Significance
Powdery mildew fungi are widespread plant pathogens with an obligate biotrophic lifestyle causing devastating damage to many crops. Blumeria graminis f. sp. hordei (Bgh) infects only barley and is engaged in an evolutionary arms race with the host immune system. Genome sequencing of Bgh isolates revealed an isolate-specific mosaic of monomorphic and polymorphic DNA blocks, suggesting a mechanism that provides a large standing genetic variation in virulence polymorphisms. Detailed Bgh transcriptome profiles during early pathogenesis on barley and immunocompromised Arabidopsis revealed a conserved Bgh transcriptional program despite ∼200 million years of reproductive isolation of these hosts.
Keywords: comparative genomics, fungal genetics, obligate biotrophy, plant–microbe interactions, effector-triggered immunity
Abstract
Barley powdery mildew, Blumeria graminis f. sp. hordei (Bgh), is an obligate biotrophic ascomycete fungal pathogen that can grow and reproduce only on living cells of wild or domesticated barley (Hordeum sp.). Domestication and deployment of resistant barley cultivars by humans selected for amplification of Bgh isolates with different virulence combinations. We sequenced the genomes of two European Bgh isolates, A6 and K1, for comparative analysis with the reference genome of isolate DH14. This revealed a mosaic genome structure consisting of large isolate-specific DNA blocks with either high or low SNP densities. Some of the highly polymorphic blocks likely accumulated SNPs for over 10,000 years, well before the domestication of barley. These isolate-specific blocks of alternating monomorphic and polymorphic regions imply an exceptionally large standing genetic variation in the Bgh population and might be generated and maintained by rare outbreeding and frequent clonal reproduction. RNA-sequencing experiments with isolates A6 and K1 during four early stages of compatible and incompatible interactions on leaves of partially immunocompromised Arabidopsis mutants revealed a conserved Bgh transcriptional program during pathogenesis compared with the natural host barley despite ∼200 million years of reproductive isolation of these hosts. Transcripts encoding candidate-secreted effector proteins are massively induced in successive waves. A specific decrease in candidate-secreted effector protein transcript abundance in the incompatible interaction follows extensive transcriptional reprogramming of the host transcriptome and coincides with the onset of localized host cell death, suggesting a host-inducible defense mechanism that targets fungal effector secretion or production.
Powdery mildew fungi (Ascomycota phylum) are widespread plant pathogens that cause devastating damage on a wide range of monocotyledonous and dicotyledonous plants, including many crops. They are obligate biotrophs that are entirely dependent on nutrient supply from living host cells for their growth and reproduction (1, 2). During pathogenesis, powdery mildews differentiate a specialized infection structure after host cell-wall penetration, the so-called haustorium, by invagination of the host plant plasma membrane. Haustoria are believed to function in nutrient uptake from host cells and in the export of effector molecules to facilitate fungal infection (3, 4).
The powdery mildew Blumeria graminis f. sp. hordei (Bgh) infects cultivated and wild barley (Hordeum vulgare ssp. vulgare and H. vulgare ssp. spontaneum), leading to substantial yield loss every year (5). During the past century intensive breeding programs have introgressed numerous race-specific resistance specificities to Bgh into modern barley cultivars. However, these monogenic resistances have been rapidly and invariably overcome by virulent Bgh isolates (6). At least 11 resistance loci in barley condition dominantly inherited isolate-specific resistance to Bgh (7). One of these, the mildew resistance locus A (Mla), has been subject to extreme functional diversification, encoding by far the largest number of Bgh recognition specificities (>30) (7). MLA resistance specificities to Bgh encode mainly allelic nucleotide binding and leucine-rich-repeat–type immune receptors (8, 9). Similarly, in bread wheat (Triticum aestivum), a close relative of barley, domestication appears to have influenced the evolution of allelic disease resistance recognition specificities at the powdery mildew resistance gene 3 (Pm3) locus against the wheat powdery mildew, Blumeria graminis f. sp. tritici (Bgt) (10, 11, 12). Recently, the barley MLA1 immune receptor was shown to be fully functional in partially immunocompromised transgenic Arabidopsis (pen2, pad4, sag101 background—in short, pps) where it mediates the specific recognition of Bgh isolates expressing the cognate avirulence effector AVRA1 and activation of effective race-specific immune responses including host cell death. This implies ∼200 million years of evolutionary conservation of the underlying MLA1-triggered immune mechanism in flowering plants and makes the interaction between Bgh and Arabidopsis a unique tractable pathosystem for studying the molecular basis of MLA-mediated powdery mildew disease resistance (13, 14).
The recent genome sequencing of the Bgh DH14 isolate, of two related powdery mildew species colonizing dicotyledonous plants, and of obligate biotrophic filamentous phytopathogens representing two other independent evolutionary lineages (oomycetes and rust fungi) revealed several convergent evolutionary features that reflect their common extreme parasitic lifestyle. These include a massive expansion of transposable elements, a reduced set of carbohydrate-active enzymes, and deficiencies in nitrate and sulfate metabolism pathways (15, 16, 17, 18). A comprehensive analysis of the gene arsenal encoding candidate-secreted effector proteins (CSEPs) identified 491 of such genes in the DH14 isolate, the majority being expressed in haustoria (15, 19). At least 43 CSEPs have been experimentally verified in the proteome of host epidermal tissue containing Bgh haustoria (20), and only 3 were detected in the proteome of isolated haustoria (21). Therefore, an active secretion mechanism has been proposed that exports these CSEPs from the haustorium (22). Furthermore, although detailed biological roles of most CSEPs have not been documented, host-induced gene silencing (23) of some CSEPs suggested their role in host-defense suppression (24, 25). Notably, the CSEP repertoire of Bgh is strikingly different from those of the distantly related Golovinomyces orontii and Erysiphe pisi powdery mildews, suggesting specific adaptations to their respective host plants (15). In Bgh, genes encoding CSEPs are often embedded in or close to transposable elements, possibly contributing to rapid diversification of the effector arsenal (19, 26, 27).
Powdery mildews form morphologically distinctive structures during asexual reproduction (haploid conidiospores) and produce fruiting bodies (cleistothecia-containing ascospores) after sexual reproduction. The latter are thought to enable overwintering of the pathogen because many of their hosts, including barley, are annuals, and airborne conidiospores rapidly loose viability after ∼24 h of release from their hosts. A major unresolved question is the relative frequency of clonal (asexual) and sexual reproduction in powdery mildew fungi and the contribution of both reproductive mechanisms to the generation and maintenance of genetic variation. This is particularly relevant after large-scale deployment of cultivated annual barley in Europe, which has largely removed seasonal bottlenecks for Bgh because the continuity of autumn- and spring-sown barley provides a “green bridge” for the fungus (6).
In this study, we sequenced the genomes and transcriptomes of two Bgh isolates, A6 and K1, and compared their respective genomic features with those of the previously sequenced DH14 isolate reference genome. Bgh isolates A6 and K1 were originally collected in Svalöf, Sweden, and Köln, Germany, respectively (28, 29). The reference Bgh isolate DH14 was collected in England (30). By investigating isolate-specific single-nucleotide polymorphisms, we detected unexpected evolutionary footprints and identified candidate virulence determinants that show hallmarks of rapid evolution. Using transgenic Arabidopsis pps plants expressing barley MLA1 as the dicotyledonous host for Bgh, we investigated early powdery mildew pathogenesis of A6 and K1 isolates by RNA sequencing (RNA-seq) experiments during compatible and MLA1-mediated incompatible interactions. This revealed that haustorially expressed CSEPs are nearly all down-regulated in the incompatible interaction and that the Bgh transcriptional program is conserved during pathogenesis on dicotyledonous Arabidopsis and monocotyledonous barley.
Results
Genome Sequencing and de Novo Genome Assembly of Bgh Isolates.
We sequenced two Bgh isolates that differ from each other and from the previously sequenced DH14 isolate by virulence patterns and geographically distant collection sites (Dataset S1, Table S1) (15). We generated both medium-long reads (∼700 bases) using 454 pyrosequencing and short reads (∼100 bases) using Illumina sequencing (Dataset S1, Table S2). Both short- and medium-long reads were used to build a de novo genome assembly for each isolate. On the basis of the depth of coverage of contigs, we estimated the genome sizes for isolates A6 and K1 at 121 ± 40 Mb and 127 ± 42 Mb, respectively, which is consistent with the genome size of ∼120 Mb estimated for the reference genome DH14 (15). However, the total assembly sizes are 60.1 and 64.6 Mb for A6 and K1, respectively, indicating that a large proportion remains unassembled. This likely reflects the high content of repetitive elements present in Bgh genomes, which could also account for the highly fragmented distribution of the assemblies over the genome (L50: 2.6 kb for A6 and 3.9 kb for K1) (Dataset S1, Table S2). Nevertheless, 94.8% and 99.2% of 248 core eukaryotic genes were detected in Bgh isolates A6 and K1, respectively, supporting an overall assembly quality (Dataset S1, Table S2). Calculation of sequence similarity among the mapped contigs between isolates revealed almost 99% of overall sequence similarity, indicating a very close relationship between the three isolates (Dataset S1, Table S3).
Distribution of SNPs Over the Reference Genome.
