Abstract
Although the mechanisms by which malaria parasites develop resistance to drugs are unclear, current knowledge suggests a main mechanism of resistance is the alteration of target enzymes by point mutation. In other organisms, defects in DNA mismatch repair have been linked to increased mutation rates and drug resistance. We have identified an unusual complement of mismatch repair genes in the Plasmodium genome. An initial functional test of two of these genes (PfMSH2-1 and PfMSH2-2) using a dominant mutator assay showed an elevation in mutation frequency with the PfMSH2-2 homolog, indirectly demonstrating a role for this gene in mismatch repair. We successfully disrupted PbMSH2-2 in the P. berghei laboratory isolate NK65, and showed that this gene is not essential for parasite growth in either the asexual (rodent) or sexual (mosquito) stages of the lifecycle. Although we observed some differences in levels of drug resistance between wild type and mutant parasites, no uniform trend emerged and preliminary evidence does not support a strong link between PbMSH2-2 disruption and dramatically increased drug resistance. We found microsatellite polymorphism in the PbMSH2-2 disrupted parasites in less than 40 life cycles post-transfection, but not in PbMap2K disrupted controls or mosquito passaged wild type parasites, which suggests a possible role for PbMSH2-2 in preventing microsatellite slippage, similar to MSH2 in other organisms. Our studies suggest that Plasmodium species may have evolved a unique variation on the highly conserved system of DNA repair compared to the mismatch repair systems in other eukaryotes.
Keywords: DNA mismatch repair, Plasmodium falciparum, Plasmodium berghei, malaria, MSH2, drug resistance
Introduction
Global resistance to multiple antimalarial drugs is becoming an increasing challenge in worldwide efforts to control malaria [1]. Identifying the mechanisms by which the parasite generates genetic changes that lead to drug resistance is critical to the understanding of drug resistance and the design of effective therapies. Current knowledge suggests a main mechanism of resistance to antimalarial drugs is alteration of target enzymes by point mutations, as has been demonstrated for pyrimethamine, sulfadoxine, and atovaquone [2–4].
While there are many mechanisms by which an organism can increase its mutation rate in response to environmental stresses such as drug pressure, defects in mismatch repair (MMR) have been found in the majority of naturally occurring, strong bacterial mutators. These defects are mainly in the mismatch recognition enzyme MutS. Mutator populations of bacteria with defects in mismatch repair have been shown to increase the frequency of drug resistance up to 1000 times more than in normal cells (reviewed in [5]).
The MMR proteins were named for their involvement in mutation avoidance and replication fidelity. They have also been linked to the regulation of homeologous recombination, gene conversion, meiotic chromosome pairing and segregation, speciation, stationary phase mutagenesis, immunoglobulin class switching and hypermutation, and responses to DNA damage including repair, tolerance, and triggering of apoptosis and cell cycle checkpoints [6–10]. MMR is highly conserved in prokaryotes and higher organisms.
The most well characterized MMR pathway is the methyl-directed MutHLS system of E. coli. Yeast, humans, and other eukaryotes have multiple homologs of bacterial MutS (MSH) and MutL (MLH), but no MutH. MMR proteins in multiple eukaryotic species have been inactivated without killing the cells or preventing reproduction [8–11].
We have found that P. falciparum has an unusual complement of putative MMR proteins based on homology to functionally characterized MMR protein sequences and motifs. We tested the function of the P. falciparum MSH2 homologs by a bacterial dominant mutator assay. We tested our hypothesis that defective DNA mismatch repair may play a role in generating genetic diversity and drug resistance in malaria parasites by disrupting the P. berghei homolog of PfMSH2-2 by gene targeting. We cloned a mutant strain with this disruption, and passaged the mutant strain through the mosquito stages of the life cycle in order to assay its role in the sexual life cycle stages. We analyzed the PbMSH2-2 mutant parasites by an in vivo drug resistance assay. We also assayed microsatellite instability, a common hallmark of the mutator phenotype.
Materials and Methods
Identification of putative Plasmodium MMR genes
We identified putative P. falciparum MMR proteins based on homology to functionally characterized MMR protein sequences and motifs, using BLAST with default settings to the P. falciparum Genome Database (PlasmoDB) at www.plasmodb.org [12]. The identity of predicted MMR proteins was confirmed by conserved domain searches using the tools available on the NCBI website (http://www.ncbi.nlm.nih.gov), PFAM (http://www.sanger.ac.uk/Software/Pfam/), and Prosite (http://us.expasy.org/prosite/), and by BLAST to the non-redundant database at NCBI. We identified the P. yoelii and P. berghei homologs of the P. falciparum MMR proteins by BLAST searches to PlasmoDB and The Institute for Genomic Research (TIGR) website at www.tigr.org (P. yoelii), and the Sanger Institute website at www.sanger.ac.uk/DataSearch (P. berghei). Predicted protein sequences in the unannotated species P. berghei and P. yoelii were confirmed by alignment to the annotated P. falciparum database sequences.
