Abstract
T-cell costimulation and coinhibition generated by engagement of the B7 family and their receptor CD28 family are of central importance in regulating the T-cell response, making these pathways very attractive therapeutic targets. Here we describe HERV–H LTR-associating protein 2 (HHLA2) as a member of the B7 family that shares 10–18% amino acid identity and 23–33% similarity to other human B7 proteins and phylogenetically forms a subfamily with B7x and B7-H3 within the family. HHLA2 is expressed in humans but not in mice, which is unique within the B7 and CD28 families. HHLA2 protein is constitutively expressed on the surface of human monocytes and is induced on B cells after stimulation with LPS and IFN-γ. HHLA2 does not interact with other known members of the CD28 family or the B7 family, but does bind a putative receptor that is constitutively expressed not only on resting and activated CD4 and CD8 T cells but also on antigen-presenting cells. HHLA2 inhibits proliferation of both CD4 and CD8 T cells in the presence of T-cell receptor signaling. In addition, HHLA2 significantly reduces cytokine production by T cells including IFN-γ, TNF-α, IL-5, IL-10, IL-13, IL-17A, and IL-22. Thus, we have identified a unique B7 pathway that is able to inhibit human CD4 and CD8 T-cell proliferation and cytokine production. This unique human T-cell coinhibitory pathway may afford unique strategies for the treatment of human cancers, autoimmune disorders, infection, and transplant rejection and may help to design better vaccines.
Interactions between members of the B7 ligand and CD28 receptor families generate positive costimulation and negative coinhibition, which are of central importance in regulating T-cell responses (1–3). B7-1/B7-2/CD28/CTLA-4 is the most extensively characterized of these pathways. Ligands B7-1 (CD80) and B7-2 (CD86) on antigen-presenting cells (APCs) bind to CD28 on naïve T cells and provide a major costimulatory signal to activate naïve T cells. After the initial activation, coinhibitory molecule cytotoxic T lymphocyte antigen-4 (CTLA-4, CD152) is induced on T cells and engages the same B7-1 and B7-2 ligands to restrain T-cell function. In contrast to the costimulatory activity of CD28, the interaction of B7-1 or B7-2 with CTLA-4 is essential for limiting the proliferative response of recently activated T cells to antigen and CD28-mediated costimulation.
During the past decade, several new pathways in the B7 and CD28 families have been identified, including B7h/ICOS, PD-L1/PD-L2/PD-1, B7-H3/receptor, and B7x/receptor. B7h (4) (also called ICOS-L, B7RP-1 (5), GL50 (6), B7H2 (7), LCOS (8), and CD275) binds to the inducible costimulator (ICOS, CD278) on activated T cells (9), which induces strong phosphatidylinositol 3-kinase activity (10, 11) and leads to the expression of transcription factors involved in follicular helper CD4 T (Tfh) differentiation (12). Therefore, the B7h/ICOS pathway provides critical T-cell help to B cells. Deficiencies in this pathway result in substantially reduced numbers of memory B cells and markedly reduced levels of serum Ig in patients with common variable immunodeficiency (13). In humans, but not in mice, B7h can bind both CD28 and CTLA-4 (14). The B7 family members PD-L1 (15) [also termed B7-H1 (16), CD274] and PD-L2 (17) [also called B7-DC (18), CD273] bind to the programmed death 1 receptor (PD-1, CD279), which ultimately decreases induction of cytokines and cell survival proteins in T cells. The PD-L/PD-1 pathway plays an important role in the control of tolerance and autoimmunity (19, 20), and contributes critically to T-cell exhaustion and viral persistence during chronic infections (21). In addition, PD-L1 can also bind to B7-1 (22, 23). Finally, B7-H3 (24) (CD276) and B7x (25) [also called B7-H4 (26) or B7S1 (27)] are recently discovered members of the B7 family, and their contributions to immune response have not yet been clearly defined. Furthermore, the receptors for B7-H3 and B7x are currently unidentified. B7-H3 binds activated T cells, but the physiological role of this pathway is unclear, as both costimulatory and coinhibitory effects have been observed (24, 28, 29). B7x binds activated T cells and inhibits T-cell functions. In addition, myeloid-derived suppressor cells (MDSCs) also express a receptor for B7x (30). Clinical data also support a coinhibitory function for B7x, as aberrant expression of this molecule is observed in many types of human cancers and is often associated with enhanced disease progression and poor clinical outcome (31). It appears that the B7x pathway is exploited as part of the immune evasion mechanisms used by many human cancers. Collectively, the regulated spatial and temporal expression of costimulatory and coinhibitory B7 molecules provides the controls that underlie T cell-mediated immune responses.
Due to their fundamental biological importance and therapeutic potential, there has been considerable interest in the identification of additional molecules with costimulatory or coinhibitory function. Here we describe the HERV–HLTR-associating 2 (HHLA2) (32) as a member of the B7 family with coinhibitory function for both human CD4 and CD8 T cells, which is comparable to other important family members. A putative receptor for HHLA2 is expressed widely on T cells and APCs. This pathway may present a unique therapeutic target.
Results
Characterization of HHLA2 as a B7 Family Member.
Through a homology search of various databases using amino acid sequences of human B7x and B7-H3, we identified HHLA2 which was shown previously to share significant homology with the B7 family (32, 33) and was also called B7H7 (33). The human HHLA2 gene is located in the q13.13 region of chromosome 3 and is near the B7-1 and B7-2 genes (q13.3-q21). We sequenced the ORF and found the deduced protein sequence of HHLA2 contained 414 amino acids (Fig. 1A), longer than most B7 members, but shorter than human B7-H3. HHLA2 shares varying levels of amino acid identity and similarity with human B7-1 (10% and 23%), B7-2 (13% and 29%), B7h (15% and 30%), PD-L1 (12% and 26%), PD-L2 (14% and 27%), B7-H3 long form (15% and 32%), and short form (16% and 33%), and B7x (18% and 30%), which are comparable to the homologies exhibited by other members of the family; for example, B7-1, the founding member of the B7 family, shares 13–21% of amino acid identity and 22–37% of similarity with other human B7 molecules.
