Abstract
The endoplasmic reticulum (ER) is a large, continuous membrane-bound organelle comprised of functionally and structurally distinct domains including the nuclear envelope, peripheral tubular ER, peripheral cisternae, and numerous membrane contact sites at the plasma membrane, mitochondria, Golgi, endosomes, and peroxisomes. These domains are required for multiple cellular processes, including synthesis of proteins and lipids, calcium level regulation, and exchange of macromolecules with various organelles at ER-membrane contact sites. The ER maintains its unique overall structure regardless of dynamics or transfer at ER-organelle contacts. In this review, we describe the numerous factors that contribute to the structure of the ER.
The ER is one continuous membrane with three morphologies: A nuclear envelope, cisternae, and tubules. Its distinct domains are used to synthesize proteins and lipids, regulate calcium levels, and interact with organelles.
The endoplasmic reticulum (ER) is a dynamic organelle responsible for many cellular functions, including the synthesis of proteins and lipids, and regulation of intracellular calcium levels. This review focuses on the distinct and complex morphology of the ER. The structure of the ER is complex because of the numerous distinct domains that exist within one continuous membrane bilayer. These domains are shaped by interactions with the cytoskeleton, by proteins that stabilize membrane shape, and by a homotypic fusion machinery that allows the ER membrane to maintain its continuity and identity. The ER also contains domains that contact the plasma membrane (PM) and other organelles including the Golgi, endosomes, mitochondria, lipid droplets, and peroxisomes. ER contact sites with other organelles and the PM are both abundant and dispersed throughout the cytoplasm, suggesting that they too could influence the overall architecture of the ER. As we will discuss here, ER shape and distribution are regulated by many intrinsic and extrinsic forces.
ER STRUCTURE AND FORMATION
Domains of the ER Are Stabilized by Membrane-Shaping Proteins
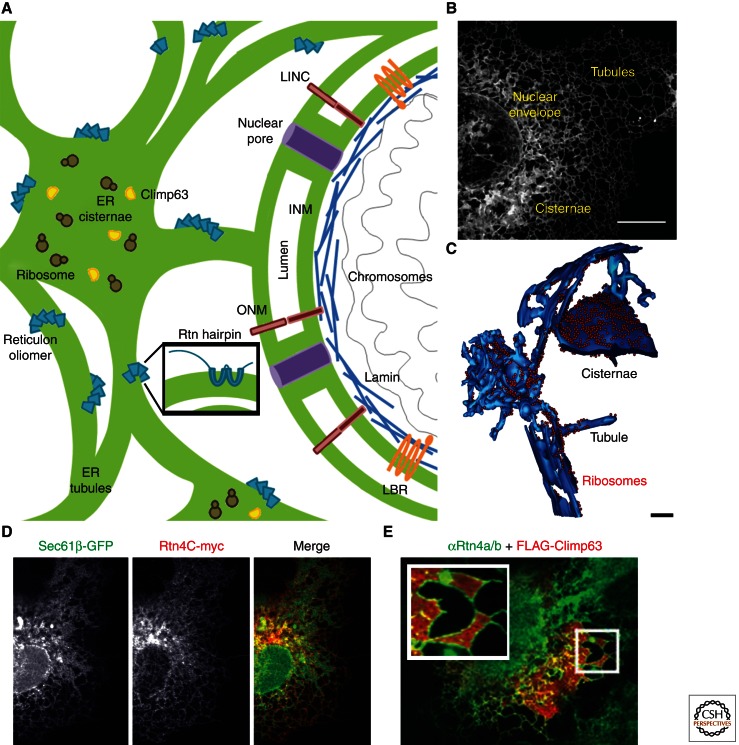
The endoplasmic reticulum (ER) is a large membrane-bound compartment spread throughout the cytoplasm of eukaryotic cells. It is divided into three major morphologies that include the nuclear envelope (NE), peripheral ER cisternae, and an interconnected tubular network (Fig. 1A,B). The ER is composed of one completely continuous membrane bilayer and has a single continuous lumen; the ability of the ER to have differently shaped domains requires some membrane proteins to segregate and form these domains through their assembly. The NE is the most visually obvious domain made from ER membrane. It surrounds the nucleus as a double membrane bilayer and acts as a barrier to selectively control transport of molecules into and out of the nucleus (Fig. 1A,B). The inner and outer nuclear membranes (INM and ONM) are large flat cisternal membranes stacked over each other, separated by the internuclear membrane space (INS) (Hetzer 2010). The INM is flattened around the nuclear contents by forming contacts with binding sites on chromatin and nuclear lamins, when present (Fig. 1A) (Zheng and Tsai 2006; Anderson and Hetzer 2007). The flat cisternae of the ONM and INM are separated by the distance of the INS (about 50 nm), which is held constant by the LINC complex (Fig. 1A) (Tzur et al. 2006; Sosa et al. 2012). The membrane bilayers of the INM and ONM cisternae are continuous with each other at nuclear pores (NPs), providing a conduit for membrane proteins to diffuse between the nuclear and cytoplasmic compartments (Fig. 1A) (Suntharalingam and Wente 2003).
Figure 1.
Domains of the ER are stabilized by membrane-shaping proteins. (A) Model depicting known ER domains (in green) and domain regulating proteins. The structure of the NE double membrane bilayer is regulated by the LINC complex (in red), nuclear pores (in purple) and lamin B-receptor (LBR) (in orange) interactions with lamin (in blue). The organization of the peripheral ER cisternae is regulated by Climp63 (in yellow) and large protein complexes such as polyribosomes (in brown). Reticulon proteins (in blue) oligomerize to control the tubular ER and curved edges of the cisternal ER. (B) Confocal fluorescence image of a Cos-7 cell expressing the ER luminal marker KDEL-venus. The continuous membrane network of the ER is comprised of the NE, peripheral cisternae and peripheral ER tubules. (C) EM tomogram of the 3D structure of ER domains shown with bound ribosomes in a yeast cell (ER in blue; ribosomes are shown as red spheres). (D) Image of a Cos-7 cell coexpressing Rtn4C-myc (red) and general ER marker mCh-Sec61β (green) shows that Rtn4 localizes preferentially to ER tubules. Note the absence of Rtn4C staining at the NE and peripheral ER cisternae (compare middle panel with left panel, and see merge). (E) Cisternal ER expands in Cos-7 cells expressing FLAG-Climp63. Expansion of ER cisternae reveals endogenous Rtn4a/b localized to the edges of ER cisternae. Scale bars, B, 10 µm; C, 100 nm. (Image in C adapted from West et al. 2011; adapted, with permission, from the Journal of Cell Biology. Image in D from Voeltz et al. 2006; reprinted, with permission, from Elsevier © 2006. Image in E from Shibata et al. 2010; reprinted, with permission, from Elsevier © 2010.)
