Summary
Nucleotide excision repair (NER) removes UV-induced DNA damage and other bulky DNA lesions thereby maintaining genomic integrity. Dr. Qingyi Wei’s group demonstrated over the last decade that NER fidelity and single nucleotide polymorphisms (SNPs) in NER genes constitute melanoma risk biomarkers. In this issue (Li et al.) they provide evidence that SNPs in NER genes may also predict melanoma survival.
Keywords: Cutaneous melanoma (CM), nucleotide excision repair (NER), xeroderma pigmentosum (XP), single nucleotide polymorphisms (SNPs)
The melanoma problem
Skin cancer is the most frequent form of cancer in Caucasians. Cutaneous melanoma is the most aggressive form of skin cancer and its frequency is increasing rapidly. In the United States, there were approximately 76,000 new melanoma cases and 56,000 melanoma in situ cases in 2012. Melanoma mortality increased in the U.S. at an annual rate of 1.5% from 1950 to 2005. More than 8,000 deaths from melanoma are estimated per year.
In the clinical setting relatively crude melanoma risk factors are applied today, including family history of melanoma, age, gender, amount of sunlight exposure (particularly intense intermittent exposure), sensitivity to sunburning, freckling, fair hair, eye and skin color and number and size of nevi (Blankenburg et al. 2005) (See also the NCI melanoma risk assessment tool http://www.cancer.gov/melanomarisktool). Discoveries reported in recent years constitute breakthroughs in the fundamental understanding of the molecular basis of melanoma and have resulted in development of novel therapeutic targets including small molecules (BRAF inhibitors) and immunotherapies (CTLA-4 antibodies). Many of the molecular pathways to melanoma appear to depend on exposure to ultraviolet radiation (UV).
UV and melanoma
The physiologically relevant UV spectrum reaching the human skin can be divided into UVA (400–320 nm) and UVB (320–280 nm). Despite constituting only 5 % of the solar spectrum, UVB is considered mainly responsible for skin carcinogenesis. UVB forms cyclobutane pyrimidine dimers (CPD), pyrimidine-6,4-pyrimidone photoproducts (6–4PP), and some minor photoproducts via direct energy transfer to the DNA molecule. If not repaired, these DNA lesions may lead to UV “fingerprint” mutations at adjacent pyrimidines. In contrast to UVB, UVA penetrates deeper into skin reaching the dermis and acts indirectly by producing reactive oxygen species leading to oxidative DNA damage. However, UVA can also generate CPD lesions. Thus both UVB and UVA appear to play a role in melanoma induction (von Thaler et al. 2010).
The prognostic markers currently used in the clinic indirectly reflect the molecular biology that drives melanoma progression. Thus, the markers included in the latest AJCC melanoma staging system are surrogates of key biological events (Spatz et al. 2010). Gene expression signatures suggest that the “primary melanoma thickness” marker may represent a quantitative surrogate of the total multi-factorial biological machinery driving melanoma progression and invasion. The biological significance of “primary melanoma ulceration” may indicate a special tumor attribute or directly indicate tumor cell dissemination, e.g. by modifying the local environment. The male gender effect on adverse melanoma outcome is clearly established (adjusted relative excess risk of death from melanoma is 1.85). One explanation, supported by microarray data, could be the existence of metastases suppressor genes located on the X chromosome. Genes involved in DNA replication as well as DNA repair may represent the molecular basis of the “mitotic activity” marker. Aggressively growing melanomas need fast replication machinery and need to repair DNA damage effectively. The fact that cutaneous melanomas are continuously exposed to UV even after tumor formation highlights the rationale for nucleotide excision repair (NER) as an important molecular pathway for tumor maintenance.
Xeroderma pigmentosum as a melanoma model disease
That the NER pathway (Fig. 1) plays a role in melanoma prevention is evident from studies of the rare autosomal recessive disease, xeroderma pigmentosum (XP) (Wang et al. 2009; Bradford et al. 2011; DiGiovanna and Kraemer 2012). XP patients have defective NER and develop freckle like lesions in sun exposed skin. XP patients under 20 years of age have a more than 1000-fold increased melanoma risk. Interestingly, the hallmark acute burning on minimal sun exposure may not be present in 30–40% of all XP patients, mainly in those patients with defects in the XPC or polymerase eta genes. The median age at diagnosis of the first cutaneous melanoma was 22 years in the XP patients compared to 50–55 years in the normal Caucasian population. The median age of death due to skin cancer was 37 years in XP (Bradford et al. 2011). Of note, the site distribution of cutaneous melanomas was similar in XP patients and normal Caucasians.
