Abstract
Proper control of mitochondrial turnover is critical for maintenance of cellular energetics under basal and stressed conditions, and for prevention of endogenous oxidative stress. Whole organelle turnover is mediated through macroautophagy, a process by which autophagosomes deliver mitochondria to the lysosome for hydrolytic degradation. While mitochondrial autophagy can occur as part of a nonselective upregulation of autophagy, selective degradation of damaged or unneeded mitochondria (mitophagy) is a rapidly growing area in development, cancer, and neurodegeneration, particularly with regard to Parkinson’s disease. Due to its dynamic nature, and the potential for regulatory perturbation by disease processes, no single technique is sufficient to evaluate mitophagy. Here, we describe several complementary techniques that include electron microscopy, single cell analysis of LC3 fluorescent puncta, and Western blot, each used in conjunction with a flux inhibitor to trap newly formed autophagosomes in order to monitor mitophagy in neuronal cells.
Keywords: Autophagy, Mitophagy, Electron microscopy, Western Blot, RFP-LC3, GFP-LC3, Immunofluorescence
1. Introduction
Mitochondria are the power generators of the cell, converting oxygen and nutrients into adenosine triphosphate (ATP). Mitochondria also play an important role in maintaining calcium homeostasis, regulating lipid metabolism and heme biogenesis (1). However, excessive or damaged mitochondria may generate ROS and release cytochrome c, AIF, or other proapoptotic proteins to promote cell death (2–4). During nutrient deprivation or chronic hypoxia, mitochondria could be recycled to provide more urgently needed molecules. In pathological situations, timely elimination of dysfunctional mitochondria represents a cytoprotective response (5, 6) while global mitochondrial elimination has been implicated as part of a regulated cell death program (7).
Mitochondrial quality control is essential for cells to maintain mitochondrial integrity and normal function (8). Even under basal conditions, continuous cycles of mitochondrial fusion and fission and of biogenesis and degradation serve to produce daughter mitochondria and remove dysfunctional or effete mitochondria. The predominant mechanism for whole organelle turnover is autophagic sequestration and delivery to the lysosome for hydrolytic degradation. Selective autophagy of mitochondria is termed mitophagy (1, 9, 10), although mitochondria are also degraded during nonselective bulk cytoplasmic degradation. Mitochondrial dynamics and mitophagy are believed to play a key role in neurodegenerative diseases and the aging process itself (10, 11).
Recent studies indicate that the efficiency of mitophagy may be genetically controlled. Two proteins whose mutations are associated with autosomal recessive parkinsonism have been implicated in mitophagy. Parkin, an E3 ubiquitin ligase, is recruited to depolarized mitochondria to promote mitophagy in cancer cells (12). While loss of PTEN-induced kinase 1 (PINK1) results in increased mitophagy (6), overexpression of full-length PINK1 enhances Parkin recruitment to depolarized mitochondria (13–15), and mutations in either protein may impair mitochondrial depolarization-induced mitophagy. Nix, a BH3-only member of the Bcl-2 family, is a mitochondrial outer membrane protein that is essential for autophagic elimination of mitochondria during red blood cell development (16). The mitochondrial protein Atg32 interacts with the autophagy machinery in yeast (17), and other yeast proteins essential for mitophagy include the phosphatase AUP1P (18) and outer membrane-localized UTH1p (19). Mitophagy is also regulated by serine/threonine kinases, such as ERK2 (20), and mitochondrial fission and fusion-related proteins, such as Drp1 and mitofusin, may play a permissive role (10). Drp1 knockdown or expression of OPA1 or dominant negative Drp1 suppresses mitophagy (6, 21), which is of course dependent upon general autophagy machinery proteins Atg5, Atg7, Ulk1, and the Atg8 homolog microtubule-associated protein 1 light chain 3 (LC3) (22–24).
To better understand the role and mechanism of mitophagy during disease processes, several protocols to monitor and visualize mitochondrial autophagy have been reported. Each has caveats and limitations; therefore, multiple complementary techniques are necessary to establish induction of mitophagy by a particular cellular condition. As with any other dynamic process in cell biology, steady-state levels of a given organelle, such as a mitochondria-containing autophagosome (“mitophagosome”), are regulated by multiple potential factors, including rate of autophagosome formation, rate of maturation and lysosomal degradation, efficacy of cargo targeting to the forming autophagosomes, and the stability of the cargo and/or its detection method to the local environment inside maturing autophagic vacuoles (AVs) and autolysosomes.
Ultrastructural evaluation of mitophagy is a direct method to provide confirmation of mitochondrial autophagy or clearance. However, maturation and lysosomal fusion is a relatively rapid process, and it may be difficult to recognize mitochondrial morphology in autophagosomes. Preventing autophagosome maturation with short-term pulses of bafilomycin A1 traps newly formed autophagosomes, facilitating identification of cargo as well as providing comparative information on rates of autophagosome formation (8).
Disappearance of fluorescent signal for individual proteins, such as translocase of the outer membrane (TOM20), cytochrome c, HtrA2/Omi, or mitochondrially targeted fluorescent proteins, are frequently used as assays for mitophagy. However, intermembrane space proteins are lost with permeability transition in damaged mitochondria, and mitochondrially targeted proteins, such as TOM20 and Mito-GFP, are degraded by the proteasome under conditions that disrupt mitochondrial import (25). GFP itself shows pH-sensitive fluorescence and can be degraded by the lysosome (26). The possibility that observed loss of an overexpressed cytosolic protein reflects nonselective bulk autophagy should also be considered. Thus, we recommend that these types of studies, which are very useful for demonstrating potential mitophagic flux, are performed in conjunction with assays that (1) directly demonstrate association of mitochondria with autophagosomes and (2) confirm the ability to reverse the loss of mitochondrial signal using specific inhibitors of the autophagolysosomal pathway.