The peripheral ER branches out from the ONM as an interconnected network comprised of cisternae and tubules (Fig. 1B,C). ER cisternae tend to be localized closer to the NE whereas tubules predominate in the periphery (Fig. 1B) (Terasaki et al. 1986; Puhka et al. 2007). The morphology of the tubular peripheral ER in many organisms, including animals, yeast, and plants, has been shown to be regulated by the Reticulon (Rtn) family of integral membrane proteins and DP1/Yop1 (De Craene et al. 2006; Voeltz et al. 2006; Audhya et al. 2007; Anderson and Hetzer 2008a; Tolley et al. 2008; West et al. 2011). Depletion of Rtn and DP1/Yop1 results in a large reduction in ER tubules; however, recent evidence suggests that Rtn and DP1/Yop1 proteins regulate the structure of multiple ER domains that contain high membrane curvature. In fact, Rtns are not only specifically localized to tubular ER domains but also to the edges of cisternae and edges of the fenestra found within cisternae (Fig. 1D,E) (Kiseleva et al. 2007; Schuck et al. 2009; Shibata et al. 2010). Consistent with this localization, yeast cells in which Rtn/Yop1 have been deleted lack tubules but also contain expansive cisternae that are no longer fenestrated (West et al. 2011).
Rtns and DP1/Yop1 are thought to stabilize the tubular network and other regions of high membrane curvature through their unique topology and by forming higher-order oligomers (Voeltz et al. 2006; Shibata et al. 2008). Rtns and DP1 each have two long transmembrane domains (TMDs) with amino- and carboxy-terminal domains, as well as the soluble domain between the TMDs, facing the cytoplasm (Fig. 1A) (Voeltz et al. 2006). These TMDs are not quite long enough to make a double pass hairpin, and are too long to span the membrane once, indicating the intriguing possibility that the TMDs of Rtns and DP1 form a wedge to increase the area of the outer leaflet compared with the inner leaflet, resulting in high membrane curvature (Fig. 1A). To test this concept, the TMDs of Rtn were lengthened to make a double pass hairpin in the membrane bilayer, resulting in the loss of Rtn-induced membrane tubules (Zurek et al. 2011). FRAP assays have shown that Rtns and DP1 organize into oligomers to establish high membrane curvature (Shibata et al. 2008). Purified Rtn or DP1/Yop1 is capable of deforming reconstituted proteoliposomes into tubules resembling the dimensions of ER tubules found in vivo, showing that these proteins are sufficient to shape membranes (Hu et al. 2008). A structure of the Rtn or DP1/Yop1 proteins in membranes has not been determined; however, the most appealing organization model posits that they would form a scaffold of C-shaped oligomers to stabilize the curvature found on tubules and at the edges of cisternae, and to accommodate passage of other ER proteins past the oligomer (Fig. 1A) (Shibata et al. 2010).
Much less is known regarding how peripheral ER cisternae are shaped. Although it is clear that ER cisternae can be propagated by the depletion of Rtns and DP1/Yop1 (De Craene et al. 2006; Voeltz et al. 2006; Anderson and Hetzer 2008a; Shibata et al. 2010; West et al. 2011), there are other factors whose overexpression can similarly generate and stabilize cisternal ER shape. Some of these are large protein complexes, in particular polyribosomes associated with translocation complexes, thought to help stabilize the flat membrane regions of the cisternae (Fig. 1A) (Shibata et al. 2006, 2010; Puhka et al. 2007). In support of this theory, addition of drugs that dislocate polyribosomes from ER membranes causes a reduction in the amount of cisternal peripheral ER (Puhka et al. 2007). Conversely, the amount of cisternal peripheral ER increases when an integral ER membrane protein that binds ribosomes, p180, is overexpressed (Benyamini et al. 2009; Shibata et al. 2010). Climp63 is another integral ER membrane protein that plays a role in regulating the shape of ER cisternae (Fig. 1E) (Shibata et al. 2010). Endogenous Climp63 preferentially localizes to ER cisternae relative to tubules and like reticulons, it is also excluded from the NE (Fig. 1E) (Vedrenne and Hauri 2006; Shibata et al. 2010). Climp63 overexpression leads to the propagation of ER cisternae (Fig. 1E), whereas its depletion alters cisternal intraluminal spacing (Shibata et al. 2010). Climp63 has a luminal domain that homo-oligomerizes, and this could explain how it directly regulates the luminal spacing of cisternae in a manner analogous to the LINC complex at the NE (Tzur et al. 2006). Climp63 also has a cytoplasmic domain that binds to MTs; however, it is unclear how MT binding would organize the shape of ER cisternae.
The purpose of differently shaped ER domains remains a subject of debate. Historically, ER cisternae have been classified as ribosome-bound, “rough” ER; by default, ER tubules are considered ribosome-free, “smooth” ER. Cisternae do have higher concentrations of ribosomes than tubules and also have a larger luminal volume to surface area than tubules, suggesting that they would be the preferred site for luminal processes like protein folding (Fig. 1C) (Shibata et al. 2010; West et al. 2011; Puhka et al. 2012). Consistent with this idea, during the ER stress response yeast peripheral ER shows increased cisternae to accommodate an increase in protein folding (Schuck et al. 2009). ER tubules have lower luminal volume to surface area than cisternae, suggesting that ER tubules could be the preferred site for the accumulation of integral membrane proteins and for processes connected to lipid synthesis (West et al. 2011). Nevertheless, there are many examples of ER tubules that do have bound ribosomes, just at a lower density than cisternae, which shows that they are not always ribosome excluded (West et al. 2011; Puhka et al. 2012).
ER during Mitosis
At the onset of mitosis in animal cells, the elaborate domain architecture of the NE and peripheral ER degenerate. During prophase, the INM proteins of the NE that tether the INM to chromatin and lamins are phosphorylated (Hetzer 2010); as a result of phosphorylation, lamin B receptor (LBR) dissociates from its nuclear contact sites and the nuclear lamin basket disassembles, leading to NE disassembly (Courvalin et al. 1992). NE disassembly does not involve vesiculation of the NE; rather, the membranes of the NE enclosure are absorbed into the peripheral ER, with which it is continuous (Fig. 2A) (Ellenberg et al. 1997; Yang et al. 1997; Anderson and Hetzer 2008b; Lu et al. 2009). At the end of mitosis, the NE is reformed when the nuclear lamins reassemble a basket around the chromatin, LBR is dephosphorylated, and the proteins located to the NE reestablish contact with the chromatin and nuclear lamin basket (Anderson and Hetzer 2007).
Figure 2.
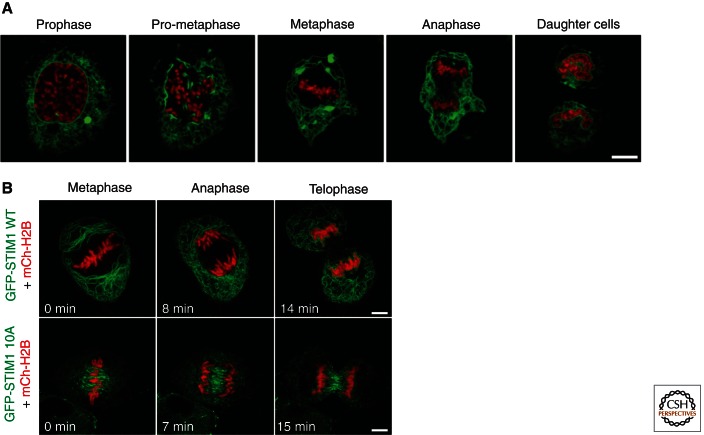
The structural organization of the ER during mitosis in mammalian cells. (A) Images of HeLa cells expressing GFP-Sec61β (ER in green) and H2B-tdTomato (chromatin in red) show the dramatic ER structural changes that occur between interphase and the indicated stages of mitosis. Note the movement of ER domains to the periphery which is particularly striking in metaphase. (B) Images of HeLa cells through mitosis expressing mCh-H2B (red) along with either GFP-STIM1 WT (top panels) or GFP-STIM1 with 10 phosphorylation sites mutated to alanine (bottom panels). Note that expression of the GFP-STIM1 10A mutant causes the ER to accumulate on the mitotic spindle. Scale bars, A, 10 µm; B, 5 µm. (Images in panel A from Anderson and Hetzer 2008b; reprinted, with permission, from the Journal of Cell Science © 2008. Images in panel B from Smyth et al. 2012; reprinted, with permission, from Elsevier © 2012.)
