Abstract
Background: Epidemiologic studies have suggested that most cases of sporadic colon cancer can be attributed to diet. The recognition that colonic microbiota have a major influence on colonic health suggests that they might mediate colonic carcinogenesis.
Objective: To examine the hypothesis that the influence of diet on colon cancer risk is mediated by the microbiota through their metabolites, we measured differences in colonic microbes and their metabolites in African Americans with a high risk and in rural native Africans with a low risk of colon cancer.
Design: Fresh fecal samples were collected from 12 healthy African Americans aged 50–65 y and from 12 age- and sex-matched native Africans. Microbiomes were analyzed with 16S ribosomal RNA gene pyrosequencing together with quantitative polymerase chain reaction of the major fermentative, butyrate-producing, and bile acid–deconjugating bacteria. Fecal short-chain fatty acids were measured by gas chromatography and bile acids by liquid chromatography–mass spectrometry.
Results: Microbial composition was fundamentally different, with a predominance of Prevotella in native Africans (enterotype 2) and of Bacteroides in African Americans (enterotype 1). Total bacteria and major butyrate-producing groups were significantly more abundant in fecal samples from native Africans. Microbial genes encoding for secondary bile acid production were more abundant in African Americans, whereas those encoding for methanogenesis and hydrogen sulfide production were higher in native Africans. Fecal secondary bile acid concentrations were higher in African Americans, whereas short-chain fatty acids were higher in native Africans.
Conclusion: Our results support the hypothesis that colon cancer risk is influenced by the balance between microbial production of health-promoting metabolites such as butyrate and potentially carcinogenic metabolites such as secondary bile acids.
INTRODUCTION
There are wide geographic variations in colorectal incidence around the world, and most of these differences have been attributed to diet (1). Within the continental United States, the African American population shoulders the major burden, with an incidence of ∼65:100,000 and a death rate of 25:100,000 (2). In sharp contrast, rural Africans rarely get the disease (3). Studies of ours have ascribed this difference to higher meat and fat intakes in Americans and to higher resistant starch intakes in Africans (4).
Colonic microbiota are dependent on dietary residues that escape small intestinal digestion and absorption. Consumption of a normal balanced diet predominantly yields carbohydrate residues such as fiber, which stimulates saccharolytic fermentation and the production of the health-promoting short-chain fatty acids (SCFAs)5 acetate, propionate, and butyrate. Butyrate is the preferred energy source for the colonic mucosa, and all 3 SCFAs have antiinflammatory and antiproliferative properties (5). Consumption of an unbalanced diet rich in meat and low in fiber increases the delivery of proteinaceous residues, which promote proteolytic fermentation with the production of ammoniac compounds and branched-chain fatty acids, which are inflammatory and may enhance colon cancer risk (5, 6).
The influence of dietary fat on cancer risk may also be determined by microbial metabolism, because it increases the hepatic synthesis of bile acids (BAs) and the quantity of BAs that escape the enterohepatic circulation and enter the colon. This provides substrate for microbes with 7α-dehydroxylating enzymes, which convert primary BA into secondary BAs, which are proinflammatory and have carcinogenic properties (7).
Digestion of food is fundamentally different in the small and large intestine. In the small intestine, the enzymic digestion rate is determined by substrate concentrations, according to the Michaelis-Menten equation. In the colon, the fermentation rate is complex, termed autocatalytic, and is determined by using both the substrate concentration and the microbe concentration. In autocatalytic reactions, the maximal rate of reaction occurs at an intermediate, rather than at the highest, reactant concentration (8). Thus, SCFA and secondary BA production is codetermined by the microbiota composition.
To test our hypothesis that the higher risk of colon cancer in African Americans than in native Africans is related to the influence of their diet on the microbiota composition and metabolic activity, we measured the differences in microbiota composition and specific bacteria known to influence SCFA and secondary BA production in fecal samples from these 2 populations.
SUBJECTS AND METHODS
Study design
The relative contents of SCFAs, BAs, and microbes of specific interest were measured in fresh fecal samples from 2 populations of varying colon cancer risk, namely high-risk African Americans (Americans) and low-risk rural Africans (Africans) (3). Middle-aged subjects were selected because colon cancer affects that group most. Microbial analysis was first untargeted, based on high-throughput 16S ribosomal RNA (rRNA) pyrosequencing, and secondly targeted based on quantitative polymerase chain reaction (qPCR) to measure numbers of microbes of specific interest, which included the major butyrate producers Faecalibacterium prausnitzii, Clostridium cluster IV, and XIVa (9); the major starch fermenters Succinivibrio spp. and Prevotella spp. (10); the Bacteroides fragilis group, lactic acid bacteria, and Lactobacillus spp. (11); and Bifdobacterium spp. (12). Finally, a functional gene analysis was performed to compare the potential for butyrogenesis, methanogenesis, hydrogen sulfide production, and secondary bile salt conversion.
Study populations
Normal healthy volunteers of either sex aged 50–65 y were selected on the basis of their medical history and results of a medical examination. African Americans rather than white Americans were chosen because their risk of colon cancer is higher and there is more genetic similarity. Subjects with a history of gastrointestinal disease or surgery were excluded, as were those with a history of antibiotic use within the past 6 wk. First-morning fecal samples were collected from 12 healthy Americans in the Pittsburgh area and from 12 age-, sex-, and BMI-matched Africans from a rural area outside the town of Empangeni in the KwaZulu-Natal Province of South Africa (Table 1). Our earlier study showed that the dietary intake patterns of these 2 populations were widely different, with Americans consuming twice as much protein and 3 times as much fat [mean (±SEM) protein: 94 ± 9 compared with 58 ± 4 g/d; mean (±SEM) fat: 114 ± 11 compared with 38 ± 3 g/d] (4). The protocol was reviewed and approved by the Institutional Review Boards of the University of Pittsburgh and the University of KwaZulu-Natal (Biomedical Research Ethics Committee).
TABLE 1.
Real-time quantitative polymerase chain reaction primers used to determine the density of specific microbes and functional microbial genes
| Target group and primer | Sequence (5′-3′) | Annealing temperature | Reference |
| °C | |||
| All bacteria | 60 | (13) | |
| Uni331F | TCCTACGGGAGGCAGCAGT | ||
| Uin797R | GGACTACCAGGGTATCTATCCTGTT | ||
| Butyrate-production gene (BcoA) | 60 | (14) | |
| BcoA-F | GCIGAICATTTCACITGGAAYWSITGGCAYATG | ||
| BcoA-R | CCTGCCTTTGCAATRTCIACRAANGC | ||
| Clostridium cluster IV1 | 50 | (15) | |
| Sg-Clept-F | GCACAAGCAGTGGAGT | ||
| Sg-Clept-R3 | CTTCCTCCGTTTTGTCAA | ||
| Costridium cluster XIVa1 | 50 | (15) | |
| g-Ccoc-F | AAATGACGGTACCTGACTAA | ||
| g-Ccoc-R | CTTTGAGTTTCATTCTTGCGAA | ||
| Bifidobacterium spp. | 60 | (16) | |
| Bif164F | GGGTGGTAATGCCGGATG | ||
| Bif662R | CCACCGTTACACCGGGAA | ||
| Succinivibrio | 60 | (17) | |
| SucDex1F | CGTCAGCTCGTGTCGTGAGA | ||
| SucDex1R | CCCGCTGGCAACAAAGG | ||
| Prevotella spp. | 60 | (17) | |
| PreGen4F | GGTTCTGAGAGGAAGGTCCCC | ||
| PreGen4R | TCCTGCACGCTACTTGGCTG | ||
| Bacteroides fragilis group | 60 | (15) | |
| g-Bfra-F | ATAGCCTTTCGAAAGRAAGAT | ||
| g-Bfra-R | CCAGTATCAACTGCAATTTTA | ||
| Lactobacillus spp. | 60 | (18) | |
| LactoF | TGGAAACAGRTGCTAATACCG | ||
| LactoR | GTCCATTGTGGAAGATTCCC | ||
| Faecalibacterium prausnitzii | 60 | (19) | |
| FpF | CCCTTCAGTGCCGCAGT | ||
| FpR | GTCGCAGGATGTCAAGAC | ||
| Functional gene for hydrogen sulfide | 60 | ||
| DSR1F+ | ACSCACTGGAAGCACGGCGG | (20) | |
| DSR-R | GTGGMRCCGTGCAKRTTHG | (20) | |
| Functional gene for methanogenesis | 60 | ||
| mcrA-F | TTCGGTGGATCDCARAGRGC | (21) | |
| mcrA-R | GBARGTCGWAWCCGTAGAATCC | (21) | |
| Functional gene for secondary bile acids | 60 | ||
| BaiCD-F | CAGCCCRCAGATGTTCTTTG | (unpublished observation)2 | |
| BaiCD-R | GCATGGAATTCWACTGCYTC |
Number indicates phylogenetic cluster of Clostridium as defined by Collins et al (22).