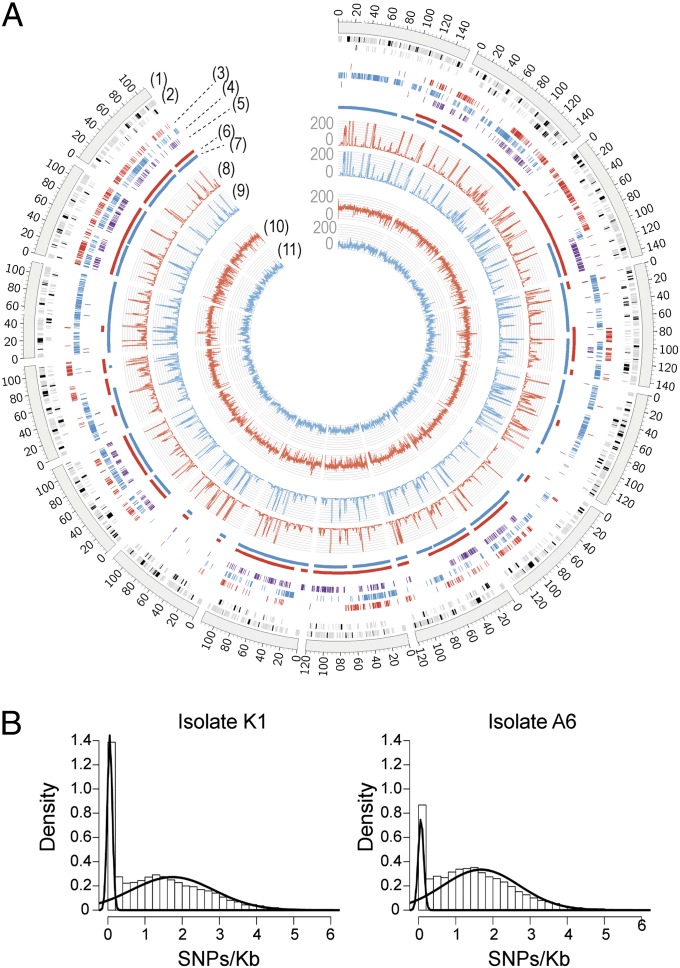
To identify single-nucleotide variations among the three Bgh genomes, we mapped Illumina short reads obtained by genome sequencing of isolates A6 and K1 onto the reference genome of isolate DH14. In total, 102,500, 87,005, and 79,186 positions could be detected with a variant nucleotide occurring specifically in one of the isolates A6, K1, or DH14. This corresponds to a frequency of isolate-specific variant nucleotides of ∼1 SNP per kilobase (1.17, 0.99, and 0.90 SNPs per kilobase for A6, K1, and DH14, respectively) (Dataset S1, Table S4). Unexpectedly, the SNP locations are not evenly distributed throughout the Bgh genomes. Instead, several large genomic stretches are fully sequence-conserved among the three isolates, whereas other regions accumulate isolate-specific SNPs (Fig. 1A). A two-component mixture model fitted to SNP densities obtained via a sliding window of 10 kb at steps of 1 kb estimated that regions with high SNP density have 1.68 ± 1.03 SNPs per kilobase whereas regions with low SNP density have 0.06 ± 0.07 SNPs per kilobase for A6, and 1.74 ± 1.11 SNPs per kilobase vs. 0.05 ± 0.06 SNPs per kilobase for K1 (Fig. 1B). The mixture model was used in combination with a hidden Markov model to predict high- and low-SNP-density blocks (Fig. 1A). In total, we estimate that 14% (A6) and 25% (K1) of the DH14 genome belongs to blocks with low SNP density. Moreover, we find that 9.4% of the largest DH14 contigs is monomorphic between all three isolates whereas 32.1% is polymorphic between all three isolates. The remaining regions are polymorphic in only one of the isolates. To estimate divergence times, we calculated SNP densities in intergenic regions and used a mutation rate of 1.3e-8 ± 2.29e-9 per site per year (31) (Materials and Methods), resulting in estimates of divergence times for low-density regions of 5.7 ± 1.2 (A6) and 4.6 ± 1.0 (K1) thousand years and for high-density regions between 137.5 ± 20.6 (A6) and 141.7 ± 21.2 (K1) thousand years.
Fig. 1.
SNP distribution in B. graminis f. sp. hordei (Bgh) isolates. (A) Circular visualization of the alignment of Bgh isolates A6 and K1 genome and transcriptome sequencing reads and SNP locations with respect to the 15 largest contigs of the Bgh isolate DH14 reference genome. Bgh K1 data are highlighted in red and Bgh A6 data in blue. The scale of the coverage plots ranges from 0 to 200 (1). The 15 largest contigs of the Bgh DH14 genome (2). Locations of annotated Bgh DH14 gene models (black bars) and repeats (gray bars). As the repeat regions and gene models can be located very close to one another, the tile track has been stacked in two layers for proper visualization (3). Bgh K1 isolate-specific genomic SNPs (4). Bgh A6 isolate-specific genomic SNPs (5). Bgh DH14 isolate-specific genomic SNPs (6). Regions with a high density of SNPs between K1 and DH14 as identified through a hidden Markov model (7). Regions with a high density of SNPs between A6 and DH14 (8). Bgh K1 RNA-Seq read coverage (average per 100 bases) (9). Bgh A6 RNA-Seq read coverage (average per 100 bases) (10). Bgh K1 genome read coverage (average per 100 bases) (11). Bgh A6 genome read coverage (average per 100 bases). (B) Histograms of SNP density distributions in Bgh isolates A6 (Right) and K1 (Left) based on a sliding window of 10 kb. The curves are based on a two-component mixture model fitted to the distributions using the expectation-maximization algorithm.
Isolate-Specific Genome Stretches and Genes Without Read Coverage.
By mapping all DNA-derived Illumina reads generated for the isolates A6 and K1 onto the DH14 reference genome, we found 69 kb (A6) and 106 kb (K1) with zero coverage in stretches of at least 1 kb. In total, five previously annotated genes are encoded in these regions (Table 1). Four of five genes are missing only in Bgh isolate K1, and two of these encode CSEPs. The other two encode a homolog of the effectors paralogous to AVRK1 and AVRA10 (EKAs) (27) and a zinc-finger FYVE domain-containing protein (32). In Bgh isolate A6, we detected only one gene with zero coverage in the DH14 reference genome, corresponding to the mating-type locus MAT1-2–1. Using the de novo assembly of isolate A6, we recovered the contig with the mating-type locus containing the cytoskeleton assembly control protein (SLA2) and MAT1-1–1 (33) (Fig. S1). Because haploid powdery mildews harbor either of these two mating-type loci to control mating compatibility for sexual reproduction (33), this validates at least one of the identified five candidate genes in the isolate-specific genome stretches as a true isolate-specific gene.
Table 1.
DH14 gene models located in regions not covered by K1/A6 genome sequencing reads
| Isolate | Bgh ID* | Description† |
| K1 | Bgh03596 | Candidate effector (CSEP0120) |
| K1 | Bgh04044 | EKA-like protein |
| K1 | Bgh04177 | FYVE zinc-finger protein |
| K1 | Bgh02875 | Candidate effector (CSEP0065) |
| A6 | Bgh03211 | MAT1-2–1 (mating type) |
Gene identification numbers retrieved via www.blugen.org.
CSEP identification numbers as described in ref. 19.
Variation of CSEP Repertoires Among the Three Bgh Isolates.
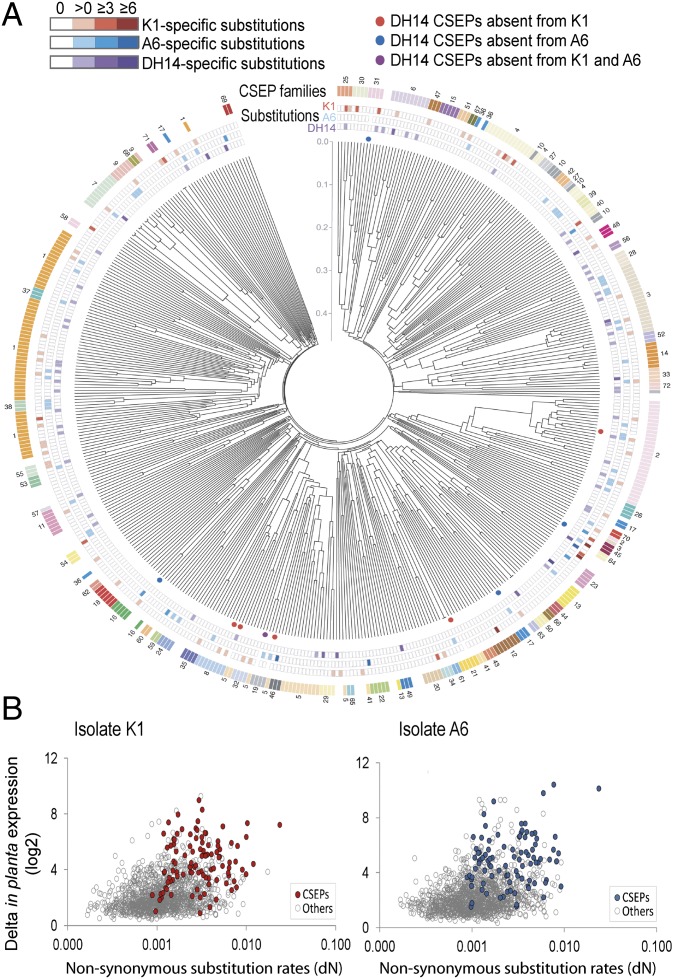
The large number of 491 previously annotated CSEP coding sequences in the DH14 Bgh reference genome and their proposed function in powdery mildew pathogenesis prompted us to examine potential isolate-specific variation in this gene family. To predict genes present in A6 and K1, we used three approaches, including a full de novo gene prediction, a de novo transcriptome assembly, and gene prediction by mapping existing CSEPs (Materials and Methods). Orthologs were identified using orthoMCL or by reciprocal best blast hits. If these three approaches did not provide conclusive evidence, we manually inspected the alignment of the A6 and K1 genome-sequencing reads to DH14. Using this strategy, nearly all CSEPs present in DH14 have an ortholog in the two other isolates (450 orthologous CSEPs in A6, 442 orthologous CSEPs in K1) (Fig. 2A). We also found a small subset of CSEP genes completely lacking in the genomes of one of the two newly sequenced Bgh genomes (three CSEPs in K1, one CSEP in A6), only partly covered by K1 or A6 Bgh sequencing reads (three CSEPs in K1, four CSEPs in A6), or with small gaps in the alignment of the K1 or A6 Bgh sequencing reads (seven CSEPs in each isolate) (Fig. 2A and Fig. S2). Furthermore, for some close paralogs in DH14, we found at least one ortholog in the other isolates, although the exact ortholog pairing could not be determined in these cases (30 CSEPs in K1, 23 CSEPs in A6). In accordance with these results, the de novo transcriptome assembly identified only one additional CSEP, which is present in isolate K1 but absent in DH14 and A6.
Fig. 2.
Comparison of CSEP repertoires between B. graminis f. sp. hordei isolates. (A) Comparison of CSEP repertoires between Bgh isolates DH14, A6, and K1. At the center of the circular plot is a dendrogram of all DH14 CSEPs obtained via neighbor-joining phylogenetic analysis. Colored circles immediately next to the dendrogram mark DH14 CSEPs that are missing in one or both of the other two isolates. For each CSEP, the next three rings of colored rectangles give the amount of nonsynonymous substitutions (per kilobase) specific to each of the three isolates DH14, A6, and K1. The rectangles in the outermost ring are color-coded according to family affiliation so that all CSEPs belonging to the same family (according to MCL analysis) have the same color. (B) XY plots between nonsynonymous substitution rates (dN) and in planta transcript induction. For each Bgh isolate (A6 and K1), only genes with at least one nonsynonymous substitution compared with the DH14 isolate reference genome were selected. The x axis corresponds to the dN rates, and the y axis corresponds to the difference between the minimal and the maximal log2-fold change values (Dataset S1, Table S6) measured between all four time points and reflects the amplitude of fungal transcript activation during in planta infection. For each isolate, genes encoding CSEPs are highlighted in color and the other genes are in gray.