Dominant mutator assay
The bacterial strain CC108, created by Cupples, et al. [13], was kindly provided by Dr. Leona Samson. The full-length PfMSH2-1 and PfMSH2-2 genes cloned into the pBlueScript SK+ vector (pBS-SK+; Stratagene) were transformed in separate experiments into the CC108 strain. Transformation with pBS-SK+ vector alone served as the negative control, while the human MSH2 gene [14] cloned into pBS-SK+ was used as a positive control (kindly provided by Dr. Richard Kolodner). The mutator assay was performed as previously described by Glassner, et al. [15], with minor modifications. Cell viability was determined by plating appropriately diluted samples of three cultures on minimal A/glucose/ampicillin plates, while the rate of lac+ reversion was determined by plating pelleted cultures on minimal A/lactose/ampicillin plates. The median lac+ reversion frequency was calculated by dividing the median number of lac+ colonies by the mean number of viable cells plated.
Analysis of synteny
Syntenic location of orthologs in different species was found by searching the available contiguous sequence for adjacent open reading frames long enough to encode genes (The Sanger Institute, PlasmoDB). Adjacent open reading frames were then used to query the P. falciparum database of annotated genes (PlasmoDB) by BLASTX, and if a single very highly significant hit resulted, this was considered to be the likely falciparum ortholog of the query. Orthology was confirmed by reciprocal BLAST. When two genes were found to be adjacent in two different species, they were determined to be located in the same synteny segment in those species.
Insertion construct design and P. berghei transfection
Insertion plasmids were constructed in order to disrupt the P. berghei MMR genes. The plasmid designed to disrupt PbMSH2-2 included a 1380 bp gene fragment (Fig. 1A), which lacked sequence encoding the first 127 amino acids of the protein, including a portion of the putative DNA binding domain, and was also missing the C-terminal 269 amino acids, including the ATP binding domain and the mutS signature motif. The gene fragment was inserted into the pDb.Dh.^Db vector upstream of a selectable marker expressing the pyramethamine-resistant human dihydrofolate reductase (hDHFR) enzyme [16]. The targeting construct was linearized with XcmI and used to transfect P. berghei strain NK65 parasites as previously described [17]. Similarly designed targeting plasmids were constructed to disrupt the genes PbMSH2-1, PbMSH6, and PbMLH1. Total genomic DNA (gDNA) was isolated from parasite pellets using the QIAamp DNA blood mini kit (Qiagen). Resistant parasite populations were assayed by Southern hybridization and PCR to check for gene disruption at the targeted locus. Southern hybridization procedures were performed according to the DIG System User’s Guide for Filter Hybridization (Boehringer Mannheim, Mannheim, Germany). Oligonucleotide primers are described in supplementary table 1.
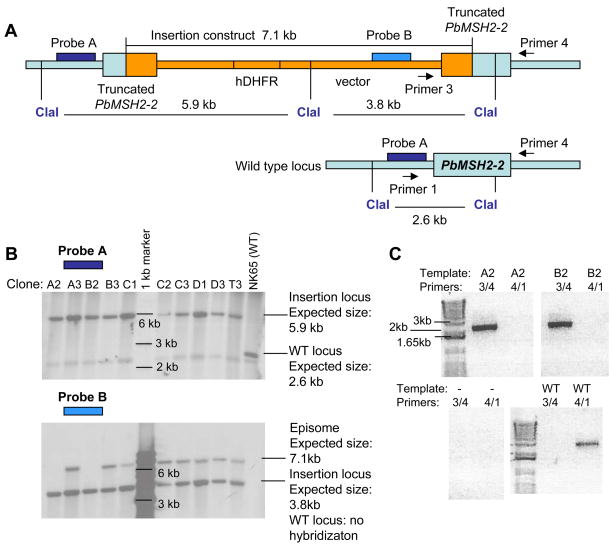
Figure 1. PbMSH2-2 gene disruption.
(A) Targeting construct designed to disrupt the P. berghei MSH2-2 gene by homologous recombination. (B) Southern hybridization verifies PbMSH2-2 disruption in parasites transfected with the targeting construct and cloned by limiting dilution. Probe A detects presence of insertion and absence of wild type locus in all clonal populations. Probe B detects presence of construct episome in all clones except A2 and B2. (C) PCR confirms presence of insertion and disruption of wild type locus in MSH2-2 mutant clones. PCR was used to amplify the PbMSH2-2 locus from gDNA isolated from clones A2 and B2, NK65 wild type gDNA (WT), and the MSH2-2 insertion construct (construct). A control reaction with no template (−) was simultaneously performed.
Passage of recombinant parasite clone A2 through mosquito vector
Anopheles stephensi mosquitoes were fed on infected Balb/c mice and dissected on day 10 post-feeding to confirm infection. Infected mosquitoes were fed on naïve Sprague-Daley (SD) rats on days 18 and 19 post-feeding. Parasite populations were cryopreserved and gDNA was isolated as previously described. NK65 wild type parasites were simultaneously passaged through the mosquito vector as a control.