J Ou, A Heather, J Riddlon, S Curry, and SJD O'Keefe, April 2012.
Materials
All BA and SCFA analytic chemicals were purchased from Sigma-Aldrich. qPCR primers for different bacteria were synthesized by Sigma-Aldrich. qPCR master mix was purchased from Applied Biosystem. An Econo-Cap EC-1000 gas chromatographic capillary column was purchased from Grace Davison Discovery Science. Millex-GS 0.22-μm syringe filters were purchased, and microconcentrators were purchased from Millipore. The Luna C18 column (3 μm, 2.0-mm internal diameter × 150 mm) was purchased from Phenomenex.
Fecal collection, preservation, and transport
Freshly voided fecal samples were collected immediately into airtight plastic vials and were transported on ice to be stored frozen at −80°C within 2 h. The samples collected in South Africa were air-couriered frozen on dry ice to Pittsburgh for analysis.
SCFA assay
Fecal samples (0.1 g) were transferred into plastic tubes; 2,2-dimethylbutyric acid was added at 1 mmol/L as internal standard. After undergoing vortex mixing and centrifugation (1900 × g, 10 min), the supernatant fluid was filtered through a Millex-GS 0.22-μm syringe filter unit (Millipore). The solution was refiltered through a microconcentrator (Ultracel YM-10; Millipore) with a molecular mass cutoff of 10,000 Da, by centrifugation (7000 × g at 4°C for 1.5 h). The filtrate was then analyzed by using an Agilent Technologies 6890N Network GC System with a flame-ionization detector for SCFA based on the method described by Scheppach et al (23). Compounds were separated on a Grace EC-1000 (15 m in length, 1.20-μm film thicknesses, 0.53 mm internal diameter) capillary column (Grace Davison Discovery Science). The oven temperature of the gas chromatograph was programmed at 5 min from 80°C to 175°C, which was held for 10 min with a total running time of 25 min. The temperatures of both the detector and injector were 200°C. The inlet was operated in a splitless mode. High-purity helium was used as carrier gas.
A mixed-SCFA standard solution was prepared by using high purity (>99%) reagents (Sigma). SCFA concentrations were computed by using a peak area ratio of the sample profile relative to the internal standard. A good linear correlation was found between the peak area ratio and the corresponding standard SCFA (r2 > 0.99 for all SCFAs). The interday and intraday CVs ranged from 2.4% to 3.9%. This method does not separate 2-methylbutyric and isovaleric acids.
BA assay
Fecal BA concentrations were measured by using the method described by Tagliacozzi et al (24), except that quantification was carried out by using liquid chromatography (LC)–mass spectrometry (MS) as opposed to LC-tandem MS. A 125-μL colonic evacuate was mixed with 400 μL acetonitrile, followed by 1 min of vortex mixing. After 15 min of centrifugation at 13,000 × g, 450 μL supernatant fluid was transferred to an autosampler vial and blown to dryness with nitrogen. The residue was dissolved with 125 μL methanol and water (1:1). Ten microliters of this solution was injected into a Shimadzu HPLC-MS (model 2010A) for quantification by using electrospray ionization in negative ion mode by monitoring the (M-H)− ion. The analytic conditions for LC-MS were as follow: column, Luna 3u, C18, 100A (2.0-mm internal diameter × 150 mm; Phenomenex); mobile phase A: 20% acetonitrile-water containing 10 mmol ammonium acetate/L; mobile phase B: 80% acetonitrile-water, gradient program; mobile phase B: 0–6 min 15%, 20 min 30%, 30 min 60%, 40 min 80%, 45 min 15%; flow rate: 0.2 mL/min; column temperature: 40°C; probe voltage: 4.5 kv; curved desolvation line temperature: 230°C. BA concentrations were calculated based on standard curves run with each sample set.
Microbial identification
DNA isolation
Fecal bacterial DNA was isolated and purified with the QIAamp DNA Stool Mini Kit (Qiagen) in combination with a bead-beating step (30 s at 30 Hz 3 times) by using the FastPrep-24 System (MP Biomedicals), as described by Zoetendal et al (25).
Microbiota composition
Samples for 454 FLX pyrosequencing were amplified with universal forward 519F (5′-Fusion A-Barcode -CAGCMGCCGCGGTAATWC-3′) and reverse 926R (5′-Fusion B-Barcode- CCGTCAATTCMTTTRAGTT-3′) primer pairs (Roche). PCR reaction mixtures were set up with the TopTaq PCR kit (Qiagen) according to the manufacturer's recommendations with 10 pmol/L each of forward and reverse primers in 25 μL reaction. Amplification was carried out with 30 cycles of thermal program (denaturation, 95°C for 30 s; annealing, 55°C for 45 s; and extension, 72°C for 60 s). All amplicons were gel-excised, concentrated, and purified with the Gel extraction kit (Qiagen), and amplicon concentrations were measured with a Qubit fluorometer (Invitrogen). A 454 FLX Titanium was used for 454 pyrosequencing (454 Life Sciences; Roche Applied Science). The paired-end pyrosequencing services were provided by Roy J Carver Biotechnology Center, University of Illinois. A total of 248,594 16S rRNA sequences (also referred to as 16S pyrotags) were obtained from the 454 Titanium pyrosequencing run. The 16S pyrotags were sorted based on their respective barcodes and handled by using the QIIME pipeline (26). RDP Classifier was used for taxonomic assignments of the aligned 16S pyrotags at the 95% confidence level (27).