Through mitosis, the spatial organization of both the NE and peripheral ER is altered in animal cells. Specifically, the interphase peripheral ER is dispersed throughout the cytoplasm, whereas the mitotic ER is located near the PM and away from the mitotic spindle (Fig. 2A) (McCullough and Lucocq 2005; Puhka et al. 2007, 2012; Anderson and Hetzer 2008b; Lu et al. 2009; Hetzer 2010; Lu et al. 2011; Smyth et al. 2012). The mechanism and proteins involved in regulating ER movement during mitosis are not well understood; however, it was recently shown that phosphorylation of STIM1 is required to prevent the ER membrane from associating with the mitotic spindle (Smyth et al. 2012). STIM1 is an integral membrane protein that binds to EB1, a microtubule (MT) plus tip binding protein, that will be further discussed in a subsequent section (Grigoriev et al. 2008). During mitosis, STIM1 is phosphorylated, resulting in dissociation from EB1. Interestingly, expression of a STIM1 mutant that cannot be phosphorylated causes the mitotic ER to accumulate around the mitotic spindle, rather than distributed close to the PM (Fig. 2B) (Smyth et al. 2012). Thus, STIM1 phosphorylation is one mechanism that regulates the position of the ER during mitosis. The factors actively involved in redistributing the ER toward the periphery during mitosis are not yet known.
What remains controversial is whether ER shape changes from having a higher degree of membrane curvature in interphase to having less membrane curvature in mitosis, or vice versa. Analysis of ER structure in HeLa cells and CHO cells, by live 3D confocal fluorescence microscopy and electron microscopy (EM), reveals an interphase ER morphology that is cisternal in the perinuclear region and mostly tubular in the periphery towards the PM, and a mitotic morphology that is almost entirely cisternal (McCullough and Lucocq 2005; Lu et al. 2009). In contrast, experiments where ER structure was analyzed by transmission EM and EM tomography of either chemically fixed or high-pressure frozen cells, reveal an ER structure that is mostly cisternal in interphase and more fenestrated and tubular in mitosis (Puhka et al. 2007, 2012). The disparity in conclusions could result from the complexity of ER domains that are reorganized as a result of NE breakdown and relocalization of the mitotic ER toward the cell periphery. Three-dimensional EM tomography reveals domains that appear cisternal by fluorescence microscopy are in fact highly fenestrated and may still contain a high degree of membrane curvature (Puhka et al. 2012). It is possible that together these data show that when the NE and ER are redistributed to the periphery during mitosis, the total amount of ER membrane curvature is accommodated through differently shaped mitotic ER domains.
ER Dynamics and Assembly
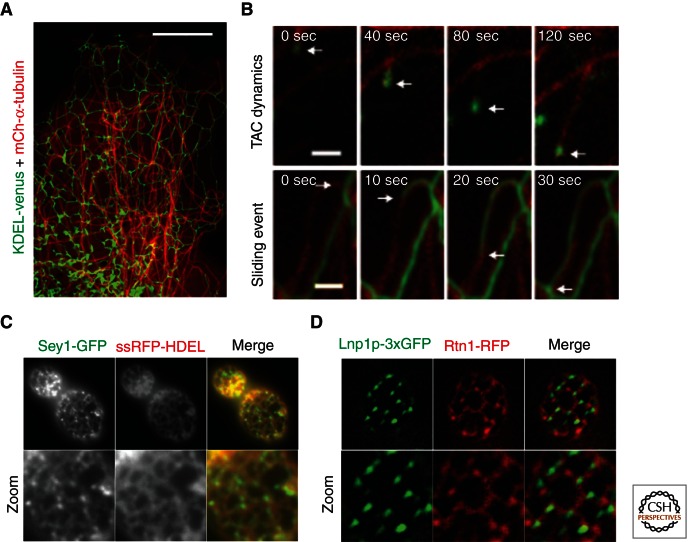
Visualization of the peripheral ER by live cell fluorescence microscopy shows a constantly changing landscape; the structure of the tubular ER network rearranges by tubule growth, retraction and homotypic fusion with adjacent ER membranes. Membrane shaping proteins like Rtns are likely to stabilize and maintain, rather than drive ER tubule formation. In the absence of Rtns and DP1/Yop1, new ER tubules are still generated, although poorly maintained (West et al. 2011). ER tubule dynamics are generated through forces exerted by molecular motors as they pull new ER tubules out from existing ER domains along the cytoskeleton. These dynamics are dependent on the MT network in animal cells (Fig. 3A) (Lee and Chen 1988; Waterman-Storer and Salmon 1998). ER dynamics on MT tracks occur by two distinct methods: the tip attachment complex (TAC) and ER sliding (Fig. 3B) (Lee and Chen 1988; Waterman-Storer and Salmon 1998). TAC dynamics are defined by the tip of a dynamic ER tubule linked to the tip of a MT, the ER grows and shrinks concurrent with MT dynamics (Waterman-Storer and Salmon 1998). TAC movements depend on tethering between the ER protein, STIM1, and an MT plus end-binding protein, EB1 (Fig. 3B, top panel) (Grigoriev et al. 2008). The ER sliding mechanism accounts for the majority of ER dynamics in the cell and occurs through a machinery involving kinesin-1 and dynein MT motors that pull ER tubules along the sides of established MTs (Wozniak et al. 2009). The movement of growing ER tubules sliding along MTs can be visualized nicely by live confocal fluorescence microscopy (Fig. 3B, bottom panel). The purpose of the constant reorganization of the tubular ER network is not known; however, it is reasonable to suggest that ER dynamics could be required for the necessary functions of the ER, such as facilitating organelle contact for exchange of proteins, lipids, and Ca2+ (Baumann and Walz 2001; English et al. 2009). To further understand the nature of ER dynamics it is important to identify the unknown factors involved, including the factors linking ER tubules to the motor proteins on MTs during sliding dynamics.
Figure 3.
ER structure is regulated by dynamics on the cytoskeleton and homotypic fusion. (A) Cos-7 cells expressing KDEL-venus (ER in green) and mCh-α-tubulin (MTs in red) illustrate the close relationship between the ER and MT networks. (B) Time lapse images of Cos-7 cells expressing YFP-STIM1 (TAC dynamics, top panel) or GFP-Sec61β (sliding event, bottom panel) to visualize ER dynamic events that are associated with MTs (labeled with mCh-α-tubulin). Arrows indicate the position of the dynamic ER tubule tip. (C) Image of a yeast cell expressing Sey1-GFP (Atl homolog, in green) and a luminal ER marker (ssRFP-HDEL, in red). Note that Sey1 is highly enriched at three-way junctions (see zoom, left panel). (D) Image of a yeast cell expressing Lnp1p-3xGFP (lunapark, in green) and the tubular ER marker Rtn1-RFP (in red). Note that Lnp1p also localizes to three-way junctions. Scale bars, A, 10 µm; B, 1 µm. (Images in panel B from Friedman et al. 2010; reprinted, with permission, from the Journal of Cell Biology. Images in panel C from Hu et al. 2009; reprinted, with permission, from Elsevier Ltd. © 2009. Images in panel D from Chen et al. 2012; reprinted, with permission, from Nature Cell Biology © 2012.)