Decreases in expression levels of mitochondrial proteins is another useful method to confirm loss of the organelle, although electron microscopy allows for a direct assessment that the entire mitochondrion is absent as opposed to more selective degradation of certain protein components by other mechanisms. Ideally, several different proteins from different mitochondrial subcompartments to include the matrix and inner membrane are monitored for parallel decreases. Changes in protein levels due to alterations in biosynthesis, epitope destruction without loss of the entire protein, or alternative degradation pathways, such as proteasomal and intramitochondrial proteases, are important factors to consider (8).
In general, inferences of autophagy that depend solely upon loss of a signal are less optimal than assays that produce a positive signal confirming delivery to autophagosomes and lysosomes. On the other hand, steady-state increases in “mitophagosomes” may reflect increased sequestration or decreased maturation, requiring specific attention to issues of flux. Tracking multiple proteins in different subcompartments of the mitochondria and reversal studies using specific methods to inhibit autophagy or proteasomal degradation should be employed to evaluate relative contributions of these major degradative pathways. Herein, we describe methods employing electron microscopy, single cell analysis of LC3 puncta and mitochondrial cofluorescence, and Western blot experiments to evaluate mitophagy and mitophagic flux in neuronal cells.
2. Materials
2.1. Cell Culture
Cell lines: SH-SY5Y, a human neuroblastoma cell line (American Type Culture Collection, Rockville, MD).
Culture medium for SH-SY5Y cells (Dulbecco’s modified Eagle’s medium, DMEM): Antibiotic-free DMEM with 4.5 g/l d-glucose (BioWhittaker, Walkerville, MD); 10% heat-inactivated fetal bovine serum (Gibco/Invitrogen, Carlsbad, CA); 10 mmol/l HEPES; 2 mmol/l glutamine.
10 mM Retinoic acid (1,000× stock solution) (Sigma, St. Louis, MO, USA), dissolved in DMSO and stored at −20°C in 20–50 µl aliquots.
Poly-d-lysine (10 mg/ml stock) (Sigma).
Culture medium for primary neurons (NB/B27): Serum-free neurobasal medium supplemented with l-gluta Max™ (2.0 mM) and B27™ (Invitrogen, Carlsbad, CA).
6-hydroxydopamine (6-OHDA; Sigma), freshly prepared in distilled water or media.
1-methyl-4-phenylpyridinium (MPP+; Sigma), prepared in distilled water, sterile filtered, and stored at −20°C.
10 mM E64-D (Calbiochem, San Diego, CA): Stock made in DMSO, stored at −20°C; use at 10 µM for SH-SY5Y cells.
25 mM Pepstatin-A (Calbiochem): Dissolved in methanol or DMSO; use at 25 µM for SH-SY5Y cells.
10 µM Bafilomycin A1 (Sigma), dissolved in DMSO as stock solution.
10 mM MG132 (Sigma), dissolved in methanol as stock solution; stored at −20°C.
Chambered Lab-tek II cover glasses (#1.5 German borosilicate; Nalge Nunc International, Naperville, IL, USA).
2.2. Electron Microscopy
0.1 M PBS, pH 7.4.
2.5% glutaraldehyde in 0.1 M PBS, pH 7.4.
1% osmium in 0.1 M PBS.
2% uranyl acetate solution, prepared in distilled water. Store at 4°C.
1% lead citrate solution: Prepared in distilled water. Store for 3–6 months at 4°C.
Polybed 812 epoxy resin (Polysciences, Warrington, PA).
JEOL JEM 1210 transmission electron microscope (JEOL, Peabody, MA).
2.3. Image (Fluoresence)-Based Analysis of Mitophagy
1 mM MitoTracker Red dye 580 (MTR; Molecular Probes, Eugene CA), dissolved in DMSO and stored at −20°C.
1 mM MitoTracker Green FM dye (Molecular Probes), dissolved in DMSO and stored at −20°C.
1 mM LysoTracker Red DND-99 (Molecular Probes), dissolved in DMSO and stored at −20°C.
FluoView 1000 (Olympus America) or Zeiss LSM 510 Meta laser-scanning confocal microscope (Carl Zeiss MicroImaging, Thornwood, NY).
2.4. Western Blot Analysis of Mitophagy
Lysis buffer: 25 mM HEPES, pH 7.5; 150 mM NaCl; 1% Triton X-100; 10% glycerol with freshly added proteinase and phosphatase inhibitors, including 100 µM E64, 1 mM sodium orthovanadate, 2 mM sodium pyrophosphate, 2 mM PMSF.
5–15% polyacrylamide gradient gel.
Immobilon-P membranes (Millipore, Bedford, MA, USA).
Blocking solution: 5% nonfat dry milk in 20 mM potassium phosphate, 150 mM potassium chloride, pH 7.4, containing 0.3% (w/v) Tween-20 (PBST).
Mouse anti-pyruvate dehydrogenase antibody (1:1000, Molecular Probes).
Mouse anti-inner mitochondrial membrane protein human mitochondrial antigen of 60 kDa antibody (1:1000, Biogenex, San Ramon, CA).
Rabbit anti-outer mitochondrial membrane protein TOM20 antibody (1:10,000, Santa Cruz Biotechnologies, Santa Cruz, CA).
