Abstract
Photosynthetic reaction centers are sensitive to high light conditions, which can cause damage because of the formation of reactive oxygen species. To prevent high-light induced damage, cyanobacteria have developed photoprotective mechanisms. One involves a photoactive carotenoid protein that decreases the transfer of excess energy to the reaction centers. This protein, the orange carotenoid protein (OCP), is present in most cyanobacterial strains; it is activated by high light conditions and able to dissipate excess energy at the site of the light-harvesting antennae, the phycobilisomes. Restoration of normal antenna capacity involves the fluorescence recovery protein (FRP). The FRP acts to dissociate the OCP from the phycobilisomes by accelerating the conversion of the active red OCP to the inactive orange form. We have determined the 3D crystal structure of the FRP at 2.5 Å resolution. Remarkably, the FRP is found in two very different conformational and oligomeric states in the same crystal. Based on amino acid conservation analysis, activity assays of FRP mutants, FRP:OCP docking simulations, and coimmunoprecipitation experiments, we conclude that the dimer is the active form. The second form, a tetramer, may be an inactive form of FRP. In addition, we have identified a surface patch of highly conserved residues and shown that those residues are essential to FRP activity.
Keywords: nonphotochemical quenching, Synechocystis
Light is vital for the survival and growth of photosynthetic organisms. In natural environments, these organisms are exposed to varying light conditions in addition to the day/night cycle. Too much exposure to light causes the formation of reactive oxygen species that damage the sensitive photochemical reaction centers, and thus, a careful regulation of energy flow is critical. Under low light conditions, an efficient energy collection by the antennae complexes is achieved, whereas under high light conditions, the excess energy has to be diverted from photosynthesis (1).
Plants and cyanobacteria have evolved different ways to deal with excess energy arriving at the reaction centers. Higher plants and green algae contain antenna complexes consisting of transmembrane proteins that sense the acidification of the thylakoid lumen and react by switching from efficient energy collection to heat dissipation (1, 2). Cyanobacteria, however, contain antenna complexes called phycobilisomes, which are membrane-anchored and consist of phycobilin proteins. Instead of sensing the effect of high light through pH changes, cyanobacterial phycobilisomes are quenched by a protein capable of directly sensing high light conditions, the orange carotenoid protein (OCP). The 35 kDa OCP consists of two distinct domains that encompass a keto-carotenoid (3′-hydroxyechinenone) in an all trans conformation (3–6). Irradiance with high light changes the OCP from an inactive orange (OCPo) to an active red form (OCPr) that is capable of binding to the phycobilisomes to prevent excess energy from flowing to the reaction centers. The low quantum yield of the OCPo to OCPr photoconversion together with instability of the OCPr form lead to effective OCP inactivity under low light conditions; therefore, the OCP acts as a switch for photoprotection triggered by a specific light level (7).
In darkness, isolated OCPr spontaneously reverts back to the OCPo form. This reversion is greatly affected by the presence of the fluorescence recovery protein (FRP) (8). In vitro, the FRP accelerates the conversion of free OCPr back to the orange form. In vivo, the FRP is essential to recover the full capacity of the antenna, presumably by playing a role in detaching OCPr from the phycobilisomes (8). Mutant strains of cyanobacteria lacking this protein are unable to recover the normal antenna capacity under low light conditions. The FRP is a 13 kDa protein that does not bind a chromophore. It is exclusively found in organisms that also contain the OCP, and the two genes are typically in close proximity in the genome. Of the currently available 130 cyanobacterial genomes, 97 genomes contain a gene for the OCP, and 71 genomes also contain a gene for the FRP (9) (Table S1). The FRP from Synechocystis sp. PCC 6803 was first characterized in a form containing an additional 25 aa compared with the FRP from almost all other strains (the other exception is the FRP encoded in the Microcystis aeruginosa genome) (8). It has recently been shown that this longer form is caused by a misidentified start site, and the active form in vivo is a shorter protein that begins with Met26 (10); we have used this form for this study.
Here, we present the 3D structure of the FRP; the protein crystallized in two different conformations and different quaternary states. We show that the active form of the FRP is a homodimer, with a cluster of highly conserved residues on one surface of the dimer. Based on these observations, we made several single amino acid mutations, analyzed the mutant forms for activity, and thereby, identified the active site of the FRP. Based on the structural information, docking, and coimmunopreciptation studies, we propose a model for the interaction between the FRP and the OCP. These results provide the foundation for additional studies on the molecular mechanism of the regulation of photoprotection in cyanobacteria.
Results
Structure of the FRP Shows That It Adopts Two Distinct Conformations.