5-Fluoroorotate (5-FOA) drug resistance assay
The 5-FOA drug resistance assay was performed 3 times with minor variations in protocol as described in the results and Figure 2. Cryopreserved parasites were thawed and injected intraperitoneally to 2 SD rats. When parasitemia reached 4–6%, infected blood was collected by cardiac puncture, and approximately 200μl was injected intraperitoneally to naïve SD rats. Infected animals were treated as indicated in Figure 2. First treatment was administered when mean parasitemia of each experimental and control group of animals was 1% (Fig. 2A), 2–3% (Fig. 2B and 2C), or 7–8% (Fig. 2D). All rats were pre-treated before the first injection with 1500 mg/kg uridine injected intraperitoneally, and 30 minutes later treated with 10 mg/kg 5-FOA, 800 mg/kg uridine in 1x PBS. Control animals were injected with 800 mg/kg uridine. At first drug treatment in Fig. 2A, the MSH2-2 mutant clone A2 had been passaged for nineteen 24-hour periods (life cycles) post-cloning. At first drug treatment in Fig. 2B–D, the MSH2-2 mutant clone A2 had been passaged for twenty-three 24-hour periods since cloning, while the mosquito passaged MSH2-2 mutant population (A2m) population was passaged for eighteen 24-hour periods prior to mosquito passage, and nineteen additional 24-hour periods post-mosquito infection. The NK65 wild type population was passaged for eighteen 24-hour periods post-mosquito infection.
Figure 2. Frequency of resistance to 5-FOA in wild type and MSH2-2 disrupted parasites.
Swiss mice were intraperitoneally injected with P. berghei infected red blood cells, and treated as indicated once or twice a day (T) with 10 mg/kg 5-FOA. Parasitemia was measured daily by thin blood smear. (A and B) Wild type parasites appear to have a higher frequency of drug resistance than PbMSH2-2 disrupted parasites. (C) PbMSH2-2 disrupted parasites which have been passaged through the sexual lifecycle stages appear to have a higher frequency of drug resistance than sexually passaged wild type parasites. (D) Sexually passaged PbMSH2-2 disrupted parasites show a higher frequency of drug resistance than asexually passaged mutant parasites.
Microsatellite identification and sequencing
Microsatellite regions in the P. berghei genome were identified using BLAST with default settings to the P. berghei database (isolate ANKA) at the Wellcome Trust Sanger Institute website (www.sanger.ac.uk). Additional microsatellite repeats were identified by placing P. berghei contigs into the Perfect Microsatellite Repeat Finder available at http://sgdp.iop.kcl.ac.uk/nikammar/repeatfinder.html (based on Tandyman written by Robert Leach). We defined microsatellite repeat as any mononucleotide repeat of 10 or more bp, and any di-nucleotide (or larger) repeat of 5 or more repeating units.
Amplification and sequencing primers were designed using Primer3 software written by S. Rozen and H.J. Skaletsky and available at http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi (supplementary table 1). Standard polymerase chain reaction (PCR) methods and Platinum Taq DNA Polymerase High Fidelity (Invitrogen) were used for amplification of the target region, and the products from 3 independent PCR reactions were either treated with Exosap (USB) and sequenced directly in the forward and reverse direction, or ligated to a bacterial vector using the TA cloning technique (Invitrogen) before transformation into E. coli. Plasmid DNA from 3 bacterial clones for each PCR product was sequenced in the forward and reverse direction. Sequencing reactions were performed by Dana-Farber/Harvard Cancer Center High-Throughput DNA Sequencing Facility. Editing of raw chromatograms and multiple sequence alignments were performed using Lasergene software (DNASTAR Inc., Madison, WI).
Results
Identification of putative Plasmodium MMR genes
Five putative MMR proteins were identified, including 2 homologs of MSH2, which we have called PfMSH2-1 (PlasmoDB identifiers PF14_0254) and PfMSH2-2 (MAL7P1.206). We also found putative homologs for MSH6 (PFE0270c), MLH1 (PF11_0184), and PMS1 (MAL7P1.145). P. berghei sequences of the putative MMR genes are located in the Sanger Institute database as follows: PbMSH2-1, contig 3793 position 1 – 2145; PbMSH2-2, contig 5351 position 4817 – 7381; PbMSH6, contig 4985 position 4579 – 8277; PbMLH1, contig 4157 position 757 –1440; PbPMS1, contig 5138 position 2298 – 5618. Although both PfMSH6 and PfPMS1 are predicted to contain introns, no intronic sequence was identified in the P. berghei or P. yoelii homologs of these genes.
As two MutS2 homologs have not yet been observed in any eukaryotic species, we confirmed their homology was closest to MSH2, and not any of the other numerous MutS homologs, by several bioinformatics methods including conserved domain and Blast searches to the NCBI, PlasmoDB, and GeneDB databases. Both proteins appear to have highest homology to the C-terminal cd03285 domain ABC_MSH2_euk domain, which is specific to the MutS2 homolog in eukaryotes. Blast P to the non-redundant database at NCBI (www.ncbi.nlm.nih.gov) confirmed that both proteins have higher homology to MSH2 in all of the organisms with homologous protein sequence data, including other eukaryotes, than to any other MutS homolog in those organisms. Prosite, Pfam, and InterProScan (http://www.ebi.ac.uk/InterProScan/) also confirmed that the ATP-binding site, the putative DNA binding domain, and the mutS family signature motif are present in both MSH2 homologs in P. falciparum, P. yoelii, and P. berghei. The full length sequence of genes encoding PbMSH2-1 and PyMSH2-1 was not available in any of the genome databases. We were not able to identify any putative functional differences between the two genes using bioinformatics methods.