Nonsynonymous Substitution Rates Among Bgh Isolates.
To identify genes showing elevated evolutionary rates among the three Bgh isolates, we focused on the SNPs detected in the coding sequence (Dataset S1, Table S5). Evolutionary pressure on proteins is often quantified by the ratio of the nonsynonymous and synonymous substitution rates (dN/dS). However, due to the restricted number of genomes analyzed (n = 3) and the low SNP frequency in the coding sequences (0.3 SNPs per kilobases), dN/dS ratio calculation did not provide enough depth for reliable statistical analyses as most genes (>70%) do not have any synonymous substitutions (Dataset S1, Table S5). Therefore, we focused our analysis on the nonsynonymous substitution rate (dN). By combining gene expression differences (see below) with the dN rates, a clear shift was observed between genes encoding CSEPs and the other gene categories, suggesting that most candidate effectors are plant-induced and have evolved faster than the other genes (Fig. 2B). Consistent with this, calculation of the ratio of all nonsynonymous substitutions vs. all synonymous substitutions also revealed a higher accumulation of nonsynonymous substitutions in CSEP-encoding genes compared with all other genes (Fig. S3). The high nonsynonymous substitution rates observed for some CSEP-encoding genes may have arisen from selection pressure exerted by introgressed race-specific resistance genes into domesticated barley (Fig. 2 A and B and Fig. S4), consistent with the model of an arms race between the innate immune system of barley and candidate-secreted Bgh effectors (34). Among the top 50 of genes with the highest dN rates in Bgh isolates A6 and K1 compared with isolate DH14 are a great number of CSEPs (13/50 and 20/50 when comparing A6 vs. DH14 and K1 vs. DH14, respectively) (Dataset S1, Table S5). Nevertheless, other genes, for example, encoding serine/threonine-protein kinases (35), are also represented among the genes that show the highest dN rates (5/50 when comparing A6 or K1 vs. DH14), suggesting that they might also interact and/or coevolve with host factors (Dataset S1, Table S5).
RNA Sequencing of Bgh Isolates A6 and K1 During Infection on Arabidopsis.
To investigate the transcriptional dynamics of A6 and K1 Bgh isolates during early stages of fungal pathogenesis, we sequenced their respective transcriptomes using Illumina RNA sequencing (Dataset S1, Table S6). Although wild-type Arabidopsis is a strict nonhost for the grass powdery mildew and fungal pathogenesis is typically terminated before haustorium differentiation (36), simultaneous inactivation of the sequentially acting immunity components PEN2, PAD4, and SAG101 (triple mutant pps: pen2, pad4, sag101) renders Arabidopsis a host for Bgh, enabling completion of the asexual life cycle by conidiospore formation (13). In addition, as the barley MLA1 immune receptor is fully functional in the Arabidopsis pps mutant background and confers isolate-specific disease resistance to the Bgh isolate K1, but not to A6, we used transgenic Arabidopsis plants in the triple-mutant background (pps-MLA1) (14) to study fungal gene expression during MLA1-specified incompatible and compatible interactions (Fig. S5). As the amount of fungal biomass is extremely low during early powdery mildew pathogenesis, a total of 2,016,225,898 reads (in 48 samples) was sequenced. More than 90% of the reads were assigned to the Arabidopsis genome whereas 1.79% could be aligned to the Bgh isolate DH14 reference genome, representing a total of 36,142,737 fungal reads (752,973 reads per sample on average) (Dataset S1, Table S7). Of the 6,477 genes previously annotated in the Bgh DH14 reference genome, 5,831 genes accumulated more than 100 reads (taking all 48 samples together) and were considered as expressed. This allowed us to analyze the transcriptional program deployed by the powdery mildew isolates A6 and K1 at intervals corresponding to conidiospore germination at 6 hours postinoculation (hpi), penetration of leaf epidermal cell walls (12 hpi), and formation of haustorial initials (18 hpi) and mature haustoria (24 hpi) (Fig. S5).
Transcriptional Dynamics of Powdery Mildew During Early Pathogenesis.
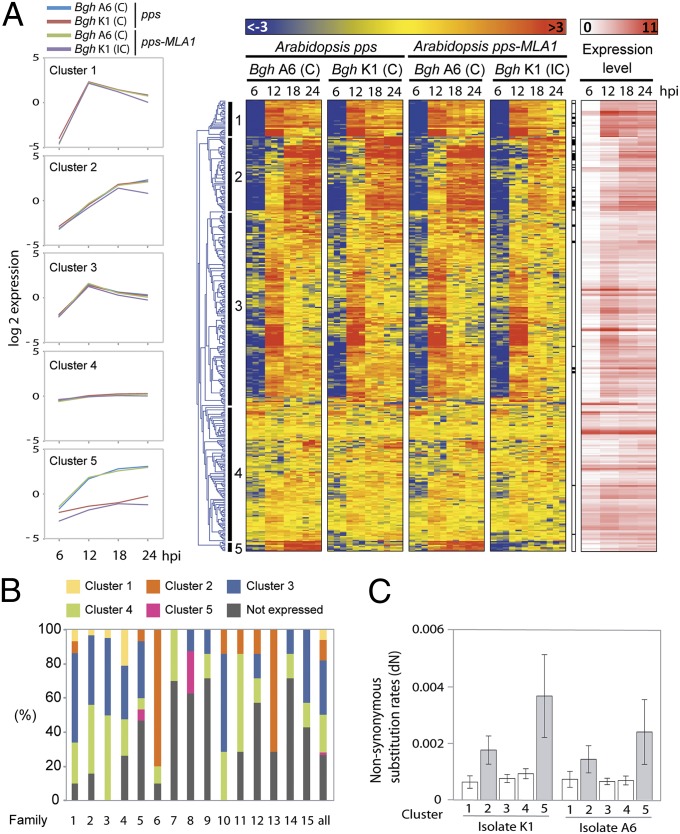
Overall, we found highly similar gene expression patterns during host colonization in both isolates, irrespective of the outcome of the interaction (Figs. 3A and 4). Gene ontology (GO) term enrichment analysis revealed that gene categories associated with DNA packaging, nucleosome organization, and chromatin structure are strongly up-regulated during conidiospore germination (6 hpi), presumably reflecting a high rate of DNA replication at this stage. In contrast, the initiation of invasive growth into host cells (12 hpi) is marked by a transient activation of gene categories related to entry and movement into the host, interaction with the host, and pathogenesis (Dataset S1, Table S8). Transcript profiling of CSEP-encoding genes revealed that most are not or are barely expressed in germinating spores, but transcripts massively accumulate during and after host cell entry (between 12 and 24 hpi), supporting a key role of effector genes during early pathogenesis (Fig. 3A). Two major sets of CSEP-encoding genes are transcribed in successive waves coinciding with penetration of the plant epidermis (clusters 1 and 3) and haustorium differentiation (clusters 2 and 5), respectively. Although most genes within a given CSEP family showed distinct expression patterns, CSEP-encoding genes belonging to families 6 and 13 are almost all induced during haustorium formation (cluster 2, Fig. 3B). These two families are expanded in Bgh compared with the closely related wheat powdery mildew Bgt. The N-terminal part of the polypeptide sequences contains the Y×C motif previously identified in the sequence of effector proteins of pathogenic fungi that form haustoria (22) (Fig. S6). Notably, clusters 2 and 5 are significantly enriched (P = 0.00183 for A6, P = 0.000168 for K1) for CSEPs having higher dN rates compared with those detected in clusters 1, 3, and 4, indicating that haustorially expressed CSEPs may become engaged in a coevolutionary arms race with the innate immune system of the host (Fig. 3C).
Fig. 3.
RNA-seq expression profiling of powdery mildew CSEPs during early stages of infection on Arabidopsis. (A) Transcriptional profiles of CSEPs. Partially immunocompromised Arabidopsis (pps: pen2, pad4, sag101 background) with and without barley MLA1 were infected with Bgh isolates A6 or K1. Samples were harvested at 6 hpi (conidiospore germination), 12 hpi (penetration), 18 hpi (haustorial initials), and 24 hpi (mature haustoria) (Fig. S5). Overrepresented (light red to dark red) and underrepresented transcripts (light blue to dark blue) are shown as log2-fold changes relative to the mean expression measured across all samples. The average expression patterns for five major gene expression clusters (from 1 to 5) are highlighted on the left side. CSEPs that are significantly regulated during the incompatible interaction (Table 2) are highlighted in black on the right side. Log2-transformed expression levels corresponding to the mean expression at each time point are depicted on the right. C, compatible interaction; IC, incompatible interaction. (B) For each of the 15 largest CSEP families, the proportion of genes detected in each gene expression cluster described in A are presented. (C) dN rates per cluster (A) and per isolate. Haustoria-induced CSEPs (clusters 2 and 5) are indicated in gray and have significantly higher nonsynonymous substitution rates than the other CSEPs (P = 0.00183 for A6 and P = 0.000168 for K1, t test).
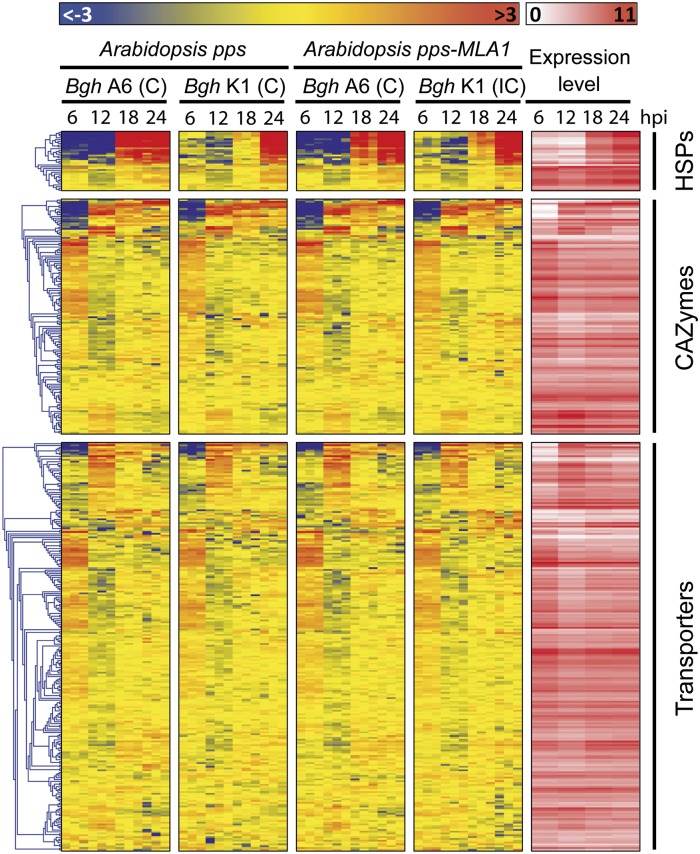
Fig. 4.