We have determined the crystal structure of Synechocystis sp. PCC 6803 FRP at 2.5 Å resolution using iodine phasing (Table S2). There are six FRP polypeptide chains in the asymmetric unit of the P41212 space group in two distinct conformations. Four (chains A–D) form dimers, with the twofold axis either in the asymmetric unit or generated by crystal symmetry. Remarkably, the other two chains (chains E and F) in the asymmetric unit exist in a very different conformation: α-helices α1 and α2 form one long extended helix α1′, making a dimer that, together with its symmetry-related chains (E′ and F′), forms a four-helix bundle (Fig. 1 A and B). Sample electron density for these regions can be found in Fig. S1 A and B. Both forms of the FRP are all helical, and they consist of an extended helical stalk of α1 and α2/α1′ (residues from 1 to 65) and a compact C-terminal head domain (residues 66–109). The stalk contains all of the contacts for the dimerization interface and likewise, forms the core of the tetramer. Chains B, D, and F show a higher flexibility, which is reflected in their temperature factor (B factor) because of fewer crystal contacts (Fig. S1C). The availability of noncrystallographically related chains enabled us to build into even weak electron density of regions of chain D, which as a result, have a high B factor (Fig. S1C, red arrow).
Fig. 1.
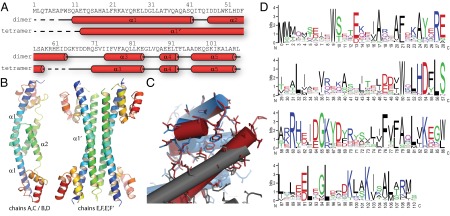
Structural overview of the FRP. (A) Primary and secondary structure of the Synechocystis sp. PCC 6803 FRP dimer and tetramer forms. Red tubes indicate α-helices, and the dashed line shows residues disordered in the structure. (B) Cartoon representation of the dimer and tetramer forms of the FRP observed in the crystals. Rainbow coloring from N to C termini from blue to red. (C) Alignment of the head domains of the dimer (red, chain B) and tetramer (blue, chain F; gray, chain E) illustrating the conservation of interactions of the head and stalk domain between the two forms. (D) Sequence conservation logo of the FRP with numbering corresponding to the Synechocystis sp. PCC 6803 protein.
Although the structure of the head domain remains essentially the same in both the dimer and tetramer, the conformational change needed for their interconversion involves a rotation of helix α1 by 180° to form one extended helix α1′, which includes a short loop and most of the residues from α2 (Fig. 1B). The interactions between the N-terminal part of helix α1/α1′ and the C-terminal head domain are intramolecular in the dimer form, and although conserved in the tetramer assembly, they are instead intermolecular (Fig. 1C).
We analyzed the interfaces between the FRP monomers using the protein interfaces, surfaces, and assemblies (PISA) tool at the European Bioinformatics Institute (11). Both the dimer and tetramer forms have a large buried surface, consistent with a native state (Tables S3 and S4). Calculation of the energy of association of the interfaces between the different protein chains (Tables S3 and S4) results in a large negative ΔG for all interfaces, indicative of strong affinity. In addition, we measured the P value for each interface; this number quantifies its specificity. Values larger than 0.5 indicate an artifact of crystal packing. Based on the P value, the dimer interface is more specific (P = 0.048 for AC, P = 0.055 for BD), which is consistent with the large number (12) of highly conserved residues that it contains. The P value close to zero also implicates the dimer as the most stable form; however, the values calculated for the tetramer (P = 0.338 for E–F, P = 0.227 for E–F′) are consistent with a viable alternative native state.
Likewise, analytical size exclusion chromatography of the purified FRP indicates that the dimer form is predominant in solution (Fig. S2A). Additionally, a time course glutaraldehyde cross-linking experiment followed by denaturing SDS/PAGE analysis confirm a preferential monomer–monomer cross-linking (Fig. S2B). When the FRP is stored on ice for more than 1 wk before characterization, a size exclusion peak corresponding to a tetramer is occasionally observed (Fig. S2A). However, we cannot exclude that this larger form is a dimer of dimers instead of the four-helix bundle observed in the crystal structure. Efforts to generate the tetramer in solution by varying pH and salt concentration were not successful.
Putative Active Site of the FRP.
In the distribution of conserved amino acids, there is a striking patch of highly conserved residues on one surface of the dimer form of the FRP (Figs. 1D and 2A). In contrast, there is very little conservation of the residues on the opposite side. In addition to highly conserved residues on the inside of the patch (L56 and H53), there is a close intermolecular (distance between planes is 3.5 Å) cation-π interaction between W50 and R60, which orients the arginine in the correct position to form an intermolecular salt bridge with D54 (Fig. 2B). All of these conserved residues are located around the twofold symmetry axis between residues H53 and S57, forming a symmetrical active site with an extended hydrogen bonding network. In addition to those absolutely conserved residues, there is a histidine residue (H61) that forms a surface bulge. In all of the available primary structures of the FRP, the amino acid at the position corresponding to H61 is an aromatic residue (Phe/Tyr/His) that is also likely to form a similar surface bulge. In the tetramer form, the group of residues that makes up the conserved patch in the dimer is buried in the interface between the two dimers (W50 and H53), involved in interactions with the head domain (D54 and L56), or located in the disordered region between residues 58 and 65 (Figs. 1A and 2A). Accordingly, we hypothesized that the region between residues 50 and 61 on one side of the FRP dimer is the active site of the FRP.
