In an interesting work published recently in PNAS, Drake et al. (1) presented a proteomic study of the skeleton from the stony coral Stylophora pistillata. This study identified proteins that are associated to the mineral phase (i.e., that potentially contribute to shape the skeleton). In other words, this set of proteins is supposed to represent the so-called “biomineralization toolkit.” Although some of the 36 proteins reported in Drake et al. (1) appear as genuine extracellular matrix (ECM) proteins related to biomineralization, such as coral acid-rich proteins or carbonic anhydrase, some others are obvious intracellular contaminants that should not be considered as skeletal organic matrix proteins (SOMPs).
Indeed, Drake et al. (1) observed proteins from the cytoskeleton, such as actins, tubulins, and myosin. These proteins are intracellular components and should not be named SOMPs: as far as we know, there is no scientific evidence that they interact directly with the growing biomineral. We consider that the integration of intracellular components to the growing list of calcifying-matrix proteins is misleading and detrimental to our understanding of biocalcification mechanisms and to the elaboration of molecular models, and this problem needs to be carefully appreciated.
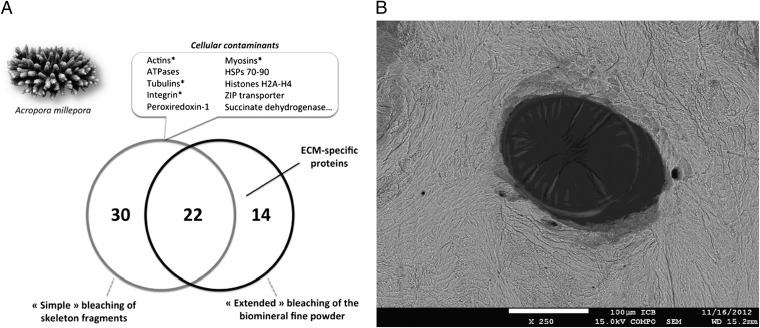
In our hands, when similarly investigating SOMPs from the coral Acropora millepora, we observed cytoskeletal proteins that were contaminants from calicoblastic cellular debris (Fig. 1 and Table 1). These contaminants could be simply removed by extensive and appropriate cleaning of the biomineral (Fig. 1). By using two types of sample treatment, we demonstrated convincingly that the presence of cytoskeletal proteins indicates an inadequate cleaning of the biomineral structures, which typically hold superficial contamination from skeleton-neighboring tissues (Table 1).
Fig. 1.
Removal of organic contamination of A. millepora’s skeleton. (A) Comparison of the proteins identified by proteomics on the skeletal organic matrix of A. millepora in two different conditions. “Simple bleaching” consisted in treating the skeletal fragments with sodium hypochlorite solution once [5% (vol/vol), 72 h], and “extended bleaching” consisted in the simple bleaching followed by cleaning the skeletal sieved powder (< 200 µm) with sodium hypochlorite solution [10% (vol/vol) 5 h]. The asterisk represents similar proteins to those reported as ECM proteins in Drake et al.’s study (1). (B) SEM image from polished transversal section of A. millepora skeleton with focusing a pore covered with residual soft tissue that remained after cleaning the fragments by simple bleaching.
Table 1.
List of the 30 proteins identified in the samples from coral skeleton treated by simple bleaching, which were further removed by extended bleaching
| Transcript references | BLASTP (above) and SwissProt reference (below) | E value | |
| 1* | >gi|379118176|gb|JT015846.1| | Actin | 0.0 |
| sp|P12716.1|ACTC_PISOC | |||
| 2 | >gi|379125045|gb|JT022715.1| | Tubulin alpha-1C chain | 0.0 |
| sp|P68365.1|TBA1C_CRIGR | |||
| 3* | >gi|379099717|gb|JR997386.1| | Tubulin beta-4 | 0.0 |
| sp|P30883.1|TBB4_XENLA | |||
| 4 | >gi|379084254|gb|JR981923.1| | Tubulin alpha-1C | 0.0 |
| sp|Q9BQE3.1|TBA1C_HUMAN | |||
| 5 | >gi|379076599|gb|JR974268.1| | Tubulin alpha | 6e-85 |
| sp|P10872.1|TBA_TETPY | |||
| 6 | >gi|379089391|gb|JR987060.1| | Tubulin alpha | 4e-161 |
| sp|P41351.1|TBA_TETTH | |||
| 7* | >gi|379122351|gb|JT020021.1| | Tubulin beta-4B | 0.0 |
| sp|P68371.1|TBB4B_HUMAN | |||
| 8 | >mf105_rep_c206 | ATP synthase beta | 0.0 |
| Ssp|Q4FP38.1|ATPB_PELUB | |||
| 9 | >gi|379098186|gb|JR995855.1| | ATP synthase alpha | 0.0 |
| sp|Q5R546.1|ATPA_PONAB | |||
| 10* | >gi|379075456|gb|JR973125.1| | Myosin heavy chain | 4e-06 |
| sp|P24733.1|MYS_AEQIR | |||
| 11 | >gi|379082904|gb|JR980573.1| | Myocilin | 7e-29 |
| Ssp|O70624.1|MYOC_MOUSE | |||