Real-time quantitative PCR
qPCR was performed with a 7900HT Fast Real-Time PCR System (Applied Biosystem). 16S rRNA gene-specific primers were used to target total and specific bacteria (F. prausnitzii, Clostridium cluster IV and XIVa, Lactobacillus spp., Succinivibrio spp., Prevotella spp., B. fragilis group, and Bifdobacterium spp.) (Table 1). Cloned 16S rRNA genes were used to construct standard curves: Clostridium leptum 29065 (representing Clostridium cluster IV), Clostridium coccoides (representing Clostridium cluster XIVa), Clostridium sindense, Bifidobacterium longum 15707, Lactobacillus delbrueckii 12315, Succinivibrio dextrinosolvens 19716, Prevotella ruminicola 19189, B. fragilis 25285, and F. prausnitzii 27766 (also a member of Clostridium cluster IV) (obtained from the American Type Culture Collection) were cultured on reinforced clostridial medium broth and incubated at 37°C in an anaerobic chamber. Genomic DNA was extracted from a 2-mL culture by using the QIAamp DNA stool mini kit (Qiagen), and bacterial 16S rRNA genes were amplified with their respective primers (Table 2). PCR products were purified by using the MinElute PCR purification Kit (Qiagen) and cloned into pCR 2.1 TOPO vector with a TOPO-TA cloning kit (Invitrogen). Plasmid DNA was isolated with a QIAprep Spin Miniprep kit (Qiagen), and plasmid DNA concentrations were measured spectrophotometrically (NanoDrop 1000; Thermol Scientific). The number of target gene copies was calculated from the mass of DNA with consideration of the size of insert and plasmid. The plasmid standard for sulfate-reducing bacteria was obtained from a previous study (28).
TABLE 2.
Demographic data of the research subjects
| Female | Male | Age | Weight | BMI | |
| y | kg | kg/m2 | |||
| Native Africans | 8 | 4 | 57 ± 1.91 | 80 ± 4.7 | 32 ± 2.4 |
| African Americans | 9 | 3 | 58 ± 2.5 | 74 ± 4.0 | 28 ± 1.8 |
Mean ± SEM (all such values).
Functional microbial genes
In the context of microbial metabolite production, analysis of functional genes rather than taxonomic groups based on the 16S rRNA gene allows better quantification (29). Thus, we also examined differences in the abundance of genes encoding the enzymes responsible for butyrate production, secondary BA synthesis, methanogenesis, and hydrogen sulfide production. The butyryl-coenzyme-A-CoA transferase (BcoA) gene was used for quantification of butyrate producers (14). For secondary BA conversion potential, we measured the baiCD gene, which encodes the enzyme that dehydroxylates the 7α-hydroxy group in primary BAs to form secondary BAs (30). To compare methanogenic potential, we measured the gene that encodes the enzyme methyl coenzyme-M reductase (mcrA), which catalyzes the crucial removal of hydrogen produced from fermentation into methane (31). Finally, we measured the gene encoding the enzyme dissimilatory (bi)sulfite reductase (dsrA) that catalyzes a step in the reduction of inorganic sulfate to hydrogen sulfide (32).
The primer sequences are listed in Table 1. All PCR experiments were done in triplicate with a reaction volume of 10 μL by using MicroAmp optical 384-well reaction plates sealed with MicroAmp optical adhesive film (Applied Biosystems). Each reaction contained 5 μL 2 Power SYBR Green PCR Master mix (Applied Biosystems), 1 μL bovine serum albumin (New England Biolabs) at 1 mg/mL (final concentration: 100 μg/mL), 0.5 μM of each primer, and 2 μL template DNA. The cycling conditions were as follows: 50°C for 2 min and 95°C for 10 min followed by 40 cycles of 95°C for 15 s, primer-specific annealing temperature (Table 1) for 20 s, and 72°C for 45 s. After amplification, a dissociation step was included to analyze the melting profile of the amplified products. Ten-fold dilution series of the plasmid standard for the respective bacterial group or species were run along with the samples. Sample DNA concentrations were calculated by using standard curves of the diluted standards containing the respective gene target for each set of primers. Data analysis was processed with SDS v2.3 software supplied by Applied Biosystems.
Power SYBR Green PCR Master mix (Applied Biosystems), 1 μL bovine serum albumin (New England Biolabs) at 1 mg/mL (final concentration: 100 μg/mL), 0.5 μM of each primer, and 2 μL template DNA. The cycling conditions were as follows: 50°C for 2 min and 95°C for 10 min followed by 40 cycles of 95°C for 15 s, primer-specific annealing temperature (Table 1) for 20 s, and 72°C for 45 s. After amplification, a dissociation step was included to analyze the melting profile of the amplified products. Ten-fold dilution series of the plasmid standard for the respective bacterial group or species were run along with the samples. Sample DNA concentrations were calculated by using standard curves of the diluted standards containing the respective gene target for each set of primers. Data analysis was processed with SDS v2.3 software supplied by Applied Biosystems.
Statistical analysis
Statistical analysis was conducted by using SPSS 16.0 (SPSS Inc). The significance of group differences for normally distributed data was assessed with Student's t test. The nonparametric data were analyzed with a Mann-Whitney U test. The significance of the association was evaluated with Spearman's rank correlation test. A level of P ≤ 0.05 was accepted as statistically significant. Data are presented as means ± SEs. Pyrosequencing data were analyzed by several multivariate ordinations (principal component analyses, nonmetric multidimensional scaling), Kruskal-Wallis independent tests, and multivariate ANOVA with Bonferroni correction.
RESULTS
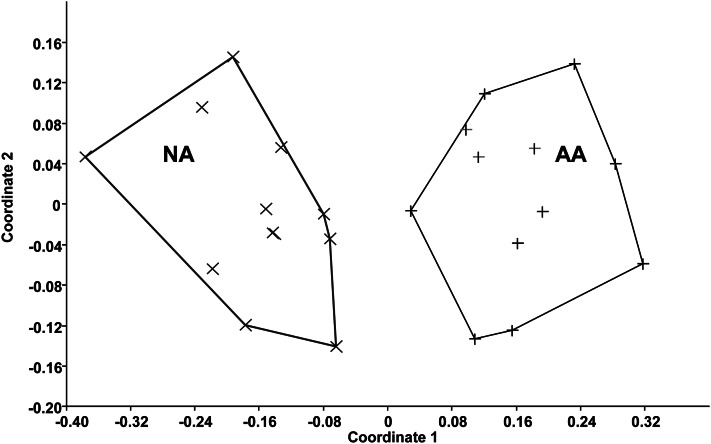
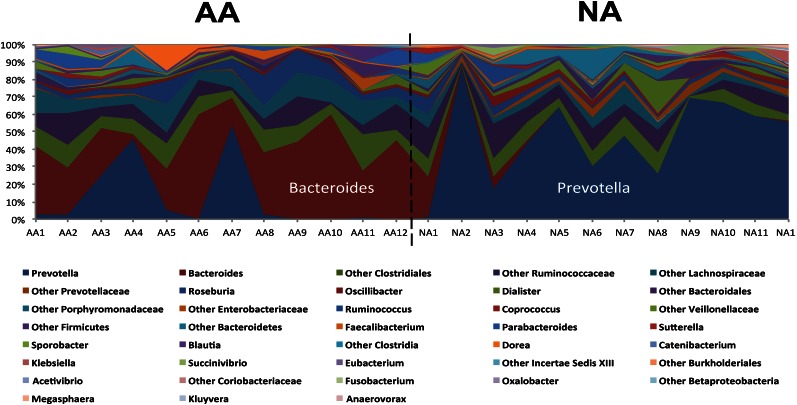
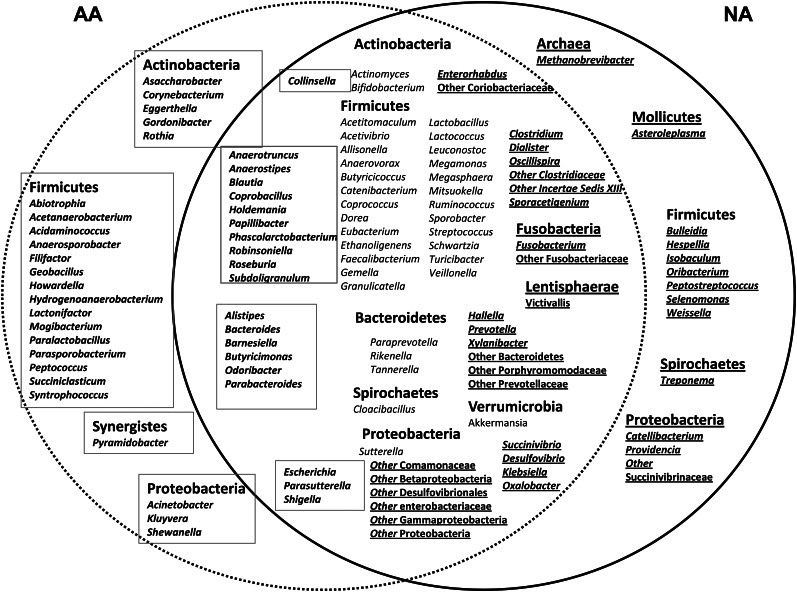
Body weight and BMI were similar between the 2 groups (Table 2). Four of the 12 Americans were obese [ie, BMI (in kg/m2) >30], as were 5 of the 12 Africans. The composition of the fecal microbiota, as shown by pyrosequencing analysis, was fundamentally different between Africans and Americans, as summarized in Figure 1. Nonmetric multidimensional scaling showed an unequivocal distinction (multivariate ANOVA: P < 0.01) between the 2 groups based on their fecal microbiota (Figure 1). This difference appeared to override interindividual differences between the groups shown in Figure 2. The most distinct feature was a predominance of Prevotella species in most Africans and Bacteroides in most Americans (Figure 2). Thus, Africans correspond to enterotype 1 and Americans to enterotype 2. Africans also had higher proportions of Succinivibrio and Oscillospira—microbes that might be involved, such as Prevotella—in starch, hemicellulose, and xylan degradation (Figure 3) (33, 34). The African gut microbiota were also characterized by a large number of sequences that could not be affiliated to reference taxa. Less than one-third of the taxa was detected in comparable abundance between the groups. The American gut microbiota were characterized by a greater abundance of potentially pathogenic proteobacteria (Escherichia, Acinetobacter). Interestingly, the American gut microbiota were more diverse (Simpson index: 0.7 compared with 0.8; P = 0.04), which presumably reflects consumption of a more diversified diet.