Fig. 2.
Amino acid conservation mapping onto the structure of the FRP. (A) FRP dimer and tetramer structures shown in surface representation are colored according to conservation (red, high; white, medium; yellow, low). (B) Close-up view of the proposed active site with stick representation of side chains. Side chain oxygen and nitrogen atoms are colored red and blue, respectively. The location of the twofold symmetry axis is indicated with a black ellipse.
Mutations in the Proposed Active Site Alter FRP Activity.
To confirm our putative identification of the FRP active site, we generated several single-point FRP mutants (W9L, W50L, W50F, H53L, D54E, D54L, R60L, and R60K) and tested their activity in an assay measuring the conversion of OCPo to OCPr. The D54L and W50L FRP mutants expressed poorly and tended to precipitate, possibly because of misfolding. These mutants also showed a rather large decrease in activity relative to WT (Fig. 3). The H53L mutant was obtained in large quantities as a soluble protein but likewise, was impaired in activity (Fig. 3). The W50F and D54E mutants were purified in large quantities, behaved as homogeneous soluble dimers, and showed only a slight reduction in activity. A mutant of a highly conserved tryptophan from the head domain (W9L) showed no reduction in activity.
Fig. 3.
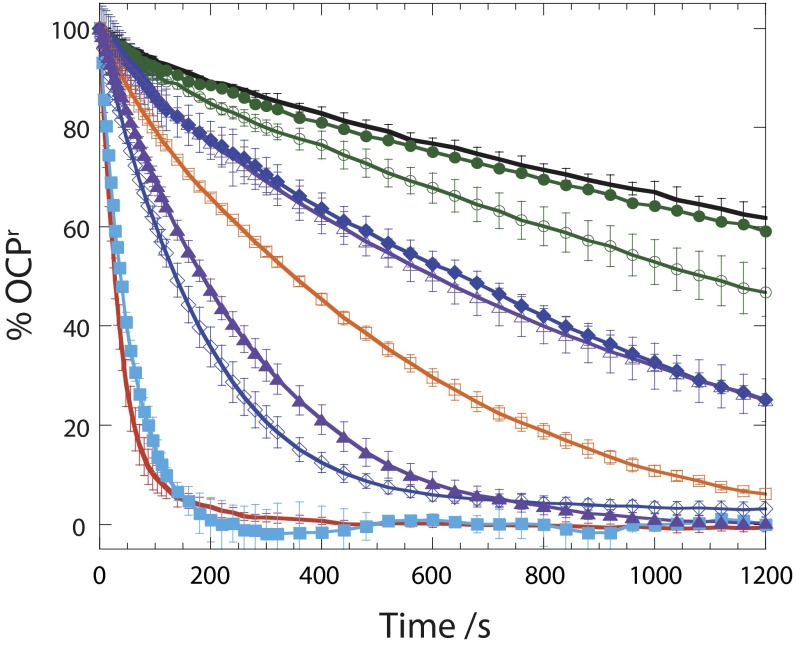
Kinetics of dark reconversion of OCPr to OCPo at 8 °C in the absence and presence of WT and mutant FRPs with a ratio of OCP to FRP of 2:1: without FRP (black), with WT (red), and with W9L (closed squares; sky blue), H53L (open squares; orange), W50F (closed triangles; violet), W50L (open triangles; violet), R60L (closed circles,; green), R60K (open circles; green), D54E (open rhomboids; blue), or D54L (closed rhomboids; blue). Average of three independent experiments. Error bars represent SD.
The most remarkable difference, however, was observed in the R60L and R60K mutants, which behaved similarly to WT during purification and analytical size exclusion chromatography but showed almost no activity, even with the relatively conservative change from arginine to lysine (Fig. 3). To exclude the possibility that the R60K mutant was disordered as a consequence of the mutation, we solved its structure. The R60K mutant FRP crystallized in the same space group, and the structure was solved by molecular replacement (Table S2 shows statistics). Although the resolution is comparatively low (3.5 Å), the electron density is clear enough to see that the overall fold of the R60K mutant of the FRP is identical to the fold of the WT (Fig. S1D). Collectively, these data show that R60 is directly involved in the conversion of OCP and that the conserved side of the dimer surface interacts with OCP in the red form.