Fig. 1.
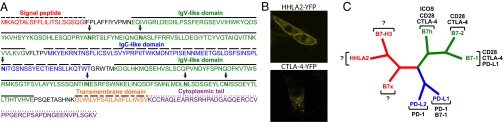
HHLA2 is a member of the B7 family and forms a subfamily with B7x and B7-H3. (A) Predicted signal peptide, IgV-like and IgC-like domains, transmembrane region, and cytoplasmic tail of human HHLA2 protein were indicated. The potential N-glycosylation sites were arrowed. (B) Confocal microscopy showed that human HHLA2-YFP protein was predominantly expressed on cell membranes of the 3T3 cells, whereas human CTLA-4-YFP fusion protein was mainly localized intracellularly in the 3T3 cells. (C) Phylogenetic tree of the human B7 family. The phylogenetic comparison of human B7 molecules was generated by PAUP version 4.0b10. The family was divided into three groups: HHLA2, B7x, and B7-H3 for group III; PD-L1 and PD-L2 for group II; and B7-1, B7-2, and B7h for group I. Receptors for human B7 molecules were also indicated.
The putative HHLA2 protein has an N-terminal signal peptide, an ectodomain composed of tandem IgV-IgC-IgV domains, six potential sites for N-linked glycosylation, a transmembrane region, and a 49-aa cytoplasmic tail (Fig. 1A). The predicted HHLA2 protein is a type I transmembrane molecule. To test this prediction, we examined the HHLA2 protein localization by expressing HHLA2-YFP fusion protein in a mouse embryonic fibroblast (3T3) cell line, which did not express endogenous HHLA2. Confocal microscopy analysis revealed that HHLA2 protein was predominantly found on cell membranes with some in the cytoplasm (Fig. 1B). In contrast, human CTLA-4–YFP fusion protein was mainly localized intracellularly in the 3T3 cell (Fig. 1B).
Evolution of HHLA2.
In a previous study we divided the B7 family of proteins into three groups by phylogenetic analysis (25). With HHLA2 added into this family, we used PAUP 4.0b10 (34) to reevaluate the relationship among human B7 proteins. As shown in Fig. 1C, a phylogenetic comparison of the family divided the human B7 molecules into three groups: group I includes B7-1, B7-2, and B7h; group II consists of PD-L1 and PD-L2; and group III contains B7x, HHLA2, and B7-H3. For group I, CD28 and CTLA-4 are receptors for all three B7 molecules and the closely related ICOS is a receptor for B7h, whereas PD-L1 can bind B7-1. For group II, PD-1 is the receptor for both PD-L1 and PD-L2. For group III, receptors have not been identified yet. The phylogenetic comparison suggests that receptors for group III would not be real homologs of receptors for groups I and II.
Based on sequence analyses, putative HHLA2 orthologs appear to be present in a wide range of species, including fish (GeneBank accession no. ACH85300), frog (NP_001122116), Heterocephalus glaber (EHB18400), giant panda (EFB27984), and monkey (EHH16036 and EHH51009), suggesting evolutionally conserved function. However, in contrast to other B7 family members, laboratory mouse and rat strains do not express HHLA2, which makes it the first B7 family member expressed in humans but not in mice.
Protein Expression Pattern of HHLA2.
The expression of HHLA2 at the protein level is completely unknown at present. To examine the protein expression, we generated a panel of monoclonal antibodies (mAbs) against HHLA2 from mice as mice do not express the HHLA2 gene. The specificity of the mAbs was demonstrated by ELISA and FACS in which mAbs reacted with HHLA2 but not with other B7 molecules (Fig. 2A and Fig. S1).
Fig. 2.
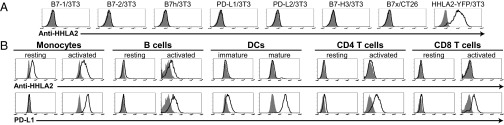
Analysis of endogenous HHLA2 protein expression by flow cytometry with specific mAb. (A) 3T3 or CT26 cells were transfected with MSCV vectors to stably express cell surface human B7-1, B7-2, B7h, PD-L1, PD-L2, B7-H3, B7x, and HHLA2-YFP. Transfectants were stained with an anti-HHLA2 mAb clone 566.1 (open histograms) or isotype control (shaded histograms) for FACS. (B) Human PBMCs were stained with biotin–anti-HHLA2 mAb/APC-streptavidin, and PE-, FITC-, or Percp-Cy5.5–conjugated anti-CD14 (monocytes), anti-CD19 (B cells), anti-CD4, anti-CD8, and anti–PD-L1. Monocytes and B cells were activated with LPS/IFN-γ for 3 d, whereas T cells were activated with anti-CD3 for 3 d. Immature DCs were generated from blood monocytes incubated with GM-CSF/IL-4 and were induced with LPS/IFN-γ to become mature DCs. Endogenous HHLA2 protein was highly detected on monocytes and induced on B cells, whereas PD-L1 was induced on activated immune cells. Anti-HHLA2 mAb (open histograms) and isotype control (shaded histograms) are shown. Results represent at least seven experiments.