Unlike other organelles, the entire ER network is completely continuous at all times, even though it constantly rearranges its structure (Lee and Chen 1988; Ellenberg et al. 1997; Dayel et al. 1999). During ER dynamics, the ER forms three-way junctions by sliding along MTs to fuse with adjacent ER regions it contacts, contributing to the overall “reticular” appearance of the ER. The factors that regulate homotypic ER membrane fusion must also be ER-specific, this is important because the ER is closely apposed and potentially tethered to nearly every membrane-bound compartment in the cell. Homotypic ER fusion is regulated by the Atlastin family of dynamin-like GTPases (Rismanchi et al. 2008; Hu et al. 2009; Orso et al. 2009; Anwar et al. 2012). Atlastin family members localize to the tubular ER and accumulate in a striking pattern at three-way junctions (Fig. 3C) (Rismanchi et al. 2008; Hu et al. 2009; Orso et al. 2009; Park et al. 2010b; Chen et al. 2012). Atlastin and its yeast homolog Sey1 do not localize to the NE or to peripheral ER cisternae, suggesting that these proteins belong to a group of proteins that partition to regions of high membrane curvature (Fig. 3C) (Rismanchi et al. 2008; Hu et al. 2009; Orso et al. 2009; Park et al. 2010b). Depletion of endogenous Homo sapiens atl2 and atl3 by siRNA or overexpression of atlastin mutants in HeLa cells results in the formation of long, unbranched ER tubules, presumably as a result of the loss of three-way junctions (Hu et al. 2009). In contrast, overexpression of wild-type atlastin in drosophila motor neurons leads to hyper fusion of ER membranes (Orso et al. 2009) and an expansion of ER cisternae in mammalian tissue culture cells (Hu et al. 2009). Proteoliposomes reconstituted with purified atlastin undergo membrane fusion in a GTP-dependent manner, providing evidence that atlastins directly regulate ER fusion (Orso et al. 2009). Atlastin clearly plays an important role in regulating ER fusion; however, Atlastin/Sey1 depletion does not cause the ER membrane to fragment in yeast or mammalian cells, suggesting that there are likely to be additional machineries involved. It is also not known whether the Atlastins/Sey1 provide a direct link between the fusion machinery and dynamics that occur on the cytoskeleton.
Recently, the highly conserved Lunapark (Lnp) family of proteins was shown to also localize at three-way junctions within the tubular ER network (Fig. 3D) (Chen et al. 2012). Deletion of Lnp1 from yeast cells results in a collapsed, but highly reticulated ER network (Chen et al. 2012). Deletion of Sey1 can suppress the ER morphology defects of the lnp1Δ mutant (Chen et al. 2012). Thus, Lnp1 appears to act in an antagonistic manner with Sey1 to regulate ER network formation; however, these proteins may also function synergistically to some extent because they are each required for the other’s ability to localize to three-way junctions (Chen et al. 2012).
In vitro systems for ER formation have suggested a role for Rab GTPases during ER vesicle fusion (Turner et al. 1997; Audhya et al. 2007). Rab proteins are GTP-binding proteins that control the fusion of target membranes through GTP hydrolysis (Cai et al. 2007; Schwartz et al. 2007). The precise combination of Rab proteins and SNAREs determine the functional identity of numerous membrane compartments and guide fusion between donor and acceptor compartments (Pfeffer 2001; Murray et al. 2002; Behnia and Munro 2005). Previous studies illustrate that the addition of recombinant Rab GDI reduces the efficiency of ER network formation in vitro, presumably by disrupting the membrane association of all Rab proteins (Turner et al. 1997; Audhya et al. 2007). Notably, no Rab protein has been identified that localizes specifically to the ER and affects ER fusion, morphology or dynamics; however, a Rab-interacting SNARE protein, yeast Ufe1, has previously been implicated in ER assembly and provides more evidence for the involvement of a Rab GTPase in ER assembly (Patel et al. 1998; Anwar et al. 2012).
ER-ORGANELLE MEMBRANE CONTACTS
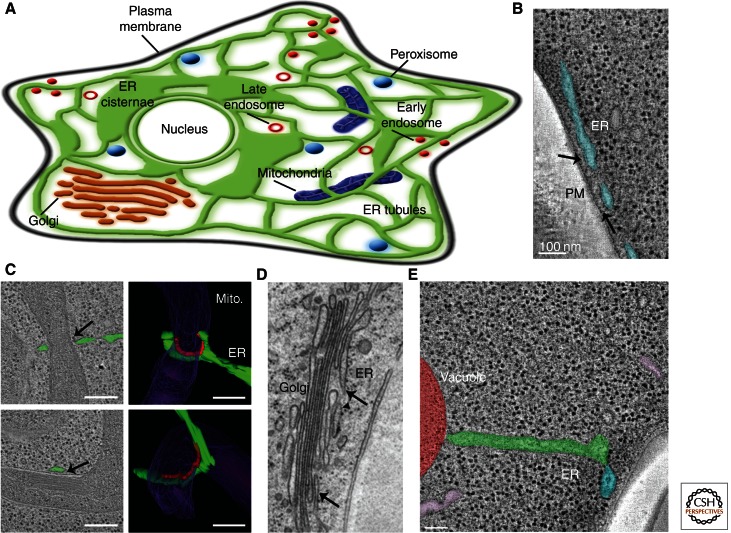
The ER network extends throughout the entire cytoplasm and the dynamic nature of the ER ensures that it probes areas where it has not yet spread. During dynamics, the ER not only forms new contacts with itself, but must also forge new contacts with organelles in the cytoplasm. The ER has been shown to be closely apposed to almost all membrane-bound organelles, including the PM, mitochondria, lipid droplets, Golgi, endosomes, and peroxisomes (Fig. 4A) (Friedman and Voeltz 2011; Toulmay and Prinz 2011). Here we discuss numerous studies that have determined these interactions to be functionally important and suggest the purpose of the ER’s wide distribution (Lebiedzinska et al. 2009; Toulmay and Prinz 2011).
Figure 4.
The ER forms membrane contact sites with the PM and other organelles. (A) Model depicting observed membrane contact sites in mammalian cells between the ER and PM, mitochondria, Golgi, endosomes and peroxisomes. (B) EM tomograph of a yeast cell illustrates the close contact between the peripheral ER (in blue) and PM (dark edge). (C) EM tomogram reveals ER tubules (in green) wrapped around mitochondria (in purple) at positions of constriction in a yeast cell. Marked in red are positions where the ER and mitochondria are closely apposed. Mito., mitochondria. (D) EM tomograph of an NRK (normal rat kidney) mammalian cell shows the close contacts between the ER and Golgi cisternae. (E) EM tomograph illustrates contacts between the ER (tubular in green, PM-associated ER in blue) and a vacuole (in red) in yeast. Note that ribosomes are excluded from the ER membrane where it apposes the PM, mitochondria, Golgi, and vacuole (in B, C, D, and E). Scale bars, B, 100 nm; C, 200 nm. (Images in B and E from West et al. 2011; reprinted, with permission, from the Journal of Cell Biology. Images in C from Friedman et al. 2011; reprinted, with permission, from the American Association for the Advancement of Science © 2011. Image in D courtesy of M. Ladinsky.)