FRP–OCP Docking Simulations.
We next performed docking simulations between combinations of full-length FRP dimer, tetramer, FRP active site region, and full-length OCP or its C- and N-terminal domains with or without carotenoid. We analyzed the solutions based on Rosetta score (12) and biological relevance considering the newly identified active site. The most convincing solution was found in a docking simulation between the active site region of the FRP and the C-terminal domain of the OCP lacking the carotenoid (Fig. 4). We then expanded the docking refinement to the full-length FRP docking to the C-terminal OCP domain containing the carotenoid based on the initial solution and found a narrow funnel in an rmsd vs. interface energy plot (Fig. S3), indicative of a good solution (12, 13).
Fig. 4.
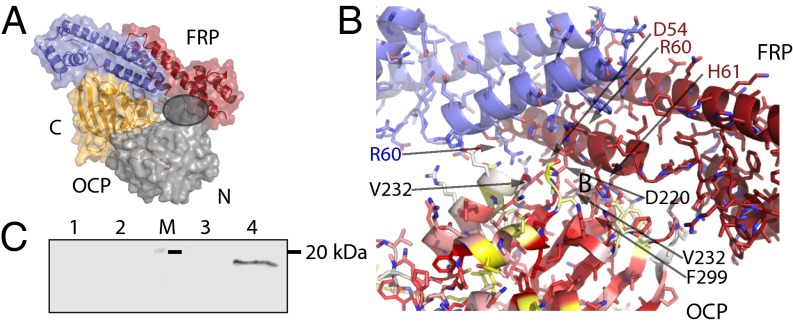
Analysis of the FRP–OCP interaction. (A) Docking solution of the FRP dimer (active site region) to the C-terminal domain of the OCP. Overview of the best solution shown as the surface with the residues involved in the docking as a cartoon and the N-terminal domain in gray, the C-terminal domain in orange. The gray circle indicates the clash between the docked FRP and the N-terminal domain of OCP. (B) Details of the docking solution interaction between OCP and FRP. Amino acids in the OCP involved in the interaction are marked in black, and amino acids of the FRP are marked in blue and red. The surface of the OCP C-terminal domain is colored according to conservation (red, high; white, medium; yellow, low). (C) Anti-OCP immunoblot of the N- (lane 2) and C-terminal domains (lane 4) after coimmunoprecipitation of each domain with the OCP. Control experiments (without FRP) are also shown: N- (lane 1) and C-terminal controls (lane 3). Molecular mass marker (20 kDa; lane M) is denoted with a black line).
The best solution based on score and surface complementarity places the FRP dimer binding to one side of the C-terminal domain (Fig. 4 A and B). This region includes a short helix, which is contributed by the N terminus of the protein, including residues T4, I5, and D6. Loops connecting the strands of the C-terminal β-sheet (T218, D220, R229 V232, N236, D262, F264, and F299) are also involved in the interaction. The binding region on the FRP is, as expected, the patch of conserved residues (around 50–62) as well as a contact from the head domain (Q80). This solution resulted from docking with the isolated C-terminal domain only; interestingly, the surface that interacts with the FRP is partially obstructed by the N-terminal domain in the full-length OCPo form (Fig. 4A, shaded circle). The FRP dimer binds to the OCP on a surface groove lined by a cluster of conserved residues (Fig. 4B). This finding is notable because although OCP homologs are, in general, very similar, the amino acid conservation is usually highest for the residues interacting with the carotenoid; also, the surface is typically less well-conserved. The electrostatics of the FRP–OCP interface are complementary (Fig. S4, Top Left, shaded circle); there is an overall negative charge around the putative active site of the FRP, which aligns with positive LY charge at the location of the docking site of the OCP (Fig. S4, Bottom Left).
To confirm the predicted interaction between the FRP and the C-terminal domain of the OCP, coimmunoprecipitation experiments were performed. The C-terminal domain of the OCP was purified from a Synechocystis PCC 6803 mutant overexpressing a construct encoding residues 170–317 of the OCP (Fig. S5A). For the N-terminal domain, we used the 16 kDa red carotenoid protein (RCP), which is an N-terminal proteolytic derivative of the OCP that binds the carotenoid (3, 14) (Fig. S5A). Interaction between the C- and N-terminal domains of the OCP with FRP was tested by coimmunoprecipitation, and OCP was detected using an antibody that recognizes both domains (Fig. S5B). As shown in Fig. 4C, only the C-terminal domain of the OCP coprecipitates with the FRP.
