E) (24). Only the BFA analogs carrying a hydroxyl group at position 7, such as B27 (a BFA diastereoisomer) and B18 (Fig. 1 D and E), supported the formation of BAC-like derivatives. Importantly, however, these new types of BAC did not react with BARS, which indicated that the structure of BFA is required for BARS ADP-ribosylation (Fig. 1E). The metabolite induced by B27 bound to GAPDH [the other less efficient substrate modified by BAC (25, 26)] to a lesser extent (Fig. 1E).
These results indicate that the hydroxyl group at position 7 of BFA reacts with the ADP-ribosyl–oxocarbenium ion intermediate generated by ADP-ribosyl cyclase, to form a BAC (Fig. 1F) that binds to BARS with high efficiency (Fig. S3). Other BFA analogs (i.e., those that lack the hydroxyl at position 4 or have an inverted configuration at C7) can induce the formation of BAC-like molecules with distinct features but do not bind to BARS.
BAC Binds Covalently into the NAD+-Binding Pocket of BARS.
Next, we focused on the binding of BAC to BARS. This binding must be covalent, as it persists under denaturing SDS/PAGE conditions (Fig. 1 A and C). We first tested whether the whole BAC molecule binds to BARS, or whether BAC acts as an ADPR donor. To address this point, we produced BAC using [3H]-labeled BFA. The HPLC-purified [3H]-BAC was incubated with recombinant BARS and examined by SDS/PAGE and a radioactivity imager. BARS incubated with [3H]-BAC was radiolabeled, indicating that the BARS-bound BAC contains BFA. No signal was detected when BARS was incubated with only [3H]-BFA (Fig. S4A). We also made use of an antibody developed against BFA to further examine whether the whole BAC molecule (i.e., both the BFA and ADPR portions) binds to BARS. If this were the case, the binding of the BFA portion of BAC should be revealed by the immunoreactivity of modified BARS to this anti-BFA antibody. Purified recombinant BARS was incubated with BFA or BAC and treated for immunoblotting. The anti-BFA antibody immunoreacted with BARS after incubation with BAC (Fig. S4 B and C), confirming the notion that the entire BAC binds to BARS (see below). Notably, this also was the case for the derivative produced using cADPR bound to BARS (Fig. S4D). In addition, we used a complementary approach based on the ability of the protein module macrodomain to recognize ADP-ribose (27). As shown in Fig. S4E, the macrodomain recognized BARS, indicating that ADP-ribose was present in the modified BARS. These collective data suggest that BARS-bound BAC contains both the BFA and ADPR moieties.
To characterize this reaction further, recombinant BARS was incubated with BAC and subjected to both MALDI-TOF mass spectrometry (MS) and MS/MS and liquid chromatography (LC)-MS analyses. The MALDI analysis showed that after binding to BAC, BARS has a molecular mass of 822 Da greater than that of the control BARS protein. This shift corresponds to the calculated mass of BAC, confirming the above predictions that BAC consists of a BFA molecule conjugated with an ADPR, presumably through its hydroxyl group at position 7 (Fig. 1F). Secondly, the fragmentation data indicated that BAC is bound to the His304 residue (Fig. S5A). In confirmation of this finding, the BARS point mutant His304Ala was not modified by BAC, whereas several other mutants in the BARS dinucleotide binding site (see below) showed covalent BAC binding levels indistinguishable from those of wild-type BARS (Fig. S5B).