FIGURE 1.
Illustration of the marked phylogenic differences in microbiota composition between AAs (n = 12) and NAs (n = 12) detected by 16S-rRNA-based taxonomic pyrosequencing. Nonmetric multidimensional scaling shows strong clustering (multivariate ANOVA: P < 0.01) according to ethnic group. AA, African American; NA, native African; rRNA, ribosomal RNA.
FIGURE 2.
Composition was dominated by Bacteroides in the 12 AAs, which indicated that they belonged to enterotype 1, and was dominated by Prevotella in the 12 NAs, which categorized them as enterotype 2 (10). AA, African American; NA, native African.
FIGURE 3.
Illustration of the similarities and differences in fecal microbial taxa between AAs and NAs. The solid circle encloses the taxa that were detected in NA and the dotted circle those identified in AA. The overlap between the 2 circles contains taxa common to both populations. Much of the shared taxa were significantly (independent Kruskal-Wallis tests) more enriched in one group than in the other, indicated by the boxed text in AAs on the left and underlined text in NAs on the right. AA, African American; NA, native African.
The targeted qPCR analysis showed that total bacteria, Prevotella spp., Succinivibrio spp. F. prausnitzii, Clostridium cluster IV, and Clostridium cluster XIVa bacterial counts were all significantly higher in the fecal samples from Africans, but Lactobacillus spp. were more abundant in Americans (Table 3). With regard to functional gene analysis, the gene-encoding secondary BA production (baiCD) was detected in greater abundance in African Americans, whereas the functional genes targeting butyrate producers (BcoA), methane producers (mcrA), and hydrogen sulfide producers (dsrA) were more abundant in native Africans.
TABLE 3.
Bacterial abundance in fecal samples from native Africans and African Americans1
| Native African | African American | P value | |
| gene copies/g feces | gene copies/g feces | ||
| Total bacteria | 7.1×1011 ± 4.5×1010* | 1.8×1011 ± 6.3×1010 | 0.046 |
| Butyrate-production gene (BcoA) | 5.4×1010 ± 1.1×1010* | 1.4×1010 ± 2.7×109 | 0.022 |
| Faecalibacterium prausnitzii | 1.6×109 ± 2.7×108* | 8.3×108 ± 2.7×108 | 0.038 |
| Clostridium cluster IV | 5.1×109 ± 8.7×108* | 2.9×109 ± 7.3×108 | 0.032 |
| Clostridium cluster XIVa | 9.5×109 ± 1.7×109* | 5.1×109 ± 9.3×108 | 0.049 |
| Succinivibrio spp. | 1.1×108 ± 6.4×107* | 4.5×106 ± 1.3×106 | 0.021 |
| Prevotella spp. | 8.2×1010 ± 1.4×1010* | 3.5×1010 ± 6.4×109 | 0.011 |
| Bacteroides fragilis group | 5.7×109 ± 1.6×106 | 4.4×1010 ± 1.3×107 | 0.177 |
| Lactobacillus spp. | 5.9×106 ± 3.4×106* | 7.1×107 ± 5.7×107 | 0.021 |
| Bifidobacterium spp. | 6.8×107 ± 2.8×107 | 3.0×108 ± 1.2×108 | 0.291 |
| Hydrogen sulfide producers | 1.1×107 ± 1.5×107* | 4.1×106 ± 1.4×106 | 0.048 |
| Methane producers | 3.1×106 ± 2.1×106 | UDL2 | |
| Secondary bile acid producers | 2.2×107 ± 5.3×106* | 4.7×108 ± 1.6×108 | 0.037 |
Values are means ± SEMs. *Mann-Whitney U test; the sample number was 12 for each group.
UDL, under detection limit.
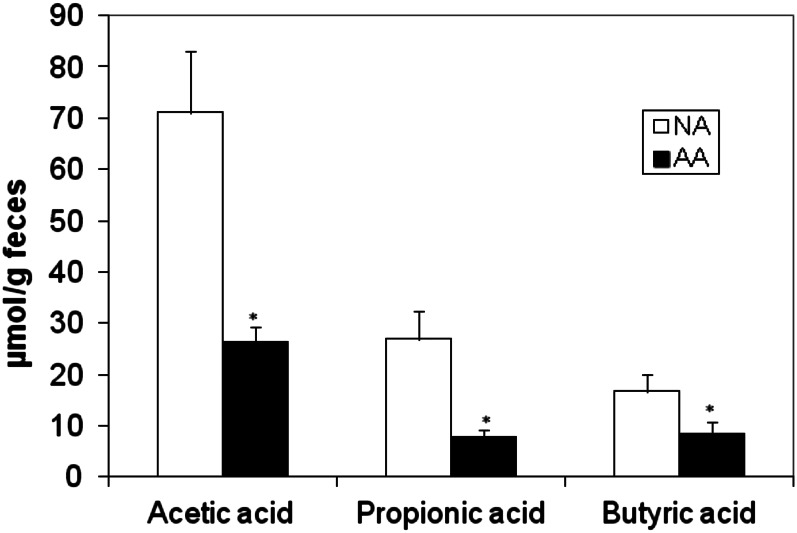
The chief SCFA products of saccharolytic fermentation, acetate, propionate, and butyrate were significantly higher in stool samples from Africans (P < 0.05; Figure 4), whereas the products of proteolytic fermentation, namely the branched SCFAs isobutyric and 2-methylbutyric/isovaleric acids, were significantly higher in Americans (isobutyrate: 1.73 ± 0.26 compared with 1.22 ± 0.24 μmol/g feces, P = 0.02; 2-methylbutyric/isovaleric acid: 1.49 ± 0.19 compared with 0.33 ± 0.19 μmol/g feces, P = 0.0002; Mann-Whitney U test).