ER and Plasma Membrane
The extensive network of the ER has multiple positions where it is closely apposed to the PM (Pichler et al. 2001; West et al. 2011). Previous studies have identified the PM-associated ER as sites of phosphatidylinositol (PI) metabolism, nonvesicular transfer of sterols, and Ca2+ level regulation (Li and Prinz 2004; Baumann et al. 2005; Carrasco and Meyer 2011; Stefan et al. 2011). EM analysis has revealed that 20%–45% of the cytoplasmic surface of the PM in budding yeast is within tethering distance of the ER and ribosome excluded (Fig. 4B) (West et al. 2011). Extensive contact between the ER and the PM may also occur in animal cells, although the extent of contact has not been characterized by EM tomography, except under conditions that activate store-operated Ca2+ release and during mitosis (McCullough and Lucocq 2005; Orci et al. 2009).
The oxysterol-binding homology (Osh) protein family and the integral ER VAP proteins regulate PI metabolism at ER-PM contact sites. Regulation of the essential lipid-signaling molecule phosphatidylinositol 4-phosphate (PI4P) is controlled by Osh and VAP mediated activation of Sac1 phosphatase (Roy and Levine 2004; Stefan et al. 2011). Deletion of Osh proteins in yeast cells results in a six- to sevenfold increase in PI4P levels; furthermore, addition of recombinant Osh3 to a microsome fraction depleted of peripherally bound proteins (including endogenous Osh proteins) was able to stimulate Sac1 phosphatase activity, suggesting that Osh proteins control PI4P levels at ER-PM contact sites (Stefan et al. 2011).
Numerous studies also implicate Osh proteins in nonvesicular sterol transportation (Raychaudhuri et al. 2006; de Saint-Jean et al. 2011; Jansen et al. 2011). By measuring the amount of free sterols converted to steryl esters (esterification), Raychaudhuri et al. showed a sevenfold decrease in total sterol transport from the PM to the ER in temperature-sensitive mutant yeast strains missing all seven Osh proteins. A similar decrease in PM-ER cholesterol transport was observed when the human Osh homologs, ORP1 and ORP2, were depleted in HeLa tissue culture cells by siRNA (Jansen et al. 2011). Structural studies have determined that Osh4 acts as a sterol and phosphoinositol phosphate exchanger by selectively binding both PI4P and sterols to transport to the PM and ER, respectively (de Saint-Jean et al. 2011). Currently, debate surrounds the function of Osh proteins in sterol transport between the PM and ER. Recent work suggests that Osh proteins regulate sterol organization at the PM and have little effect on nonvesicular transport of sterols between the PM and ER (Georgiev et al. 2011). These studies showed that transportation of a fluorescent sterol from the PM to ER was not inhibited in a yeast mutant lacking functional copies of all seven Osh proteins, contrary to previous data (Georgiev et al. 2011). It will be interesting to watch this exciting field surrounding nonvesicular transport of sterols between the PM and ER as it develops.
In animal cells, the junction between the ER and the PM has also been shown to be important for regulating proper Ca2+ levels (Carrasco and Meyer 2011). During times of depleted Ca2+ in the ER, Ca2+ is transported from the PM, using the process of store-operated Ca2+ entry (SOCE). At ER-PM junctions, STIM1 and STIM2 act as resident Ca2+ sensors and interact with the calcium channel protein Orai1 to form a Ca2+-released-activated-Ca2+ influx (CRAC) channel to facilitate the entry of Ca2+ into the ER (Liou et al. 2005; Roos et al. 2005; Feske et al. 2006; Park et al. 2010a). As mentioned previously, STIM1 is important for tethering the ER to MTs during TAC-regulated ER dynamics; however, inhibition of TAC movements does not affect SOCE (Grigoriev et al. 2008). It is currently unclear whether STIM1’s roles in SOCE and TAC movements are related.
ER and Mitochondria
Contact sites between the ER and mitochondria are a highly conserved feature of eukaryotic cells and have been associated with several important functions including Ca2+ signaling, lipid biosynthesis, organelle inheritance, and mitochondrial division (Vance 1990; Rizzuto et al. 1998; Boldogh and Pon 2007; Friedman et al. 2011). EM tomography has shown that the two apposing membranes come within 30 nm of each other (Csordas et al. 2006; West et al. 2011); in some striking examples of ER-mitochondria contacts, electron dense structures can be seen by EM that resemble proteinaceous tethers (Csordas et al. 2006; Hayashi et al. 2009). Contact between these two organelles can also be visualized by live confocal fluorescence microscopy, time-lapsed images reveal contact sites that are so securely tethered that the two organelles remain attached even as they move along the cytoskeleton (Friedman et al. 2010).
The influx of Ca2+ into the intermembrane space and matrix of the mitochondria has been observed at ER contact sites (Rizzuto et al. 1993, 1998; Csordas et al. 2006). Calcium uptake into mitochondria requires IP3 receptor (IP3R) interaction with the voltage-dependent anion selective channel protein 1 (VDAC-1) on the OMM (Rizzuto et al. 1998; Szabadkai et al. 2006; Hayashi et al. 2009). Both deletion of IP3R or ligand induced inhibition of IP3R blocks activities associated with mitochondrial Ca2+ influx (Rizzuto et al. 1993, 1998; Khan et al. 1996; Jayaraman and Marks 1997). Elegant experiments have shown that Ca2+ signaling from the ER to the mitochondria was prevented by altering the spacing between the ER and mitochondria by artificially tethering the two membranes too closely (Csordas et al. 2006, 2010). The close spacing was designed so that it would prevent the large IP3R channel from being able to fit between the ER and mitochondria (Csordas et al. 2006, 2010). These experiments suggest a model whereby the ER channel would need to release Ca2+ directly to the mitochondrial membrane rather than from nearby. There are several functions associated with localized positions of Ca2+ exchange on the mitochondria; changes in Ca2+ levels has been shown to affect apoptosis, mitochondrial division and motility, and to regulate the activity of Ca2+-binding proteins on the mitochondria that can only bind Ca2+ at high local concentrations (Rizzuto et al. 1993, 1998; Berridge 2002; Scorrano et al. 2003).
Contact sites between the ER and mitochondria are also likely to be required for lipid flipping between the apposed membranes during lipid biosynthesis (Vance 1990; Achleitner et al. 1999; Voelker 2000; Holthuis and Levine 2005; van Meer et al. 2008; Kornmann et al. 2009). Resident proteins of both the ER and the mitochondria are required for the biosynthesis of one of the cells most abundant phospholipids, phosphatidylcholine (PC). During this process, phosphatidylserine (PS) is first synthesized from phosphatidylalanine on the ER membrane; PS conversion to phosphatidylethanolamine (PE) requires proteins on the mitochondria, whereas PE conversion to PC requires ER localized enzymes (van Meer et al. 2008; Osman et al. 2011). Therefore, before each of these conversion steps, the phospholipid must be moved back and forth between the apposing membranes. At the same time, the steady state levels of each of these phospholipids are likely to be maintained for each organelle. Many enzymes involved in lipid biosynthesis are found to be biochemically enriched in the membrane fraction that constitutes mitochondrial associated membranes (MAM) (Vance 1990; Stone and Vance 2000; Voelker 2000); however, the factors involved in transferring of phospholipids between the ER and mitochondrial membranes during conversion are not known.