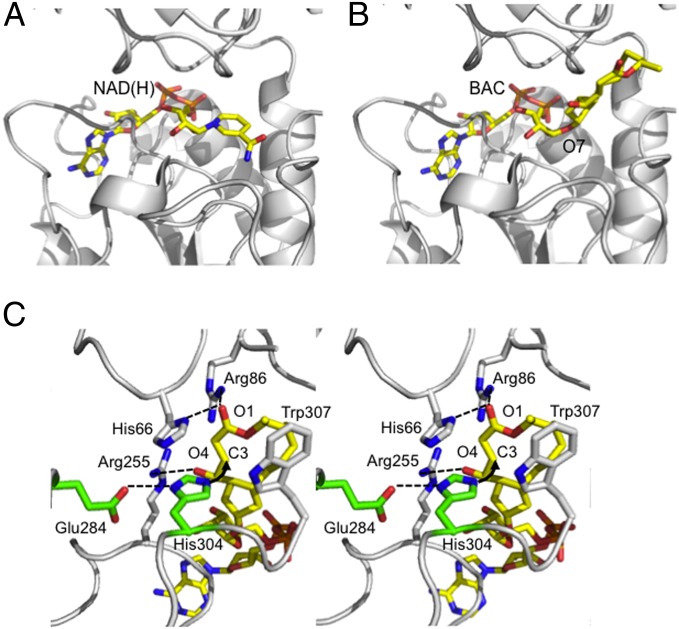
Then, to investigate the structural details of the covalent binding of BAC to BARS, we used a molecular modeling approach and our previous knowledge of the BARS crystal structure to fit a BAC molecule into the well-characterized NAD(H) Rossmann fold in BARS (28), based on the common ADP molecular framework shared by NAD(H) and BAC (Fig. 2 A and B). In the crystal structure of the BARS–NAD(H) complex [Protein Data Bank (PDB) ID code 1HKU], a protein cavity lined by residues Tyr65, His66, Arg86, Gly88, Ser89, Gly90, Asp92, Ser113, Thr117, Arg255, His304, Trp307, Ser313, and Met316 is occupied by the NAD(H) nicotinamide moiety (28). This cavity may host the BFA moiety of BAC (Fig. 2). Analysis of the polarity/hydrophobicity side-chain distribution within the cavity suggests that the orientation of the BFA moiety of BAC would be with its C3 atom close to the imidazole ring of His304 (Fig. 2C). According to this model, the His304 side chain might be hydrogen bonded to Glu284 [as found in the crystal structure of the BARS-NAD(H) complex], which would assist His304 during a nucleophilic attack on the BFA C3 atom. Furthermore, the BFA carbonylic O1 and hydroxylic O4 atoms would be located in two positively charged pockets lined by residues His66/Arg86 and Arg86/Arg255, respectively (Fig. 2C). In particular, the Arg86 side chain is positioned to form a hydrogen bond with the carbonyl group of BFA, thus polarizing the carbon–oxygen bond, whereas Arg255 binds the carboxylate moiety of BFA, helping position the BFA moiety of BAC correctly in the active site. The BFA plane is further kept in the correct orientation for catalysis by a stacking interaction with the Trp307 side chain. As the C3 atom of BFA is strongly polarized as a result of the electron-attractor effect of the nearby conjugated electrophilic carbonyl (lactone) group and of the 4-hydroxy group (29), a rational explanation of the strong and specific binding of BAC to BARS is that the ADPR portion of BAC is involved in recognition and binding to the BARS nucleotide-binding cleft, whereas the C3 atom of the BFA portion is involved in covalent binding to His304, which may act as an electron donor in a nucleophilic reaction (Michael addition). This hypothesis is supported by the observation that the BAC formed using B18, which is characterized by a diffused conjugated double bond that ranges from C1 to C4, did not bind covalently to BARS (Fig. 1F). In addition, molecules with a coumarinic moiety, which we previously showed to act as specific inhibitors of ADP-ribosylation (30), and which are known to bind in the Rossmann fold (31), behaved as competitive inhibitors for the binding of BAC to BARS (Fig. S6).
Fig. 2.
Model of BAC binding to BARS. View of the BARS nucleotide binding site with (A) the NAD(H) cofactor bound (PDB ID code 1HKU) and (B) the modeled BAC molecule. Position 7 of BFA, where the conjugation between ADPR and BFA takes place to form BAC, is indicated. (C) Stereoview of the BAC binding site. Residues relevant to BAC interaction and to catalysis are shown in stick representation (white and green, respectively) and labeled. Hydrogen bonds are indicated as dashed lines, whereas the nucleophilic attack between His304 and the C3 atom of BAC is indicated by an arrow. Hydrogen atoms are not shown.
Collectively, these findings and the MS data in Fig. S5 A and B support our computational model of the BARS modification by BAC and indicate that (i) BAC fits into the BARS Rossmann fold with high affinity and specificity and (ii) its BFA portion is involved in covalent binding to His304 through a nucleophilic reaction.
BAC Is Synthesized in Living Cells by the ADP-Ribosyl Cyclase CD38.
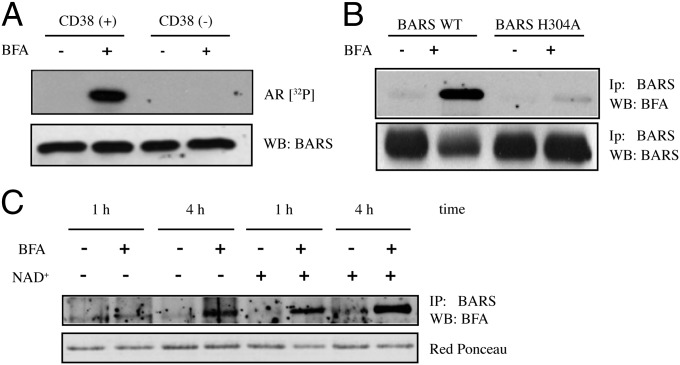
As the above data suggest that BAC is synthesized by an ADP-ribosyl cyclase, we focused on the membrane-bound ADP-ribosyl cyclase CD38, a mammalian enzyme responsible for synthesizing the Ca2+-releasing signaling metabolite cADPR (32). To test for a role of CD38, we performed the in vitro assay for BAC formation using whole membrane fractions prepared from control HeLa cells that do not express CD38 (CD38−) and from HeLa cells stably transfected with a vector for CD38 expression (CD38+) (33), with analysis by SDS/PAGE and autoradiography. As shown in Fig. 3A, only the membranes obtained from CD38+ HeLa cells supported the formation of BAC, which demonstrates that this ADP-ribosyl cyclase is indeed involved in the synthesis of BAC. Whether the mechanism of BAC formation involves a covalent or a noncovalent CD38–ADP-ribosyl–oxocarbenium ion intermediate remains undetermined. According to some authors, this intermediate is covalently bound to CD38 (21), whereas others have suggested that it is not covalently attached (22).