FIGURE 4.
Summary of the differences in mean (±SE) group concentrations of the major short-chain fatty acids in fecal samples. Concentrations were significantly greater in NAs (n = 12) than in AAs (n = 12): Mann-Whitney U test for acetate (P = 0.001), propionate (P = 0.003), and butyrate (P = 0.049). AA, African American; NA, native African.
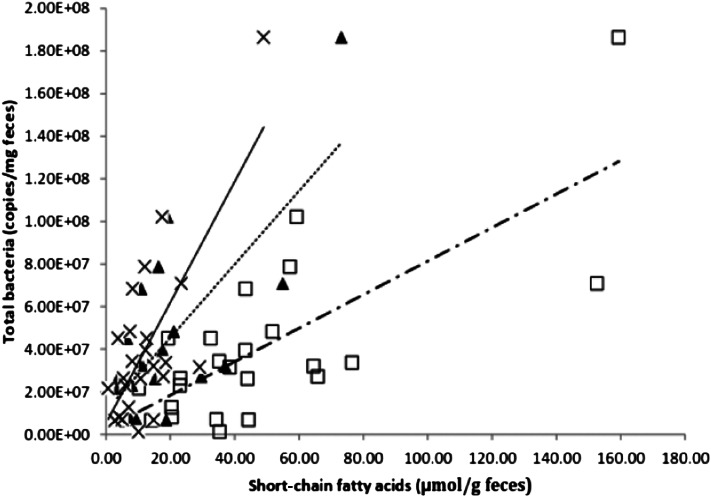
To assess the relation between production of SCFAs and the abundance of specific microbial groups, correlation analyses of fecal components were performed. The results showed that the total bacterial abundance was significantly correlated (P < 0.05; Table 4) with concentrations of stool acetate (r2 = 0.38), propionate (r2 = 0.33), and butyrate (r2 = 0.37) (Figure 5). Second, stool butyrate concentrations were significantly correlated with the abundance of the butyrate producers Clostridium cluster IV (r2 = 0.29, P = 0.047) and Clostridium cluster XIVa (r2 = 0.29, P = 0.047) (Table 4).
TABLE 4.
Nonparametric Spearman correlations (R) between fecal short-chain fatty acid concentrations and copy numbers of bacteria1
| Total SCFAs |
Acetic acid |
Propionic acid |
Butyric acid |
|||||
| R | P | R | P | R | P | R | P | |
| Total bacteria | 0.37 | 0.011 | 0.38 | 0.010 | 0.33 | 0.026 | 0.37 | 0.042 |
| Bifidobacterium spp. | −0.23 | 0.112 | −0.27 | 0.066 | −0.17 | 0.234 | −0.19 | 0.197 |
| Faecalibacterium prausnitzii | −0.28 | 0.215 | −0.17 | 0.105 | −0.27 | 0.265 | −0.20 | 0.246 |
| Clostridium IV | 0.26 | 0.074 | 0.25 | 0.083 | 0.17 | 0.234 | 0.29 | 0.047 |
| Clostridium XIVa | 0.26 | 0.074 | 0.27 | 0.066 | 0.19 | 0.197 | 0.29 | 0.047 |
Sample number = 24. SCFA, short-chain fatty acid.
FIGURE 5.
The abundance of total bacteria correlated significantly (Spearman's test) with the concentrations of acetate (□, dashed and dotted line; r2 = 0.38, P = 0.033), propionate (▴, dashed line; r2 = 0.33, P = 0.039), and butyrate (×, continuous line; r2 = 0.37, P = 0.042) in fecal samples from African Americans and native Africans (n = 24).
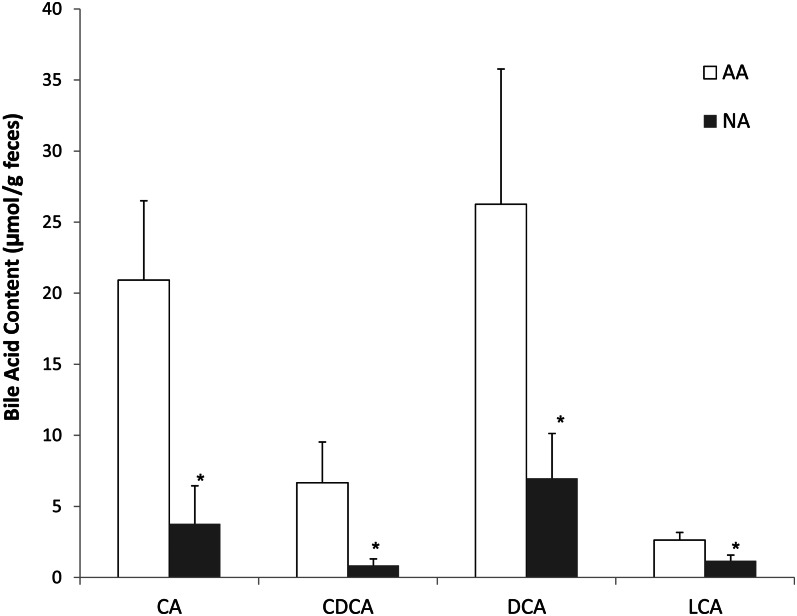
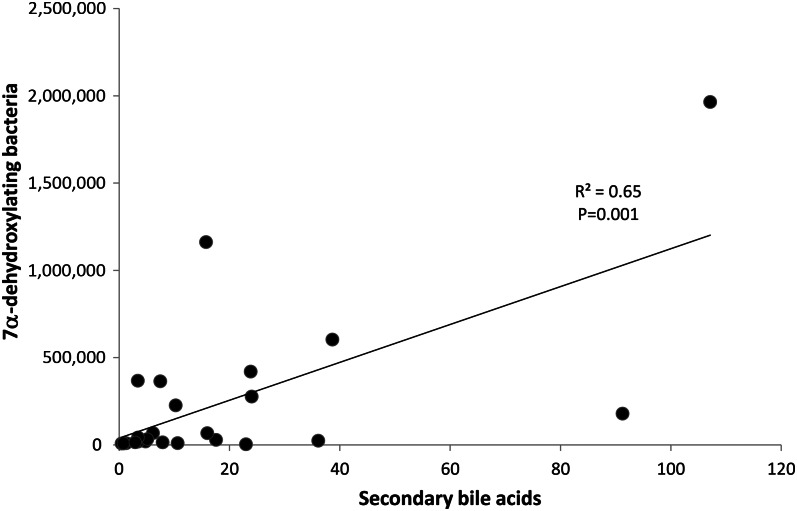
Amounts of the 4 major BAs in feces were significantly higher (P < 0.05) in Americans than in Africans (Figure 6). The fecal primary BAs cholic acid (CA) and chenodeoxycholic acid and the secondary BAs deoxycholic acid (formed from CA) and lithocholic acid (LCA, formed from CA and chenodeoxycholic acid) were significantly higher (P < 0.05) in Americans than in Africans (Figure 6). The ratio of butyrate to LCA was significantly higher in Africans (39.2 ± 19.0 compared with 6.0 ± 3.2; P = 0.047). A significant correlation was also found between 7α-dehydroxylating bacteria and secondary BA concentrations in stool (r2 = 0.65, P = 0.01; Spearman's test) as well (Figure 7), which likely indicated a higher microbial 7α-dehydroxylating capacity for colonic BAs in the Americans.