Discussion
We have determined the structure of the FRP, which regulates photoprotection in cyanobacteria. The FRP is present in two distinct forms in the crystal: a dimer that we identify as the active state and a tetramer that may be an inactive form. We were not able to purify the tetramer, but the structure shows that the active site that we identified in the dimer is disrupted in the tetramer. The difference between the dimer and tetramer is reminiscent of the conformational change observed in hemagglutinin of the influenza A virus on fusion at low pH, where a loop and an α-helix are drastically rearranged to become a long extended α-helix in the form of a triple-stranded coiled coil (15). Despite secondary structure consisting only of α-helices, the tertiary structure of either the active dimer or the tetramer form seems to be unique, because there is no close structural homolog to either in the Protein Data Bank (PDB).
Highly conserved residues forming an intricate network of hydrogen bonds are concentrated on one surface of the FRP dimer (Fig. 2A). Mutation of the absolutely conserved amino acids in this region greatly affected the activity of the FRP. Among them, the R60K mutant is almost completely inactive. The dimer form is the predominant form in solution and, based on activity assays, we propose that it is the active form of the FRP.
Based on structural evidence, the tetramer form seems to be a viable alternate conformation of the FRP and could represent an inactive state. The active site residues in the tetramer are either buried or too flexible (e.g., R60) to show ordered electron density. It seems that the crystallization conditions generate the right environment for the tetramer to form. In solution, there is a small fraction of apparent tetramer present, but it is unclear whether this larger form is a dimer of dimers or a tetramer with an extended α1′ helix. Whether the inactive tetramer can be formed in a cellular environment is an open question. Previous results indicate that the FRP is constitutively active in vitro and in vivo. In vitro the presence of the FRP abolishes/reduces the accumulation of the OCPr and the phycobilisome fluorescence quenching by the OCP (16). In Synechocystis mutant cells containing a high concentration of the FRP (10 times more than in WT cells), no fluorescence quenching was observed, indicating that, in vivo, the FRP is also active under high light conditions (9). Nevertheless, it is possible that, in vivo, a tetramerization could result in the inactivation of some of the FRP. In addition to the hypothesis that the FRP is inactivated by change in conformation and oligomeric state, there is also the possibility that changes in either pH or redox state coupled to light intensity could perturb the interactions among residues in the active site and thus, reduce its activity. This mechanism would be similar to the regulation of photoprotection in plants and allow a more dynamic regulation in contrast to relying on a difference in expression level of the FRP.
We have identified a patch of residues (W50, D54, H53, and R60), contributed by both chains on the surface of the FRP dimer to be important for the acceleration of the OCPr to OCPo conversion. In contrast, a mutation of a highly conserved residue from the head domain (W9L) did not influence activity at all (Fig. 3). The most striking inactivating mutation was changing R60 to lysine. This finding suggests that not only is the positive charge important but also, the intermolecular distance between the FRP and the OCP is critical. Moreover, the observation that mutation of W50 or D54, amino acids interacting with R60, decreases FRP activity also suggested that the exact position of the positive charge of the arginine is fundamental for FRP activity.
Docking simulations suggest that the FRP interacts with the C terminus of the OCP. A recent kinetic study (17) suggests that the transition from OCPo to OCPr could be limited by cis–trans proline isomerization of residues Q224–P225 or P225–P226. Our docking solution places the FRP close to that loop, and therefore, it is possible that it could facilitate that isomerization (Fig. 4B). Recently, the OCP has been proposed to interact with the phycobilisome through its N-terminal domain, specifically through the surface surrounding Arg155, which is typically buried in interaction with the C-terminal domain in the OCPo (18). We have confirmed the interaction of the FRP with the C-terminal domain of the OCP by coimmunoprecipitation. The FRP specifically interacts with the C-terminal domain, whereas no interaction was observed with the N-terminal domain of the OCP (Fig. 4C). It has also been proposed that the OCP might partially open up to expose part of the carotenoid (6) and that the FRP could act to drive the two domains back together. Alternatively, the binding of the FRP to the C-terminal domain could trigger the conformational change back to the orange form of the OCP (Fig. 5). The position of the FRP in the most plausible docking model of FRP–OCP clashes with the N-terminal domain (in the structure of the closed orange form), which is consistent with this hypothesis. The binding of the FRP to OCPr could trigger the conversion, and the following conformational change could cause the dissociation of the FRP.