Fig. 3.
CD38 can support BAC synthesis in intact cells. (A) Total membrane fractions from control (CD38−) and CD38+ HeLa cells were incubated for 2 h at 37 °C with recombinant His-BARS and 30 µM total NAD+ (spiked with 5 µCi [32P]-NAD+), in the absence and presence of BFA (80 µg/mL). The samples were analyzed by SDS/PAGE and autoradiography (AR [32P]); total BARS levels were analyzed by Western blotting. (B) CD38+ HeLa cells were transfected with wild-type YFP-BARS (BARS WT) or YFP-BARS with the His304 point mutation (BARS H304A). After 24 h, the cells were treated with 80 µg/mL BFA for 4 h at 37 °C, in the presence of 5 mM extracellular NAD+. YFP-BARS was immunoprecipitated using an anti-BARS antibody, and the modified protein was revealed using an anti–BFA-specific antibody. (B, Lower) Total BARS levels are shown. (C) CD38+ HeLa cells were transfected with YFP-BARS. After 24 h, the cells were treated with 80 µg/mL BFA for 1 h or 4 h at 37 °C, in the presence or absence of exogenously added 5 mM NAD+, as indicated. YFP-BARS was immunoprecipitated from total lysates using an anti-BARS antibody, and the modified protein was revealed using an anti–BFA-specific antibody. (C, Lower) Total levels of BARS are shown.
We then tested whether BAC formation and binding to BARS by CD38 also occurs in living cells. For this, the CD38+ HeLa cells were transfected with YFP-BARS or the YFP-BARS His304Ala point mutant, and incubated with NAD+ in the absence or presence of BFA. These treatments did not induce major alterations in the subcellular localization of CD38, which remained prevalently localized at the plasma membrane (Fig. S7). The cells were lysed, YFP-BARS was immunoprecipitated with an anti-BARS antibody, and the samples were subjected to SDS/PAGE and immunoblotting with an anti-BFA antibody. Strikingly, BAC bound to YFP-BARS in CD38+ HeLa cells, whereas the YFP-BARS His304Ala mutant was not modified, as expected (Fig. 3B). The reaction was very efficient, as more than 90% of the overexpressed wild-type BARS was modified by BAC after 4 h of incubation. BAC formation also was induced by addition of cADPR to the medium (Fig. S8). Importantly, BARS expressed in the CD38+ HeLa cells incubated in the absence of exogenously added NAD+ (and of other possible sources of NAD+, e.g., serum) also showed BAC binding to BARS, although the fraction of the modified BARS and the rate of the reaction were lower than in cells supplemented with NAD+ (Fig. 3C). Together, these findings show that CD38 also can catalyze the formation of BAC in intact cells in the absence of exogenously added NAD+.
CD38 is an ectoenzyme, and its catalytic domain is localized extracellularly (32–34). It has been proposed that the conversion of extracellular NAD+ to cADPR and the subsequent cADPR influx is mediated by the juxtaposition of two CD38 monomers, which results in a catalytically active channel (35). Although the extracellular NAD+ concentration is low in cell culture, this concentration may be increased locally by connexin 43 hemichannels, which translocate NAD+ to the extracellular space (36, 37). To test whether BAC can be produced outside the cells, control CD38− and CD38+ HeLa cells were incubated with BFA in the presence of NAD+. Then, the various media were collected and incubated with recombinant BARS, and protein modification was monitored with the anti-BFA antibody. BARS showed BAC binding when incubated with the medium from the BFA-treated CD38+ HeLa cells (Fig. S9A). Thus, BAC can be produced extracellularly. To investigate whether extracellularly generated BAC can cross the plasma membrane, CD38− HeLa cells were transfected with YFP-BARS and treated with BFA and NAD+, as well as with a recombinant catalytically active soluble portion of CD38, to generate BAC in the medium. As expected, the addition of recombinant CD38 supported the extracellular formation of BAC (Fig. S9B), but YFP-BARS was not modified (Fig. S9C), which indicates that the BAC generated in the medium cannot cross the plasma membrane. As a further test, CD38+ cells were incubated with medium containing purified BAC. Again, no modification of BARS was observed (Fig. S9D), indicating that extracellular BAC cannot cross the plasma membrane, even in cells that express CD38.