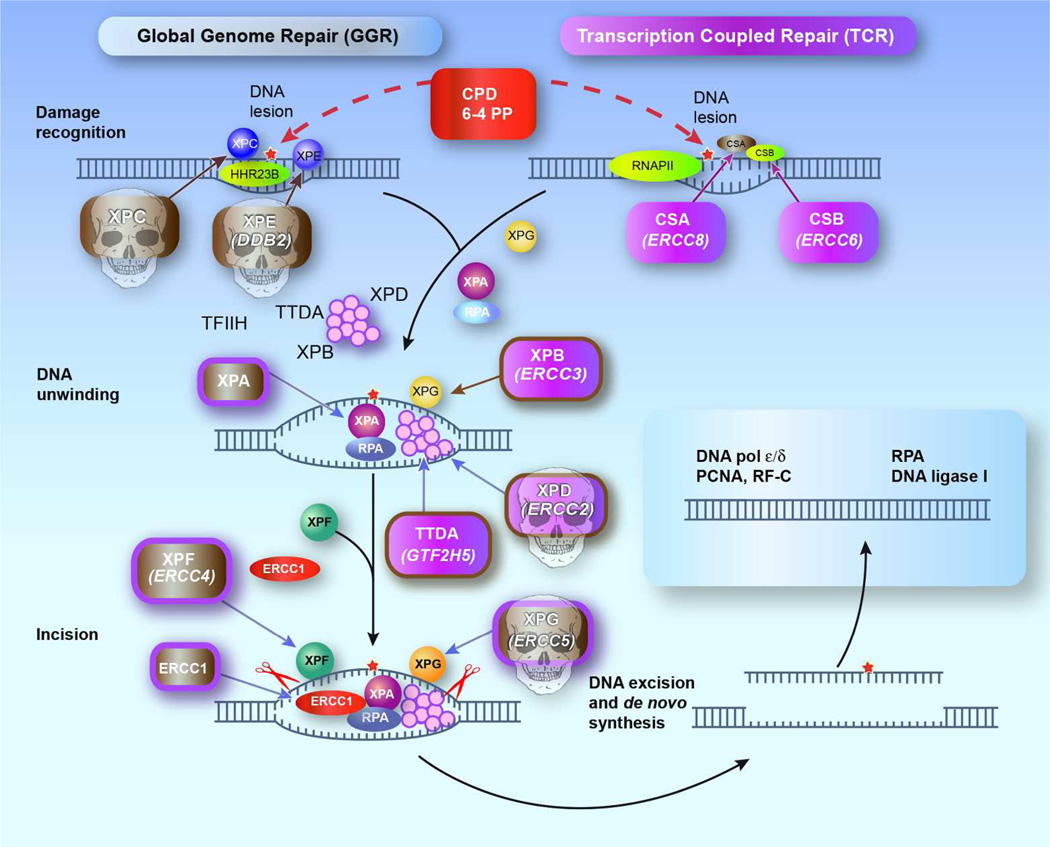
Figure 1.