Recently, ER tubules were also shown to mark the position of mitochondrial division in yeast and mammalian cells (Friedman et al. 2011). Using high-resolution EM tomography and live-confocal fluorescence microscopy, ER tubules were shown to define the position of division machinery recruitment by wrapping around the mitochondria before division in both yeast and mammalian cells (Fig. 4C). ER tubule contact was observed at positions where mitochondria were constricted, even when the mitochondrial division machinery was depleted (Friedman et al. 2011). These data suggest that ER tubule contact may play an early role in constricting the mitochondria to allow division machinery recruitment. It is currently unknown whether the ER provides a structural role in mitochondrial constriction and division, or whether the ER provides a scaffold upon which other required factors are recruited. It also remains to be determined how the mitochondria fusion and fission machinery interact with one another at sites of ER-mitochondria contacts. Regardless, this work shows a new and unexpected role for ER tubules during mitochondrial biogenesis.
There are many functions that occur at ER-mitochondria contact sites; therefore, it is possible and reasonable to expect multiple tethers. Alternatively, a single tethering complex could allow all processes to be synergized at the same position. Recently, the ER-mitochondria encounter structures (ERMES) complex was identified in yeast as a tether between the two organelles. In yeast cells, ERMES is comprised of MMM1, MDM10, MDM12, and MDM34. The components of the ERMES complex consists of membrane proteins that localize to both the ER (Mmm1) and mitochondrial membrane (Mdm10 and Mdm34) (Kornmann et al. 2009). Furthermore, mutation of ERMES components can be rescued when coexpressed with a synthetic ER-mitochondrial linker complex (Kornmann et al. 2009). Thus, ERMES makes a compelling tether candidate for ER-mitochondrial contacts. It is not known whether there are other tethers in yeast besides ERMES; mammalian homologs of the ERMES complex have not been identified, giving further indication that there are other tethers. In mammalian cells, Mitofusin2 (MFN2) has been shown to tether the ER to the mitochondria (de Brito and Scorrano 2008). ER-localized MFN2 is thought to homo- or hetero-complex with MFN2 or MFN1 on the outer mitochondrial membrane and act as an ER-mitochondria tether (de Brito and Scorrano 2008). Depletion of MFN2 in vivo decreased the amount of ER-mitochondria contact, this effect was rescued by expression of an ER localized MFN2 protein (de Brito and Scorrano 2008); however, a role for MFN2 in mitochondrial tethering to the ER in yeast has not been shown. In addition, MFN2 does not appear to be required for contact between ER tubules and mitochondria at constriction/fission sites (Friedman et al. 2011). Together, these data suggest that there could be multiple complexes involved in tethering between these organelles and that contact site formation may be highly regulated during different ER-mitochondria functions.
ER and Golgi
The Golgi forms membrane contacts with the ER to regulate the transfer of secreted proteins and lipids (Glick and Nakano 2009). Trafficking between the ER and Golgi involves anterograde (COPII mediated), retrograde (COPI mediated), and direct nonvesicular transport between the apposed membranes (Malhotra et al. 1989; Barlowe et al. 1994; Ladinsky et al. 1999; Hanada 2010). High resolution EM has shown the close relationship between the ER and Golgi at membrane contact sites thought to facilitate nonvesicular lipid transport (Fig. 4D) (Ladinsky et al. 1999; Mogelsvang et al. 2004). Depletion of integral ER membrane proteins VAP-A and VAP-B, resulted in mislocalization of Golgi lipid sensing and transport proteins (Nir2, OSBP, and CERT), and altered Golgi structure; furthermore, Nir2 was shown to directly transfer PI from the ER to the Golgi (Peretti et al. 2008). Additional work shows the role of ceramide transport protein (CERT) in the nonvesicular transport of ceramide (a sphingomyelin precursor) from the ER to the trans-Golgi (Hanada 2010). Mutations in the FFAT motif of CERT were shown to disrupt CERT-mediated transport of ceramide from the ER to the Golgi (Kawano et al. 2006). Nonvesicular ceramide transport at ER-Golgi contact sites regulated by CERT is thought to be facilitated by a conformational change in the START domain of CERT (Hanada 2010). Thus, these results show a direct connection between the ER and Golgi at membrane contacts. The proteins regulating tethering between the ER and Golgi membranes at contact sites have not yet been identified.
ER and Endosomes
A growing collection of evidence revolves around the characterization of ER-endosome contacts and the function of these contacts in lipid and sterol exchange. In the first reported example of ER-organelle membrane contact sites, the nucleus-vacuole junction in yeast cells was identified; these membrane contact sites are mediated by the direct interaction between the ER membrane protein Nvj1 and the vacuole protein Vac8 (Pan et al. 2000). The nucleus-vacuole junction is not present in mammalian cells, but other endosome compartments have been observed interacting with the ER; high-resolution immuno-EM studies showed the colocalization of the ER localized phophatase PTP1B with the endocytic cargo EGFR (Fig. 4E) (Eden et al. 2010). Additionally, sustained contact can be observed by live confocal fluorescence microscopy between Rab5-labeled early endosomes and the ER in mammalian cell culture (Friedman et al. 2010). One proposed function of ER-endosomes contacts may be to monitor and regulate cholesterol levels in the endocytic pathway (Rocha et al. 2009). Cholesterol levels effect the direction of late endosome (LE) trafficking; under cholesterol-rich conditions, LEs accumulate at the MTOC after dissociation from the dynein complex (Rocha et al. 2009). The dissociation from the dynein complex is thought to be regulated by the cholesterol sensor oxysterol-binding protein-related protein 1L (ORP1L), mutants in this protein can alter the response to cholesterol levels and association with dynein (Rocha et al. 2009). Recently, the integral ER protein VAP-A was shown to play a role in regulating trafficking of LEs in response to cholesterol levels (Rocha et al. 2009). VAP-A depletion alters the ability of LEs to traffic away from the MTOC (Rocha et al. 2009). Thin section EM under control conditions showed some interaction between LEs and the ER; this interaction can be increased or decreased by decreasing or increasing cholesterol levels, respectively (Rocha et al. 2009). The proteins regulating tethering between the ER and different endosomal membrane compartments are currently unknown.
ER and Peroxisomes
The peroxisome is a unique organelle that receives much of its starting material, including lipids and peroxisomal membrane proteins, from the ER for de novo biogenesis (Kim et al. 2006; Lam et al. 2010). Past results show that both vesicular and nonvesicular transport of lipids and proteins from the ER are required for proper peroxisomal formation and maintenance (Raychaudhuri and Prinz 2008; Lam et al. 2010; Agrawal et al. 2011; van der Zand et al. 2012). In vitro ER-peroxisome lipid transfer assays also show nonvesicular transport between the membrane compartments; however, the proteins mediating this process remain to be identified (Raychaudhuri and Prinz 2008). Recent live cell image analysis and in vitro cell-free ER budding assays identified a distinct pool of vesicles rich in peroxisomal membrane proteins budding from the ER at peroxisomal ER exit sites, these vesicles then underwent heterotypic fusion with other ER-derived peroxisomal membrane protein vesicles to form a new peroxisome compartment (Agrawal et al. 2011; van der Zand et al. 2012). The ER-derived peroxisome remains a striking example of the ER’s ability to generate an organelle distinct from itself in both morphology and function.