Together, these data indicate that CD38 is required for the intracellular translocation of BAC, although it might not necessarily transport BAC itself. The exact mechanism of this translocation remains to be determined.
BAC Affects the Oligomerization/Conformation of BARS and Inhibits the Binding of BARS with Interactors Involved in Fission.
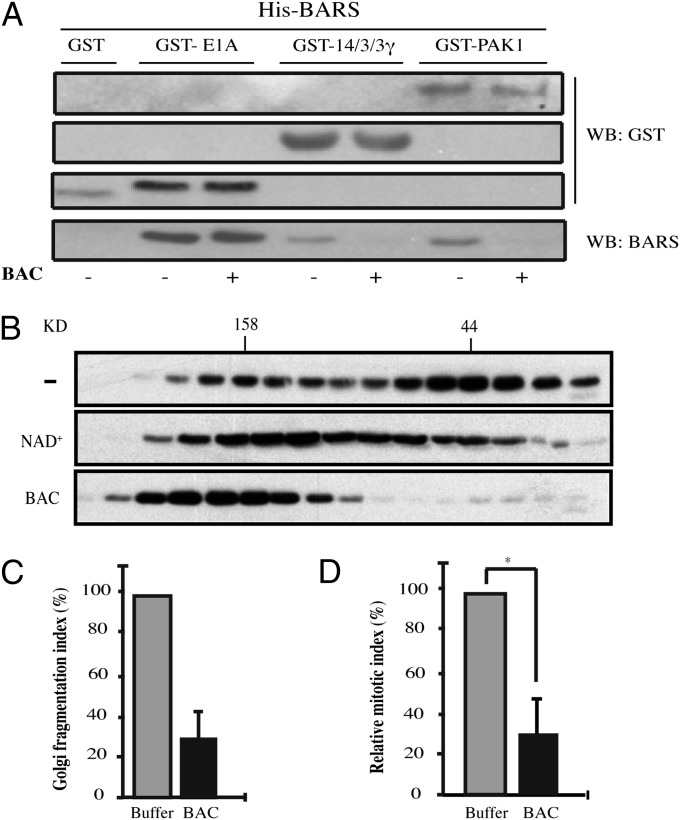
Next, we investigated the mechanism through which the binding of BAC to BARS affects the activity of BARS (20, 25). It has been proposed that BARS can switch between its nuclear corepression activity and its membrane-fission activity depending on its binding with two cofactors, NAD(H) and acyl-CoA. Under this model, NAD(H) promotes a “closed dimeric/tetrameric conformation” and enhances the binding of BARS to cellular and viral transcriptional repressors (28, 38), whereas the binding of palmitoyl-CoA to BARS promotes an “open monomeric conformation” of BARS, which appears to promote membrane fission (11). Recently, 14-3-3γ and p21-activated kinase 1 (PAK1) were shown to be essential BARS interactors involved in the fission of post-Golgi carriers and of macropinosomes (9, 10). We investigated whether BAC binding can selectively inhibit BARS interactions with molecular partners involved in membrane fission. To test this hypothesis, we performed an in vitro pull-down assay (9). The preincubation of immobilized, His-tagged BARS with BAC strongly impaired the ability of BARS to bind to GST-tagged 14-3-3γ and GST-tagged PAK1 (Fig. 4A) (9, 10). Instead, the interaction with E1A, a known cofactor in transcription corepression (28), was not affected by BAC (Fig. 4A). Based on these findings, we postulated that the covalent binding of BAC to BARS would irreversibly lock BARS in a closed dimeric/tetrameric conformation, which would be inactive in fission (11). We tested this hypothesis by first examining whether the covalent binding of BAC to BARS can alter the oligomerization state of BARS. Rat brain cytosol was incubated with control buffer, or with NAD+ or BAC, and subjected to gel-filtration chromatography. The native protein eluted in two main peaks, which approximately corresponded to the 50-kDa and 170-kDa molecular weight markers, suggesting a BARS conformation compatible with an equilibrium between the open monomeric and closed dimeric (and/or tetrameric) states (25). The incubation with NAD+ increased the proportion of BARS detected at a molecular mass of 158 kDa (Fig. 4B); after incubation with BAC, BARS was found exclusively in fractions corresponding to an apparent molecular mass >158 kDa (Fig. 4B). This indicates that the covalent binding of BAC to BARS alters its oligomerization/interaction state by favoring the tetramer, most probably as a result of changes in the BARS conformation (28).