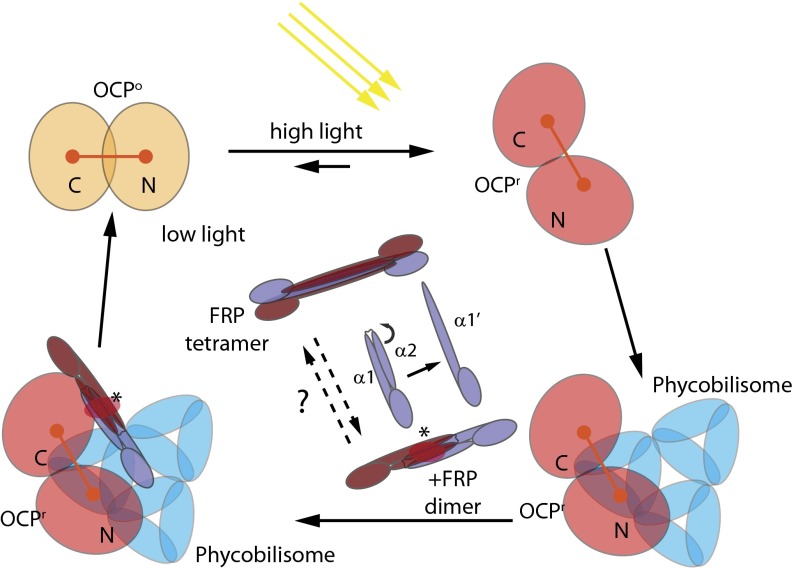
Fig. 5.
Model of the interplay between the FRP, OCP, and phycobilisome under high and low light conditions. The OCPo is converted to OCPr under high light conditions, undergoes a conformational change that exposes part of the carotenoid and R155, and binds to the phycobilisomes to quench excess energy. The FRP can bind to unattached or phycobilisome-attached OCPr, converting the OCPr into OCPo and dissociating it from the phycobilisomes. Under low light conditions, the decrease of OCPr concentration leads to a situation in which all of the phycobilisomes will be free of OCP and unquenched. The dimer FRP is the active form. The state of the FRP could be regulated by exterior factors that convert it to an inactive tetramer form under high light conditions by rearranging helix α2 to form an extended α1′. The FRP dissociates from the OCPo after conversion. The red ellipse and asterisk indicate the position of the FRP active site.
Additional studies are needed to probe the residues in the OCP involved in the interaction and confirm the site of the interaction of the FRP with the C-terminal domain of the OCP, which is challenging because of the transient nature of the active red form of the OCP. A structure of the complex of the active red form of the OCP and the FRP would provide the information needed to explain how the newly identified active site residues of the FRP can act to convert OCP back to its orange form.
Methods
Site-Directed Mutagenesis.
To obtain a plasmid containing the sequence of the gene encoding the short Synechocystis sp. PCC 6803 His-tagged FRP, a deletion of the thrombin sequence and the nucleotides coding for the first 25 aa of FRP was done in the plasmid pcB9 (8) using the synthetic primers petM26Hisfor/petM26hisrev. The construction of the pCB9 plasmid, in which the frp gene (slr1964) is under control of a T7 promoter, was described in ref. 8. The point mutations were added in the short FRP gene by site-directed mutagenesis using the Quickchange XL site-directed mutagenesis kit (Stratagene) and synthetic mutagenic oligonucleotides (all of the oligonucleotides used in this work are described in Table S5). Oligonucleotides used to construct site-directed mutants were synthesized by Eurofins MWG. Incorporated mutations were confirmed by DNA sequencing. DNA containing the desired mutations was transformed into Escherichia coli strain BL21 (DE3) for protein expression.
Protein Purification for Crystallization.
E. coli cells containing the frp pcB9-derived plasmid were grown to an OD600 of 0.8 at 37 °C, at which point they were cooled to room temperature followed by induction with 0.05 mM isopropylthio-β-d-galactoside and incubation overnight. Harvested cells were resuspended in buffer A (20 mM Tris, pH 8.0, at room temperature, 300 mM NaCl) and lysed using a French Press. Cleared lysate was applied on a 5-mL HisTrap HP column (Amersham) and washed with buffer A containing 20 mM imidazole. His-FRP was eluted with 2 column volumes buffer A containing 300 mM imidazole, concentrated, and then applied on a HiLoad 26/60 Superdex 75 (GE Healthcare) column equilibrated in 20 mM Tris, pH 7.4, at room temperature and 50 mM NaCl for final cleanup. Protein was then concentrated to 10–20 mg/mL for crystallization. Crystals were obtained from sitting drop experiments mixing 3–5 µL protein solution with 1–5 µL reservoir solution containing 5–10% (wt/vol) PEG-3350, 100 mM Na-citrate, pH 5.5–5.6, and 2% (vol/vol) tacsimate, pH 5.0 (Hampton Research). Crystals were stabilized by adding an 80% (vol/vol) glycerol solution to the drop for a final concentration of 25% (vol/vol) glycerol, frozen in liquid nitrogen, and measured at beam lines 5.0.1 and 5.0.2 of the Advanced Light Source at Lawrence Berkeley National Laboratory. For iodine soaks, in addition to glycerol, 1 µL 4 M potassium iodide solution was added to the drop, and crystals were frozen within 5 min.