3. Methods
3.1. Ultrastructural Assay of Mitophagy
SH-SY5Y cells are maintained in antibiotic-free media and used at passages 30–45. Cells are plated at a density of 3 × 104/cm2 in six-well culture plates. Parallel cultures of the experimental condition being tested (such as molecular or pharmacologic manipulation) are treated with 5–10 nM bafilomycin A1, an inhibitor of vacuolar-type H(+)-ATPase and autophagosome–lysosome fusion, or its vehicle for 2–4 h prior to fixation (see Note 1).
Cells are rinsed three times with 0.1 M PBS, fixed for at least 60 min in 2.5% glutaraldehyde at room temperature, or overnight at 4°C.
After fixation, cell monolayers are washed three times in PBS, and then postfixed in aqueous 1% OsO4 and 1% K3Fe(CN)6 for 1 h.
After three PBS washes, the cultures are dehydrated through a graded series of 30–100% ethanol solutions, infiltrated, and then embedded in Polybed 812 epoxy resin.
Ultrathin (60 nm) sections are collected on copper grids and stained with 2% uranyl acetate in 50% methanol for 10 min, followed by an incubation in 1% lead citrate for 7 min.
Sections are photographed using a JEOL JEM 1210 transmission electron microscope at 80 kV.
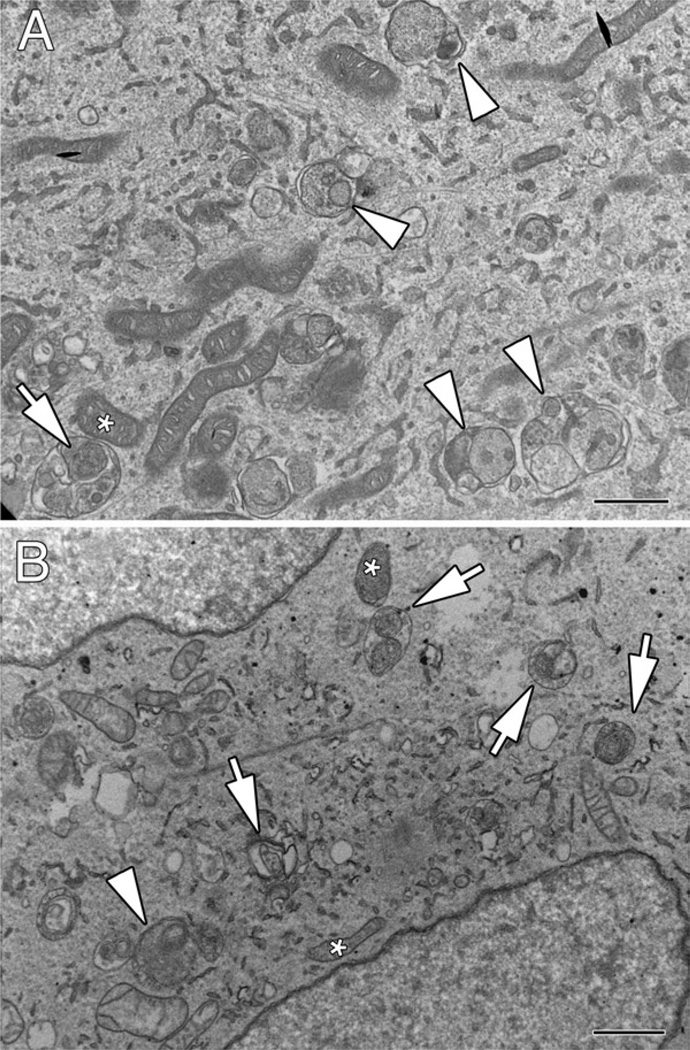
As an example of ultrastructural analysis of mitophagy, Fig. 1 shows a comparison of early autophagosomes (AVis) trapped by bafilomycin A1 treatment of control SH-SY5Y cells (Fig. 1a) versus the PINK1 shRNA line A14 (Fig. 1b). Because of the low rate of autophagy induction in control cells, a 24 h treatment with bafilomycin A1 was used to derive sufficient numbers of AVis for analysis of their content, although such a lengthy pulse is not recommended for stressed cells or for flux analysis, due to saturation. For comparative analysis of sequestration rates, pulses of 2–6 h are ideal (8) (see Note 2). While the majority of AVis trapped by bafilomycin A1 in control cells contain a variety of organellar and membranous structures (Fig. 1a, arrowheads) with only rare mitochondria identified amid other structures (Fig. 1a, arrow), multiple AVis containing mitochondrial profiles (Fig. 1b, arrows) are identified in PINK1 shRNA cells. Stress and injury can change the appearance of mitochondria in cells. Comparison of putative mitochondrial profiles within the AVi with nearby mitochondria in the cytoplasm (Fig. 1, asterisks) facilitates their identification.
Fig. 1.
Analysis of AVi content by electron microscopy using an inhibitor of autophagosome–lysosome fusion. (a) Control SH-SY5Y cells were treated with 5 nM bafilomycin A1 × 24 h prior to fixation and analysis by electron microscopy. Note the accumulation of several AVis containing heterogeneous cytoplasmic material (arrowheads). An occasional AVi exhibits a mitochondrial profile (arrow) similar to an adjacent mitochondrion in the cytoplasm (asterisk). (b) The PINK1 shRNA line A14 was treated with 10 nM bafilomycin A1 × 2 h prior to fixation and analysis by electron microscopy. Mitochondrial profiles are readily identified in the bafilomycin-trapped AVis (arrows), although generalized cargo was also observed in some (arrowhead). Note heterogeneity in electron density of mitochondria within AVis that are similar to heterogeneity observed among free mitochondria in the cytoplasm (asterisks). In the absence of bafilomycin A1, cargo is typically altered to the extent that it can no longer be recognized (see, for example, published images of the A14 cell line in (6, 8)). Scale bars: 1 µm.