RNA-seq expression profiling of powdery mildew pathogenicity-related genes during early stages of infection on Arabidopsis. The heatmaps show gene expression patterns of three gene families (encoding heat-shock proteins, carbohydrate-active enzymes, and transporters). Partially immunocompromised Arabidopsis (pps: pen2, pad4, sag101 background) with and without barley MLA1 were infected with Bgh isolates A6 or K1, and samples were harvested at 6, 12, 18, and 24 hpi (Fig. S5). Overrepresented (light red to dark red) and underrepresented transcripts (light blue to dark blue) are shown as log2-fold changes as described for Fig. 3A. C, compatible interaction; IC, incompatible interaction.
Analysis of three additional pathogenicity-related gene categories (encoding carbohydrate-active enzymes, transporters, and heat-shock proteins) revealed strikingly distinct patterns of gene activation (Fig. 4). Similar to CSEP-encoding genes, transcripts encoding small heat-shock proteins of 30 kDa, which can contribute to the maintenance of membrane integrity (37), were also massively induced during haustorium formation. In contrast to the strong modulation of gene expression observed for the above-mentioned gene categories, expression changes were less pronounced for genes encoding carbohydrate-active enzymes and transporters (Fig. 4). Nevertheless, some of these genes showed remarkable expression patterns. For example, transcripts of two genes encoding cutinases (Bgh00226 and Bgh00227) were specifically detected at 6 and 12 hpi, respectively (Fig. 4 and Fig. S7), suggesting that they might be engaged in enzymatic degradation of the leaf cuticle during the switch from extracellular to invasive growth into the host. Notably, although only a small subset of genes (seven in total) encoding transporters were markedly induced during or after fungal entry (Fig. 4 and Fig. S7), five of these encode sugar (Bgh00499, Bgh00500) or amino acid (Bgh02324, Bgh00557, Bgh06082) transporters. This implies that uptake of host nutrients likely takes place before full differentiation of haustoria.
Modulation of Bgh and Arabidopsis Gene Expression in the Incompatible Interaction.
We found that 215 genes showed significant [f-test, false discovery rate (fdr) < 0.05] overall differences in gene regulation between the compatible (Bgh A6 and K1 on Arabidopsis pps, Bgh A6 on Arabidopsis pps-MLA1) and the incompatible interaction (Bgh K1 on Arabidopsis pps-MLA1) (Dataset S1, Table S6). To test when the expression differences are most prominent, we compared gene expression between compatible and incompatible interactions at each of the four time points separately while correcting for isolate and host genotype effects. Notably, we found significant expression differences almost exclusively at 24 hpi (Table 2). At this time point, out of 76 differentially expressed genes [fdr < 0.05, |log2 fold change (FC)| ≥1], 65 genes are down-regulated (Table 2). Of these down-regulated genes, the majority (43 genes) are CSEPs, suggesting that primarily expression of effector genes is affected in the incompatible interaction (Table 2). Moreover, these down-regulated CSEPs are strongly induced during haustorium formation (cluster 2, Fig. 3A) and show an underrepresentation of the largest families but an overrepresentation of small families and family 6. On the other hand, simultaneous analysis of the corresponding Arabidopsis host transcriptional profiles showed extensive transcriptional reprogramming in the incompatible interaction from 18 hpi (Table 2), well before the transcriptomic profiles of Bgh isolates differ between the compatible and the incompatible interactions (14).
Table 2.
Simultaneous analysis of host (Arabidopsis) and pathogen (Blumeria) gene expression profiles
| Species | Regulation | 6 hpi | 12 hpi | 18 hpi | 24 hpi |
| Arabidopsis | Up | 2 | 0 | 763* | 269 |
| Down | 0 | 0 | 69 | 219 | |
| Blumeria | Up | 5 | 0 | 0 | 11 |
| Down | 0 | 0 | 0 | 65† |
Numbers indicate genes differentially expressed (fdr < 0.05, |log2FC| ≥1) comparing incompatible vs. compatible interactions.
Enriched with chitin response genes (14).
Forty-three (66%) encode CSEPs.
Isolate-Specific Gene Expression Patterns.
To identify major transcriptional differences between Bgh isolates A6 and K1 during host colonization, we selected genes showing significant differences in transcript accumulation for at least three of the four sampling time points tested. In total, we identified 225 genes with significant differences (fdr < 0.05) between the two isolates (Dataset S1, Table S6). Among these, we identified some of the aforementioned “missing genes” such as those encoding two CSEPs (CSEP0128 and CSEP0058, Fig. S8) and a FYVE zinc-finger protein (Table 1), thereby supporting their Bgh isolate specificity. The other missing CSEP-encoding genes identified before (Fig. S2 and Table 1) were not considered as expressed and are closely associated with repeat sequences (Dataset S1, Table S9), indicating that these CSEP-encoding genes might be pseudogenes or are expressed at different time points. Furthermore, genes encoding 11 additional CSEPs were preferentially expressed in Bgh isolate A6 whereas 10 others were preferentially expressed in Bgh isolate K1 (Fig. S8 and Dataset S1, Table S9), suggesting that isolate-specific regulation of effector transcription could contribute to the avoidance of host recognition. In addition, other genes such as those encoding the GAL10/UDP epimerase (Bgh01641) or a conserved protein (Bgh04330) were exclusively expressed in the Bgh isolate K1 (Fig. S8).
Influence of the Host Species on Bgh Gene Expression.
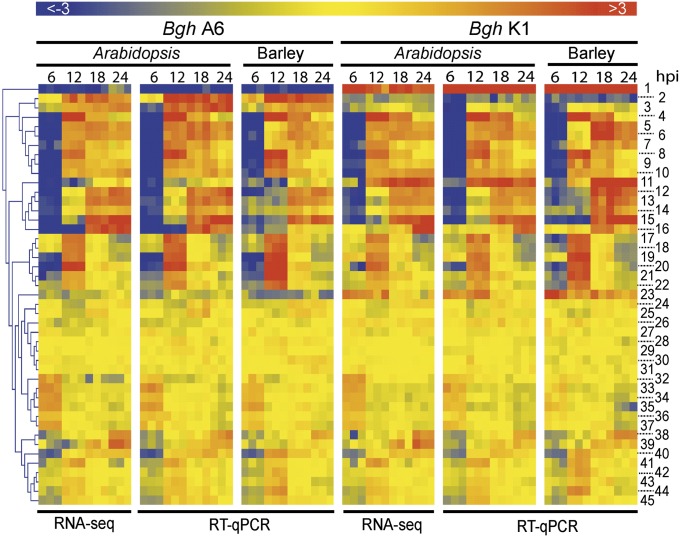
Because Bgh is a host-adapted pathogen of monocotyledonous barley but is nonadapted for growth and reproduction on Arabidopsis, we tested whether the complex stage-specific powdery mildew gene expression profiles on Arabidopsis leaves are conserved during colonization of barley leaves. We selected 45 genes based on the RNA sequencing (RNA-seq) data that showed distinctive expression patterns during the interaction with immunocompromised Arabidopsis (highly expressed, differentially regulated during infection in planta or differentially expressed/regulated between the two isolates). For these genes, we compared the expression profiles during Bgh infection on Arabidopsis pps and its natural host barley using RT-quantitative PCR (RT-qPCR) (Fig. 5). To validate this approach, gene expression levels measured during Arabidopsis infection using RNA-seq were first compared with those determined by RT-qPCR. A near-perfect correlation was observed (y = 1.02×–0.011; R2 = 0.88; Fig. S9A), indicating minimal variations between the two experimental approaches (Fig. 5). We then compared fungal gene expression during infection of barley and Arabidopsis, which revealed remarkably similar expression patterns (Fig. 5). Unexpectedly, the observed linear correlation (y = 0.89 + 9E-19; R2 = 0.82; Fig. S9B) suggests that during powdery mildew pathogenesis the fungal gene expression program is essentially robust against exposure to vastly divergent host-encoded molecules in monocots and dicots, including cell-wall constituents. These data corroborate that the dicotyledonous model plant Arabidopsis can be used as a host mimetic instead of the natural host barley to explore the molecular basis of Bgh pathogenesis.
Fig. 5.
Comparison of powdery mildew gene expression profiles during infection on Arabidopsis and barley. Expression profiles of 45 selected powdery mildew genes were monitored during the compatible interaction on Arabidopsis and barley using RT-qPCR. Arabidopsis pps (pen2, pad4, sag101 background) and barley cultivar “Ingrid” leaves were inoculated with Bgh isolate A6 and K1. Samples were harvested at 6 hpi (conidiospore germination), 12 hpi (penetration), 18 hpi (haustorial initials), and 24 hpi (mature haustoria). Gene expression levels were normalized to the transcript levels of the two reference genes actin and β-tubulin. Overrepresented (light red to dark red) and underrepresented transcripts (light blue to dark blue) are shown as log2-fold changes relative to the mean expression measured across all samples. The gene numbers on the right side refer to the corresponding gene names in Dataset S1, Table S10.