Using the anti-HHLA2 mAb clone 566.1, HHLA2 expression was examined on APCs by FACS. CD14 positive monocytes in human peripheral blood mononuclear cells (PBMCs) expressed significant levels of HHLA2 and the expression was further up-regulated by stimulation with LPS and IFN-γ (Fig. 2B). Resting CD19 positive B cells did not express HHLA2, but the expression was induced by LPS and IFN-γ stimulation (Fig. 2B). No clear HHLA2 protein could be detected on blood monocyte-derived immature dendritic cells (DCs) or LPS/IFN-γ–induced mature DCs (Fig. 2B). For T cells, both CD4 and CD8 T cells in PBMCs did not express HHLA2; both were still negative after stimulation with plate-bound anti-CD3 (Fig. 2B). As a control, PD-L1, another B7 molecule, was induced on monocytes, B cells, and DCs after LPS/IFN-γ stimulation and was induced on CD4 and CD8 T cells after stimulation with anti-CD3 (Fig. 2B). Collectively, these results demonstrate that endogenous HHLA2 is an integral cell surface protein constitutively expressed on monocytes and induced on B cells.
HHLA2 Does Not Bind Other Known Members of the CD28 and B7 Families.
It is well documented that all previously characterized B7 family members can act as ligands and regulate T-cell function by binding to receptors. The receptor for HHLA2 is unknown, therefore we first asked whether any of the known CD28 family members are the receptor for HHLA2. To this end, we first generated an HHLA2-Ig fusion protein consisting of the extracellular portion of human HHLA2 and the Fc portion of IgG1 and three other controls including B7x-Ig, B7-H3-Ig, and only the Fc portion of human IgG1 (Ig control). The HHLA2-Ig fusion protein and the controls, produced in the same system and purified in the same way, were used to search for the interactions between HHLA2 and the CD28 family members using FACS analysis. To do this, we established 3T3 lines expressing cell surface human CD28, ICOS, and PD-1 (Fig. 3A). CTLA-4 is not primarily a cell surface protein (Fig. 1B), mainly due to the fact that it contains an intracellular localization motif (TTGVYVKMPPT) in its cytoplasmic tail (35). We established a 3T3 line expressing cell surface CTLA-4, which did not contain the cytoplasmic tail (Fig. 3A). In FACS experiments, HHLA2-Ig, like the other control Igs (Ig, B7x-Ig, and B7-H3-Ig), did not bind CD28, CTLA-4, ICOS, and PD-1 on the cell surface of 3T3 cells (Fig. 3B). Within the B7 family, B7-1 can bind to another B7 molecule PD-L1 (22). To test whether HHLA2 binds to any of the known B7 family members, we established 3T3 and mouse colon carcinoma (CT26) lines that expressed cell surface human B7-1, B7-2, B7h, PD-L1, PD-L2, B7-H3, and B7x, and found that neither HHLA2-Ig nor B7x-Ig bound the other B7 molecules (Fig. 3B). As a positive control, PD-1–Ig bound 3T3 cells expressing PD-L1 or PD-L2 (Fig. 3C). These results reveal that neither the known members of the CD28 family nor those of the B7 family interact with HHLA2.
Fig. 3.

HHLA2 does not bind other known members of the CD28 and B7 families. (A) 3T3 or CT26 cells were transfected with MSCV vectors to stably express cell surface human CD28, CTLA-4 without cytoplasmic tail, ICOS, PD-1, B7-1, B7-2, B7h, PD-L1, PD-L2, B7-H3, and B7x. All transfectants were stained with specific mAbs (open histograms) or control Abs (shaded histograms). (B) Transfectants were stained with HHLA2-Ig fusion protein (open histograms) or control fusion proteins Ig or B7x-Ig (shaded histograms) and then stained with a PE-conjugate antihuman IgG Fc. (C) As positive controls, 3T3 cells expressing PD-L1 or PD-L2 were stained with PD-1–Ig (open histograms) or control Ig (shaded histograms).
Constitutive Expression of an HHLA2 Putative Receptor on T Cells and Other Immune Cells.
T cells express receptors for members of the B7 family. To test whether T cells have an HHLA2 receptor, we used HHLA2-Ig and control Ig to stain T cells from PBMCs. FACS analyses showed that HHLA2 bound freshly isolated, resting CD4 and CD8 T cells (Fig. 4A). After stimulation with plate-bound anti-CD3 for 3 d, activated CD4 and CD8 T cells still expressed a receptor for HHLA2 (Fig. 4A). In contrast, ICOS was not expressed on resting CD4 and CD8 T cells but was induced after stimulation (Fig. 4A). HHLA2 receptor positive cells and ICOS positive cells were partially overlapping. PD-1, another CD28 family member, was recently reported to be expressed on human B cells (36). We therefore examined whether APCs have a receptor for HHLA2. HHLA2-Ig bound freshly isolated B cells and monocytes, suggesting these cells express a receptor for HHLA2 (Fig. 4A). We further stimulated PBMCs with LPS/IFN-γ for 3 d and found B cells and monocytes were activated, as evidenced by induced expression of PD-L1. Both activated B cells and monocytes were stained by HHLA2-Ig (Fig. 4A), suggesting resting and activated B cells as well as monocytes have a putative HHLA receptor. Finally, we examined DCs. We found that HHLA2 bound blood monocyte-derived immature DCs as well as LSP/IFN-γ–induced mature DCs (Fig. 4A). In contrast to immune cells, HHLA2 did not bind human HeLa cells and mouse 3T3 cells (Fig. 4B), suggesting these cells did not have an HHLA2 receptor. Taken together, these results indicate that a putative HHLA2 receptor is constitutively expressed on T cells, B cells, monocytes, and DCs.
Fig. 4.