3.2. Fluorescence Methods for Analyzing Mitophagy
Confocal or epifluorescence microscopy using GFP-LC3 as a marker of autophagy can be used to monitor mitophagy induction in primary neurons or SH-SY5Y cells.
-
SH-SY5Y cells are seeded in complete DMEM medium on uncoated chambered Lab-tek II cover glasses. SH-SY5Y cells can be studied in their undifferentiated state or following retinoic acid treatment (10 µM for at least 3 days as judged by the extension of neurites of at least two soma lengths) to induce neuronal differentiation.
Chambered cover glasses (four well) are coated with 100 µg/ml per well of poly-d-lysine for at least 4 h prior to seeding with primary cortical or midbrain neurons (100,000 neurons per well) in NB/B27.
Cells are transiently transfected with GFP-LC3 (1 µg DNA diluted in OPTIMEM in 0.10% Lipofectamine 2000). Four to six hours following transfection, one volume of complete DMEM medium or NB/B27 medium is added to the SH-SY5Y or primary neurons, respectively, and the cultures incubated overnight in a 5% CO2 incubator at 37°C. The day following transfection, approximately two-thirds of the media is replenished with fresh media.
To visualize the “colocalization” of mitochondria with autophagosomes, GFP-LC3-expressing cultures are loaded with 100 nM MTR dye 580 (diluted from 1 mM DMSO stocks directly into media overlying cells). This loading should be done prior to experimental treatments that may affect mitochondrial membrane potential (see Note 3). The cultures are washed once with warm media to remove unbound/cytosolic background staining prior to performing image analysis using an epifluorescence or a laser confocal microscope.
In order to achieve statistical significance for measuring mitophagy, it is necessary to analyze high-quality images captured at a high magnification (60× or 90× if a 10× amplifier is used), imaging at least 25–30 cells per condition (27). We use an inverted FluoView 1000 or Zeiss LSM 510 Meta laser-scanning confocal microscope (excitation/emission filter, 488/510 nm; 561/592 nm) to image for mitophagy. Optimization experiments to compare the results of live imaging with imaging after fixation in a positive control condition are advised for each cell type, as fewer GFP-LC3 puncta are detected in fixed cells under basal conditions, but the background is also decreased with formaldehyde fixation (27). Glutaraldehyde fixation may produce autofluorescent puncta (possibly related to lipofuscin) as an artifact of fixation, and use of this fixative is not recommended for fluorescence studies.
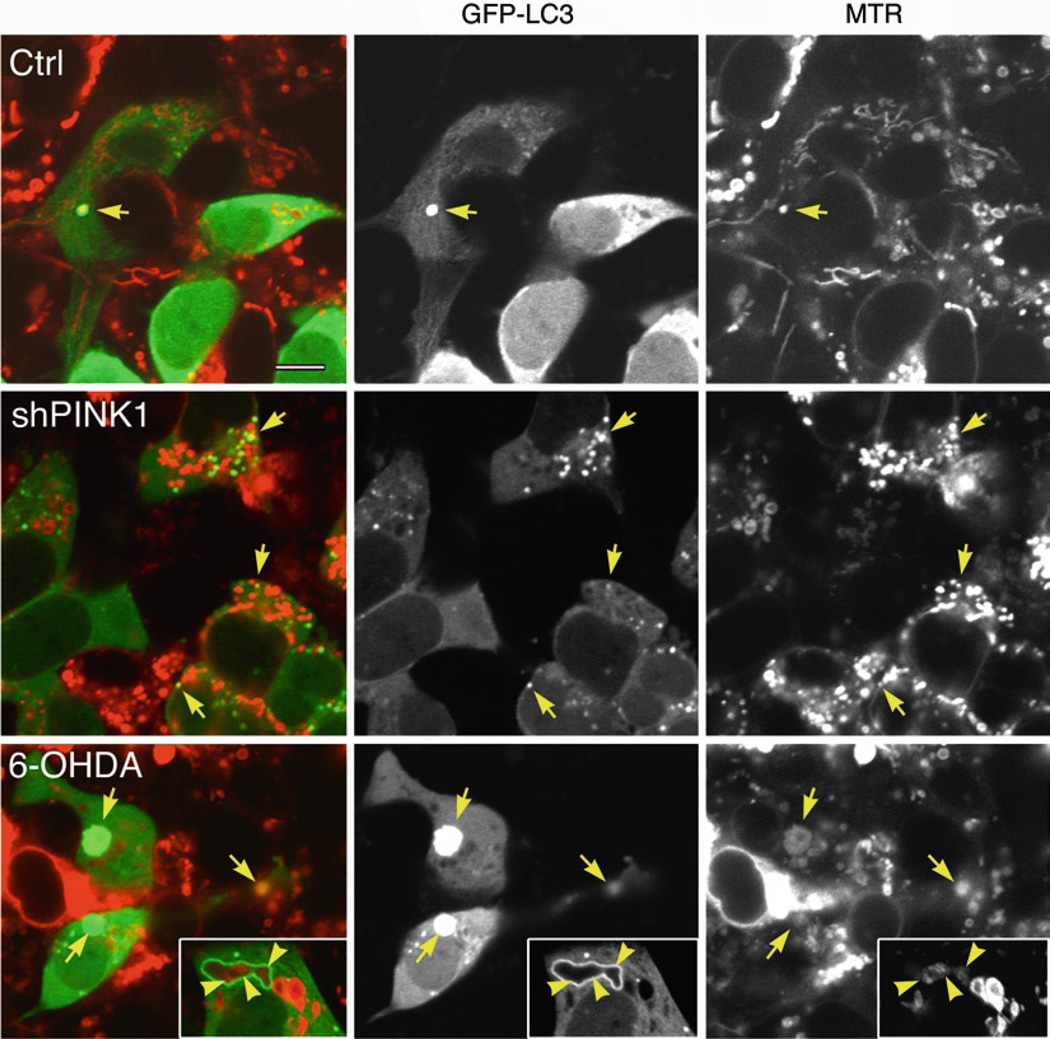
There are two patterns of fluorescence association reflective of mitophagy. First, a straightforward colocalization of LC3 and mitochondrial signals in a punctate form (Fig. 2, arrows) is most often observed. Notably, in merged images, colocalizing LC3 and mitochondria do not always appear yellow due to differences in intensity of staining. For example, a strong GFP-LC3 signal may overwhelm a weaker MTR signal exhibited by marginally polarized mitochondria or a dim mito-RFP signal in low-expressing cells (see Note 4). Conversely, yellow pixels due to overlay of a GFP-LC3 puncta over a larger region of MTR signal most likely do not represent mitophagy. The second pattern that can be observed is of ring-like GFP-LC3 structures surrounding discrete mitochondrial fragments (Fig. 2, bottom row, inset); while there is technically no colocalization of the fluorescent pixels, this is counted as a “mitophagosome.”