Discussion
Agricultural ecosystems, characterized by a low diversity of plant species, a low intraspecies genetic diversity, but high host density, have facilitated the emergence and propagation of aggressive host-specialized pathogens (38). Extensive pan-European surveys of Bgh powdery mildew conidiospores over successive years have suggested that the virulence patterns of these conidiospores are influenced by the cultivation of domesticated barley cultivars with Bgh resistance specificities introgressed from wild barley (H. vulgare ssp. spontaneum) (6). A study comprising 9,000 Bgh isolates, collected over 3 consecutive years across Europe, detected 609 virulence phenotypes (39). This and similar studies have led to the proposal that the barley mildew pathogen has established a single enormous pool of genetic variation across Europe in which local selection maintains distinct subpopulations (6). Our study suggests how this standing genetic variation at the population level has been generated and might be maintained. The comparative genome analysis of isolates A6, K1, and DH14 revealed for each strain a characteristic mosaic of relatively large isolate-specific blocks of varying SNP densities (Fig. 1). Because the collection sites of these three isolates are each geographically 500–1,000 km apart from each other, the isolate-specific mosaic Bgh genomes could reflect sexual Bgh reproduction events at each site, followed by a selective sweep in the Bgh population in response to locally grown barley accessions carrying different combinations of powdery mildew recognition specificities. However, we estimate the divergence time of the monomorphic and polymorphic blocks to be ∼5,000 and ∼140,000 y ago, respectively. Even though divergence time estimates are error-prone due to indirect estimates of the mutation rate in the powdery mildews (Materials and Methods), at least the regions with high SNP density very likely existed before barley domestication ∼10,000 y ago. Similar peculiar genome diversity patterns alternating between fully conserved and variable regions were observed in a genome alignment of five individuals of Zymoseptoria pseudotritici, a close relative of the wheat pathogen Z. tritici (40). However, in contrast to our findings, the blocks are 10-fold smaller, 54% of the Z. tritici genome is monomorphic between all isolates, and the variable regions showed only two haplotypes. These differences can be explained as the Z. pseudotritici species likely result from a very recent hybridization event between two closely related Zymoseptoria species. Sexual reproduction is relatively frequent in this species and recombination has contributed to the generation of a mosaic genome structure.
The detection of large “ancient” sequence blocks in extant Bgh isolates in this study implies that sexual recombination is likely rare in the Bgh population. If sexual recombination were frequent, one would expect a largely homogenous SNP distribution and smaller block sizes. Asexual reproduction could potentially enable indefinite clonal maintenance in the population and preservation of particular virulence arsenals. Consequently, the combined sexual and asexual reproduction mode of Bgh could have resulted in a gradual buildup of an enormous diversity of clonal lines across Europe. Because of an extraordinary large number of up to 1014 airborne conidia were released and dispersed during powdery mildew epidemics over Europe (6), even the rarest virulence haplotype could be locally selected from this standing genetic variation in the Bgh population.
A limitation of our study is the number of resequenced isolates, and therefore we cannot exclude inbreeding by frequent sexual reproduction within a local population. Sequencing the genomes of dozens of randomized but geographically stratified Bgh isolates is needed to estimate frequencies of geographically bounded sexual reproduction and clonal reproduction. The observed Bgh genomic signatures could reflect the obligate biotrophic lifestyle of powdery mildews, as (near) clonal reproduction enables maintenance of the virulence necessary for survival whereas maintenance of sexual reproduction both facilitates the elimination of deleterious mutations and generates genetic diversity in the arms race with the host.
Effector proteins have been described in many plant pathogens including bacteria, nematodes, fungi, and oomycetes (41), and a subset are known to manipulate host innate immune mechanisms, thereby facilitating pathogenesis. A comprehensive analysis of the 491 genes encoding candidate-secreted effector proteins in the Bgh isolate DH14 revealed the presence of large gene families, some of them showing hallmarks of diversifying selection (19). The Bgh isolates differing in avirulence gene combinations that were resequenced in this study permitted us to examine polymorphisms between CSEP orthologs. The massive proliferation of repetitive elements in the genome of the Bgh isolate DH14, accounting for ∼64% of the fungal genome (15, 27), suggests that CSEP arsenals could have been impacted by the activity of these elements as recently demonstrated by resequencing of three isolates of Pyrenophora tritici-repentis, a necrotrophic fungus responsible for tan spot disease of wheat (42). Consistent with this, the flanking regions of most of the Bgh isolate-specific CSEPs show similarity to repeat sequences. Our study also revealed largely conserved CSEP repertoires in the three Bgh isolates. Accordingly, we found only five and six CSEPs, respectively, in isolate DH14 that are likely absent from the genomes of isolate A6 and K1 (Fig. 2) and one potential unique CSEP in isolate K1. It remains unclear, however, whether sexual recombination between parental clones of the tested isolates or clonal variation within the isolates or both contributed to these isolate-specific polymorphisms in CSEP repertoires. Notably, single nonsynonymous substitutions in orthologous CSEPs can also be sufficient to escape or to trigger host immune responses. Similar to other plant pathogens (43–45), we found that CSEPs accumulate higher nonsynonymous substitution rates compared with other gene categories. Despite the fact that we could not quantify the evolutionary pressure acting on these genes by calculating the dN/dS ratio, most of them accumulate only nonsynonymous substitutions, suggesting that they are under strong diversifying selection. Interestingly, we found that, on average, CSEPs that are specifically induced in haustorial initials accumulate higher nonsynonymous substitution rates compared with the other CSEPs (Fig. 3). Haustoria have been shown to establish an intimate interface with host cells and to mediate effector delivery into host cells (4). Thus, it is tempting to speculate that this specific set of CSEPs plays an important role during early pathogenesis in the establishment of compatibility by interfering with the host innate immune system, potentially triggering iterated cycles of pathogen effector and innate immune receptor adaptations (arms race). Accordingly, CSEPs expressed at late stages of powdery mildew pathogenesis (mature haustoria) might have a role in biotrophy maintenance, and their structure or action might be less vulnerable to host innate immune receptors due to an already defeated immune system.
We found a few other Bgh genes that are specific to a particular isolate. For example, a gene encoding a FYVE zinc-finger protein that targets proteins to a membrane lipid via interaction with phosphatidylinositol-3-phosphate (46) is not present in the genome of the Bgh isolate K1, suggesting isolate-specific variation in vesicular trafficking. In Bgh isolate A6, we found only the mating-type locus MAT1-2–1 to be absent, indicating that A6 has an opposite mating type compared with isolates DH14 and K1. Therefore, mating is likely possible between Bgh isolates A6 and DH14 or K1 but not between K1 and DH14. The DH14 region absent from A6 spans at least 11 kb until the end of the contig, suggesting a much larger mating-type locus in the powdery mildews compared with other Leotiomycetes, which are typically between 3 and 5 kb in size (33). Using the de novo genome assembly of Bgh isolate A6, we retrieved the opposite mating-type locus gene MAT1-1–1. Similar to the grapevine powdery mildew and other Leotiomycetes, MAT1-1–1 is flanked by SLA2, which encodes a cytoskeleton assembly protein (47). In addition, we found two CSEPs and one EKA homolog with our genome mapping approach that are completely absent in the Bgh isolate K1 compared with the isolate DH14. However, our transcript data show that these two CSEPs are not expressed in A6 (<100 reads in total), suggesting that these might be pseudogenes. This underlines the importance of integrating transcriptome data into comparative genome analysis.
By taking advantage of the established pathosystem between immunocompromised Arabidopsis pps lines expressing or lacking the barley MLA1 immune receptor and the Bgh isolates expressing or lacking the cognate AVRA1 effector (13, 14), we monitored gene expression patterns in compatible and incompatible interactions. Using Arabidopsis as a host plant allowed in silico elimination of host-derived transcripts from the RNA sequencing analysis, thereby enabling a de novo assembly of pathogen-derived transcripts. In addition, host gene expression profiles of the Arabidopsis pps lines upon Bgh challenge have been analyzed before and reveal sustained expression of a large MLA1-dependent gene cluster enriched in chitin responsive genes at 18 hpi (14). This enables us to directly correlate fungal effector gene induction with plant defense gene activation.
Gene expression profiles of Bgh have been previously monitored for a restricted set of genes using a custom microarray and have revealed coordinate shifts in the expression patterns of genes encoding key enzymes of the primary metabolism (48) as well as a cluster of genes encoding potential virulence determinants (49). In the present study, the complete gene repertoire of Bgh has been analyzed, and we have focused on gene categories often associated with fungal pathogenicity such as those encoding carbohydrate-active enzymes, transporters, CSEPs, and heat-shock proteins (50, 51). Transcriptional profiles of CSEPs revealed the existence of two major expression waves associated with penetration of the plant epidermis (12 hpi) and the differentiation of haustoria (18 and 24 hpi) (Fig. 3). Similar waves of expression have been described for candidate effector genes in other filamentous pathogens including rust fungi (50, 52), Colletotrichum fungi (51, 53), and oomycetes (54), suggesting that different suites of effector proteins are sequentially deployed by plant pathogens for successful invasion. Unexpectedly, CSEP-encoding genes belonging to the same family mostly showed differential expression patterns, indicating that Bgh sequentially delivers structurally related effectors during colonization. This might be advantageous to the fungus to escape host recognition without altering the biological function of the effectors. Many CSEP-encoding genes are induced during appressorial penetration and the formation of haustorium initials, suggesting that those structures are key interfaces for effector secretion. Thus, powdery mildew CSEPs expressed in penetrating appressoria may suppress early plant defense responses to prepare subsequent invasive fungal growth and haustorium formation. In transgenic Arabidopsis pps plants, barley MLA1-mediated immunity to Bgh is accompanied by a sustained expression of a large cluster of genes at 18 hpi (coincident with the formation of haustorium initials) of which many are known to be responsive to chitin, one of the well-known microbe-associated molecular patterns commonly found in fungal cell walls (Table 2) (14). Consistent with this, we found that CSEP genes induced in haustorium initials (18 hpi) evolved faster than the other CSEPs, and we identified two CSEP gene families in which almost all members are induced at that stage (family 6 and family 13). For members of family 6, the N terminus of the mature protein is highly conserved, possibly indicating a conserved folding structure, whereas the C terminus is under diversifying selection (19). Interestingly, these two families are also among the most expanded families in Bgh compared with Bgt, which suggests species-specific expansion. Taken together, these results indicate that members of these families may encode candidate avirulence effectors that are recognized by barley race-specific immune receptors. Notably, haustoria-induced CSEPs are nearly all down-regulated after 24 hpi in the MLA1-mediated incompatible interaction. This result is striking and suggests that primarily the expression of effectors is affected during the incompatible interaction. Importantly, the host Arabidopsis responds with clear differential transcriptional activities between MLA1-triggered incompatible and compatible interactions at an earlier time point at 18 hpi (Table 2) (14). These results indicate either that the host activated a generic mechanism to reduce the transcription of small secreted proteins or that the fungus senses unsuccessful invasion and consequently shuts down transcription of a particular set of CSEPs.