FIGURE 6.
Contents of major bile acids in feces were significantly lower in NAs (n = 12) than in AAs (n = 12): Mann-Whitney U test for CA (P = 0.027), CDCA (P = 0.016), DCA (P = 0.043), and LCA (P = 0.031). *P < 0.05 compared with AA. AA, African American; CA, cholic acid; CDCA, chenodeoxycholic acid; DCA, deoxycholic acid; LCA, lithocholic acid; NA, native African.
FIGURE 7.
The abundance of 7α-dehydroxylating bacteria, estimated from detection of their functional genes by polymerase chain reaction, correlated significantly with the concentration of secondary bile acids in fecal samples (n = 24). r2 = 0.65, P = 0.001 (Spearman's test).
DISCUSSION
The findings of the current study support the hypothesis that the balance between health-promoting and inflammatory microbial metabolites may determine colon cancer risk and that their production is dependent on both the composition of food and composition of the microbiota, as in an autocatalytic system. The data provide evidence that saccharolytic fermentation was lower, proteolytic fermentation was higher, and secondary BA production was higher in African Americans than in Africans. From our earlier studies in the same populations, it is reasonable to postulate that these differences can be ascribed to differences in diet, with African Americans eating more dietary meat and fat and less complex carbohydrate and fiber (4).
Fecal microbial composition was shown to be very different in Africans and African Americans, with the pattern in the former corresponding to enterotype 2 and the latter to enterotype 1. Three enterotypes were recently described by a European consortium based on their pooled analysis of fecal samples obtained from healthy adults from 4 European countries, the United States, and Japan (10). From this, they proposed that all human populations could be categorized into 1 of 3 enterotypes depending on their common networks of co- and anticorrelating genera, which are driven by the genus Bacteroides (type 1), Prevotella (type 2), or Ruminococcus (type 3). Interestingly, the composition in our African sample was remarkably similar to that shown in Central African children from Burkina Faso by De Filippo et al (35), dominated by Prevotella (type 2) and showing an overabundance of microbes that are likely involved in starch and cellulose degradation. They related these differences to differences in diet, with African children consuming a diet rich in coarse grains and vegetables that was somewhat similar to what Burkitt described as the “traditional African diet” that was associated with a low risk of colonic chronic diseases and colon cancer (36). Importantly, butyrate and the other SCFAs were also higher in the African children—as in our study; the authors proposed that this might help reduce the risk of enteric infections.
Interestingly in the current study, whereas the higher proportions of Prevotella, Succinivibrio, and Oscillospira in the African gut microbiome also represented higher numbers of these microbes in stool samples, as measured by qPCR, the higher compositional representation of bacteroides in African Americans did not appear to reflect higher numbers. It is possible that the total numbers of bacteroides were underestimated because the qPCR assay used targeted the B. fragilis group and not all human Bacteroides strains. On the other hand, the assay does include the 2 major human gut species, namely B. fragilis and Bacteroides thetaiotamicron (15). Presumably, the greater numbers of total bacteria in the Africans was a consequence of the expansion of populations of microbes that degrade starches, hemicelluloses, and xylans—notably Prevotella, Succinivibrio, and Oscillospira—and those that ferment their products.
Our targeted microbe analysis showed that both the butyrate production gene and the recognized major butyrate-producing bacteria, including F. prausnitzii, and those contained within Clostridium cluster IV and cluster XIVa were more abundant in stool samples from native Africans. Evidence that these differences were of functional significance was provided by our observation of positive correlations between the abundance of butyrate producers and butyrate concentrations in stool samples. Early cultural and molecular studies from Flint's group (37, 38) in Aberdeen, Scotland, indicated that the most numerous butyrate-producing bacteria found in human feces were highly oxygen-sensitive anaerobes belonging to the Clostridium clusters IV and XIVa. Their studies also highlighted cross-feeding between bacteria, with most butyrate producers consuming acetate produced by other microbes. Higher rates of fermentation in Africans might also have been facilitated by the higher numbers of the hydrogen-utilizing microbes, ie, sulfate reducers and methanogens, measured in stool samples, because it has been shown that the removal of the end product hydrogen increases fermentation potential (39).
Butyrate, propionate, and acetate have all have been shown in experimental models to have antineoplastic properties, but butyrate appears to be the most potent (5). Butyrate has a unique role in the maintenance of colonic mucosal health: first, because of its position as the preferred energy source, and second, because of its protean antineoplastic properties. A wealth of experimental evidence, recently reviewed by ourselves (40, 41), indicates the inhibitory effect of butyrate on tumorigenesis, possibly mediated by its antiinflammatory and immunomodulatory effects and downregulation of the key canonical Wnt-signaling pathway linked to colonic carcinogenesis (42). Butyrate may also play a role in primary prevention through the activation of different drug-metabolizing enzymes. This can reduce the burden of carcinogens, such as BAs, and therefore decrease the number of mutations, which reduces cancer risk. Experimental studies have shown that it regulates colonic epithelial growth and protects against carcinogenesis by inhibiting the proliferation and migration of neoplastic cells, restricting tumor angiogenesis, inducing apoptosis, and promoting differentiation of the neoplastic colonocytes (43–49). F. prausnitzii, a member of Clostridium cluster IV, is one of the most dominant butyrate producers, but also may have independent antiinflammatory properties related to secreted metabolites, which have been shown to block nuclear factor κB activation and IL-8 production (50).
Food residues from an unbalanced diet deficient in fiber and high in meat promotes proteolytic rather than saccharolytic fermentation, with generation of the branched-chain SCFAs isobutyrate, isovaleric, and 2-methylbutyric acid (51, 52) and nitrogenous metabolites. Whereas little is known about the functional significance of branched SCFAs, proteolytic products such as hippurate, p-cresyl, ammonia, and phenols as well as sulfide metabolites have been shown to be inflammatory and carcinogenic in experimental models (53, 54). Thus, the lower risk of colon cancer in Africans could also be a consequence of lower proteolytic fermentation. We have also hypothesized that the high meat content of Westernized diets may increase colon cancer risk because of its stimulatory effect on sulfate-reducing bacteria, which use the sulfur residues of meat to release hydrogen sulfide, which has been shown to be genotoxic in experimental models (54). Surprisingly, the opposite was observed, with higher numbers of sulfate-reducing bacteria in Africans, which suggests that increased saccharolytic fermentation with increased hydrogen production may be more stimulatory to these hydrogenotrophs.
Strong epidemiologic evidence links high fat consumption to increased colon cancer risk (55, 56). Whereas experimental evidence indicates that fat, particularly fat high in n−6 fatty acids, is proinflammatory and that a Westernized diet high in fat and deficient in vitamin D increases colonic tumors in normal mice (57), fat digestion and absorption are very efficient and little dietary fat enters the colon, which suggests that other mechanisms may be involved. Perhaps the strongest contender is BAs. Increased fat consumption stimulates the liver to synthesize more BAs. This in turn increases the quantity of BAs that escape enterohepatic recirculation and enter the colon. Once in the colon, primary BAs, notably CA and chenodeoxycholic acid, are converted to secondary BAs, namely deoxycholic acid and LCA, by specific bacteria that contain 7α-dehydroxylating enzymes. Our studies showed that not only were microbes containing this enzyme more common in high-risk African Americans, but so also were fecal secondary BAs. Human studies have associated increased fecal secondary BAs with colon polyps (58) and colon cancer (59), and there is substantial experimental evidence that they have carcinogenic properties (7). A further potential mechanism recently reported in a mouse model was the ability of a high-fat diet to stimulate the delivery of sulfur-rich taurine conjugates of BAs to the colon, where they produced a blossom of Biophila wadsworthia, which released hydrogen sulfide from taurine that lead to acute inflammation and colitis (60).