Fig. 2.
Analysis of mitophagic sequestration by dual fluorescence. Stable cell lines that express an empty vector (Ctrl) or that stably knock down endogenous PINK1 (shPINK1) were transfected with GFP-LC3 for 2 days. Mitochondria were labeled by loading cells with 100 nM MTR 580 dye for 30 min at 37°C prior to treating some wells with 6-hydroxydopamine (6-OHDA, 120 µM × 4 h). Confocal images show a significant increase in GFP-LC3 puncta that colocalize with mitochondria in shPINK1 or 6-OHDA-treated cells (arrows). Note that 6-OHDA increases the average size of AVs and causes mitochondrial swelling, often associated with decreased MTR staining intensity, while shPINK1 results in fragmentation of the mitochondrial network with smaller autophagosomes consistent with the EM image of Fig. 21.1b. The inset shows a large, irregular GFP-LC3 ring encircling three distinct mitochondrial profiles (arrowheads); typically, the GFP-LC3 rings are rounder, encircling only one mitochondrial profile. Scale bar: 10 µm.
In addition to analyzing for mitophagy (see Note 5), changes in the general level of macroautophagy can be analyzed within the same experiment by quantifying the average number of GFP-LC3 puncta per cell in the green channel (27).
3.3. Flux Analysis
For each of the image-based methods of analyzing mitochondria contained in autophagosomes (EM and dual fluorescence), an estimate of flux can be derived from comparisons in the presence and absence of bafilomycin A1 (see Note 1) using the following formula:
Mitophagic Flux = (mean mitophagosome #/ cell with Baf − mean mitophagosome #/ cell without Baf)/ time in Baf.
Alternatively, a qualitative assessment can be made through monitoring lysosomal delivery of mitochondria. Primary cortical or SH-SY5Y neuroblastoma cells are colabeled with 250 nM MitoTracker Green FM dye, which is relatively independent of mitochondrial membrane potential in many cell types, and loaded with LysoTracker Red DND-99 (100 nM) to label lysosomes, followed by a wash with warmed complete DME media (6) (see Note 6). RFP-LC3, an AV reporter construct that is more stable in the acidic environment of lysosomes compared to GFP-LC3 (26), can also be used to study both early and late AVs (28). The cells can be live-cell imaged immediately or they can be fixed in 4% paraformaldehyde (15 min at room temperature).
Finally, disappearance of mitochondrial fluorescence relative to diffuse GFP fluorescence in transfected cells (percent cellular pixels occupied by mitochondria) can be used to quantify mitochondrial loss, particularly at later time points (see Note 7).
3.4. Western Blot Analysis of Mitophagy
Cells are treated with a possible inducer of mitophagy in the presence or absence of 10 nM bafilomycin A1 (see Note 1) for at least 4 h, which blocks the fusion of AVs with lysosomes, or with the cell-permeable lysosomal protease inhibitors E64-D and pepstatin-A. Alternatively, RNAi against autophagy-related genes Atg7 and Atg8 can be employed 2–3 days prior (20, 29). Western blots (see Note 8) are probed for the total cellular levels of multiple mitochondrial proteins (e.g., manganese superoxide dismutase, porin, adenine nucleotide translocase, pyruvate dehydrogenase, or the complex IV human mitochondrial antigen of 60 kDa) (see Notes 9 and 10). Western blots are also probed for nonmitochondrial proteins, such as cytochrome P450 of the endoplasmic reticulum, to determine selectivity for mitochondria.