CONCLUDING REMARKS
Thorough investigation of the ER, its structure and contacts with other organelles has illuminated many of the factors and forces involved in regulation of the ER and its numerous processes. High-resolution fluorescence microscopy, 3D EM tomographs and numerous biochemical assays have revealed ER structures, membrane contacts and functions that were previously unknown and uncharacterized. Much work has surrounded understanding how the distinct domains of the ER are regulated; the culmination of quality work has identified numerous proteins and protein complexes as being responsible. Dramatic ER shape changes during mitosis have been observed, but the proteins or processes involved are currently unknown. Although we know that the ER is extremely dynamic on the MT cytoskeleton and contacts numerous organelles, only a handful of regulating proteins have been identified and even fewer mechanisms are currently known. Future work addressing these questions will further our understanding of the ER and its capabilities.
ACKNOWLEDGMENTS
We thank S. Ferro-Novick, J. Friedman, M. Hetzer, M. Ladinsky, W. Prinz, T. Rapoport, N. Rusan, Y. Shibata, and M. West for providing images, and C. English for helpful suggestions. This work is supported by grant RO1GM083977 to G.K.V. from the National Institutes of Health (NIH). A.R.E. is supported by an NIH predoctoral training grant GM07135.
Footnotes
Editors: Susan Ferro-Novick, Tom A. Rapoport, and Randy Schekman
Additional Perspectives on The Endoplasmic Reticulum available at www.cshperspectives.org
REFERENCES
- Achleitner G, Gaigg B, Krasser A, Kainersdorfer E, Kohlwein SD, Perktold A, Zellnig G, Daum G 1999. Association between the endoplasmic reticulum and mitochondria of yeast facilitates interorganelle transport of phospholipids through membrane contact. Eur J Biochem 264: 545–553 [DOI] [PubMed] [Google Scholar]
- Agrawal G, Joshi S, Subramani S 2011. Cell-free sorting of peroxisomal membrane proteins from the endoplasmic reticulum. Proc Natl Acad Sci 108: 9113–9118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DJ, Hetzer MW 2007. Nuclear envelope formation by chromatin-mediated reorganization of the endoplasmic reticulum. Nat Cell Biol 9: 1160–1166 [DOI] [PubMed] [Google Scholar]
- Anderson DJ, Hetzer MW 2008a. Reshaping of the endoplasmic reticulum limits the rate for nuclear envelope formation. J Cell Biol 182: 911–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DJ, Hetzer MW 2008b. Shaping the endoplasmic reticulum into the nuclear envelope. J Cell Sci 121: 137–142 [DOI] [PubMed] [Google Scholar]
- Anwar K, Klemm RW, Condon A, Severin KN, Zhang M, Ghirlando R, Hu J, Rapoport TA, Prinz WA 2012. The dynamin-like GTPase Sey1p mediates homotypic ER fusion in S. cerevisiae. J Cell Biol 197: 209–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audhya A, Desai A, Oegema K 2007. A role for Rab5 in structuring the endoplasmic reticulum. J Cell Biol 178: 43–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe C, Orci L, Yeung T, Hosobuchi M, Hamamoto S, Salama N, Rexach MF, Ravazzola M, Amherdt M, Schekman R 1994. COPII: A membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell 77: 895–907 [DOI] [PubMed] [Google Scholar]
- Baumann O, Walz B 2001. Endoplasmic reticulum of animal cells and its organization into structural and functional domains. Int Rev Cytol 205: 149–214 [DOI] [PubMed] [Google Scholar]
- Baumann NA, Sullivan DP, Ohvo-Rekila H, Simonot C, Pottekat A, Klaassen Z, Beh CT, Menon AK 2005. Transport of newly synthesized sterol to the sterol-enriched plasma membrane occurs via nonvesicular equilibration. Biochemistry 44: 5816–5826 [DOI] [PubMed] [Google Scholar]
- Behnia R, Munro S 2005. Organelle identity and the signposts for membrane traffic. Nature 438: 597–604 [DOI] [PubMed] [Google Scholar]
- Benyamini P, Webster P, Meyer DI 2009. Knockdown of p180 eliminates the terminal differentiation of a secretory cell line. Mol Biol Cell 20: 732–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge MJ 2002. The endoplasmic reticulum: A multifunctional signaling organelle. Cell Calcium 32: 235–249 [DOI] [PubMed] [Google Scholar]
- Boldogh IR, Pon LA 2007. Mitochondria on the move. Trends Cell Biol 17: 502–510 [DOI] [PubMed] [Google Scholar]
- Cai H, Reinisch K, Ferro-Novick S 2007. Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev Cell 12: 671–682 [DOI] [PubMed] [Google Scholar]
- Carrasco S, Meyer T 2011. STIM proteins and the endoplasmic reticulum-plasma membrane junctions. Annu Rev Biochem 80: 973–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Novick P, Ferro-Novick S 2012. ER network formation requires a balance of the dynamin-like GTPase Sey1p and the Lunapark family member Lnp1p. Nat Cell Biol 30: 328–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courvalin JC, Segil N, Blobel G, Worman HJ 1992. The lamin B receptor of the inner nuclear membrane undergoes mitosis-specific phosphorylation and is a substrate for p34cdc2–type protein kinase. J Biol Chem 267: 19035–19038 [PubMed] [Google Scholar]
- Csordas G, Renken C, Varnai P, Walter L, Weaver D, Buttle KF, Balla T, Mannella CA, Hajnoczky G 2006. Structural and functional features and significance of the physical linkage between ER and mitochondria. J Cell Biol 174: 915–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csordas G, Varnai P, Golenar T, Roy S, Purkins G, Schneider TG, Balla T, Hajnoczky G 2010. Imaging interorganelle contacts and local calcium dynamics at the ER-mitochondrial interface. Mol Cell 39: 121–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayel MJ, Hom EF, Verkman AS 1999. Diffusion of green fluorescent protein in the aqueous-phase lumen of endoplasmic reticulum. Biophys J 76: 2843–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Brito OM, Scorrano L 2008. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature 456: 605–610 [DOI] [PubMed] [Google Scholar]
- De Craene JO, Coleman J, Estrada de Martin P, Pypaert M, Anderson S, Yates JR 3rd, Ferro-Novick S, Novick P 2006. Rtn1p is involved in structuring the cortical endoplasmic reticulum. Mol Biol Cell 17: 3009–3020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Saint-Jean M, Delfosse V, Douguet D, Chicanne G, Payrastre B, Bourguet W, Antonny B, Drin G 2011. Osh4p exchanges sterols for phosphatidylinositol 4-phosphate between lipid bilayers. J Cell Biol 195: 965–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden ER, White IJ, Tsapara A, Futter CE 2010. Membrane contacts between endosomes and ER provide sites for PTP1B-epidermal growth factor receptor interaction. Nat Cell Biol 12: 267–272 [DOI] [PubMed] [Google Scholar]