T cells and other immune cells constitutively express a putative receptor for HHLA2. (A) T cells, B cells, and monocytes from PBMCs and DCs derived from blood monocytes were stained with HHLA2-Ig fusion protein (open histograms) or control Ig (shaded histograms) and then stained with a PE-conjugate antihuman IgG Fc. CD4 and CD8 T cells were stimulated with anti-CD3 for 3 d, whereas B cells and monocytes were stimulated with LPS/IFN-γ for 3 d. Immature DCs were generated from blood monocytes and were induced with LPS/IFN-γ to be mature DCs. HHLA2 bound T cells, B cells, monocytes, and DCs. ICOS was induced on activated CD4 and CD8 T cells, whereas PD-L1 was induced on APCs. (B) In contrast to these immune cells, HHLA2 bound neither human HeLa cells nor mouse 3T3 cells. HHLA2-Ig fusion protein (open histograms) and control Ig (shaded histograms) are shown. Results represent at least five experiments.
HHLA2 Inhibits TCR-Mediated CD4 and CD8 T-Cell Proliferation.
Based on our data showing that HHLA2 protein was detected on APCs and a putative receptor was constitutively expressed on T cells, we examined whether HHLA2 was able to regulate T-cell function using a system modified from our previous studies (25). In this system, purified T cells were activated with plate-bound mAb to human CD3 and the activation of T cells was determined on days 3 and 5. We first performed a dose titration of anti-CD3 and found that T cells from different normal donors needed different concentrations of anti-CD3 to achieve the middle level of proliferation. Therefore, different suitable concentrations of anti-CD3 were used for T-cell experiments from each donor. We used the MTT assay to quantify anti-CD3–induced T-cell activation in the presence of immobilized HHLA2-Ig, control Ig, or B7x-Ig (Fig. 5A). HHLA2-Ig significantly decreased T-cell activation induced by anti-CD3 (Fig. 5A). As a control, B7x-Ig also inhibited T-cell activation in the same system, consistent with our previous report (25). As both CD4 and CD8 T cells constitutively express an HHLA2 receptor, we next examined whether HHLA2 was able to inhibit both CD4 and CD8 T cells. Purified T cells from PBMCs were labeled with carboxyfluorescein diacetate succinimidyl ester (CFSE), and stimulated with anti-CD3 in the presence of immobilized HHLA2-Ig, control Ig, or B7x-Ig for 5 d. These cells were then analyzed by FACS with anti-CD4 and anti-CD8. HHLA2-mediated inhibition was determined by gating on CD4 and CD8 T-cell populations and measuring CFSE fluorescence intensity. As expected, both CD4 and CD8 T cells proliferated vigorously when incubated with anti-CD3 and control Ig (Fig. 5 B and C), with more than 55% of CD4 and 69% of CD8 T cells dividing. However, when T cells were incubated with anti-CD3 and HHLA2-Ig, significantly fewer CD4 and CD8 T cells proliferated, with less than 38% of CD4 and less than 52% of CD8 T cells dividing (Fig. 5 B and C). Similarly, B7x-Ig also inhibited both CD4 and CD8 T-cell proliferation, with less than 44% of CD4 and less than 57% of CD8 T cells dividing (Fig. 5). Therefore, these findings from two functional assays demonstrate that HHLA2 inhibits TCR-mediated proliferation of both human CD4 and CD8 T cells to a similar degree as B7x.
Fig. 5.

Coinhibition of HHLA2 on TCR-mediated CD4 and CD8 T-cell proliferation. (A) T cells purified from PBMCs were activated with a combination of plate-bound anti-CD3 and either plate-bound HHLA2-Ig (4 μg/mL), control Ig (4 μg/mL), or B7x-Ig (4 μg/mL) for 3 d. Metabolic activity was then determined by MTT assay. (B and C) CFSE-labeled T cells were stimulated with a combination of plate-bound anti-CD3 and either plate-bound HHLA2-Ig (10 μg/mL), control Ig (10 μg/mL), or B7x-Ig (10 μg/mL) for 5 d. T cells were then stained with anti-CD4 and anti-CD8 and analyzed by flow cytometry. Representative FACS plots showed CFSE dilution among CD4 and CD8 T cells (B). The percentages of proliferating CD4 and CD8 T cells were calculated by CFSE dilution (C). n = 9–12, *P < 0.05; **P < 0.01, ***P < 0.001.
HHLA2 Inhibits Cytokine Production from T Cells.
We next tested the effect of HHLA2 on cytokine production from T cells. Purified T cells from PBMCs were stimulated with anti-CD3 in the presence of immobilized HHLA2-Ig or control Ig for 3 d and cytokines in the supernatants were measured using Th1/Th2/Th9/Th17/Th22 flowcytomix. Among the 13 T cell-derived cytokines tested, we found HHLA2 significantly reduced production of 7 cytokines from T cells: IFN-γ (21% reduction), TNF-α (30% reduction), IL-5 (39% reduction), IL-10 (56% reduction), IL-13 (39% reduction), IL-17A (36% reduction), and IL-22 (35% reduction) (Fig. 6). HHLA2 reduced production of IL-2 and IL-9, but the differences did not reach statistical significance. In addition, there was no effect of HHLA2 on cytokine production of IL-1β, IL-4, IL-6, or IL-12p70. These results suggest that HHLA2 is able to suppress certain cytokines produced from T cells induced by TCR signaling.
Fig. 6.
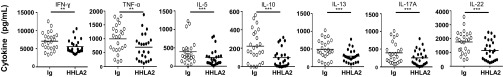
Inhibition of HHLA2 on cytokine production from T cells. Purified T cells were stimulated with a combination of plate-bound anti-CD3 and either plate-bound HHLA2-Ig (4 μg/mL) or control Ig (4 μg/mL) for 3 d. The cytokine levels of the supernatants were measured using Th1/Th2/Th9/Th17/Th22 flowcytomix. HHLA2 significantly reduced production of seven cytokines from T cells including IFN-γ, TNF-α, IL-5, IL-10, IL-13, IL-17A, and IL-22. n = 24, **P < 0.01, ***P < 0.001.