Reprobing the Western blot membranes for LC3, and for β-actin as a normalizing protein, is used to verify efficacy of the autophagy inhibitors (bafilomycin A1 or RNAi). The lipidated form of LC3 (LC3-II) exhibits a faster electrophoretic migration by SDS-PAGE compared to LC3-I, and the LC3-II/β-actin ratio is a well-accepted measure of autophagosome content (reviewed in (30)). For example, an increase in the LC3-II-to-β-actin ratio in cells pretreated with bafilomycin A1 (4 h at 10 nM for SH-SY5Y cells) is indicative of successful pharmacological suppression of autophagosome maturation. Likewise, a significant decrease in LC3-II induced in the presence of RNAi targeting essential Atg proteins is indicative of successful suppression of autophagy induction (20, 29, 31).
3.5. Conclusion
To summarize, no single technique is sufficient to establish an effect of a given experimental manipulation on mitophagy. Employing several complementary techniques that include (1) direct electron microscopy visualization of “mitophagosomes,” (2) flux analysis of fluorescent puncta exhibiting colocalization of cargo and markers of early and late AVs, and (3) carefully controlled Western blot experiments should be sufficient for evaluating effects of a particular, gene product, treatment, or small compound on mitophagy (6, 20, 29).
Acknowledgements
This work was supported in part by the National Institutes of Health (AG026389, NS065789). CTC is recipient of an AFAR/ Ellison Medical Foundation Julie Martin Mid-Career Award in Aging Research.
Footnotes
The optimal bafilomycin A1 concentrations and treatment times are likely to be cell type-dependent. The ideal concentration and time course should be optimized to minimize toxicity and prevent saturation of the observed LC3 II accumulation. LysoTracker Red staining should be titrated to establish the minimum dose that is effective in increasing lysosomal pH for a particular cell type (32). In our hands, a concentration of bafilomycin A1 of 10 nM for more than 6 h leads to saturation of AV content in SH-SY5Y cells while a 20 nM concentration leads to cytotoxicity. Even though 10 nM does not cause toxicity in this time frame, there are ultrastructural effects on mitochondria (pallor, cristae separation) in control cells; thus, 5 nM may be optimal for ultrastructural studies.
At the EM level, the decrease of mitochondria and increase of autophagy/mitophagy could be quantitatively analyzed. Comparisons in the presence or absence of bafilomycin A1 provide information about whether neurotoxin or other experimental treatments increase or decrease lysosome-dependent flux using the formula indicated above.
The disappearance of MTR or tetramethylrhodamine methyl ester (TMRM) signal from cells by flow cytometry or fluores-cence microscopy has been used to infer autophagic clearance of mitochondria. However, the loading and accumulation of MTR and TMRM in mitochondria are dependent on trans-membrane potential. Loss of TMRM signal indicates a membrane potential change that could initiate mitophagy, but does not necessarily reflect mitophagy (33). Even the advertised membrane potential-independent dye Mitotracker Green may show cell type- and concentration-dependent sensitivity to oxidative and depolarizing treatments (34), necessitating optimization experiments for each cellular context. In short, these probes are ideally loaded into cells prior to treatments that may dissipate mitochondrial membrane potential.
Cells may be cotransfected with monomeric RFP (mito-RFP) targeted to mitochondria via the leader sequences of various cytochrome c oxidase (COX) subunits to visualize colocalization of mitochondria with GFP-LC3 (6, 27). A word of caution is that transient overexpression of RFP targeted into mitochondria by COX subunit presequences may cause increased basal autophagy/mitophagy in neurons for several days after transfection. Also, proper mitochondrial import is dependent upon healthy mitochondrial membrane potentials, and an overexpressed protein may compete with other proteins at the level of the outer mitochondrial membrane translocase system (Dagda & Chu, unpublished observations).
An increase in the percentage of GFP-LC3 puncta, that colocalize with MTR or mito-RFP-labeled mitochondria, or the percentage of RFP-LC3 puncta that colocalize with mitochondria labeled with mito-GFP suggests induction of mitophagy, rather than simply increased bulk autophagy.
Bafilomycin A1 cannot be used with LysoTracker Red, as it quenches lysosomal-specific fluorescence by disrupting pH (27, 35).
Given that GFP-LC3 and RFP-LC3 show dual cytosolic and punctate distributions, the percentage of the cellular area occupied by mitochondria in GFP-LC3 or RFP-LC3 transfected cells can be measured using NIH Image J (Bethesda, MD) to quantify the extent of mitochondrial loss induced by a specific treatment (27, 29). A low threshold level is set for the green channel of the RGB image containing the GFP-LC3 fluores-cence in order to detect faint diffuse LC3 labeling that highlights the entire cell area (as opposed to detecting only bright spots >1.5 standard deviations above the background for detection of autophagic puncta). A decrease in the percentage of cellular area occupied by mitochondria, induced by a specific treatment compared to untreated cells, suggests mitochondrial degradation. However, reversing this mitochondrial loss by cotreating cells with bafilomycin A1, lysosomal protease inhibitors, or Atg7/Atg8 RNAi is needed to confirm induction of mitophagy (6). Rates of mitochondrial biogenesis is another factor to be considered.
Depending upon the mitochondrial protein of interest, a zwit-terionic detergent, such as CHAPS, may be added to the lysis buffer for better extraction of membrane proteins.
Interpretation of immunoblots for apoptogenic factors of the intermembrane space, which can be released by different toxic insults, can be difficult (2). Complex I proteins frequently show disproportionate decreases compared to other mitochondrial proteins, which may reflect mechanisms other than whole organelle turnover (selective degradation or impaired biosynthesis). It is important to keep in mind that mitochondrially targeted proteins can undergo proteasomal degradation under conditions that impair potential-dependent import (25).