Nucleotide excision repair (NER) pathway. In global genome repair (GGR) the XPC and XPE(DDB2) gene products recognize the DNA damage such as cyclobutane pyrimidine dimers (CPDs) or 6–4 photoproducts (6–4 PP) and initiate the NER cascade. In transcription coupled repair (TCR) in actively transcribed genes, the stalled RNA polymerase II in concert with CSA(ERCC8) and CSB(ERCC6) gene products initiate the NER cascade. XPB(ERCC3) and XPD(ERCC2) gene products are components of the 10 subunit multiprotein complex TFIIH (transcription factor II H), and demarcate the damage due to their helicase activity. These helicases along with the XPA and XPG(ERCC5) gene products and replication protein A (RPA) unwind the DNA surrounding the lesion. The TTD-A(GTF2H5) gene product is also part of the TFIIH complex. XPF(ERCC4) and XPG(ERCC5) gene products are endonucleases that cut the damage containing DNA strand removing the lesion along with a fragment of about 30 nucleotides. The resulting gap is filled using polymerases and ligases and the complementary DNA strand as a template. Mutations in the genes in rectangles have been associated with clinical disease. TagSNPs in XPC, XPE(DDB2), XPD(ERCC2) and XPG(ERCC5) genes (Skulls) are associated with increased risk of death from melanoma (See paper by Li et al in this issue). PCNA – proliferating cell nuclear antigen; RF-C replication factor C. (Figure modified from (DiGiovanna and Kraemer 2012)).
Seven different NER genes that correct seven distinct genetic XP complementation groups (XP-A to XP-G) have been identified (DiGiovanna and Kraemer 2012). However, the NER pathway consists of at least 23 genes/proteins that act in a well-defined sequential manner to repair bulky and UV-induced DNA damage (Fig. 1). Several steps constitute the NER process: First the DNA lesion is recognized by the XPE(DDB2) and XPC gene products, then the strand containing the lesion is unwound by XPB(ERCC3) and XPD(ERCC2) gene product helicases and incised on both sides of the lesion by XPG(ERCC5) and XPF(ERCC4) gene product endonucleases with damaged strand displacement. Finally, the resulting gap is filled by DNA polymerases using the opposite strand as a template followed by strand ligation. Two sub-pathways of NER, global genome repair (GGR) and transcription-coupled repair (TCR) can be discerned. These sub-pathways only differ in the first damage recognition step. TCR eliminates damage in actively transcribed genes while GGR removes DNA damage throughout the remainder of the genome. Unrepaired DNA damage may lead to somatic mutations that play a role in cancer induction and progression.
Because the clinical findings in XP are associated with cellular defects, including sensitivity of killing and mutagenic effects of UV and the inability of XP cells to repair UV-induced DNA damage, XP provides a powerful model for the study of melanoma in humans. Multiple melanomas with the same exposure history and genetic background can be studied in few patients where the effects of UV-damage are amplified due to the NER deficiency. For example, it was shown that more than 90% of melanomas in XP patients carried UV-type “fingerprint” mutations in the PTEN melanoma suppressor gene (Wang et al. 2009). This provides direct molecular evidence of UV involvement in melanoma disease in humans.
Markers for melanoma risk
The group of Dr. Qingyi Wei (Li et al., this issue) has studied molecular melanoma marker analysis for more than a decade. Originating from a laboratory headed by the late Dr. Larry Grossman in Baltimore they started out assessing functional cellular NER capacity measured in peripheral blood lymphocytes. Such studies necessitate the establishment of small scale functional assays due to the very limited lymphocyte cell numbers available per patient. Wei and others successfully applied host cell reactivation for that purpose and in 2003 demonstrated that a reduced NER activity is an independent risk factor for the development of cutaneous melanoma in the general population (Wei et al. 2003). Others have confirmed these findings and shown that even a slight reduction in DNA repair capacity was associated with a significantly increased cancer risk in patients with lung cancer, with head and neck squamous cell cancer, or with squamous cell carcinomas of the skin. That defective mechanisms for handling of UV-induced DNA damage constitute a melanoma risk marker was also demonstrated in patients with dysplastic nevus syndrome / familial melanoma. Their lymphocytes exhibited spontaneous as well as post-UV plasmid hypermutability using a shuttle vector assay (See (Blankenburg et al. 2005) and references therein).
Markers for melanoma survival
Treatment efficacy and melanoma resistance to certain chemotherapeutic regimens may depend on DNA repair. For example, applying functional assays, it was shown that increased base excision repair conferred resistance to fotemustine and cisplatin in melanoma cells in vitro and diminished mismatch repair conferred cellular fotemustine and etoposide resistance. The efficacy of the “standard” chemotherapeutic treatment of metastasized melanoma with dacarbazine or temozolomide depends on low O-6-methylguanine-DNA-methyltransferase (MGMT) repair and on high mismatch repair.