Discussion
Here we provide evidence for HHLA2 as a unique member of the B7 family that inhibits proliferation and cytokine production of both human CD4 and CD8 T cells. HHLA2 was originally cloned as a gene that was polyadenylated within a long terminal repeat (LTR) of the HERV-H endogenous retrovirus family (32), exhibiting homology with B7 (32, 33). We found that HHLA2 has all of the characteristics of a B7 family member. Similar to other members of the B7 family, HHLA2 shared 10–18% of amino acid identity and 23–33% of similarity to other human B7 molecules. It is already demonstrated that the IgV domain is the receptor-binding domain for B7-1 (37), B7-2 (38), PD-L1 (39), and PD-L2 (40). Like other B7s, HHLA2 has extracellular IgV and IgC domains. With the phylogenetic analyses, we found that HHLA2 formed the third subfamily with B7x and B7-H3 within the B7 family. Indeed, the highest homologous sequences to HHLA2 are B7-H3 and B7x. Our bioinformatic analyses and other results (32, 33) reveal that HHLA2 is found in various species including human, monkey, frog, and fish, but is not expressed in mouse and rat. Mouse and rat have only HHLA2 pseudogenes (32, 33). This is unique, as all other known members of the B7 family and of the CD28 family are found in both humans and mice.
Compared with other B7s, human HHLA2 has a different expression pattern. HHLA2 protein was expressed highly and constitutively on monocytes, whereas its expression on human B cells was induced by inflammatory stimulation. However, blood monocyte-derived DCs and T cells were HHLA2 negative even after activation with LPS/IFN-γ and anti-CD3, respectively. Future investigation is warranted to determine whether other stimuli are able to induce HHLA2 on DCs and T cells. Further study will also be required to determine whether HHLA2 is expressed on nonhematopoietic cells and tumors. Different from HHLA2, we found that the other B7 molecule, PD-L1, was not expressed in human resting T cells and APCs but was induced on these immune cells after activation. B7-2 is expressed at very low levels on human resting B cells and immature DCs and is induced to high levels with stimuli (41, 42), whereas B7x is hardly detected on normal human T cells and APCs (41). These studies highlight the dramatic differences in the spatial and temporal expression of the individual members of the B7 family. These findings have important implications for understanding the in vivo functions of previously characterized and newly identified B7s.
HHLA2 appears to have a counter receptor that is distinct from CD28, CTLA-4, ICOS, PD-1, and all B7 molecules. CD28 is constitutively expressed on T cells, whereas CTLA-4, ICOS, and PD-1 are induced after T-cell activation (1–3). HHLA2-Ig fusion protein did not interact with any known members of the CD28 and B7 families, demonstrating that these molecules are not the receptor for HHLA2. These results are consistent with the phylogenetic analyses, which suggest the receptors for group III (HHLA2, B7x, and B7-H3) may be distinct from the receptors for groups I and II. Interestingly, HHLA2 bound not only to activated human CD4 and CD8 T cells but also to resting CD4 and CD8 T cells. Therefore, HHLA2 joins B7-1 and B7-2 to recognize receptors expressed on both resting and activated T cells.
HHLA2 is able to function as a negative regulator of human T cells. In the presence of TCR signaling, immobilized HHLA2 protein suppressed proliferation of both human CD4 and CD8 T cells as effectively as B7x in the same experimental system. The second line of evidence that supported an inhibitory role for HHLA2 in T-cell regulation is its effect on cytokine production. Among 13 cytokines from T cells induced by TCR signaling, HHLA2 significantly reduced the production of 7 cytokines including IFN-γ, TNF-α, IL-5, IL-10, IL-13, IL-17A, and IL-22, indicating that HHLA2 is able to inhibit T-cell cytokine production. We found that human T cells from different donors had considerable variation in cytokine production, which may reflect an interesting heterogeneity in the human response. In group III of the B7 family, B7x suppresses T cells and is widely overexpressed in many human solid tumors; B7-H3 is reported to have costimulatory and coinhibitory effects (28), although clinical observations suggest that it may function in tumor immune evasion. We have now expanded this group to include HHLA2 as a T cell coinhibitor. In addition to T cells, human APCs also express a receptor for HHLA2. The potential function of this pathway in APCs remains to be clarified.
In summary, we have characterized a member of the B7 family that serves as an attenuator of T-cell responses. It is a unique B7 family member in that it exists in humans but not in mice. Its putative receptor is constitutively expressed on human T cells and APCs. The expression patterns of HHLA2 and its putative counter receptor coupled with its coinhibitory function suggest that this pathway may be a potent regulator of human immune responses at both the very early and late stages. In the clinic, CTLA-4–Ig fusion proteins (Abatacept and Belatacept) inhibit T-cell functions and have already been used to treat adult rheumatoid arthritis and to prevent acute kidney transplant rejection (43, 44), respectively. A mAb that blocks CTLA-4 functions (Ipilimumab) was recently approved for treatment of metastatic melanoma (45, 46). Some mAbs against PD-1 and PD-L1 are currently in clinical trials with cancer patients (47, 48). Clearly, further studies on this inhibitory HHLA2 pathway may lead to new therapies for human cancers, autoimmune disorders, infection, and transplant rejection.
Materials and Methods
Bioinformatic Analysis.
BLAST was used to search public databases with protein sequences. Sequence alignment and homology comparison were done with MacVector 10.6. The phylogenetic tree was generated by PAUP (4.0b10) using sequence alignment by removal of significant inserts and trimming C- and N-terminal extensions (34). Motifs and domains were analyzed with EMBL-EBI tools, SMART, and CBS Prediction.
Production and Purification of Fusion Proteins.