In summary, our study supports the hypothesis that colon cancer risk is determined by the interaction between diet and gut microbiota (61, 62) and that the higher risk in African Americans could be attributed to their chronically lower consumption of fiber and resistant starch and their higher consumption of dietary fat. Further studies are needed to assess the functional significance of other differences noted in the pyrosequencing analysis (eg, Bacteroides) and the responsiveness of the identified microbial and metabolomic differences to dietary change.
Supplementary Material
Acknowledgments
The authors’ responsibilities were as follows—JO, SJDO, KN, and HRG: participated in the design and conduct of the study; JPD: conducted the BA and SCFA analyses; FC, EGZ, and MW: assisted with the microbiota analysis; and JO, FC, and SJDO: performed the data analysis. All authors were involved in finalizing the manuscript. None of the authors had any conflicts of interest to declare.
Footnotes
Abbreviations used: BA, bile acid; CA, cholic acid; LC, liquid chromatography; LCA, lithocholic acid; MS, mass spectrometry; qPCR, quantitative polymerase chain reaction; rRNA, ribosomal RNA; SCFA, short-chain fatty acid.
REFERENCES
- 1.Doll R, Peto R. The causes of cancer: quantitative estimates of avoidable risks of cancer in the United States today. J Natl Cancer Inst 1981;66:1191–308. [PubMed] [Google Scholar]
- 2.Sharma S, O'Keefe SJD. Environmental influences on the high mortality from colorectal cancer in African Americans. Postgrad Med J 2007;83:583–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O'Keefe SJD, Kidd M, Noel G, Owira P. The rarity of colon cancer in Africans is associated with low animal product consumption, not fiber. Am J Gastroenterol 1999;94:1373–80. [DOI] [PubMed] [Google Scholar]
- 4.O'Keefe SJD, Chung D, Mahmoud N, Sepulveda AR, Manafe M. Why Do African Americans get more colon cancer than Native Africans? J Nutr 2007;137:175S–82S. [DOI] [PubMed] [Google Scholar]
- 5.Waldecker M, Kautenburger T, Daumann H, Busch C, Schrenk D. Inhibition of histone-deacetylase activity by short-chain fatty acids and some polyphenol metabolites formed in the colon. J Nutr Biochem 2008;19:587–93. [DOI] [PubMed] [Google Scholar]
- 6.Windey K, De Preter V, Verbeke K. Relevance of protein fermentation to gut health. Mol Nutr Food Res 2012;56:184–96. [DOI] [PubMed] [Google Scholar]
- 7.Bernstein C, Holubec H, Bhattacharyya AK, Nguyen H, Payne CM, Zaitlin B, Bernstein H. Carcinogenicity of deoxycholate, a secondary bile acid. Arch Toxicol 2011;85:863–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Penry DL, Jumars P. Modeling animal guts as chemical reactors. Am Nat 1987;129:69–96. [Google Scholar]
- 9.Louis P, Flint HJ. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett 2009;294:1–8. [DOI] [PubMed] [Google Scholar]
- 10.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, et al. Enterotypes of the human gut microbiome. Nature 2011;473:174–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fooks LJ, Gibson GR. Probiotics as modulators of the gut flora. Br J Nutr 2002;88:S39–49. [DOI] [PubMed] [Google Scholar]
- 12.Arunachalam KD. Role of Bifidobacteria in nutrition, medicine and technology. Nutr Res 1999;19:1559–97. [Google Scholar]
- 13.Nadkarni MA, Martin FE, Jacques NA, Hunter N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 2002;148:257–66. [DOI] [PubMed] [Google Scholar]
- 14.Louis P, Flint HJ. Development of a semiquantitative degenerate real-time pcr-based assay for estimation of numbers of butyryl-coenzyme A (CoA) CoA transferase genes in complex bacterial samples. Appl Environ Microbiol 2007;73:2009–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsuki T, Watanabe K, Fujimoto J, Tanaka R. Development of 16S rRNA-gene-targeted group-specific primers for the detection and identification of predominant bacteria in human feces. Appl Environ Microbiol 2002;68:5445–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kok RG, de Waal A, Schut F, Hellingwerf K. Specific detection and analysis of a probiotic Bifidobacterium strain in infant feces. Appl Environ Microbiol 1996;62:3668–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stevenson DM, Weimer P. Dominance of Prevotella and low abundance of classical ruminal bacterial species in the bovine rumen revealed by relative quantification real-time PCR. Appl Microbiol Biotechnol 2007;75:165–74. [DOI] [PubMed] [Google Scholar]
- 18.Byun R, Nadkarni MA, Chhour KL, Martin FE, Jacques NA, Hunter N. Quantitative analysis of diverse Lactobacillus species present in advanced dental caries. J Clin Microbiol 2004;42:3128–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rinttilä T, Kassinen A, Malinen E, Krogius L, Palva A. Development of an extensive set of 16S rDNA-targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. J Appl Microbiol 2004;97:1166–77. [DOI] [PubMed] [Google Scholar]
- 20.Spence C, Whitehead TR, Cotta MA. Development and comparison of SYBR Green quantitative real-time PCR assays for detection and enumeration of sulfate-reducing bacteria in stored swine manure. J Appl Microbiol 2008;105:2143–52. [DOI] [PubMed] [Google Scholar]
- 21.Denman SE, Tomkins NW, McSweeney CS. Quantitation and diversity analysis of ruminal methanogenic populations in response to the antimethanogenic compound bromochloromethane. FEMS Microbiol Ecol 2007;62:313–22. [DOI] [PubMed] [Google Scholar]
- 22.Collins MD, Lawson PA, Willems A, Cordoba JJ, Fernandez-Garayzabal J, Garcia P, Cai J, Hippe H, Farrow JA. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol 1994;44:812–26. [DOI] [PubMed] [Google Scholar]
- 23.Scheppach W, Fabian C, Kasper H. Fecal short-chain fatty acid (SCFA) analysis by capillary gas-liquid chromatography. Am J Clin Nutr 1987;46:641–6. [DOI] [PubMed] [Google Scholar]
- 24.Tagliacozzi D, Mozzi AF, Casetta B, Bertucci P, Bernardini S, Ilio CD, Urbani A, Federici G. Quantitative analysis of bile acids in human plasma by liquid chromatography-electrospray tandem mass spectrometry: a simple and rapid one-step method. Clin Chem Lab Med 2003;41:1633–41. [DOI] [PubMed] [Google Scholar]
- 25.Zoetendal EG, Heilig HG, Klaassens ES, Booijink CC, Kleerebezem M, Smidt H, de Vos WM. Isolation of DNA from bacterial samples of the human gastrointestinal tract. Nat Protoc 2006;1:870–3. [DOI] [PubMed] [Google Scholar]
- 26.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010;7:335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 2009;37:D141–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakamura N, Leigh SR, Mackie RI, Gaskins HR. Microbial community analysis of rectal methanogens and sulfate reducing bacteria in two non-human primate species. J Med Primatol 2009;38:360–70. [DOI] [PubMed] [Google Scholar]
- 29.Nava GM, Carbonero F, Croix JA, Greenberg E, Gaskins HR. Abundance and diversity of mucosa-associated hydrogenotrophic microbes in the healthy human colon. ISME J 2012;6:57–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wells JE, Berr F, Thomas LA, Dowling RH, Hylemon PB. Isolation and characterization of cholic acid 7α-dehydroxylating fecal bacteria from cholesterol gallstone patients. J Hepatol 2000;32:4–10. [DOI] [PubMed] [Google Scholar]