Fig. 4.
BAC affects the interactions of BARS with its partners involved in fission. (A) In vitro GST pull-down assay. His-BARS was preincubated with HPLC-purified BAC to induce BAC binding. After 3 h, equimolar amounts of GST, GST-E1A, GST-14/3/3γ or GST-PAK1 were added to the mixture and incubated for another 2 h at 4 °C. The GST-tagged proteins were recovered and eluted. The samples were subjected to SDS/PAGE and immunoblotted with antibodies directed against GST or BARS. (B) Gel-filtration patterns of rat brain cytosol were treated as indicated. The fractions were separated by SDS/PAGE and immunoblotted with an anti-BARS antibody. (C) Golgi fragmentation assay. Digitonin-permeabilized normal rat kidney cells were incubated with mitotic cytosol preincubated with HPLC-purified BAC (BAC) or buffer alone. Golgi fragmentation was evaluated and quantified by immunofluorescence using an antibody against giantin (13). (D) Quantification of the mitotic index of cells grown on coverslips and arrested in G2 phase of the cell cycle, as described in SI Materials and Methods. One hour before the G2 block release, the cells were microinjected with BAC or buffer alone (both mixed previously with dextran-FITC as a tracer of microinjection). The cells were fixed 40 min after the G2 block release and stained with the DNA dye Hoechst 33342. Quantification data are means ± SD from three independent experiments, each carried out in duplicate. More than 200 cells were microinjected for each condition. *P = 0.02.
As BAC cannot cross the plasma membrane, we investigated the effects of BAC on the fission-inducing activity of BARS in an in vitro assay in permeabilized cells that reconstitutes the BARS-dependent mitotic fission and fragmentation of the Golgi complex, a process required for entry into mitosis (13). Incubation of permeabilized cells with mitotic cytosol induced fission of the Golgi complex into dispersed fragments, as already reported (Fig. 4C) (13). When BAC was added to mitotic cytosol under conditions resulting in exhaustive binding of BARS to BAC (Fig. S3), Golgi fission/fragmentation was strongly inhibited (Fig. 4C), indicating that the covalent binding of BAC to BARS inhibits the ability of BARS to induce mitotic Golgi fragmentation. Then, to test the effect of BAC in living cells, we induced the ADP-ribosylation of BARS by microinjecting purified BAC in G2-blocked HeLa cells and monitored the effect of this treatment on mitotic entry, which depends tightly on Golgi fragmentation (13). As shown in Fig. 4D, the injection of BAC caused a strong impairment of entry into mitosis, in line with the expected effect of the inhibition of BARS on Golgi fragmentation.
Together, these data indicate that the binding of BAC to BARS favors an oligomeric conformation of BARS whereby BARS cannot interact with the proteins necessary for BARS-induced fission, and that this reaction therefore inhibits the ability of BARS to support mitotic fission of the Golgi complex and mitotic entry.
Discussion
In this study, we describe a two-step mechanism that underlies the modification of BARS by BFA, and we test the functional role of this reaction. The first reaction step is catalyzed by the ADP-ribosyl cyclase CD38 and leads to the formation of BAC, a BFA–ADP-ribose conjugate. The mechanism of BAC formation is based on the catalytic mechanism of conversion of NAD+ to cADPR. This involves cleavage of the NAD+ nicotinamide–ribose bond and the subsequent formation of an enzyme-stabilized ADP-ribosyl–oxocarbenium ion intermediate with good electrophilic properties, which reacts with the hydroxyl groups in position 7 of BFA to form BAC (Fig. S1). Notably, again in common with the mechanism of conversion of NAD+ to cADPR, the synthesis of BAC occurs at the external cell surface, where BAC is translated during synthesis into the cell cytosol, as has been proposed to occur for cADPR synthesis and influx (35).
The second step is the covalent binding of BAC into the BARS NAD(H)-binding pocket (the Rossmann fold) (28). As shown in the model in Fig. 2, a compelling explanation of this reaction is that the C3 atom of BFA in BAC is positioned close to the imidazole ring of His304 of the BARS-binding pocket, and it undergoes nucleophilic attack by His304. The reaction is assisted by the hydrogen bond between His304 and Glu284 (Fig. 2 B and C). Notably, this binding mechanism is in agreement with the known similarities between BARS and D2-hydroxy acid dehydrogenases (28), in which the structurally equivalent His/Glu(Asp)/Arg triad functions as the center for substrate binding and dehydrogenase activity. Here, the His residue is postulated to be the acid/base catalyst, with the Glu/Asp residue helping to lower the His pKa to stabilize it in an unprotonated state. The Arg residue is proposed to polarize the substrate 2-hydroxyl group for catalysis (28).