Materials and Methods
Biological Material.
Bgh isolates A6 and K1 were originally collected in Svalöf, Sweden, and Köln, Germany, respectively (28, 29). Plant growth (barley, Arabidopsis), conidia spore inoculation of Bgh, and RNA purification were performed as described previously (9, 14).
Genome Resequencing.
Library construction and shotgun sequencing were performed by the Max Planck Genome Centre Cologne. Long reads were obtained using Roche 454 GS FLX titanium with the XL+ chemistry for standard shotgun sequencing (rapid key). For both isolates (K1 and A6) we obtained 1.98 million reads, with an average read length of 529 and 505 for K1 and A6, respectively. Short paired-end reads (100 bases) were obtained using Illumina HiSeq2000 sequencing and TruSeq DNA library preparation protocols and an insert size of 250 bases. In total, 135 and 77 million paired end reads were generated for K1 and A6, respectively. The whole genome shotgun projects have been deposited in the GenBank database [accession nos. AOLT00000000 (isolate A6) and AOIY01000000 (isolate K1)].
Preprocessing and Mapping of Short Reads.
All short reads were trimmed using the fastx toolkit function fastq_quality_trimmer with minimum quality score set to 20 and minimum read length to 50. Nearly all reads (>98%) were of sufficient quality and length. After trimming, reads were mapped to the genome of Bgh isolate DH14 (15) using Bowtie with the default settings and –best. After mapping, duplicate reads were removed using function rmdups from the Samtools toolkit (default setting). In total, 70 and 184 million reads mapped to the DH14 reference genome, corresponding to 35 and 77 million unique reads for A6 and K1, respectively. To identify regions in the reference genome with zero coverage, we used a sliding window of 1 kb that moved 100 bp at each step and summed the amount of bases with mapped reads.
De Novo Genome Assembly.
For each isolate we made a de novo genome assembly using CLC Genomics software (version 5.5.1). We used the long reads (sff files) and trimmed short reads (fastq files) as input. We noted a contamination with yeast involving ∼2% of the short reads and excluded all assembled contigs that aligned to the yeast genome using MUMmer with default settings [identity (%IDY) > 95% or coverage (COV Q) > 95%]. To assess the quality of the final genome assemblies, we calculated the L50 and estimated the gene coverage using CEGMA (version 2.0) (55) (Dataset S1, Table S2). To estimate the genome size, we divided the amount of high-quality bases by the average coverage per contig (>1,000 bp) using a separate assembly of 454 data only (Newbler V2.6). For A6, we generated 995 million high-quality bases in long reads, and for K1 we generated 1,004 million high-quality bases. In the assembly this resulted in an average coverage of 7.8 ± 3.9 Mb and 8.3 ± 4.1 Mb, respectively. Hence the genome sizes are estimated to be 127 ± 42 Mb and 121 ± 40 Mb, respectively.
RNA Sequencing.
RNA purification, library construction, and sequencing were performed as described previously (14) and resulted in 20–60 million 100-base long reads per sample (total 2 billion). Reads were mapped to the Bgh DH14 BluGen (www.blugen.org/) genome assembly version 3.0 using Tophat (56) with settings -a 10 -g 5 and splice sites based on BluGen gene models from July 2012 (gff-file can be obtained via www.mpipz.mpg.de/23693/Powdery_Mildews). Likewise, reads were mapped to the genome of the host plant Arabidopsis (tair10) (14). The RNA-seq data used in this study are deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database and available via accession nos. GSE39463 (Arabidopsis thaliana) and GSE43163 (Blumeria graminis f.sp. hordei). The raw reads are available via the Sequence Read Archive project no. SRP014482.
SNP Analysis.
SNP calling was performed separately for the mapped (see above) Illumina genome, and transcriptome sequencing reads of A6 and K1 using the function mpileup of SAMtools version 0.1.18 (57) with options -ug. The obtained SNP sets were filtered by applying the bcftools script vcfutils.pl varFilter (SAMtools) with default read depth settings for the transcriptome data and adjusted read depth settings for the genome data according to the respective sequencing coverage to -d 20 and -D 200 for K1 and to -d 10 and -D 100 for A6, respectively. Subsequent variant annotation and effect prediction was performed using snpEff version 2.0.5 (default settings) (58) based on the DH14 gene models. Additionally, we computed the dN (number of observed nonsynonymous substitutions/number of possible nonsynonymous sites) and dS (number of observed synonymous substitutions/number of possible synonymous sites) rates for A6 vs. DH14 and K1 vs. DH14 using the function kaks from the R package seqinr (version 3.0–6). To identify SNP density as a function of the genomic location, we used a 10-kb sliding window that moved 1 kb at each step for all DH14 contigs larger than 50 kb. To estimate the distributions of the low and high SNP densities, we fitted a two-component mixture model to the observed SNP densities using the expectation-maximization (EM) algorithm (function normalmixEM, R-package mixtools). To identify regions with high or low SNP densities, a two-state hidden Markov model (HMM) was fitted on the 10-kb SNP densities using the EM algorithm (function depmix, R-package depmixS4), and subsequently the posterior state sequence was computed via the Viterbi algorithm (function fit, package depmixS4). Three isolate-specific sets of genome sequence variants were extracted for DH14 (genome SNPs identified in both K1 and A6), K1 (genome SNPs identified only in K1), and A6 (genome SNPs identified only in A6). Isolate-specific polymorphic regions were calculated using the same EM/HMM approach as mentioned above, but on all isolate-specific SNPs. From these predicted segments we derived the percentages of the 15 largest contigs where all three isolates are monomorphic (i.e., low SNP density) or polymorphic (i.e., all three isolates have a high isolate-specific SNP density).
To obtain high-confidence SNPs for the protein-coding regions, we extracted common polymorphisms detected in both the genome and the transcriptome SNP analysis. As for the genomic SNPs, three high confidence sets of isolate-specific coding SNPs were obtained for DH14, K1, and A6. To estimate divergence times, we used the mutation rate for intergenic regions of 1.3e-08 previously used to estimate the divergence time between Bgh and Bgt (31). The intergenic-based estimate is in line with divergence estimates for the powdery mildews based on the internal transcribed spacer (ITS) regions (59). The accuracy and confidence interval of the intergenic mutation rate in the powdery mildews have never been directly estimated and might be dependent on mode and frequency of reproduction.
Circular Visualizations.
Circos version 0.62.1 (60) was used to construct the circular visualization of the alignment of the A6 and K1 genome and transcriptome sequencing reads and genomic SNP locations with respect to the 15 largest contigs of the DH14 reference genome shown in Fig. 1A. For visualization of the CSEP repertoire in the three Bgh isolates in Fig. 2A, a multiple sequence alignment of the DH14 CSEP protein sequences and subsequent phylogenetic analysis (with the neighbor-joining algorithm) was performed using ClustalW (default settings) (61). To plot this tree, the obtained Newick tree file was fed into MEGA5 (62). The plotted tree was subsequently combined with a circular plot of the amount of nonsynonymous isolate-specific substitutions (per kilobase) for each DH14 CSEP created with Circos.
Comparison of CSEP Repertoires Between Isolates.
For the comparison of CSEP repertoires between isolates, a combination of different tools was used to search for orthologs of the previously annotated DH14 CSEPs in the isolates K1 and A6. The analysis included 485 of the 491 previously annotated CSEPs (19) that were included in the BluGen gene models from July 2012 and the respective cDNA fasta files. First, we performed de novo gene prediction on the K1 and A6 genome assemblies using Augustus version 2.6.1 (63) and providing the RNA sequencing data and DH14 cDNA information as hints in the gene prediction (for workflow and used parameter settings, see protocols at http://bioinf.uni-greifswald.de/bioinf/wiki/pmwiki.php?n=IncorporatingRNAseq.Tophat and http://bioinf.uni-greifswald.de/bioinf/wiki/pmwiki.php?n=Augustus.IncorporateESTs). To identify orthologs of the 485 analyzed DH14 CSEPs among the predicted genes of the other two isolates, we used orthoMCL version 2.0.3 (64) on the predicted A6 and K1 proteins and previously annotated DH14 proteins with default settings according to the orthoMCL user guide. In a second, independent approach, we extracted all RNA sequencing reads that did not align to the Arabidopsis reference genome and used these for a transcriptome de novo assembly. The transcriptome assembly was performed using Trinity (65) with default parameter settings for single-end reads (see protocol at http://trinityrnaseq.sourceforge.net/). The obtained transcript set was subsequently filtered in several steps to obtain a high-confidence transcript set. The filtering included removal of redundant transcripts, low-coverage transcripts (with less than 10× coverage), transcripts that aligned to the Arabidopsis or yeast genomes (using MUMmer version 3.22 with default parameters) (66), transcripts for which no good ORF was predicted by Trinity, and transcripts that did not align to the respective genome assemblies (BLASTN, e-value < 1e-05). The filtered sets contained 6,084 transcripts for A6 and 5,828 transcripts for K1. To identify orthologous pairs between the DH14 CSEPs and the assembled A6/K1 transcripts, we performed alignments (BLASTP, e-value < 1e-06) and extracted pairs of reciprocal best hits. As a third approach, we aligned the coding sequences of the previously annotated DH14 CSEPs to the A6 and K1 genome assemblies using Gmap version 2012–11-09 (67) with default parameters and extracted all DH14 CSEPs with a Gmap alignment in which at least 99% (high/medium confidence orthologs) or 90% (low-confidence orthologs) of the coding sequence aligned to the A6/K1 genome with at least 99% (high/medium confidence orthologs) or 90% (low-confidence orthologs) similarity. Finally, in cases where these three tools did not provide conclusive evidence, we manually inspected the alignment of the A6 and K1 genome sequencing reads to the DH14 reference genome using Integrative Genomics Viewer (IGV) version 2.1 (68) and checked whether the read coverage was sufficiently deep and even across the complete DH14 gene model.
CSEP Family Analysis Between Wheat and Barley Powdery Mildews.
To examine family expansions of CSEP families between Bgh (19) and Bgt, we used an all-versus-all BLASTP (-e 1e-10) of the predicted CSEPs in both species. Afterward we used MCL (with option -I 2) to cluster the sequences based on their similarity. To create phylogenetic trees of identified families we used Jalview (ClustalW for multiple alignments and “average distance using % identity” to construct a tree).
Statistical Analysis of RNA-seq.