To rule out that the effects of a particular treatment on mitochondrial protein levels are not contributed by (a) the ubiquitin proteasome system, (b) localized degradation of mitochondrial proteins (8), or (c) decreased efficiency of biosynthesis, import, or assembly of complex proteins, additional studies are required. In addition to proteasome inhibitors (i.e., MG132 and clasto-lactacystin-lactone), quantitative RT-PCR or reprobing the membrane for TFAM, a nuclear-encoded mitochondrial-specific transcription factor, may be used to implicate alterations in biogenesis (36, 37).
References
- 1.Bueler H. Mitochondrial dynamics, cell death and the pathogenesis of Parkinson’s disease. Apoptosis. 2010;15:1336–1353. doi: 10.1007/s10495-010-0465-0. [DOI] [PubMed] [Google Scholar]
- 2.Chu CT, Zhu JH, Cao G, Signore A, Wang S, Chen J. Apoptosis inducing factor mediates caspase-independent 1-methyl-4-phenylpyridinium toxicity in dopaminergic cells. Journal of neurochemistry. 2005;94:1685–1695. doi: 10.1111/j.1471-4159.2005.03329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jennings JJ, Jr., Zhu JH, Rbaibi Y, Luo X, Chu CT, Kiselyov K. Mitochondrial aberrations in mucolipidosis Type IV. The Journal of biological chemistry. 2006;281:39041–39050. doi: 10.1074/jbc.M607982200. [DOI] [PubMed] [Google Scholar]
- 4.Gomez-Lazaro M, Bonekamp NA, Galindo MF, Jordan J, Schrader M. 6-Hydroxydopamine (6-OHDA) induces Drp1-dependent mitochondrial fragmentation in SH-SY5Y cells. Free radical biology & medicine. 2008;44:1960–1969. doi: 10.1016/j.freeradbiomed.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 5.Suen DF, Narendra DP, Tanaka A, Manfredi G, Youle RJ. Parkin overexpression selects against a deleterious mtDNA mutation in heteroplasmic cybrid cells. Proceedings of the National Academy of Sciences of the United States of America. 107:11835–11840. doi: 10.1073/pnas.0914569107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dagda RK, Cherra SJ, 3rd, Kulich SM, Tandon A, Park D, Chu CT. Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. The Journal of biological chemistry. 2009;284:13843–13855. doi: 10.1074/jbc.M808515200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tolkovsky AM, Xue L, Fletcher GC, Borutaite V. Mitochondrial disappearance from cells: a clue to the role of autophagy in programmed cell death and disease? Biochimie. 2002;84:233–240. doi: 10.1016/s0300-9084(02)01371-8. [DOI] [PubMed] [Google Scholar]
- 8.Chu CT. A pivotal role for PINK1 and autophagy in mitochondrial quality control: implications for Parkinson disease. Human molecular genetics. 2010;19:R28–R37. doi: 10.1093/hmg/ddq143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knott AB, Bossy-Wetzel E. Impairing the mitochondrial fission and fusion balance: a new mechanism of neurodegeneration. Annals of the New York Academy of Sciences. 2008;1147:283–292. doi: 10.1196/annals.1427.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen H, Chan DC. Mitochondrial dynamics--fusion, fission, movement, and mitophagy--in neurodegenerative diseases. Human molecular genetics. 2009;18:R169–R176. doi: 10.1093/hmg/ddp326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weber TA, Reichert AS. Impaired quality control of mitochondria: aging from a new perspective. Experimental gerontology. 2010;45:503–511. doi: 10.1016/j.exger.2010.03.018. [DOI] [PubMed] [Google Scholar]
- 12.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. The Journal of cell biology. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ. PINK1 Is Selectively Stabilized on Impaired Mitochondria to Activate Parkin. PLoS biology. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ziviani E, Tao RN, Whitworth AJ. Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:5018–5023. doi: 10.1073/pnas.0913485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawajiri S, Saiki S, Sato S, Sato F, Hatano T, Eguchi H, Hattori N. PINK1 is recruited to mitochondria with parkin and associates with LC3 in mitophagy. FEBS letters. 2010;584:1073–1079. doi: 10.1016/j.febslet.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 16.Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, Rogov V, Lohr F, Popovic D, Occhipinti A, Reichert AS, Terzic J, Dotsch V, Ney PA, Dikic I. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO reports. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Developmental cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tal R, Winter G, Ecker N, Klionsky DJ, Abeliovich H. Aup1p, a yeast mitochondrial protein phosphatase homolog, is required for efficient stationary phase mitophagy and cell survival. The Journal of biological chemistry. 2007;282:5617–5624. doi: 10.1074/jbc.M605940200. [DOI] [PubMed] [Google Scholar]
- 19.Kissova I, Deffieu M, Manon S, Camougrand N. Uth1p is involved in the autophagic degradation of mitochondria. The Journal of biological chemistry. 2004;279:39068–39074. doi: 10.1074/jbc.M406960200. [DOI] [PubMed] [Google Scholar]