This reaction between BAC and BARS is exquisitely selective, as BAC binds covalently exclusively to the Rossmann fold of BARS, but not to that of other dehydrogenases, with the partial exception of GAPDH, in which the reaction is orders of magnitudes less efficient. Such remarkable selectivity suggests that this reaction might have a role in the toxicity of BFA, perhaps in cell types or organisms expressing high levels of CD38 or other similar ADP-ribosyl cyclases. Indeed, the modification of BARS by BAC may impair the fission-inducing activity of BARS required for mitotic Golgi fragmentation, an effect that may result in a potent and prolonged block in G2 of the cell cycle, and eventually in cell apoptosis (12, 13).
Perhaps more important, the fact that BFA may lead to the covalent binding of BAC to BARS has implications for cancer treatment. Based on the structural data now available on the mechanism of this binding (this study) and on the binding of BFA to the ARF–GTPase exchange factor (39), it now is possible to design BFA analogs with increased selectivity toward the formation of BAC-modified BARS, and with no or strongly reduced effects on the ARF–GTPase exchange factor. This provides a strategy for generating BFA analogs with selective pharmacological effects on the cell cycle. Such analogs would be relevant for the treatment of tumors characterized by high levels of CD38 expression and, hence, high rates of BAC synthesis, such as multiple myelomas (40, 41).
Materials and Methods
Unless otherwise specified, all reagents were from Sigma–Aldrich. [32P]-β-NAD+ was from PerkinElmer. The anti-BARS antibody (BC3) was produced as described previously (9). The anti-CD38 antibody (IB4) was kindly provided by Fabio Malavasi (University of Turin, Turin, Italy). Keyhole limpet hemocyanin and cyclin dependent kinase 1 inhibitor RO-3306 were from Calbiochem. Cell culture reagents were from Gibco/Invitrogen. The TransIT-LT1 reagent was from Mirus Bio LLC. Additional materials and methods are presented in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank all colleagues who kindly provided antibodies and reagents; Dr. J. Donaldson (National Institutes of Health) for BFA analogs; Dr. C. P. Berrie for editorial assistance; and Drs. C. Limina, A. Tamburro, M. G. Silletta, R. Weigert, and S. Spanò (Negri Sud Institute) for performing initial experiments. We also acknowledge financial support from Italian Association for Cancer Research (AIRC) through the Grants IG4664 and IG10341 (to D.C.), IG4700 (to A.L.), and IG6074 (to A.C.); and from the Liguria Region and the Ministry of Education, University, and Research (Fund for Investments in Basic Research Project; A.D.F.). G.G. and C.V. received fellowships from AIRC (Italian Foundation for Cancer Research). Financial support from Technological Innovation Fund DM 24/09/2009, Legge 46/82-MEF, and Project “FaReBio di Qualità” also is acknowledged.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1222413110/-/DCSupplemental.
References
- 1.Corda D, Di Girolamo M. Functional aspects of protein mono-ADP-ribosylation. EMBO J. 2003;22(9):1953–1958. doi: 10.1093/emboj/cdg209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hassa PO, Haenni SS, Elser M, Hottiger MO. Nuclear ADP-ribosylation reactions in mammalian cells: Where are we today and where are we going? Microbiol Mol Biol Rev. 2006;70(3):789–829. doi: 10.1128/MMBR.00040-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koch-Nolte F, Kernstock S, Mueller-Dieckmann C, Weiss MS, Haag F. Mammalian ADP-ribosyltransferases and ADP-ribosylhydrolases. Front Biosci. 2008;13:6716–6729. doi: 10.2741/3184. [DOI] [PubMed] [Google Scholar]
- 4.Di Girolamo M, Dani N, Stilla A, Corda D. Physiological relevance of the endogenous mono(ADP-ribosyl)ation of cellular proteins. FEBS J. 2005;272(18):4565–4575. doi: 10.1111/j.1742-4658.2005.04876.x. [DOI] [PubMed] [Google Scholar]