Structure Determination and Analysis.
Even at 1.0 Å wavelength, the anomalous signal from the iodine was large enough to locate four sites using hkl2map/SHELX C/D/E (19). Initial density was obtained using phenix.autosol, and buccaneer successfully built an initial model of 595 residues, which was then manually corrected in Coot combined with refinement runs with phenix.refine (20–22). The sequence logo was generated at http://weblogo.berkeley.edu/logo.cgi (23) from an alignment of 71 known FRP sequences.
Protein Purification and Activity Assays.
The isolation of the short His-tagged FRP from E. coli strain BL21 (DE3) to activity assays is described in ref. 8. Briefly, protein expression was induced at an OD600 of 0.4 by adding 1 mM isopropylthio-β-d-galactoside and 2% (vol/vol) ethanol. After 12 h of growth at 20 °C, the cells were harvested and lysed by French Press. The supernatant was loaded on a column of Ni-ProBond resin (Invitrogen). The FRP was eluted with 300 mM imidazole and dialyzed during 48 h against 40 mM Tris⋅HCl at pH 8.0. Purity of FRP was checked by SDS/PAGE on 17% (wt/vol) polyacrylamide/2 M urea in a Tris/Mes system.
The OCPr to OCPo conversion in the absence or presence of FRP was monitored in a Specord S600 (Analytik Jena) spectrophotometer at 8 °C. The OCP was previously photoconverted to the red form by 5 min illumination with 5,000 μmol photons m−2 s−1 white light at 8 °C.
Construction of the Synechocystis PCC 6803 Strain Overexpressing the His-Tagged C-Terminal Domain of the OCP.
To obtain a strain overexpressing a His6-tagged C-terminal domain of the OCP, a plasmid containing the slr1963 gene under the control of the psbA2 promoter was used. The construction of this plasmid is described in ref. 24. The deletion of the nucleotides coding for the first 169 aa (N-terminal domain) of the OCP was obtained by site-directed mutagenesis using the Quickchange XL site-directed mutagenesis kit (Stratagene) and synthetic primers (Table S5). The plasmid obtained was used to transform WT cells. In the strains overexpressing the C-terminal domain, the original WT OCP was suppressed by partial deletion of the slr1963 gene as described in ref. 18. The His-tagged C-terminal domain was isolated by Nickel affinity as described in ref. 24. The isolated protein did not bind any chromophore.
Coimmunoprecipitation.
Anti-FRP antibodies (Covalab), FRP proteins, and C- or N-terminal OCP domains were incubated at 4 °C overnight. The Sepharose-protein A beads (Sigma) were blocked by incubating them in the presence of 5% BSA overnight. The mixed proteins were then incubated with the blocked Sepharose beads for 2.5 h. The beads were washed with a buffer containing 1% N-β-dodecyl maltoside to remove unbound proteins. The proteins attached to the anti-FRP antibodies were eluted with 2% (wt/vol) SDS buffer and loaded on an SDS/PAGE gel. In the control samples, the FRP protein is absent. Western blot using anti-OCP antibody was performed.
Docking Calculations.
The protein structure of the OCP was modified to include the loop region from amino acids T164 to K170, which is missing in the available structure (PDB ID code 3MG1). Protein structures (OCP: 3MG1_mod; FRP: this structure) were energy-minimized using the rosetta-relax application before the docking step. Then, docking was performed using Rosetta dock (12). Unconstrained docking simulations were performed for 100,000 runs, whereas refinement runs were executed for 20,000 runs. The best solutions were chosen based on their Rosetta score and surface complementarity (scripts are in SI Methods) and analyzed for biological significance by hand.
PDB ID Codes.
PDB ID codes are 4JDX for the WT FRP protein and 4JDQ for the R60K mutant.
Supplementary Material
Acknowledgments
We thank the entire staff at the Advanced Light Source, Lawrence Berkeley National Laboratory, which is supported by the Director, Office of Science, Office of Basic Energy Sciences of the US Department of Energy under Contract DE-AC02-05CH11231, and Peter Zwart in particular for excellent support during data collection. Modeling simulations were run on the Genepool cluster at the National Energy Research Scientific Computing Center (NERSC). M.S. was supported by a Swiss National Science Foundation Postdoctoral Fellowship. R.L.L. and C.A.K. were supported by National Science Foundation Grant MCB0851094. A.W., R.L.-I., A.T. and D.K. have been supported by grants from Agence Nationale de la Recherche (ANR, Cyanoprotect Project), Centre Nationale de Recherche Scientifique (CNRS), Commisariat à l'énergie atomique et aux energies alternatives (CEA), and HARVEST EU FP7 Marie Curie Research Training Network.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4JDX and 4JDQ).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1303673110/-/DCSupplemental.