| 12 | >gi|222798399|gb|EZ026787.1| | Histone H2A | 1e-26 |
| sp|P35061.2|H2A_ACRFO | |||
| 13 | >gi|379114242|gb|JT011912.1| | Histone H2B | 2e-76 |
| sp|P35067.1|H2B_ACRFO | |||
| 14 | >gi|379095792|gb|JR993461.1| | Histone H4; | 2e-65 |
| sp|P35059.2|H4_ACRFO | |||
| 15 | >kb8_rep_c51392 | Heat shock protein 90; | 0.0 |
| sp|O44001.1|HSP90_EIMTE | |||
| 16 | >kb8_rep_c29387 | Heat shock protein 90 | 0.0 |
| Ssp|O44001.1|HSP90_EIMTE | |||
| 17 | >gi|379104815|gb|JT002485.1| | Heat shock protein 90 | 0.0 |
| sp|O57521.2|HS90B_DANRE | |||
| 18 | >kb8_rep_c63048 | Heat shock protein 70 | 3e-66 |
| sp|Q9S9N1.1|HSP7E_ARATH | |||
| 19 | >gi|379073448|gb|JR971117.1| | Heat shock protein 70 | 0.0 |
| sp|P63018.1|HSP7C_RAT | |||
| 20 | >kb8_c48899 | Heat shock protein 70 | 0.0 |
| sp|P11144.2|HSP70_PLAFA | |||
| 21 | >gi|379105500|gb|JT003170.1| | Zinc transporter ZIP14 | 1e-75 |
| sp|Q75N73.1|S39AE_MOUSE | |||
| 22 | >gi|379096620|gb|JR994289.1| | Calpain-9 | 0.0 |
| sp|O35920.1|CAN9_RAT | |||
| 23 | >gi|379108785|gb|JT006455.1| | Photosystem II precursor | 0.0 |
| sp|P49472.1|PSBC_ODOSI | |||
| 24 | >gi|222803727|gb|EZ032115.1| | Voltage-dep. channel protein 2 | 5e-122 |
| Ssp|P81004.1|VDAC2_XENLA | |||
| 25 | >gi|379104892|gb|JT002562.1| | Peroxiredoxin-1 | 9e-100 |
| sp|P0CB50.1|PRDX1_CHICK | |||
| 26 | >gi|222782586|gb|EZ011257.1| | Succinate Dehydrogenase | 2e-65 |
| sp|Q7ZVF3.2|DHSA_DANRE | |||
| 27 | >gi|379122454|gb|JT020124.1| | Endoplasmin | 0.0 |
| sp|Q66HD0.2|ENPL_RAT | |||
| 28* | >gi|379079965|gb|JR977634.1| | Integrin | 0.49 |
| sp|P16144.5|ITB4_HUMAN | |||
| 29 | >gi|222799407|gb|EZ027795.1| | Transaldolase | 5.4 |
| sp|B6JNZ3.1|TAL_HELP2 | |||
| 30 | >kb8_c30860_frame-3 | No hit | — |
Similar proteins to those reported as ECM proteins in Drake et al.’s study (1).
According to the most commonly accepted view, the formation of metazoan calcified skeletons results from the secretion of an acellular matrix that remains occluded within the biomineral phase once precipitated. During this extracellular process, cellular contaminants can be entrapped in void structures (such as the microcavities present inside all the aragonitic skeleton of stony corals), and need to be removed by thorough incubation of skeleton fine powder (< 200 μm) in concentrated sodium hypochlorite [10% (vol/vol), 5 h] before extraction and further proteomic analysis of the biomineralization proteins. This simple treatment removes most—if not all—cellular debris, leaving intact the skeleton-associated proteins, the true SOMPs that are part of the biomineralization toolkit.
We are convinced by our previous experiments (2–4)—which are reproducible and coherent with the current understanding of biomineralization processes—that a careful and appropriate cleaning of biominerals is crucial for generating accurate proteomic data and further correctly interpreting the results.
Footnotes
The authors declare no conflict of interest.
References
- 1.Drake JL, et al. Proteomic analysis of skeletal organic matrix from the stony coral Stylophora pistillata. Proc Natl Acad Sci USA. 2013;110(10):3788–3793. doi: 10.1073/pnas.1301419110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Joubert C, et al. Transcriptome and proteome analysis of Pinctada margaritifera calcifying mantle and shell: focus on biomineralization. BMC Genomics. 2010;11:613. doi: 10.1186/1471-2164-11-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marie B, et al. The shell-forming proteome of Lottia gigantea reveals both deep conservations and lineage-specific novelties. FEBS J. 2013;280(1):214–232. doi: 10.1111/febs.12062. [DOI] [PubMed] [Google Scholar]
- 4.Marie B, et al. Different secretory repertoires control the biomineralization processes of prism and nacre deposition of the pearl oyster shell. Proc Natl Acad Sci USA. 2012;109(51):20986–20991. doi: 10.1073/pnas.1210552109. [DOI] [PMC free article] [PubMed] [Google Scholar]