- 5.De Matteis MA, et al. Stimulation of endogenous ADP-ribosylation by brefeldin A. Proc Natl Acad Sci USA. 1994;91(3):1114–1118. doi: 10.1073/pnas.91.3.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Di Girolamo M, et al. Evidence that the 50-kDa substrate of brefeldin A-dependent ADP-ribosylation binds GTP and is modulated by the G-protein beta gamma subunit complex. Proc Natl Acad Sci USA. 1995;92(15):7065–7069. doi: 10.1073/pnas.92.15.7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corda D, Colanzi A, Luini A. The multiple activities of CtBP/BARS proteins: The Golgi view. Trends Cell Biol. 2006;16(3):167–173. doi: 10.1016/j.tcb.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 8.Bonazzi M, et al. CtBP3/BARS drives membrane fission in dynamin-independent transport pathways. Nat Cell Biol. 2005;7(6):570–580. doi: 10.1038/ncb1260. [DOI] [PubMed] [Google Scholar]
- 9.Valente C, et al. A 14-3-3γ dimer-based scaffold bridges CtBP1-S/BARS to PI(4)KIIIβ to regulate post-Golgi carrier formation. Nat Cell Biol. 2012;14(4):343–354. doi: 10.1038/ncb2445. [DOI] [PubMed] [Google Scholar]
- 10.Liberali P, et al. The closure of Pak1-dependent macropinosomes requires the phosphorylation of CtBP1/BARS. EMBO J. 2008;27(7):970–981. doi: 10.1038/emboj.2008.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang JS, et al. A role for BARS at the fission step of COPI vesicle formation from Golgi membrane. EMBO J. 2005;24(23):4133–4143. doi: 10.1038/sj.emboj.7600873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colanzi A, et al. The Golgi mitotic checkpoint is controlled by BARS-dependent fission of the Golgi ribbon into separate stacks in G2. EMBO J. 2007;26(10):2465–2476. doi: 10.1038/sj.emboj.7601686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hidalgo Carcedo C, et al. Mitotic Golgi partitioning is driven by the membrane-fissioning protein CtBP3/BARS. Science. 2004;305(5680):93–96. doi: 10.1126/science.1097775. [DOI] [PubMed] [Google Scholar]
- 14.Chinnadurai G. Transcriptional regulation by C-terminal binding proteins. Int J Biochem Cell Biol. 2007;39(9):1593–1607. doi: 10.1016/j.biocel.2007.01.025. [DOI] [PubMed] [Google Scholar]
- 15.Chinnadurai G. The transcriptional corepressor CtBP: A foe of multiple tumor suppressors. Cancer Res. 2009;69(3):731–734. doi: 10.1158/0008-5472.CAN-08-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Driouich A, Jauneau A, Staehelin LA. 7-Dehydrobrefeldin A, a naturally occurring brefeldin A derivative, inhibits secretion and causes a cis-to-trans breakdown of Golgi stacks in plant cells. Plant Physiol. 1997;113(2):487–492. doi: 10.1104/pp.113.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujiwara T, Oda K, Yokota S, Takatsuki A, Ikehara Y. Brefeldin A causes disassembly of the Golgi complex and accumulation of secretory proteins in the endoplasmic reticulum. J Biol Chem. 1988;263(34):18545–18552. [PubMed] [Google Scholar]
- 18.Donaldson JG, Finazzi D, Klausner RD. Brefeldin A inhibits Golgi membrane-catalysed exchange of guanine nucleotide onto ARF protein. Nature. 1992;360(6402):350–352. doi: 10.1038/360350a0. [DOI] [PubMed] [Google Scholar]
- 19.Mironov A, et al. Role of NAD+ and ADP-ribosylation in the maintenance of the Golgi structure. J Cell Biol. 1997;139(5):1109–1118. doi: 10.1083/jcb.139.5.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weigert R, et al. CtBP/BARS induces fission of Golgi membranes by acylating lysophosphatidic acid. Nature. 1999;402(6760):429–433. doi: 10.1038/46587. [DOI] [PubMed] [Google Scholar]
- 21.Sauve AA, Munshi C, Lee HC, Schramm VL. The reaction mechanism for CD38. A single intermediate is responsible for cyclization, hydrolysis, and base-exchange chemistries. Biochemistry. 1998;37(38):13239–13249. doi: 10.1021/bi981248s. [DOI] [PubMed] [Google Scholar]
- 22.Egea PF, et al. Insights into the mechanism of bovine CD38/NAD+glycohydrolase from the X-ray structures of its Michaelis complex and covalently-trapped intermediates. PLoS ONE. 2012;7(4):e34918. doi: 10.1371/journal.pone.0034918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kotaka M, et al. Structural studies of intermediates along the cyclization pathway of Aplysia ADP-ribosyl cyclase. J Mol Biol. 2012;415(3):514–526. doi: 10.1016/j.jmb.2011.11.022. [DOI] [PubMed] [Google Scholar]