- 20.Zhu JH, Horbinski C, Guo F, Watkins S, Uchiyama Y, Chu CT. Regulation of autophagy by extracellular signal-regulated protein kinases during 1-methyl-4-phenylpyridinium-induced cell death. The American journal of pathology. 2007;170:75–86. doi: 10.2353/ajpath.2007.060524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, Alroy J, Wu M, Py BF, Yuan J, Deeney JT, Corkey BE, Shirihai OS. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. The EMBO journal. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kundu M, Lindsten T, Yang CY, Wu J, Zhao F, Zhang J, Selak MA, Ney PA, Thompson CB. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood. 2008;112:1493–1502. doi: 10.1182/blood-2008-02-137398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chu CT, Zhu J, Dagda R. Beclin 1-independent pathway of damage-induced mitophagy and autophagic stress: implications for neurodegeneration and cell death. Autophagy. 2007;3:663–666. doi: 10.4161/auto.4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang J, Randall MS, Loyd MR, Dorsey FC, Kundu M, Cleveland JL, Ney PA. Mitochondrial clearance is regulated by Atg7-dependent and -independent mechanisms during reticulocyte maturation. Blood. 2009;114:157–164. doi: 10.1182/blood-2008-04-151639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wright G, Terada K, Yano M, Sergeev I, Mori M. Oxidative stress inhibits the mitochondrial import of preproteins and leads to their degradation. Experimental cell research. 2001;263:107–117. doi: 10.1006/excr.2000.5096. [DOI] [PubMed] [Google Scholar]
- 26.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 27.Chu CT, Plowey ED, Dagda RK, Hickey RW, Cherra SJ, 3rd, Clark RS. Autophagy in neurite injury and neurodegeneration: in vitro and in vivo models. Methods in enzymology. 2009;453:217–249. doi: 10.1016/S0076-6879(08)04011-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cherra SJ, Kulich SM, Uechi G, Balasubramani M, Mountzouris J, Day BW, Chu CT. Regulation of the autophagy protein LC3 by phosphorylation. J. Cell. Biol. 2010;190:533–539. doi: 10.1083/jcb.201002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dagda RK, Zhu J, Kulich SM, Chu CT. Mitochondrially localized ERK2 regulates mitophagy and autophagic cell stress: implications for Parkinson’s disease. Autophagy. 2008;4:770–782. doi: 10.4161/auto.6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, Bamber BA, Bassham DC, Bergamini E, Bi X, Biard-Piechaczyk M, Blum JS, Bredesen DE, Brodsky JL, Brumell JH, Brunk UT, Bursch W, Camougrand N, Cebollero E, Cecconi F, Chen Y, Chin LS, Choi A, Chu CT, Chung J, Clarke PG, Clark RS, Clarke SG, Clave C, Cleveland JL, Codogno P, Colombo MI, Coto-Montes A, Cregg JM, Cuervo AM, Debnath J, Demarchi F, Dennis PB, Dennis PA, Deretic V, Devenish RJ, Sano FDi, Dice JF, Difiglia M, Dinesh-Kumar S, Distelhorst CW, Djavaheri-Mergny M, Dorsey FC, Droge W, Dron M, Dunn WA, Jr., Duszenko M, Eissa NT, Elazar Z, Esclatine A, Eskelinen EL, Fesus L, Finley KD, Fuentes JM, Fueyo J, Fujisaki K, Galliot B, Gao FB, Gewirtz DA, Gibson SB, Gohla A, Goldberg AL, Gonzalez R, Gonzalez-Estevez C, Gorski S, Gottlieb RA, Haussinger D, He YW, Heidenreich K, Hill JA, Hoyer-Hansen M, Hu X, Huang WP, Iwasaki A, Jaattela M, Jackson WT, Jiang X, Jin S, Johansen T, Jung JU, Kadowaki M, Kang C, Kelekar A, Kessel DH, Kiel JA, Kim HP, Kimchi A, Kinsella TJ, Kiselyov K, Kitamoto K, Knecht E, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Plowey ED, Cherra SJ, 3rd, Liu YJ, Chu CT. Role of autophagy in G2019S–LRRK2-associated neurite shortening in differentiated SH-SY5Y cells. Journal of neurochemistry. 2008;105:1048–1056. doi: 10.1111/j.1471-4159.2008.05217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shacka JJ, Klocke BJ, Shibata M, Uchiyama Y, Datta G, Schmidt RE, Roth KA. Bafilomycin A1 inhibits chloro-quine-induced death of cerebellar granule neurons. Molecular pharmacology. 2006;69:1125–1136. doi: 10.1124/mol.105.018408. [DOI] [PubMed] [Google Scholar]
- 33.Tolkovsky AM. Mitophagy. Biochimica et biophysica acta. 2009;1793:1508–1515. doi: 10.1016/j.bbamcr.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 34.Buckman JF, Hernandez H, Kress GJ, Votyakova TV, Pal S, Reynolds IJ. MitoTracker labeling in primary neuronal and astrocytic cultures: influence of mitochondrial membrane potential and oxidants. Journal of neuroscience methods. 2001;104:165–176. doi: 10.1016/s0165-0270(00)00340-x. [DOI] [PubMed] [Google Scholar]
- 35.Lang-Rollin IC, Rideout HJ, Noticewala M, Stefanis L. Mechanisms of caspase-independent neuronal death: energy depletion and free radical generation. J Neurosci. 2003;23:11015–11025. doi: 10.1523/JNEUROSCI.23-35-11015.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rantanen A, Jansson M, Oldfors A, Larsson NG. Downregulation of Tfam and mtDNA copy number during mammalian spermatogenesis. Mamm Genome. 2001;12:787–792. doi: 10.1007/s00335-001-2052-8. [DOI] [PubMed] [Google Scholar]
- 37.Fisher RP, Topper JN, Clayton DA. Promoter selection in human mitochondria involves binding of a transcription factor to orientation-independent upstream regulatory elements. Cell. 1987;50:247–258. doi: 10.1016/0092-8674(87)90220-0. [DOI] [PubMed] [Google Scholar]