A ClustalW amino acid alignment suggests that PbMSH2-1 shares 83% pairwise identity with PfMSH2-1, excluding the first 278 amino acids of PfMSH2-1 for which homologous P. berghei sequence was not available. PbMSH2-2 shares 61% amino acid identity to the full length PfMSH2-2 protein, and truncated portions of the sequence homologous to the PbMSH2-1 sequence share 64% pairwise identity with each other. The P. falciparum MSH2-1 and MSH2-2 proteins share 43% identity. The truncated portions of the P. berghei MSH2-1 and MSH2-2 proteins are 50% identical.
Dominant mutator assay
In order to test the involvement of the P. falciparum MSH2 homologs in classical mismatch repair, we determined the rate of spontaneous mutations in mismatch repair proficient E. coli in the presence of the P. falciparum MSH2 proteins. We hypothesized that functional PfMSH2-1 or PfMSH2-2 would compete with E. coli MutS for binding to mutation sites, but would be unable to interact with the other components of the bacterial MMR pathway. Thus this non-productive association with mutated sites would lead to an increase in mutation rate.
The CC108 bacterial strain was transfected with the pBS-SK+ vector expressing the hMSH2, PfMSH2-1, or PfMSH2-2 gene. The pBS-SK+ vector alone served as the negative control. Because the CC108 bacterial strain has a lacZ− genotype conferred by a +1G frameshift mutation near active site codons of β-galactosidase, a reversion mutation is necessary to restore the lacZ+ genotype. Induction of a mutator phenotype associated with the introduction of either the PfMSH2-1 or PfMSH2-2 gene was determined by measuring the lac+ reversion rate.
The hMSH2 positive control showed a 5.7-fold higher reversion frequency to lac+ compared to vector alone (Table 1). We did not detect any biological activity of PfMSH2-1, possibly due to a lack of expression of the PfMSH2-1 protein. PfMSH2-2 showed a 2.3 to 5.6-fold higher lac+ reversion rate compared to pBS-SK+ alone. The 5.6-fold difference observed for PfMSH2-2 was comparable to that seen for the hMSH2 positive control.
Table 1.
Summary of PfMSH2-1 and PfMSH2-2 dominant mutator assays using the lacZ− CC108 strain.
| Experiment no. | Constructa | Lac+ median reversion frequency | Fold Increaseb |
|---|---|---|---|
| pBS | 1.6 × 10−9 | ||
| 1 | hMSH2 | 8.9 × 10−9 | 5.7 |
| pBS | 2.9 × 10−8 | ||
| 2 | PfMutS2-2 | 6.7 × 10−8 | 2.3 |
| pBS | 4.3 × 10−8 | ||
| 3 | PfMutS2-2 | 2.4 × 10−7 | 5.6 |
Thirteen cultures were assayed for each construct.
Relative to pBS control.
Disruption of PbMSH2-2
In order to disrupt the PbMSH2-2 gene, we constructed an insertion plasmid designed to integrate via a single crossover event, resulting in two truncated copies of the gene (Fig. 1a). Transfected parasites were cloned by limiting dilution, and two clonal populations, A2 and B2, were obtained with the disrupted PbMSH2-2 locus, and with no wild type or episomal construct (Fig. 1). PbMSH2-2 disrupted parasite populations had similar growth rates in rats and mice as wild type, and similar morphology by light microscopy and giemsa staining.
We constructed similarly designed insertion plasmids in order to disrupt the genes PbMSH2-1, PbMSH6, and PbMLH1. We attempted several P. berghei transfections with these constructs, but genomic integration was not successful (data not shown). Simultaneous control transfections with the PbTRAP targeting construct pINT [17] and the PbMSH2-2 targeting construct resulted in successful integration events.
Because PfMSH2-2 seems to be maximally expressed in the mosquito stages of the life cycle [18], we reasoned that PbMSH2-2 may play a role in the sexual life cycle stages of the parasite. In order to test this hypothesis, female Anopheles stephensi mosquitoes were fed on mice infected with mutant clone A2 or NK65 wild type parasites. The PbMSH2-2 mutant parasites had similar infectivity to mice and mosquitoes and similar oocyst and sporozoite development as wild type (data not shown). No changes in parasite morphology compared to wild type were detected by light microscopy at any life cycle stage.
Drug resistance assay
In order to test our hypothesis that MSH2-2 gene disruption may result in an increased rate of forward mutation to drug resistance, we treated infected rats with 5-fluoroorotate (5-FOA), and compared rates of acquisition of parasite drug resistance (Fig. 2). While no dramatic difference was observed between wild type and mutant parasites in our first experiment involving 25 Swiss mice (Fig. 2A), there seemed to be a trend towards a higher frequency of drug resistance in the wild type population, as measured by a higher percent of animals with detectable parasitemia after drug treatment. A second drug assay comparing wild type and mutant parasite populations in 18 Swiss mice had a similar result, and detected no evidence for a mutator phenotype in the MSH2-2 mutant population, while the wild type population of parasites seemed to show a higher frequency of drug resistance (Fig. 2B).
Interestingly, in mosquito-passaged mutant and wild type parasites tested in 18 Swiss mice this trend was reversed, with the MSH2-2 disrupted parasites showing higher frequency of resistance after drug treatment than wild type parasites (Fig. 2C), although again no dramatic differences were seen between populations. In order to confirm this result, we compared asexually to sexually passaged MSH2-2 mutant parasites. In this experiment the MSH2-2 disrupted sexually passaged mutant parasites had a slightly higher frequency of drug resistance than the MSH2-2 disrupted asexually passaged mutants, confirming our observation that while trends may be detectable in these data, the differences between the two populations were not dramatic (Fig. 2D). Higher parasitemia at first drug treatment and more intense drug treatment did not seem to increase the difference in drug resistance phenotypes observed in this assay.