- 24.Klausner RD, Donaldson JG, Lippincott-Schwartz J. Brefeldin A: Insights into the control of membrane traffic and organelle structure. J Cell Biol. 1992;116(5):1071–1080. doi: 10.1083/jcb.116.5.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spanò S, et al. Molecular cloning and functional characterization of brefeldin A-ADP-ribosylated substrate. A novel protein involved in the maintenance of the Golgi structure. J Biol Chem. 1999;274(25):17705–17710. doi: 10.1074/jbc.274.25.17705. [DOI] [PubMed] [Google Scholar]
- 26.Valente C, Spanò S, Luini A, Corda D. Purification and functional properties of the membrane fissioning protein CtBP3/BARS. Methods Enzymol. 2005;404:296–316. doi: 10.1016/S0076-6879(05)04027-9. [DOI] [PubMed] [Google Scholar]
- 27.Dani N, et al. Combining affinity purification by ADP-ribose-binding macro domains with mass spectrometry to define the mammalian ADP-ribosyl proteome. Proc Natl Acad Sci USA. 2009;106(11):4243–4248. doi: 10.1073/pnas.0900066106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nardini M, et al. CtBP/BARS: A dual-function protein involved in transcription co-repression and Golgi membrane fission. EMBO J. 2003;22(12):3122–3130. doi: 10.1093/emboj/cdg283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brüning A, et al. Brefeldin A binds to glutathione S-transferase and is secreted as glutathione and cysteine conjugates by Chinese hamster ovary cells. J Biol Chem. 1992;267(11):7726–7732. [PubMed] [Google Scholar]
- 30.Weigert R, et al. Characterization of chemical inhibitors of brefeldin A-activated mono-ADP-ribosylation. J Biol Chem. 1997;272(22):14200–14207. doi: 10.1074/jbc.272.22.14200. [DOI] [PubMed] [Google Scholar]
- 31.Ma Q, Wang R, Yang CS, Lu AY. Expression of mammalian DT-diaphorase in Escherichia coli: Purification and characterization of the expressed protein. Arch Biochem Biophys. 1990;283(2):311–317. doi: 10.1016/0003-9861(90)90648-i. [DOI] [PubMed] [Google Scholar]
- 32.De Flora A, Zocchi E, Guida L, Franco L, Bruzzone S. Autocrine and paracrine calcium signaling by the CD38/NAD+/cyclic ADP-ribose system. Ann N Y Acad Sci. 2004;1028:176–191. doi: 10.1196/annals.1322.021. [DOI] [PubMed] [Google Scholar]
- 33.Zocchi E, et al. Expression of CD38 increases intracellular calcium concentration and reduces doubling time in HeLa and 3T3 cells. J Biol Chem. 1998;273(14):8017–8024. doi: 10.1074/jbc.273.14.8017. [DOI] [PubMed] [Google Scholar]
- 34.Zocchi E, et al. Ligand-induced internalization of CD38 results in intracellular Ca2+ mobilization: role of NAD+ transport across cell membranes. FASEB J. 1999;13(2):273–283. doi: 10.1096/fasebj.13.2.273. [DOI] [PubMed] [Google Scholar]
- 35.Franco L, et al. The transmembrane glycoprotein CD38 is a catalytically active transporter responsible for generation and influx of the second messenger cyclic ADP-ribose across membranes. FASEB J. 1998;12(14):1507–1520. doi: 10.1096/fasebj.12.14.1507. [DOI] [PubMed] [Google Scholar]
- 36.Bruzzone S, Guida L, Zocchi E, Franco L, De Flora A. De Flora A Connexin 43 hemi channels mediate Ca2+-regulated transmembrane NAD+ fluxes in intact cells. FASEB J. 2001;15(1):10–12. doi: 10.1096/fj.00-0566fje. [DOI] [PubMed] [Google Scholar]
- 37.Bruzzone S, et al. A self-restricted CD38-connexin 43 cross-talk affects NAD+ and cyclic ADP-ribose metabolism and regulates intracellular calcium in 3T3 fibroblasts. J Biol Chem. 2001;276(51):48300–48308. doi: 10.1074/jbc.M107308200. [DOI] [PubMed] [Google Scholar]
- 38.Barnes CJ, et al. Functional inactivation of a transcriptional corepressor by a signaling kinase. Nat Struct Biol. 2003;10(8):622–628. doi: 10.1038/nsb957. [DOI] [PubMed] [Google Scholar]
- 39.Kremer W, Steiner G, Béraud-Dufour S, Kalbitzer HR. Conformational states of the small G protein Arf-1 in complex with the guanine nucleotide exchange factor ARNO-Sec7. J Biol Chem. 2004;279(17):17004–17012. doi: 10.1074/jbc.M312780200. [DOI] [PubMed] [Google Scholar]
- 40.van der Veer MS, et al. The therapeutic human CD38 antibody daratumumab improves the anti-myeloma effect of newly emerging multi-drug therapies. Blood Cancer J. 2011;1(10):e41. doi: 10.1038/bcj.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hersher R. Companies wager high on CD38-targeting drugs for blood cancer. Nat Med. 2012;18(10):1446. doi: 10.1038/nm1012-1446a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.