To analyze differential gene expression, a 2 × 2 × 4 factorial design was used with host genotypes (MLA1-HA+/−), pathogen isolates (A6 and K1 isolates), and time (6, 12, 18, 24 hpi) as factors. Each condition contained three biological replicates totaling 48 samples. The mapped RNA-seq reads (see above) were transformed into a count per gene per sample using the function coverageBed of the bedTools suite (69). Genes with less than 100 reads in all samples were discarded, and subsequently the count data of the remaining genes were log-transformed and normalized by the function voom from the R package limma version 3.10.3 (70) to yield log2 counts per million. To analyze various aspects of differential gene expression, we fitted three distinct linear models using the function lmFit (R package limma). Subsequently, we performed a moderated f-test or t test over various specific contrasts of interest. In all cases, the resulting P values were adjusted for false discoveries due to multiple hypothesis testing via the Benjamini–Hochberg procedure.
First, we looked for common changes in gene expression during the time course of fungal infection. For this purpose, we used the following linear model: “time + fungal_isolate + host_genotype + virulence.” To obtain genes showing differential gene expression during the infection time course, we compared the gene expression between subsequent time points; i.e., we applied moderated t tests to the following three contrasts: “12 vs. 6 hpi,” “18 vs. 12 hpi,” and “24 vs. 18 hpi.” The resulting sets of significantly up- and down-regulated genes between subsequent time points (fdr < 0.01, |log2FC| ≥ 2) were examined for GO term enrichment. The enrichment analysis and the previous assignment of molecular functions to the BluGen gene models were performed using Blast2GO (71) with default settings.
In a next step, to look for virulence-associated differences, we fitted the following linear model: “virulence_time + fungal_Isolate + host_genotype” to each gene. Subsequently, we used a moderated f-test over seven contrasts: Six contrasts define the interaction between two time points and virulent and avirulent samples and the remaining contrast defines the pair-wise comparison between virulent and avirulent samples at time point 6 (72). In total, 215 genes have a significant virulence–time interaction as defined by the f-test (fdr < 0.05). We also used the same linear model to identify genes differentially expressed between virulent and avirulent conditions at each of the four time points separately and examined the resulting gene sets (fdr < 0.05, |log2FC | ≥ 1) for GO term enrichment with Blast2GO (71).
Differences in gene-expression patterns between the two isolates A6 and K1 were found by fitting the following linear model: “isolate_time + host_genotype + virulence.” To identify genes showing isolate-specific differences, we identified differentially expressed genes between A6 and K1 at each of the four analyzed time points. The resulting groups of differentially expressed genes (fdr < 0.05, |log2FC| ≥ 1) were examined for GO term enrichment with Blast2GO (71). Moreover, for CSEPs showing isolate-specific gene expression (Dataset S1, Table S9), we manually inspected the read alignment in the IGV browser to ensure that the observed differential expression was not due to any major sequence differences.
Heatmaps of Gene Expression Patterns.
Heatmaps of gene expression profiles were generated using the Genesis expression analysis package (73). To derive expression patterns of genes during powdery mildew infection, log2 expression ratios were calculated between the normalized number of reads detected for a given gene at a given fungal developmental stage and the geometrical mean of the number of reads calculated across all fungal developmental stages (50). Bgh transporters were retrieved from the Transporter Classification database (www.tcdb.org). Bgh carbohydrate-active enzymes were identified using the CAZy database (www.cazy.org) as previously reported (51). The 491 Bgh DH14 candidate-secreted effector proteins as well as their ID numbers are similar to those recently described (19).
RT-qPCR Analysis
First-strand cDNA was synthesized from 2 μg DNase-treated total RNA using the iScript cDNA synthesis kit (Bio-Rad) and PCR amplification was performed as previously described (53) using the iQ5 real-time PCR detection system (Bio-Rad). For each gene, specific primers were designed with the Primer 3 and Amplify 3× programs (Dataset S1, Table S10). BLASTN searches against the Bgh and A. thaliana genomes were performed to rule out cross-annealing artifacts. GeNorm (74) was used to assess the expression stability of six commonly used reference genes (75) encoding tubulin α-chain (Bgh01972), β-tubulin (Bgh00079), elongation factor 1-α (Bgh01866), actin (Bgh00992), ubiquitin-conjugating enzyme E2 (Bgh03777), and glyceraldehyde 3-phosphate dehydrogenase (Bgh00075), of which actin and β-tubulin were the most stable (stability value 0.032 and 0.033, respectively) and used to normalize gene expression using Pfaffl calculation (76).
Supplementary Material
Acknowledgments
We thank Sabine Haigis for collecting material, Pietro Spanu for the latest annotation files, Ralph Panstruga and Thomas Wicker for comments, and the Max Planck Genome Centre for sequencing support. This work was supported by the Max-Planck Society (S.H., E.V.L.v.T., and P.S.-L.) and the German Research Foundation in the Collaborative Research Centre SFB670 (B.K., T.M., S.V., and P.S.-L.).
Footnotes
The authors declare no conflict of interest.
Data deposition: RNA-sequencing data used in this study have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo [nos. GSE39463 (Arabidopsis thaliana) and GSE43163 (Blumeria graminis f. sp. hordei)]. The raw reads are available via Sequence Read Archive project no. SRP014482 (accession no. SRX160966). The whole genome shotgun projects have been deposited in the GenBank database [accession nos. AOLT00000000 (isolate A6) and AOIY01000000 (isolate K1)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1306807110/-/DCSupplemental.
References
- 1.Kemen E, Jones JD. Obligate biotroph parasitism: Can we link genomes to lifestyles? Trends Plant Sci. 2012;17(8):448–457. doi: 10.1016/j.tplants.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 2.O’Connell RJ, Panstruga R. Tête à tête inside a plant cell: Establishing compatibility between plants and biotrophic fungi and oomycetes. New Phytol. 2006;171(4):699–718. doi: 10.1111/j.1469-8137.2006.01829.x. [DOI] [PubMed] [Google Scholar]
- 3.Vogele RT, Mendgen K. Rust haustorium: Nutrient uptake and beyond. New Phytol. 2003;159(1):93–100. doi: 10.1046/j.1469-8137.2003.00761.x. [DOI] [PubMed] [Google Scholar]
- 4.Panstruga R, Dodds PN. Terrific protein traffic: The mystery of effector protein delivery by filamentous plant pathogens. Science. 2009;324(5928):748–750. doi: 10.1126/science.1171652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Czembor JH. Resistance to powdery mildew in selections from Moroccan barley land races. Euphytica. 2002;125:397–409. [Google Scholar]
- 6.Wolfe MS, McDermott JM. Population genetics of plant pathogen interactions: The example of the Erysiphe graminis-Hordeum vulgare pathosystem. Annu Rev Phytopathol. 1994;32:89–113. [Google Scholar]
- 7.Jørgensen JH,Wolfe M. Genetics of powdery mildew resistance in barley. Crit Rev Plant Sci. 1994;13(1):97–119. [Google Scholar]
- 8.Seeholzer S, et al. Diversity at the Mla powdery mildew resistance locus from cultivated barley reveals sites of positive selection. Mol Plant Microbe Interact. 2010;23(4):497–509. doi: 10.1094/MPMI-23-4-0497. [DOI] [PubMed] [Google Scholar]
- 9.Maekawa T, et al. Coiled-coil domain-dependent homodimerization of intracellular barley immune receptors defines a minimal functional module for triggering cell death. Cell Host Microbe. 2011;9(3):187–199. doi: 10.1016/j.chom.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 10.Srichumpa P, Brunner S, Keller B, Yahiaoui N. Allelic series of four powdery mildew resistance genes at the Pm3 locus in hexaploid bread wheat. Plant Physiol. 2005;139(2):885–895. doi: 10.1104/pp.105.062406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yahiaoui N, Brunner S, Keller B. Rapid generation of new powdery mildew resistance genes after wheat domestication. Plant J. 2006;47(1):85–98. doi: 10.1111/j.1365-313X.2006.02772.x. [DOI] [PubMed] [Google Scholar]
- 12.Yahiaoui N, Kaur N, Keller B. Independent evolution of functional Pm3 resistance genes in wild tetraploid wheat and domesticated bread wheat. Plant J. 2009;57(5):846–856. doi: 10.1111/j.1365-313X.2008.03731.x. [DOI] [PubMed] [Google Scholar]
- 13.Lipka V, et al. Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science. 2005;310(5751):1180–1183. doi: 10.1126/science.1119409. [DOI] [PubMed] [Google Scholar]
- 14.Maekawa T, Kracher B, Vernaldi S, Ver Loren van Themaat E, Schulze-Lefert P. Conservation of NLR-triggered immunity across plant lineages. Proc Natl Acad Sci USA. 2012;109(49):20119–20123. doi: 10.1073/pnas.1218059109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spanu PD, et al. Genome expansion and gene loss in powdery mildew fungi reveal tradeoffs in extreme parasitism. Science. 2010;330(6010):1543–1546. doi: 10.1126/science.1194573. [DOI] [PubMed] [Google Scholar]
- 16.Baxter L, et al. Signatures of adaptation to obligate biotrophy in the Hyaloperonospora arabidopsidis genome. Science. 2010;330(6010):1549–1551. doi: 10.1126/science.1195203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duplessis S, et al. Obligate biotrophy features unraveled by the genomic analysis of rust fungi. Proc Natl Acad Sci USA. 2011;108(22):9166–9171. doi: 10.1073/pnas.1019315108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kemen E, et al. Gene gain and loss during evolution of obligate parasitism in the white rust pathogen of Arabidopsis thaliana. PLoS Biol. 2011;9(7):e1001094. doi: 10.1371/journal.pbio.1001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pedersen C, et al. Structure and evolution of barley powdery mildew effector candidates. BMC Genomics. 2012;13:694. doi: 10.1186/1471-2164-13-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bindschedler LV, McGuffin LJ, Burgis TA, Spanu PD, Cramer R. Proteogenomics and in silico structural and functional annotation of the barley powdery mildew Blumeria graminis f. sp. hordei. Methods. 2011;54(4):432–441. doi: 10.1016/j.ymeth.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 21.Godfrey D, Zhang Z, Saalbach G, Thordal-Christensen H. A proteomics study of barley powdery mildew haustoria. Proteomics. 2009;9:3222–3232. doi: 10.1002/pmic.200800645. [DOI] [PubMed] [Google Scholar]