In order to test if the parasites that reappeared in the peripheral blood after drug treatment were truly resistant, or just persistent or slow growing parasites, we infected and treated naive Swiss mice (Fig. 3). All 3 populations that had reappeared in the peripheral blood after drug treatment appeared to be resistant to 5-FOA. The control untreated population from experiment 2 and the wild type population were susceptible to the drug.
Figure 3. Confirmation of drug resistance in parasite populations.
Three putative resistant parasite populations cryopreserved after the second drug assay, a control untreated MSH2-2 mutant population from the same assay, and a previously untreated wild type NK65 parasite population were used to infect naïve swiss mice and treated with 12.2 mg/kg 5-FOA as indicated (T).
Microsatellite instability assay
We assayed for microsatellite instability, a classical hallmark of defective MMR, by sequencing PbMSH2-2 mutant clones, wild type NK65 parasites, wild type ANKA parasites, mosquito passaged PbMSH2-2 disrupted and NK65 parasites, and a control clone disrupted at the PbMap2 kinase locus [19]. We analyzed 15 regions of genomic DNA isolated from NK65, ANKA, MSH2-2 disrupted NK65 (A2a, A2b, and B2), mosquito-passaged MSH2-2 disrupted NK65 (A2c and A2d), mosquito-passaged NK65 (NKm), and Map2K disrupted NK65 (C2) parasites (Table 2). Strains A2a, A2b, A2c, and A2d are all derived from the MSH2-2 mutant clone A2, although the populations were expanded separately for the final 14 days of passaging prior to gDNA extraction. Nine regions were identified with detectable microsatellite polymorphism between ANKA and NK65, and 2 regions were found to be polymorphic between the mutant parasite populations and the parent strain NK65. A tri-nucleotide repeat, (AAT)13 in the NK65 and ANKA strains, was contracted to (AAT)11 in both mosquito-passaged populations derived from the MSH2-2 disrupted clone A2. An additional tri-nucleotide repeat, (ATA)8, had expanded to (ATA)9 in all of the MSH2-2 mutant clones, suggesting the expansion may have occurred prior to cloning of the mutant parasites. No differences were found between the NK65 parent and mosquito-passaged NK65, or in the Map2K-disrupted NK65 clone, which had been passaged in rodents for a similar length of time post-cloning as the MSH2-2 mutant clones A2 and B2, and had presumably undergone a comparable number of DNA replications.
Table 2.
Microsatellite length polymorphism detected by sequencing P. berghei wild type and PbMSH2-2 disrupted gDNA
| Primer set | Laboratory strains | Differences in mutant and control strains relative to NK65 parent strain | |||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|||||||||
| ANKA | NK65 | MSH2-2 mutant clones | Mosquito-passaged parasites | ||||||
|
|
|||||||||
| A2a | A2b | B2 | NKm | A2c | A2d | C2 | |||
| MS7a | (ATA)24 | (ATA)22 | NC | NC | NC | NC | NC | NC | NC |
| MS8 | (ATA)24 | (ATA)25 | NC | NC | NC | NC | NC | NC | NC |
| MS11 | (TAT)29 | (TAT)27 | NC | NC | NC | NC | NC | NC | NC |
| MS13 | (TTA)24 | (TTA)20 | NC | NC | NC | NC | NC | NC | NC |
| MS18a | (TTA)14 | (TTA)16 | NC | NC | NC | NC | NC | NC | NC |
| (TAG)6 | (TAG)6 | ||||||||
| MS21 | (AAT)13 | (AAT)13 | NC | NC | NC | NC | (AAT)11 | (AAT)11 | NC |
| MS23 | (ATA)8 | (ATA)8 | (ATA)9 | (ATA)9 | (ATA)9 | NC | (ATA)9 | (ATA)9 | NC |
| (ATA)5 | (ATA)5 | (ATA)5 | (ATA)5 | (ATA)5 | (ATA)5 | (ATA)5 | |||
| CG8 | (AAT)22 | (AAT)23 | NC | NC | NC | NC | NC | NC | NC |
| (A)13 | (A)13 | ||||||||
| CG9 | (TTA)24 | (TTA)20 | NC | NC | NC | NC | NC | NC | NC |
| CG11 | (TAT)29 | (TAT)27 | NC | NC | NC | NC | NC | NC | NC |
| CG12 | (ATA)6 | (ATA)6 | NC | NC | NC | NC | NC | NC | NC |
| (TAA)5 | (TAA)5 | ||||||||
| (TAA)5 | (TAA)5 | ||||||||
| (TAA)5 | (TAA)5 | ||||||||
| CG13 | (ATTT)40 | (ATTT)40 | NC | NC | NC | NC | NC | NC | NC |
| CG16 | (T)10 | (T)10 | NC | NC | NC | NC | NC | NC | NC |
| (AT)5 | (AT)5 | ||||||||
| (TTAT)21 | (TTAT)20 | ||||||||
| CG17 | (A)18 | (A)18 | NC | NC | NC | NC | NC | NC | NC |
| (GAT)5 | (GAT)5 | ||||||||
| (GAT)5 | (GAT)5 | ||||||||
| (GAT)6 | (GAT)6 | ||||||||
| CG25 | (AT)22 | (AT)22 | NC | NC | NC | NC | NC | NC | NC |
A2a–d: populations of MSH2-2 mutant clone A2, isolated from separate rodent infections; B2: MSH2-2 mutant clone B2; NKm: NK65 wild type parasites passaged through the mosquito stages of the life cycle; C2: Map2K mutant control clone; NC: no change
Microsatellite region Blasts to putative hypothetical protein in the PlasmoDB database
Discussion
We have found three MutS and two MutL homologs in the genomes of Plasmodium species. Despite the high level of evolutionary conservation observed in the MMR systems of eukaryotes, the complement of MMR genes identified in Plasmodium species appears to be unusual.