HHLA2-Ig and B7x-Ig proteins were prepared by fusing the coding region of the extracellular domain without signal peptide of human HHLA2 or B7x to a human IgG1 Fc tag of plasmid pMT/BiP as described (25). The pMT/BiP construct itself produced human IgG1 Fc tag as a control. All constructs were cotransfected into Drosophila Schneider 2 (S2) cells with a hygromycin resistance plasmid, and the stable transfected cell lines were induced to secrete fusion proteins in Express Five serum-free medium (LifeTechnologies). Proteins were purified using Protein G Plus Agarose columns (Pierce) followed by FPLC. The purity and identity of fusion proteins were confirmed by SDS/PAGE, Western blotting, and protein sequencing with MALDI-TOF-MS/MS (Fig. S1).
Retrovirus Constructs and Cell Line Transfectants.
HHLA2-YFP fusion protein construct was generated by using PCR to amplify the coding sequence of HHLA2 without the stop codon and then cloned into the Bgl II site of the L50-YFP/MSCV vector. CTLA-4 in L50-YFP/MSCV vector was reported previously (49). The coding sequences of human CD28, PD-1, ICOS, B7-1, B7-2, B7h, PD-L1, PD-L2, B7-H3, and B7x were cloned into XhoI/NotI or XhoI/EcoRI sites of MSCV vector. The coding sequence of human CTLA-4 without a cytoplasmic tail was cloned into an MSCV vector as well. All vectors were used to generate retrovirus and then transfected into cell lines 3T3 or CT26. Positive cell line transfectants were sorted out by FACS using specific mAbs or YFP.
Generation of Monoclonal Antibodies to Human HHLA2.
Hybridomas producing mAbs to human HHLA2 were generated by standard techniques from splenocytes of HHLA2-Ig–immunized BALB/c mice fused to NSO myeloma cells. Four independent clones, 566.1 (IgG1), 351.7 (IgG1), 457.23 (IgG1), and 205.1 (IgG1) were selected by ELISA as their mAbs recognized HHLA2-Ig, but not controls including B7x-Ig, B7-H3-Ig, and normal human IgG. After this preliminary screening, specificity of mAbs were further determined by FACS positive staining of a 3T3 transfectant expressing HHLA2-YFP but negative staining of transfectants expressing other human B7s (B7-1, B7-2, B7h, PD-L1, PD-L2, B7-H3, and B7x) and human CD28 family members (CD28, CTLA-4, ICOS, and PD-1). mAbs were purified by Protein G Plus Agarose columns and biotinylated with the EZ-Link Sulfo-NHS-Biotin kit (Thermo Scientific).
Human Antigen-Presenting Cells and Activation.
Human CD19+ B cells and CD14+ monocytes in PBMCs were activated as previously described (41). B cells were activated by IFN-γ (100 ng/mL; eBioscience) and LPS (60 μg/mL; Sigma) for 3 d, and monocytes were stimulated by IFN-γ (100 ng/mL) and LPS (100 ng/mL) for 3 d. DCs were generated from human blood monocytes (50). Monocytes from PBMCs were incubated with completed RPMI1640 containing 10% human serum AB (Atlanta Biological), human GM-CSF (100 ng/mL; R&D), and human IL-4 (50 ng/mL) for 6 d to generate immature DCs. These immature DCs were further stimulated with LPS (1 μg/mL) and IFN-γ (100 ng/mL) for 2 d to generate mature DCs.
Human T-Cell Coinhibition Assay.
Human T cells were purified from PBMCs with CD2 Microbeads (Miltenyi Biotec) and incubated (2 × 105 per well) with different concentrations (0.1–10 μg/mL) of plate-bound anti-CD3 (OKT3; eBioscience) for 3 d. The T-cell proliferation was determined by 3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide (MTT) assay and plates were read at 570 nm. T cells from different donors needed different concentrations of anti-CD3 to achieve the middle level of proliferation. After determining the suitable anti-CD3 concentration for each donor’s T cells, 96-well flat-bottom plates were precoated with anti-CD3, HHLA-2-Ig, control Ig, or B7x-Ig in PBS at 4 °C overnight. Wells were washed and incubated with purified T cells for 3 d. T-cell proliferation was then measured with MTT assay. For the CFSE (Sigma)-labeled proliferation assay, CFSE-labeled human T cells were incubated with plate-bound anti-CD3, HHLA2-Ig, control Ig, or B7x-Ig for 5 d, and stained with anti-CD4 and anti-CD8 for flow cytometry.
Statistics.
Statistical significance was calculated with the paired- or unpaired t test using Prism software version 4.0b (GraphPad). A P value of <0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank Jun Sik Lee, Vladimir Vigdorovich, Kimberly Hofmeyer, Yael Abadi, and Eszter Lazar-Molnar for help at the beginning of this project; Tsvetelina Pentcheva-Hoang and James Allison (Memorial Sloan-Kettering Cancer Center) for providing CTLA-4–YFP plasmid; and Steven Almo for comments on the manuscript. This work was supported by National Institutes of Health (NIH) Grant DP2Dk083076 and Department of Defense PC094137 (to X.Z.). R.Z. was partially supported by the China Scholarship Council. J.M.C., K.C.O., and K.G. were partially supported by NIH Training Grant T32GM007288 and T32DK007218. The Albert Einstein Cancer Center, the Diabetes Center, and the Center for AIDS Research are supported by NIH Grants P30CA013330, P60DK020541, and AI-51519. The Hybridoma Facility is supported by the Albert Einstein College of Medicine.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1303524110/-/DCSupplemental.