More recently, the use of DNA repair as an independent marker for melanoma progression and disease survival has come into focus. Gene expression profiles of primary cutaneous melanomas associated with metastases to distant sites and with poor prognosis revealed DNA repair being among the most significant pathways. Four out of 44 genes over-expressed in primary melanomas with poor prognosis belonged to the NER pathway (Kauffmann et al. 2008).
SNPs and melanoma risk / survival
Single nucleotide polymorphisms (SNPs) are variations in the DNA sequence that are present in the normal population, for example replacing a nucleotide C (cytosine) with a T (thymine). There is about one SNP per 300 nucleotides with about 10 million SNPs in the entire human genome. The human genome has 23 pairs of chromosomes. Each chromosome has its own pattern of SNPs. The genotype of the same SNP on each chromosome of a pair is indicated by two letters, for example AG means “A” on one chromosome (allele) and “G” on the other chromosome of the pair.
Most SNPs are located between genes and do not have known functions. However, some SNPs are located in or near genes and can act as markers for inheritance of a large group of nucleotides that frequently appear together on the same allele (haplotype). These marker SNPs may be associated with differences in drug responses, disease states, or cancer risk. Genome wide association studies (GWAS) are a method to determine possible associations of SNPs with disease. DNA from an affected group and a control population is collected and tested for the presence of a large number of SNPs. A smaller number of representative (tag) SNPs are often used as markers of linked groups of SNPs.
SNPs in the XPC NER gene have been shown by Dr. Wei’s group to confer an increased risk for melanoma development (Li et al. 2006). SNPs in other genes and pathways have also been implicated in an increased melanoma risk. These include the regulation of skin pigmentation, such as the melanocortin-1 receptor gene, the production of proteins of the steroid and thyroid hormone superfamily, including vitamin D and the peroxisome proliferators-activated receptor genes, the detoxification of oxidative stress metabolites, such as the gluthation S-transferase genes, or cytokine genes like the interleukin-10 gene. With modern high throughput techniques even more loci will most likely be defined.
The paper by Li et al. is a follow-up on their earlier GWAS study of SNPs and melanoma susceptibility in a cohort of 1804 melanoma cases and 1026 controls (Amos et al. 2011). The GWAS study examined about 1 million SNPs and found increased melanoma risk in association with loci on several different chromosomes. They identified novel chromosomal loci predisposing to cutaneous melanoma including 15q13.1 (HERC2/OCA2 region), 16q24.3 (MC1R region), 9p21.3 (p16/ARF region), and 1q21.3 (ARNT/LASS2/ANXA9 region).
The current paper examines survival of 1042 patients from this cohort after 3 years in relation to 74 tag SNPs representing 8 NER genes. They found that 4 of the SNPs were associated with reduced survival. Two of the SNPs were rare – XPE AG genotype and ERCC5/XPG AG genotype was present in about 2% of the melanoma patients. However, the other two SNPs were much more common – XPC AA genotype in 62.1% and ERCC2/XPD AA genotype in 27.4%. The XPE and ERCC5/XPG polymorphisms were associated with 4- to11- fold increased hazard of early death while the XPC and ERCC2/XPD polymorphisms were associated with about a doubling of risk of death. The risk of death increased with increasing numbers of these SNPs up to 34-fold if 3 of these adverse SNP genotypes were present. Presence of these variant SNPs was associated with additional risk of death in melanoma patients with unfavorable histopathological risk factors such as increased tumor thickness, involvement of lymph nodes, increased mitotic rate, presence of ulceration, and stage III or IV.
This is an intriguing exploratory study. While some of these results are similar to several smaller studies of association of SNPs and melanoma survival, they should be repeated with larger cohorts with different genetic backgrounds to determine the extent of applicability of these findings. These tag SNPs have not been reported as having biological function and this can be followed-up. The rare genetic disorder, XP, has mutations in these same NER genes resulting in greatly increased melanoma risk. Molecular medicine based on studies such as that reported by Li et al. may soon be in clinical use for melanoma risk assessment.
Clinical relevance.