- 31.Carbonero F, Benefiel AC, Gaskins HR. Contributions of the microbial hydrogen economy to colonic homeostasis. Nat Rev Gastroenterol Hepatol 2012;9:504–18. [DOI] [PubMed] [Google Scholar]
- 32.Carbonero F, Benefiel AC, Alizadeh-Ghamsari AH, Gaskins HR. Microbial pathways in colonic sulfur metabolism and links with health and disease. Front Physiol 2012;3:448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tims S, Derom C, Jonkers DM, Vlietinck R, Saris WH, Kleerebezem M, de Vos WM, Zoetendal EG. Microbiota conservation and BMI signatures in adult monozygotic twins. ISME J 2013;7:707–17. [DOI] [PMC free article] [PubMed]
- 34.Walker AW, Ince J, Duncan SH, Webster LM, Holtrop G, Ze X, Brown D, Stares MD, Scott P, Bergerat A, et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J 2011;5:220–30. [DOI] [PMC free article] [PubMed]
- 35.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci USA 2010;107:14691–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burkitt DP. Diseases of the alimentary tract and western diets. Pathol Microbiol (Basel) 1973;39:177–86. [DOI] [PubMed] [Google Scholar]
- 37.Pryde SE, Duncan SH, Hold GL, Stewart CS, Flint HJ. The microbiology of butyrate formation in the human colon. FEMS Microbiol Lett 2002;217:133–9. [DOI] [PubMed] [Google Scholar]
- 38.Barcenilla A, Pryde SE, Martin JC, Duncan SH, Stewart CS, Henderson C, Flint HJ. Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl Environ Microbiol 2000;66:1654–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolin M. Interactions between H2-producing and methaneproducing species. : Schlegel HG, Gottschalk G, Pfennig N, Microbial formation and utilization of gases. Göttingen, Germany: Goltze Press, 1976:141–50. [Google Scholar]
- 40.Greer JB, O'Keefe SJ. Microbial induction of immunity, inflammation and cancer. Front Physiol 2011;1:168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vipperla K, O'Keefe SJ. The microbiota and its metabolites in colonic mucosal health and cancer risk. Nutr Clin Pract 2012;27:624–35. [DOI] [PubMed] [Google Scholar]
- 42.Bordonaro M, Lazarova DL, Sartorelli AC. Butyrate and Wnt signaling: a possible solution to the puzzle of dietary fiber and colon cancer risk? Cell Cycle 2008;7:1178–83. [DOI] [PubMed] [Google Scholar]
- 43.Chirakkal H, Leech SH, Brookes KE, Prais AL, Waby JS, Corfe BM. Upregulation of BAK by butyrate in the colon is associated with increased Sp3 binding. Oncogene 2006;25:7192–200. [DOI] [PubMed] [Google Scholar]
- 44.Hinnebusch BF, Meng S, Wu JT, Archer SY, Hodin RA. The effects of short-chain fatty acids on human colon cancer cell phenotype are associated with histone hyperacetylation. J Nutr 2002;132:1012–7. [DOI] [PubMed] [Google Scholar]
- 45.Comalada M, Bailon E, de Haro O, Lara-Villoslada F, Xaus J, Zarzuelo A, Galvez J. The effects of short-chain fatty acids on colon epithelial proliferation and survival depend on the cellular phenotype. J Cancer Res Clin Oncol 2006;132:487–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Andoh A, Shimada M, Araki Y, Fujiyama Y, Bamba T. Sodium butyrate enhances complement-mediated cell injury via down-regulation of decay-accelerating factor expression in colonic cancer cells. Cancer Immunol Immunother 2002;50:663–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodríguez-Salvador J, Armas-Pineda C, Perezpena-Diazconti M, Chico-Ponce de Leon F, Sosa-Sainz G, Lezama P, Recillas-Targa F, Arenas-Huertero F. Effect of sodium butyrate on pro-matrix metalloproteinase-9 and -2 differential secretion in pediatric tumors and cell lines. J Exp Clin Cancer Res 2005;24:463–73. [PubMed] [Google Scholar]
- 48.Zeng H, Briske-Anderson M. Prolonged butyrate treatment inhibits the migration and invasion potential of HT1080 tumor cells. J Nutr 2005;135:291–5. [DOI] [PubMed] [Google Scholar]
- 49.Zgouras D, Wachtershauser A, Frings D, Stein J. Butyrate impairs intestinal tumor cell-induced angiogenesis by inhibiting HIF-1alpha nuclear translocation. Biochem Biophys Res Commun 2003;300:832–8. [DOI] [PubMed] [Google Scholar]
- 50.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA 2008;105:16731–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dehority BA, Johnson RR, Bentley OG, Moxon AL. Studies on the metabolism of valine, proline, leucine and isoleucine by rumen microorganisms in vitro. Arch Biochem Biophys 1958;78:15–27. [DOI] [PubMed] [Google Scholar]
- 52.Allison MJ. Production of branched-chain volatile fatty acids by certain anaerobic bacteria. Appl Environ Microbiol 1978;35:872–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Russell WR, Gratz SW, Duncan SH, Holtrop G, Ince J, Scobbie L, Duncan G, Johnstone AM, Lobley GE, Wallace RJ, et al. High-protein, reduced-carbohydrate weight-loss diets promote metabolite profiles likely to be detrimental to colonic health. Am J Clin Nutr 2011;93:1062–72. [DOI] [PubMed] [Google Scholar]
- 54.Attene-Ramos MS, Wagner ED, Plewa MJ, Gaskins HR. Evidence that hydrogen sulfide is a genotoxic agent. Mol Cancer Res 2006;4:9–14. [DOI] [PubMed] [Google Scholar]
- 55.NIH. Based on November 2005 SEER data submission, posted to the SEER web site 2006. Available from: http://seer.cancer.gov/csr/1975_2003/ (cited 18 December 2012).
- 56.Stadler J, Stern HS, Yeung KS, McGuire V, Furrer R, Marcon N, Bruce WR. Effect of high fat consumption on cell proliferation activity of colorectal mucosa and on soluble faecal bile acids. Gut 1988;29:1326–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Newmark HL, Yang K, Kurihara N, Fan K, Augenlicht LH, Lipkin M. Western-style diet-induced colonic tumors and their modulation by calcium and vitamin D in C57Bl/6 mice: a preclinical model for human sporadic colon cancer. Carcinogenesis 2009;30:88–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.de Kok TM, van Faassen A, Glinghammar B, Pachen DM, Eng M, Rafter JJ, Baeten CG, Engels LG, Kleinjans JC. Bile acid concentrations, cytotoxicity, and pH of fecal water from patients with colorectal adenomas. Dig Dis Sci 1999;44:2218–25. [DOI] [PubMed] [Google Scholar]
- 59.Bernstein H, Bernstein C, Payne CM, Dvorakova K, Garewal H. Bile acids as carcinogens in human gastrointestinal cancers. Mutat Res 2005;589:47–65. [DOI] [PubMed] [Google Scholar]
- 60.Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, Antonopoulos DA, Jabri B, Chang EB. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature 2012;487:104–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dethlefsen L, Eckburg PB, Bik EM, Relman DA. Assembly of the human intestinal microbiota. Trends Ecol Evol 2006;21:517–23. [DOI] [PubMed] [Google Scholar]
- 62.O'Keefe SJD. Nutrition and colonic health: the critical role of the microbiota. Curr Opin Gastroenterol 2008;24:51–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.