References
- 1.Chen L. Co-inhibitory molecules of the B7-CD28 family in the control of T-cell immunity. Nat Rev Immunol. 2004;4(5):336–347. doi: 10.1038/nri1349. [DOI] [PubMed] [Google Scholar]
- 2.Greenwald RJ, Freeman GJ, Sharpe AH. The B7 family revisited. Annu Rev Immunol. 2005;23:515–548. doi: 10.1146/annurev.immunol.23.021704.115611. [DOI] [PubMed] [Google Scholar]
- 3.Zang X, Allison JP. The B7 family and cancer therapy: Costimulation and coinhibition. Clin Cancer Res. 2007;13(18 Pt 1):5271–5279. doi: 10.1158/1078-0432.CCR-07-1030. [DOI] [PubMed] [Google Scholar]
- 4.Swallow MM, Wallin JJ, Sha WC. B7h, a novel costimulatory homolog of B7.1 and B7.2, is induced by TNFalpha. Immunity. 1999;11(4):423–432. doi: 10.1016/s1074-7613(00)80117-x. [DOI] [PubMed] [Google Scholar]
- 5.Yoshinaga SK, et al. T-cell co-stimulation through B7RP-1 and ICOS. Nature. 1999;402(6763):827–832. doi: 10.1038/45582. [DOI] [PubMed] [Google Scholar]
- 6.Ling V, et al. Cutting edge: Identification of GL50, a novel B7-like protein that functionally binds to ICOS receptor. J Immunol. 2000;164(4):1653–1657. doi: 10.4049/jimmunol.164.4.1653. [DOI] [PubMed] [Google Scholar]
- 7.Wang S, et al. Costimulation of T cells by B7-H2, a B7-like molecule that binds ICOS. Blood. 2000;96(8):2808–2813. [PubMed] [Google Scholar]
- 8.Brodie D, et al. LICOS, a primordial costimulatory ligand? Curr Biol. 2000;10(6):333–336. doi: 10.1016/s0960-9822(00)00383-3. [DOI] [PubMed] [Google Scholar]
- 9.Hutloff A, et al. ICOS is an inducible T-cell co-stimulator structurally and functionally related to CD28. Nature. 1999;397(6716):263–266. doi: 10.1038/16717. [DOI] [PubMed] [Google Scholar]
- 10.Parry RV, Rumbley CA, Vandenberghe LH, June CH, Riley JL. CD28 and inducible costimulatory protein Src homology 2 binding domains show distinct regulation of phosphatidylinositol 3-kinase, Bcl-xL, and IL-2 expression in primary human CD4 T lymphocytes. J Immunol. 2003;171(1):166–174. doi: 10.4049/jimmunol.171.1.166. [DOI] [PubMed] [Google Scholar]
- 11.Zang X, et al. A genetic library screen for signaling proteins that interact with phosphorylated T cell costimulatory receptors. Genomics. 2006;88(6):841–845. doi: 10.1016/j.ygeno.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 12.Crotty S. Follicular helper CD4 T cells (TFH) Annu Rev Immunol. 2011;29:621–663. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- 13.Yong PF, Salzer U, Grimbacher B. The role of costimulation in antibody deficiencies: ICOS and common variable immunodeficiency. Immunol Rev. 2009;229(1):101–113. doi: 10.1111/j.1600-065X.2009.00764.x. [DOI] [PubMed] [Google Scholar]
- 14.Yao S, et al. B7-h2 is a costimulatory ligand for CD28 in human. Immunity. 2011;34(5):729–740. doi: 10.1016/j.immuni.2011.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freeman GJ, et al. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J Exp Med. 2000;192(7):1027–1034. doi: 10.1084/jem.192.7.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong H, Zhu G, Tamada K, Chen L. B7-H1, a third member of the B7 family, co-stimulates T-cell proliferation and interleukin-10 secretion. Nat Med. 1999;5(12):1365–1369. doi: 10.1038/70932. [DOI] [PubMed] [Google Scholar]
- 17.Latchman Y, et al. PD-L2 is a second ligand for PD-1 and inhibits T cell activation. Nat Immunol. 2001;2(3):261–268. doi: 10.1038/85330. [DOI] [PubMed] [Google Scholar]
- 18.Tseng SY, et al. B7-DC, a new dendritic cell molecule with potent costimulatory properties for T cells. J Exp Med. 2001;193(7):839–846. doi: 10.1084/jem.193.7.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okazaki T, Honjo T. PD-1 and PD-1 ligands: From discovery to clinical application. Int Immunol. 2007;19(7):813–824. doi: 10.1093/intimm/dxm057. [DOI] [PubMed] [Google Scholar]
- 20.Francisco LM, Sage PT, Sharpe AH. The PD-1 pathway in tolerance and autoimmunity. Immunol Rev. 2010;236:219–242. doi: 10.1111/j.1600-065X.2010.00923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hofmeyer KA, Jeon H, Zang X. The PD-1/PD-L1 (B7-H1) pathway in chronic infection-induced cytotoxic T lymphocyte exhaustion. J Biomed Biotechnol. 2011;2011:451694. doi: 10.1155/2011/451694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Butte MJ, Keir ME, Phamduy TB, Sharpe AH, Freeman GJ. Programmed death-1 ligand 1 interacts specifically with the B7-1 costimulatory molecule to inhibit T cell responses. Immunity. 2007;27(1):111–122. doi: 10.1016/j.immuni.2007.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paterson AM, et al. The programmed death-1 ligand 1:B7-1 pathway restrains diabetogenic effector T cells in vivo. J Immunol. 2011;187(3):1097–1105. doi: 10.4049/jimmunol.1003496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chapoval AI, et al. B7-H3: A costimulatory molecule for T cell activation and IFN-gamma production. Nat Immunol. 2001;2(3):269–274. doi: 10.1038/85339. [DOI] [PubMed] [Google Scholar]
- 25.Zang X, et al. B7x: A widely expressed B7 family member that inhibits T cell activation. Proc Natl Acad Sci USA. 2003;100(18):10388–10392. doi: 10.1073/pnas.1434299100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sica GL, et al. B7-H4, a molecule of the B7 family, negatively regulates T cell immunity. Immunity. 2003;18(6):849–861. doi: 10.1016/s1074-7613(03)00152-3. [DOI] [PubMed] [Google Scholar]