References
- 1.Niyogi KK. Photoprotection revisited: Genetic and molecular approaches. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:333–359. doi: 10.1146/annurev.arplant.50.1.333. [DOI] [PubMed] [Google Scholar]
- 2.Pascal AA, et al. Molecular basis of photoprotection and control of photosynthetic light-harvesting. Nature. 2005;436(7047):134–137. doi: 10.1038/nature03795. [DOI] [PubMed] [Google Scholar]
- 3.Kerfeld CA. Structure and function of the water-soluble carotenoid-binding proteins of cyanobacteria. Photosynth Res. 2004;81(3):215–225. doi: 10.1023/B:PRES.0000036886.60187.c8. [DOI] [PubMed] [Google Scholar]
- 4.Kerfeld CA. Water-soluble carotenoid proteins of cyanobacteria. Arch Biochem Biophys. 2004;430(1):2–9. doi: 10.1016/j.abb.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 5.Kerfeld CA, et al. The crystal structure of a cyanobacterial water-soluble carotenoid binding protein. Structure. 2003;11(1):55–65. doi: 10.1016/s0969-2126(02)00936-x. [DOI] [PubMed] [Google Scholar]
- 6.Wilson A, et al. Structural determinants underlying photoprotection in the photoactive orange carotenoid protein of cyanobacteria. J Biol Chem. 2010;285(24):18364–18375. doi: 10.1074/jbc.M110.115709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilson A, et al. A soluble carotenoid protein involved in phycobilisome-related energy dissipation in cyanobacteria. Plant Cell. 2006;18(4):992–1007. doi: 10.1105/tpc.105.040121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boulay C, Wilson A, D’Haene S, Kirilovsky D. Identification of a protein required for recovery of full antenna capacity in OCP-related photoprotective mechanism in cyanobacteria. Proc Natl Acad Sci USA. 2010;107(25):11620–11625. doi: 10.1073/pnas.1002912107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kerfeld CA, Kirilovsky D. Structure, function and evolution of OCP-based photoprotection in cyanobacteria. In: Chauvat F, Chauvat CC, editors. Genomics of Cyanobacteria. San Diego: Elsevier; 2013. pp. 1–26. [Google Scholar]
- 10.Gwizdala M, Wilson A, Omairi-Nasser A, Kirilovsky D. Characterization of the Synechocystis PCC 6803 Fluorescence Recovery Protein involved in photoprotection. Biochim Biophys Acta. 2013;1827(3):348–354. doi: 10.1016/j.bbabio.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 11.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372(3):774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 12.Wang C, Bradley P, Baker D. Protein-protein docking with backbone flexibility. J Mol Biol. 2007;373(2):503–519. doi: 10.1016/j.jmb.2007.07.050. [DOI] [PubMed] [Google Scholar]
- 13.Chaudhury S, et al. Benchmarking and analysis of protein docking performance in Rosetta v3.2. PLoS One. 2011;6(8):e22477. doi: 10.1371/journal.pone.0022477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chábera P, Durchan M, Shih PM, Kerfeld CA, Polívka T. Excited-state properties of the 16kDa red carotenoid protein from Arthrospira maxima. Biochim Biophys Acta. 2011;1807(1):30–35. doi: 10.1016/j.bbabio.2010.08.013. [DOI] [PubMed] [Google Scholar]
- 15.Wiley DC, Skehel JJ. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu Rev Biochem. 1987;56:365–394. doi: 10.1146/annurev.bi.56.070187.002053. [DOI] [PubMed] [Google Scholar]
- 16.Gwizdala M, Wilson A, Kirilovsky D. In vitro reconstitution of the cyanobacterial photoprotective mechanism mediated by the Orange Carotenoid Protein in Synechocystis PCC 6803. Plant Cell. 2011;23(7):2631–2643. doi: 10.1105/tpc.111.086884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorbunov MY, Kuzminov FI, Fadeev VV, Kim JD, Falkowski PG. A kinetic model of non-photochemical quenching in cyanobacteria. Biochim Biophys Acta. 2011;1807(12):1591–1599. doi: 10.1016/j.bbabio.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 18.Wilson A, et al. The essential role of the N-terminal domain of the orange carotenoid protein in cyanobacterial photoprotection: Importance of a positive charge for phycobilisome binding. Plant Cell. 2012;24(5):1972–1983. doi: 10.1105/tpc.112.096909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64(Pt 1):112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 20.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68(Pt 4):352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 22.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schneider TD, Stephens RM. Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 1990;18(20):6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson A, et al. A photoactive carotenoid protein acting as light intensity sensor. Proc Natl Acad Sci USA. 2008;105(33):12075–12080. doi: 10.1073/pnas.0804636105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.