Nucleotide excision repair (NER) removes UV-induced and other DNA damage and thereby prevents the development of a cellular “mutator” phenotype by maintaining genomic integrity.
The relevance of NER to cutaneous melanoma is demonstrated by NER-defective diseases such as xeroderma pigmentosum.
Common single nucleotide polymorphisms (SNPs) in NER genes may be associated with increased melanoma risk and decreased melanoma survival in the general population.
Acknowledgements
This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research (KHK) and partly by the Deutsche Forschungsgemeinschaft DFG, the German Cancer Aid, the Bundesministerium für Wirtschaft und Technologie, the Niedersächsische Krebsgesellschaft e.V., and the Forschungsförderungsprogramm der UMG (SE).
Abbreviations
- NER
nucleotide excision repair
- XP
xeroderma pigmentosum
- CM
cutaneous melanoma
- SNP
single nucleotide polymorphism
Footnotes
Conflicts of interest
The authors state no conflict of interest.
REFERENCES
- Amos CI, Wang LE, Lee JE, Gershenwald JE, Chen WV, Fang S, et al. Genome-wide association study identifies novel loci predisposing to cutaneous melanoma. Hum.Mol.Genet. 2011;20(24):5012–5023. doi: 10.1093/hmg/ddr415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenburg S, Konig IR, Moessner R, Laspe P, Thoms KM, Krueger U, et al. Assessment of 3 xeroderma pigmentosum group C gene polymorphisms and risk of cutaneous melanoma: a case-control study. Carcinogenesis. 2005;26(6):1085–1090. doi: 10.1093/carcin/bgi055. [DOI] [PubMed] [Google Scholar]
- Bradford PT, Goldstein AM, Tamura D, Khan SG, Ueda T, Boyle J, et al. Cancer and neurologic degeneration in xeroderma pigmentosum: long term follow-up characterises the role of DNA repair. J.Med.Genet. 2011;48(3):168–176. doi: 10.1136/jmg.2010.083022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGiovanna JJ, Kraemer KH, et al. Shining a light on xeroderma pigmentosum. J Invest Dermatol. 2012;132(3 Pt 2):785–796. doi: 10.1038/jid.2011.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffmann A, Rosselli F, Lazar V, Winnepenninckx V, Mansuet-Lupo A, Dessen P, et al. High expression of DNA repair pathways is associated with metastasis in melanoma patients. Oncogene. 2008;27(5):565–573. doi: 10.1038/sj.onc.1210700. [DOI] [PubMed] [Google Scholar]
- Li C, Hu Z, Liu Z, Wang LE, Strom SS, Gershenwald JE, et al. Polymorphisms in the DNA repair genes XPC, XPD, and XPG and risk of cutaneous melanoma: a case-control analysis. Cancer Epidemiol.Biomarkers Prev. 2006;15(12):2526–2532. doi: 10.1158/1055-9965.EPI-06-0672. [DOI] [PubMed] [Google Scholar]
- Spatz A, Batist G, Eggermont AM, et al. The biology behind prognostic factors of cutaneous melanoma. Curr.Opin.Oncol. 2010;22(3):163–168. doi: 10.1097/CCO.0b013e328337fe8f. [DOI] [PubMed] [Google Scholar]
- von Thaler AK, Kamenisch Y, Berneburg M, et al. The role of ultraviolet radiation in melanomagenesis. Exp.Dermatol. 2010;19(2):81–88. doi: 10.1111/j.1600-0625.2009.01025.x. [DOI] [PubMed] [Google Scholar]
- Wang Y, DiGiovanna JJ, Stern JB, Hornyak TJ, Raffeld M, Khan SG, et al. Evidence of ultraviolet type mutations in xeroderma pigmentosum melanomas. Proc.Natl.Acad.Sci.U.S.A. 2009;106(15):6279–6284. doi: 10.1073/pnas.0812401106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q, Lee JE, Gershenwald JE, Ross MI, Mansfield PF, Strom SS, et al. Repair of UV light-induced DNA damage and risk of cutaneous malignant melanoma. J.Natl.Cancer Inst. 2003;95(4):308–315. doi: 10.1093/jnci/95.4.308. [DOI] [PubMed] [Google Scholar]