Nuclear MMR in eukaryotes is initiated by a heterodimeric complex of either MSH2-MSH3 or MSH2-MSH6 (reviewed in [6–10]). Two of the Plasmodium falciparum MutS homologs, called PfMSH2-1 and PfMSH2-2, have closest homology to MSH2 in other organisms. The third has good homology to MSH6, and no MSH3 ortholog has yet been identified in the genome of any Plasmodium species. While MSH3 is also absent in C. elegans and Drosophila melanogaster, duplicate MSH2 genes have not yet been observed in any other eukaryotic species. While it is possible that one of the Plasmodium MSH2 genes plays a role in mitochondrial DNA repair, our bioinformatics analysis of putative mitochondrial genes in P. falciparum does not provide any evidence for this hypothesis. The two MutL homologs are closest in sequence identity to MLH1 and PMS1. MLH1-PMS1 is the major nuclear heterodimer in S. cerevisiae, and can interact with either MutS heterodimer. Two copies of another gene implicated in MMR, proliferating cell nuclear antigen (PCNA), have also been identified in the Plasmodium genome [23,24].
The unusual complement of genes in the Plasmodium MMR system may reflect the parasites’ particular needs for genetic diversity and replication fidelity. The Plasmodium cell cycle is not well characterized (reviewed in [25]), but it is clearly quite different from that of most other eukaryotes, and includes at least five DNA synthesis phases. The mechanisms used to regulate genome fidelity in these circumstances may be somewhat different from those in a classical cell cycle.
Our initial functional test of the MSH2 genes in P. falciparum using the dominant mutator assay shows an elevation in mutation frequency with the PfMSH2-2 homolog, not unlike the levels reported for other MutS homologues including A. thaliana MutS (3 to 5-fold; [28]) or hMSH2 (~8.5-fold; [14]). It is possible that the absence of highly elevated levels of forward mutation in this assay was due to low-level expression of the falciparum genes in the bacterial system. Although variation existed between experiments in the lac+ reversion frequencies observed, this was well within the range seen in assays of this nature [15,28].
We successfully disrupted the PbMSH2-2 locus in the P. berghei laboratory isolate NK65, and we showed that this gene is not essential for parasite growth in either the asexual (rodent) or sexual (mosquito) stages of the lifecycle. We did not find that PbMSH2-2 disruption had a dramatic effect on drug resistance, although we observed a trend towards a difference in frequency of drug resistance in mosquito-passaged PbMSH2-2 disrupted parasites.
We found microsatellite polymorphism in the PbMSH2-2 disrupted parasites, but not in the PbMap2K disrupted or mosquito passaged NK65 controls, with one change arising within 16 asexual life cycles (assuming a 24 hour life cycle). Although the level of microsatellite polymorphism observed is low, its appearance is rapid, and suggests that MSH2-2 may play a role in maintaining stability at microsatellite regions. Our results are consistent with those reported for other MSH2 disruptions. For example, a recent study of MSH2 and MSH6 knockout strains in C. elegans showed that mutation could be detected in an assay of 15 microsatellites after only 18 generations [29].
As expression and activity of MMR genes in other eukaryotic organisms may be regulated by the expansion/deletion of microsatellite repeats in and around the genes [20], we also assayed microsatellite stability in and around the MMR genes in P. berghei wild type and MSH2-2 mutant strains (Supplementary table 2). We found no polymorphism in any of the microsatellites sequenced, in any strain.
It has been proposed that neighboring repeat sequences may increase the recombination frequencies in MMR proteins, thus increasing the loss and regaining of a mutator phenotype (reviewed in [21]). We examined the chromosomal context, or synteny, of the database sequences of the P. falciparum, P. berghei, P. yoelii, and P. vivax MMR genes in order to detect recombination events near these genes. We found a synteny breakpoint immediately upstream of the coding sequence of PMS1 in P. falciparum compared to the other species. We then performed Southern blots of the PMS1 upstream region on genomic DNA from ten geographically diverse laboratory isolates, but we did not detect any significant changes in restriction fragment length (Supplementary figure 1).
Based on these experiments, we do not believe that PbMSH2-2 disruption results in a strong mutator phenotype. It is possible that PbMSH2-2 is a minor MMR enzyme in this system, or that its function overlaps with that of PbMSH2-1 or other Plasmodium proteins. PbMSH2-2 may play a larger role in one of the other MMR system functions, such as recombination.