- 22.Godfrey D, et al. Powdery mildew fungal effector candidates share N-terminal Y/F/WxC-motif. BMC Genomics. 2010;11:317. doi: 10.1186/1471-2164-11-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nowara D, et al. HIGS: Host-induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis. Plant Cell. 2010;22(9):3130–3141. doi: 10.1105/tpc.110.077040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang WJ, et al. Interaction of barley powdery mildew effector candidate CSEP0055 with the defence protein PR17c. Mol Plant Pathol. 2012;13(9):1110–1119. doi: 10.1111/j.1364-3703.2012.00820.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pliego C, et al. Host-induced gene silencing in barley powdery mildew reveals a class of ribonuclease-like effectors. Mol Plant Microbe Interact. 2013 doi: 10.1094/MPMI-01-13-0005-R. [DOI] [PubMed] [Google Scholar]
- 26.Ridout CJ, et al. Multiple avirulence paralogues in cereal powdery mildew fungi may contribute to parasite fitness and defeat of plant resistance. Plant Cell. 2006;18(9):2402–2414. doi: 10.1105/tpc.106.043307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sacristán S, et al. Coevolution between a family of parasite virulence effectors and a class of LINE-1 retrotransposons. PLoS ONE. 2009;4(10):e7463. doi: 10.1371/journal.pone.0007463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wiberg A. Genetical studies of spontaneous sources of resistance to powdery mildew in barley. Hereditas. 1974;77(1):89–148. doi: 10.1111/j.1601-5223.1974.tb01357.x. [DOI] [PubMed] [Google Scholar]
- 29.Hinze K, Thompson RD, Ritter E, Salamini F, Schulze-Lefert P. Restriction fragment length polymorphism-mediated targeting of the ml-o resistance locus in barley (Hordeum vulgare) Proc Natl Acad Sci USA. 1991;88(9):3691–3695. doi: 10.1073/pnas.88.9.3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hollomon DW. Genetic control of ethirimol resistance in a natural population of Erysiphe graminis f.sp. hordei. Phytopathology. 1981;71:536–540. [Google Scholar]
- 31.Oberhaensli S, et al. Comparative sequence analysis of wheat and barley powdery mildew fungi reveals gene colinearity, dates divergence and indicates host-pathogen co-evolution. Fungal Genet Biol. 2011;48(3):327–334. doi: 10.1016/j.fgb.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 32.Stenmark H, Aasland R, Driscoll PC. The phosphatidylinositol 3-phosphate-binding FYVE finger. FEBS Lett. 2002;513(1):77–84. doi: 10.1016/s0014-5793(01)03308-7. [DOI] [PubMed] [Google Scholar]
- 33.Brewer MT, Cadle-Davidson L, Cortesi P, Spanu PD, Milgroom MG. Identification and structure of the mating-type locus and development of PCR-based markers for mating type in powdery mildew fungi. Fungal Genet Biol. 2011;48(7):704–713. doi: 10.1016/j.fgb.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 34.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444(7117):323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 35.Dickman MB, Yarden O. Serine/threonine protein kinases and phosphatases in filamentious fungi. Fungal Genet Biol. 1999;26(2):99–117. doi: 10.1006/fgbi.1999.1118. [DOI] [PubMed] [Google Scholar]
- 36.Collins NC, et al. SNARE-protein-mediated disease resistance at the plant cell wall. Nature. 2003;425(6961):973–977. doi: 10.1038/nature02076. [DOI] [PubMed] [Google Scholar]
- 37.Nakamoto H, Vígh L. The small heat shock proteins and their clients. Cell Mol Life Sci. 2007;64(3):294–306. doi: 10.1007/s00018-006-6321-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stukenbrock EH, McDonald BA. The origins of plant pathogens in agro-ecosystems. Annu Rev Phytopathol. 2008;46:75–100. doi: 10.1146/annurev.phyto.010708.154114. [DOI] [PubMed] [Google Scholar]
- 39. Müller K (1993) Virulenzstruktur und Dynamik grossriiumiger Populationen des Gerstenmehltaues Erysiphe graminis DC. fsp. hordei Marchal in Europa. PhD thesis (Swiss Federal Institute of Technology, Zurich)
- 40.Stukenbrock EH, Christiansen FB, Hansen TT, Dutheil JY, Schierup MH. Fusion of two divergent fungal individuals led to the recent emergence of a unique widespread pathogen species. Proc Natl Acad Sci USA. 2012;109(27):10954–10959. doi: 10.1073/pnas.1201403109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Win J, et al. (2012) Effector biology of plant-associated organisms: Concepts and perspectives. Cold Spring Harb Symp Quant Biol, 10.1101/sqb.2012.77.015933. [DOI] [PubMed]
- 42. Manning VA, et al. (2013) Comparative Genomics of a plant-pathogenic fungus, Pyrenophora tritici-repentis, reveals transduplication and the impact of repeat elements on pathogenicity and population divergence. G3 (Bethesda) 3(1):41–63. [DOI] [PMC free article] [PubMed]
- 43.Jiang RH, Tripathy S, Govers F, Tyler BM. RXLR effector reservoir in two Phytophthora species is dominated by a single rapidly evolving superfamily with more than 700 members. Proc Natl Acad Sci USA. 2008;105(12):4874–4879. doi: 10.1073/pnas.0709303105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Joly DL, Feau N, Tanguay P, Hamelin RC. Comparative analysis of secreted protein evolution using expressed sequence tags from four poplar leaf rusts (Melampsora spp.) BMC Genomics. 2010;11:422. doi: 10.1186/1471-2164-11-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raffaele S, et al. Genome evolution following host jumps in the Irish potato famine pathogen lineage. Science. 2010;330(6010):1540–1543. doi: 10.1126/science.1193070. [DOI] [PubMed] [Google Scholar]
- 46.Gaullier JM, Ronning E, Gillooly DJ, Stenmark H. Interaction of the EEA1 FYVE finger with phosphatidylinositol 3-phosphate and early endosomes. Role of conserved residues. J Biol Chem. 2000;275(32):24595–24600. doi: 10.1074/jbc.M906554199. [DOI] [PubMed] [Google Scholar]
- 47. Debuchy R, Turgeon BG (2006) Mating-type structure, function, and evolution in Euascomycetes. The Mycota I Growth, Differentiation, and Sexuality, eds Kües U, Fischer R (Springer, Berlin/Heidelberg, Germany), pp. 293–323.
- 48.Both M, Csukai M, Stumpf MP, Spanu PD. Gene expression profiles of Blumeria graminis indicate dynamic changes to primary metabolism during development of an obligate biotrophic pathogen. Plant Cell. 2005;17(7):2107–2122. doi: 10.1105/tpc.105.032631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Both M, et al. Transcript profiles of Blumeria graminis development during infection reveal a cluster of genes that are potential virulence determinants. Mol Plant Microbe Interact. 2005;18(2):125–133. doi: 10.1094/MPMI-18-0125. [DOI] [PubMed] [Google Scholar]
- 50.Duplessis S, et al. Melampsora larici-populina transcript profiling during germination and timecourse infection of poplar leaves reveals dynamic expression patterns associated with virulence and biotrophy. Mol Plant Microbe Interact. 2011;24(7):808–818. doi: 10.1094/MPMI-01-11-0006. [DOI] [PubMed] [Google Scholar]
- 51.O’Connell RJ, et al. Lifestyle transitions in plant pathogenic Colletotrichum fungi deciphered by genome and transcriptome analyses. Nat Genet. 2012;44(9):1060–1065. doi: 10.1038/ng.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hacquard S, et al. A comprehensive analysis of genes encoding small secreted proteins identifies candidate effectors in Melampsora larici-populina (poplar leaf rust) Mol Plant Microbe Interact. 2012;25(3):279–293. doi: 10.1094/MPMI-09-11-0238. [DOI] [PubMed] [Google Scholar]
- 53.Kleemann J, et al. Sequential delivery of host-induced virulence effectors by appressoria and intracellular hyphae of the phytopathogen Colletotrichum higginsianum. PLoS Pathog. 2012;8(4):e1002643. doi: 10.1371/journal.ppat.1002643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schornack S, et al. Ten things to know about oomycete effectors. Mol Plant Pathol. 2009;10(6):795–803. doi: 10.1111/j.1364-3703.2009.00593.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parra G, Bradnam K, Korf I. CEGMA: A pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23(9):1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- 56.Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li H, et al. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6(2):80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takamatsu S. Origin and evolution of the powdery mildews (Ascomycota, Erysiphales) Mycoscience. 2013;54:75–86. [Google Scholar]
- 60.Krzywinski M, et al. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chenna R, et al. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31(13):3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tamura K, et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Stanke M, et al. (2006) AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res 34(Web Server issue):W435–W39. [DOI] [PMC free article] [PubMed]
- 64. Fischer S, et al. (2011) Using OrthoMCL to assign proteins to OrthoMCL-DB groups or to cluster proteomes into new ortholog groups. Curr Protoc Bioinformatics Chapter 6:Unit 6.12.1–19. [DOI] [PMC free article] [PubMed]
- 65.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29(7):644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kurtz S, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5(2):R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu TD, Watanabe CK. GMAP: A genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics. 2005;21(9):1859–1875. doi: 10.1093/bioinformatics/bti310. [DOI] [PubMed] [Google Scholar]
- 68.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Quinlan AR, Hall IM. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Smyth GK, Michaud J, Scott HS. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 2005;21(9):2067–2075. doi: 10.1093/bioinformatics/bti270. [DOI] [PubMed] [Google Scholar]
- 71.Conesa A, et al. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21(18):3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 72.Conesa A, Nueda MJ, Ferrer A, Talón M. maSigPro: A method to identify significantly differential expression profiles in time-course microarray experiments. Bioinformatics. 2006;22(9):1096–1102. doi: 10.1093/bioinformatics/btl056. [DOI] [PubMed] [Google Scholar]
- 73.Sturn A, Quackenbush J, Trajanoski Z. Genesis: Cluster analysis of microarray data. Bioinformatics. 2002;18(1):207–208. doi: 10.1093/bioinformatics/18.1.207. [DOI] [PubMed] [Google Scholar]
- 74. Vandesompele J, et al. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3(7):RESEARCH0034. [DOI] [PMC free article] [PubMed]
- 75.Hacquard S, Delaruelle C, Frey P, Martin F, Duplessis S. Validation of Melampsora larici-populina reference genes for in planta RT-quantitative PCR expression profiling during time-course infection of poplar leaves. Physiol Mol Plant Pathol. 2011;75(3):106–112. [Google Scholar]
- 76.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.