- 27.Prasad DV, Richards S, Mai XM, Dong C. B7S1, a novel B7 family member that negatively regulates T cell activation. Immunity. 2003;18(6):863–873. doi: 10.1016/s1074-7613(03)00147-x. [DOI] [PubMed] [Google Scholar]
- 28.Hofmeyer KA, Ray A, Zang X. The contrasting role of B7-H3. Proc Natl Acad Sci USA. 2008;105(30):10277–10278. doi: 10.1073/pnas.0805458105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suh WK, et al. The B7 family member B7-H3 preferentially down-regulates T helper type 1-mediated immune responses. Nat Immunol. 2003;4(9):899–906. doi: 10.1038/ni967. [DOI] [PubMed] [Google Scholar]
- 30.Abadi YM, et al. Host b7x promotes pulmonary metastasis of breast cancer. J Immunol. 2013;190(7):3806–3814. doi: 10.4049/jimmunol.1202439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barach YS, Lee JS, Zang X. T cell coinhibition in prostate cancer: New immune evasion pathways and emerging therapeutics. Trends Mol Med. 2010;17(1):47–55. doi: 10.1016/j.molmed.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mager DL, Hunter DG, Schertzer M, Freeman JD. Endogenous retroviruses provide the primary polyadenylation signal for two new human genes (HHLA2 and HHLA3) Genomics. 1999;59(3):255–263. doi: 10.1006/geno.1999.5877. [DOI] [PubMed] [Google Scholar]
- 33.Flajnik MF, Tlapakova T, Criscitiello MF, Krylov V, Ohta Y. Evolution of the B7 family: Co-evolution of B7H6 and NKp30, identification of a new B7 family member, B7H7, and of B7’s historical relationship with the MHC. Immunogenetics. 2012;64(8):571–590. doi: 10.1007/s00251-012-0616-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swofford DL. PAUP. Phylogenetic Analysis Using Parsimony and Other Methods. Version 4. Sunderland, MA: Sinauer; 2000. [Google Scholar]
- 35.Leung HT, Bradshaw J, Cleaveland JS, Linsley PS. Cytotoxic T lymphocyte-associated molecule-4, a high-avidity receptor for CD80 and CD86, contains an intracellular localization motif in its cytoplasmic tail. J Biol Chem. 1995;270(42):25107–25114. doi: 10.1074/jbc.270.42.25107. [DOI] [PubMed] [Google Scholar]
- 36.Thibult ML, et al. PD-1 is a novel regulator of human B-cell activation. Int Immunol. 2013;25(2):129–137. doi: 10.1093/intimm/dxs098. [DOI] [PubMed] [Google Scholar]
- 37.Stamper CC, et al. Crystal structure of the B7-1/CTLA-4 complex that inhibits human immune responses. Nature. 2001;410(6828):608–611. doi: 10.1038/35069118. [DOI] [PubMed] [Google Scholar]
- 38.Schwartz JC, Zhang X, Fedorov AA, Nathenson SG, Almo SC. Structural basis for co-stimulation by the human CTLA-4/B7-2 complex. Nature. 2001;410(6828):604–608. doi: 10.1038/35069112. [DOI] [PubMed] [Google Scholar]
- 39.Lin DY, et al. The PD-1/PD-L1 complex resembles the antigen-binding Fv domains of antibodies and T cell receptors. Proc Natl Acad Sci USA. 2008;105(8):3011–3016. doi: 10.1073/pnas.0712278105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lázár-Molnár E, et al. Crystal structure of the complex between programmed death-1 (PD-1) and its ligand PD-L2. Proc Natl Acad Sci USA. 2008;105(30):10483–10488. doi: 10.1073/pnas.0804453105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee JS, et al. B7x in the periphery abrogates pancreas-specific damage mediated by self-reactive CD8 T cells. J Immunol. 2012;189(8):4165–4174. doi: 10.4049/jimmunol.1201241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Azuma M, et al. B70 antigen is a second ligand for CTLA-4 and CD28. Nature. 1993;366(6450):76–79. doi: 10.1038/366076a0. [DOI] [PubMed] [Google Scholar]
- 43.Vincenti F, Dritselis A, Kirkpatrick P. Belatacept. Nat Rev Drug Discov. 2011;10(9):655–656. doi: 10.1038/nrd3536. [DOI] [PubMed] [Google Scholar]
- 44.Fiocco U, et al. Co-stimulatory modulation in rheumatoid arthritis: the role of (CTLA4-Ig) abatacept. Autoimmun Rev. 2008;8(1):76–82. doi: 10.1016/j.autrev.2008.07.035. [DOI] [PubMed] [Google Scholar]
- 45.Sharma P, Wagner K, Wolchok JD, Allison JP. Novel cancer immunotherapy agents with survival benefit: Recent successes and next steps. Nat Rev Cancer. 2011;11(11):805–812. doi: 10.1038/nrc3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hodi FS, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363(8):711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Topalian SL, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366(26):2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brahmer JR, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366(26):2455–2465. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pentcheva-Hoang T, Egen JG, Wojnoonski K, Allison JP. B7-1 and B7-2 selectively recruit CTLA-4 and CD28 to the immunological synapse. Immunity. 2004;21(3):401–413. doi: 10.1016/j.immuni.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 50.O'Neill DW, Bhardwaj N. 2005. Differentiation of peripheral blood monocytes into dendritic cells. Curr Protoc Immunol Chapter 22:Unit 22F 24. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