Supplementary Material
Supplementary Table 1 – Oligonucleotide primers
Supplementary Table 2 – Repeat regions sequenced in P. berghei MMR gene flanking and coding regions
(A) Signals from hybridization of probe PMS1-s5 to XmnI gDNA digestion, with a predicted fragment size 7.69 kb (B) Signals from hybridization of probe PMS1-s10 to PacI gDNA digestion, predicted restriction fragment size 5.17 kb
Acknowledgments
We thank Timothy Lepore for maintaining mosquito colonies and for assisting with passage of mutant and wild type parasite populations in A. stephensi mosquitoes. We thank Amy Bei, Julia Fisher, and Dr. Kerri Mello for assistance with rodent handling. We would also like to thank Dr. Leona Samson, Dr. Mark Ambrose and Dr. Brian Glassner for their help with setting up the dominant mutator assays. This worked was supported by the NIH (R01-AI050689-03) (A.S.) and R01-GM061351 (D.W.).
Abbreviations
- MMR
mismatch repair
- MSH
mutS homolog
- MLH
mutL homolog
- PlasmoDB
P. falciparum genome database
- pBS-SK+
pBlueScript SK+ vector
- gDNA
genomic DNA
- SD
Sprague-daley
- 5-FOA
5-fluoroorotate
- PCNA
proliferating cell nuclear antigen
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Farooq U, Mahajan RC. Drug resistance in malaria. J Vector Borne Dis. 2004;41(3–4):45–53. [PubMed] [Google Scholar]
- 2.Brooks DR, Wang P, Read M, Watkins WM, Sims PF, Hyde JE. Sequence variation of the hydroxymethyldihydropterin pyrophosphokinase: dihydropteroate synthase gene in lines of the human malaria parasite, Plasmodium falciparum, with differing resistance to sulfadoxine. Eur J Biochem. 1994;224(2):397–405. doi: 10.1111/j.1432-1033.1994.00397.x. [DOI] [PubMed] [Google Scholar]
- 3.Cowman AF, Morry MJ, Biggs BA, Cross GA, Foote SJ. Amino acid changes linked to pyrimethamine resistance in the dihydrofolate reductase-thymidylate synthase gene of Plasmodium falciparum. Proc Natl Acad Sci U S A. 1988;85(23):9109–9113. doi: 10.1073/pnas.85.23.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Korsinczky M, Chen N, Kotecka B, Saul A, Rieckmann K, Cheng Q. Mutations in Plasmodium falciparum cytochrome b that are associated with atovaquone resistance are located at a putative drug-binding site. Antimicrob Agents Chemother. 2000;44(8):2100–2108. doi: 10.1128/aac.44.8.2100-2108.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chopra I, O’Neill AJ, Miller K. The role of mutators in the emergence of antibiotic-resistant bacteria. Drug Resist Updat. 2003;6(3):137–145. doi: 10.1016/s1368-7646(03)00041-4. [DOI] [PubMed] [Google Scholar]
- 6.Harfe BD, Jinks-Robertson S. DNA mismatch repair and genetic instability. Annu Rev Genet. 2000;34:359–399. doi: 10.1146/annurev.genet.34.1.359. [DOI] [PubMed] [Google Scholar]
- 7.Jiricny J. Eukaryotic mismatch repair: an update. Mutat Res. 1998;409(3):107–121. doi: 10.1016/s0921-8777(98)00056-1. [DOI] [PubMed] [Google Scholar]
- 8.Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 9.Marti TM, Kunz C, Fleck O. DNA mismatch repair and mutation avoidance pathways. J Cell Physiol. 2002;191(1):28–41. doi: 10.1002/jcp.10077. [DOI] [PubMed] [Google Scholar]
- 10.Schofield MJ, Hsieh P. DNA mismatch repair: molecular mechanisms and biological function. Annu Rev Microbiol. 2003;57:579–608. doi: 10.1146/annurev.micro.57.030502.090847. [DOI] [PubMed] [Google Scholar]
- 11.Harfe BD, Jinks-Robertson S. Mismatch repair proteins and mitotic genome stability. Mutat Res. 2000;451(1–2):151–167. doi: 10.1016/s0027-5107(00)00047-6. [DOI] [PubMed] [Google Scholar]
- 12.Kissinger JC, Brunk BP, Crabtree J, et al. The Plasmodium genome database. Nature. 2002;419(6906):490–492. doi: 10.1038/419490a. [DOI] [PubMed] [Google Scholar]
- 13.Cupples CG, Cabrera M, Cruz C, Miller JH. A set of lacZ mutations in Escherichia coli that allow rapid detection of specific frameshift mutations. Genetics. 1990;125(2):275–280. doi: 10.1093/genetics/125.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fishel R, Lescoe MK, Rao MR, et al. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993;75(5):1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- 15.Glassner BJ, Rasmussen LJ, Najarian MT, Posnick LM, Samson LD. Generation of a strong mutator phenotype in yeast by imbalanced base excision repair. Proc Natl Acad Sci U S A. 1998;95(17):9997–10002. doi: 10.1073/pnas.95.17.9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Koning-Ward TF, Fidock DA, Thathy V, et al. The selectable marker human dihydrofolate reductase enables sequential genetic manipulation of the Plasmodium berghei genome. Mol Biochem Parasitol. 2000;106(2):199–212. doi: 10.1016/s0166-6851(99)00189-9. [DOI] [PubMed] [Google Scholar]
- 17.Sultan AA, Thathy V, Frevert U, et al. TRAP is necessary for gliding motility and infectivity of plasmodium sporozoites. Cell. 1997;90(3):511–522. doi: 10.1016/s0092-8674(00)80511-5. [DOI] [PubMed] [Google Scholar]
- 18.Le Roch KG, Zhou Y, Blair PL, et al. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science. 2003;301(5639):1503–1508. doi: 10.1126/science.1087025. [DOI] [PubMed] [Google Scholar]
- 19.Rangarajan R, Bei AK, Jethwaney D, et al. A mitogen-activated protein kinase regulates male gametogenesis and transmission of the malaria parasite Plasmodium berghei. EMBO Rep. 2005;6(5):464–469. doi: 10.1038/sj.embor.7400404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang DK, Metzgar D, Wills C, Boland CR. Microsatellites in the eukaryotic DNA mismatch repair genes as modulators of evolutionary mutation rate. Genome Res. 2001;11(7):1145–1146. doi: 10.1101/gr.186301. [DOI] [PubMed] [Google Scholar]
- 21.Caporale LH. Natural selection and the emergence of a mutation phenotype: an update of the evolutionary synthesis considering mechanisms that affect genome variation. Annu Rev Microbiol. 2003;57:467–485. doi: 10.1146/annurev.micro.57.030502.090855. [DOI] [PubMed] [Google Scholar]
- 22.Tornier C, Bessone S, Varlet I, Rudolph C, Darmon M, Fleck O. Requirement for Msh6, but not for Swi4 (Msh3), in Msh2-dependent repair of base-base mismatches and mononucleotide loops in Schizosaccharomyces pombe. Genetics. 2001;158(1):65–75. doi: 10.1093/genetics/158.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li JL, Warren AV, Cox LS. Identification of a second proliferating cell nuclear antigen in the human malarial pathogen Plasmodium falciparum. Int J Parasitol. 2002;32(14):1683–1692. doi: 10.1016/s0020-7519(02)00162-5. [DOI] [PubMed] [Google Scholar]
- 24.Patterson S, Whittle C, Robert C, Chakrabarti D. Molecular characterization and expression of an alternate proliferating cell nuclear antigen homologue, PfPCNA2, in Plasmodium falciparum. Biochem Biophys Res Commun. 2002;298(3):371–376. doi: 10.1016/s0006-291x(02)02436-1. [DOI] [PubMed] [Google Scholar]
- 25.Arnot DE, Gull K. The Plasmodium cell-cycle: facts and questions. Ann Trop Med Parasitol. 1998;92(4):361–365. doi: 10.1080/00034989859357. [DOI] [PubMed] [Google Scholar]
- 26.Janse CJ, Van der Klooster PF, Van der Kaay HJ, Van der Ploeg M, Overdulve JP. Rapid repeated DNA replication during microgametogenesis and DNA synthesis in young zygotes of Plasmodium berghei. Trans R Soc Trop Med Hyg. 1986;80(1):154–157. doi: 10.1016/0035-9203(86)90219-1. [DOI] [PubMed] [Google Scholar]
- 27.Baton LA, Ranford-Cartwright LC. Spreading the seeds of million-murdering death: metamorphoses of malaria in the mosquito. Trends Parasitol. 2005;21(12):573–580. doi: 10.1016/j.pt.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 28.Ade J, Haffani Y, Beizile FJ. Functional analysis of the Arabidopsis thaliana mismatch repair gene MSH2. Genome. 2001;44(4):651–657. [PubMed] [Google Scholar]
- 29.Denver DR, Feinberg S, Estes S, Thomas WK, Lynch M. Mutation rates, spectra and hotspots in mismatch repair-deficient Caenorhabditis elegans. Genetics. 2005;170(1):107–113. doi: 10.1534/genetics.104.038521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Machado CR, Augusto-Pinto L, McCulloch R, Teixeira SM. DNA metabolism and genetic diversity in Trypanosomes. Mutat Res. 2005 doi: 10.1016/j.mrrev.2005.05.001. [DOI] [PubMed] [Google Scholar]
Website References
- The Plasmodium Genome Database [http://www.plasmoDB.org]
- The Institute for Genomic Research (TIGR) [www.tigr.org]
- The Sanger Institute website [www.sanger.ac.uk/DataSearch]
- National Center for Biotechnology Information (NCBI) [http://www.ncbi.nlm.nih.gov]
- Protein families database alignments and HMM (PFAM) [http://www.sanger.ac.uk/Software/Pfam/]
- Prosite database of protein families and domains [http://us.expasy.org/prosite/]
- European bioinformatics institute InterProScan [http://www.ebi.ac.uk/InterProScan/]
- Primer3 [http://www-genome.wi.mit.edu/cgi-bin/primer/primer3]
- Perfect microsatellite repeat finder [http://sgdp.iop.kcl.ac.uk/nikammar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1 – Oligonucleotide primers
Supplementary Table 2 – Repeat regions sequenced in P. berghei MMR gene flanking and coding regions
(A) Signals from hybridization of probe PMS1-s5 to XmnI gDNA digestion, with a predicted fragment size 7.69 kb (B) Signals from hybridization of probe PMS1-s10 to PacI gDNA digestion, predicted restriction fragment size 5.17 kb