- Ellenberg J, Siggia ED, Moreira JE, Smith CL, Presley JF, Worman HJ, Lippincott-Schwartz J 1997. Nuclear membrane dynamics and reassembly in living cells: Targeting of an inner nuclear membrane protein in interphase and mitosis. J Cell Biol 138: 1193–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- English AR, Zurek N, Voeltz GK 2009. Peripheral ER structure and function. Curr Opin Cell Biol 21: 596–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feske S, Gwack Y, Prakriya M, Srikanth S, Puppel SH, Tanasa B, Hogan PG, Lewis RS, Daly M, Rao A 2006. A mutation in Orai1 causes immune deficiency by abrogating CRAC channel function. Nature 441: 179–185 [DOI] [PubMed] [Google Scholar]
- Friedman JR, Voeltz GK 2011. The ER in 3D: A multifunctional dynamic membrane network. Trends Cell Biol 21: 709–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JR, Webster BM, Mastronarde DN, Verhey KJ, Voeltz GK 2010. ER sliding dynamics and ER-mitochondrial contacts occur on acetylated microtubules. J Cell Biol 190: 363–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JR, Lackner LL, West M, Dibenedetto JR, Nunnari J, Voeltz GK 2011. ER tubules mark sites of mitochondrial division. Science 334: 358–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev AG, Sullivan DP, Kersting MC, Dittman JS, Beh CT, Menon AK 2011. Osh proteins regulate membrane sterol organization but are not required for sterol movement between the ER and PM. Traffic 12: 1341–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick BS, Nakano A 2009. Membrane traffic within the Golgi apparatus. Annu Rev Cell Dev Biol 25: 113–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriev I, Gouveia SM, van der Vaart B, Demmers J, Smyth JT, Honnappa S, Splinter D, Steinmetz MO, Putney JW Jr, Hoogenraad CC, et al. 2008. STIM1 is a MT-plus-end-tracking protein involved in remodeling of the ER. Curr Biol 18: 177–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K 2010. Intracellular trafficking of ceramide by ceramide transfer protein. Proc Jpn Acad Ser B Phys Biol Sci 86: 426–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Rizzuto R, Hajnoczky G, Su TP 2009. MAM: More than just a housekeeper. Trends Cell Biol 19: 81–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzer MW 2010. The nuclear envelope. Cold Spring Harb Perspect Biol 2: a000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holthuis JC, Levine TP 2005. Lipid traffic: Floppy drives and a superhighway. Nat Rev Mol Cell Biol 6: 209–220 [DOI] [PubMed] [Google Scholar]
- Hu J, Shibata Y, Voss C, Shemesh T, Li Z, Coughlin M, Kozlov MM, Rapoport TA, Prinz WA 2008. Membrane proteins of the endoplasmic reticulum induce high-curvature tubules. Science 319: 1247–1250 [DOI] [PubMed] [Google Scholar]
- Hu J, Shibata Y, Zhu PP, Voss C, Rismanchi N, Prinz WA, Rapoport TA, Blackstone C 2009. A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell 138: 549–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen M, Ohsaki Y, Rita Rega L, Bittman R, Olkkonen VM, Ikonen E 2011. Role of ORPs in sterol transport from plasma membrane to ER and lipid droplets in mammalian cells. Traffic 12: 218–231 [DOI] [PubMed] [Google Scholar]
- Jayaraman T, Marks AR 1997. T cells deficient in inositol 1,4,5-trisphosphate receptor are resistant to apoptosis. Mol Cell Biol 17: 3005–3012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano M, Kumagai K, Nishijima M, Hanada K 2006. Efficient trafficking of ceramide from the endoplasmic reticulum to the Golgi apparatus requires a VAMP-associated protein-interacting FFAT motif of CERT. J Biol Chem 281: 30279–30288 [DOI] [PubMed] [Google Scholar]
- Khan AA, Soloski MJ, Sharp AH, Schilling G, Sabatini DM, Li SH, Ross CA, Snyder SH 1996. Lymphocyte apoptosis: Mediation by increased type 3 inositol 1,4,5-trisphosphate receptor. Science 273: 503–507 [DOI] [PubMed] [Google Scholar]
- Kim PK, Mullen RT, Schumann U, Lippincott-Schwartz J 2006. The origin and maintenance of mammalian peroxisomes involves a de novo PEX16-dependent pathway from the ER. J Cell Biol 173: 521–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiseleva E, Morozova KN, Voeltz GK, Allen TD, Goldberg MW 2007. Reticulon 4a/NogoA locates to regions of high membrane curvature and may have a role in nuclear envelope growth. J Struct Biol 160: 224–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B, Currie E, Collins SR, Schuldiner M, Nunnari J, Weissman JS, Walter P 2009. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science 325: 477–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladinsky MS, Mastronarde DN, McIntosh JR, Howell KE, Staehelin LA 1999. Golgi structure in three dimensions: Functional insights from the normal rat kidney cell. J Cell Biol 144: 1135–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam SK, Yoda N, Schekman R 2010. A vesicle carrier that mediates peroxisome protein traffic from the endoplasmic reticulum. Proc Natl Acad Sci 107: 21523–21528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebiedzinska M, Szabadkai G, Jones AW, Duszynski J, Wieckowski MR 2009. Interactions between the endoplasmic reticulum, mitochondria, plasma membrane and other subcellular organelles. Int J Biochem Cell Biol 41: 1805–1816 [DOI] [PubMed] [Google Scholar]
- Lee C, Chen LB 1988. Dynamic behavior of endoplasmic reticulum in living cells. Cell 54: 37–46 [DOI] [PubMed] [Google Scholar]
- Li Y, Prinz WA 2004. ATP-binding cassette (ABC) transporters mediate nonvesicular, raft-modulated sterol movement from the plasma membrane to the endoplasmic reticulum. J Biol Chem 279: 45226–45234 [DOI] [PubMed] [Google Scholar]
- Liou J, Kim ML, Heo WD, Jones JT, Myers JW, Ferrell JE Jr, Meyer T 2005. STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr Biol 15: 1235–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Ladinsky MS, Kirchhausen T 2009. Cisternal organization of the endoplasmic reticulum during mitosis. Mol Biol Cell 20: 3471–3480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Ladinsky MS, Kirchhausen T 2011. Formation of the postmitotic nuclear envelope from extended ER cisternae precedes nuclear pore assembly. J Cell Biol 194: 425–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra V, Serafini T, Orci L, Shepherd JC, Rothman JE 1989. Purification of a novel class of coated vesicles mediating biosynthetic protein transport through the Golgi stack. Cell 58: 329–336 [DOI] [PubMed] [Google Scholar]
- McCullough S, Lucocq J 2005. Endoplasmic reticulum positioning and partitioning in mitotic HeLa cells. J Anat 206: 415–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogelsvang S, Marsh BJ, Ladinsky MS, Howell KE 2004. Predicting function from structure: 3D structure studies of the mammalian Golgi complex. Traffic 5: 338–345 [DOI] [PubMed] [Google Scholar]
- Murray JT, Panaretou C, Stenmark H, Miaczynska M, Backer JM 2002. Role of Rab5 in the recruitment of hVps34/p150 to the early endosome. Traffic 3: 416–427 [DOI] [PubMed] [Google Scholar]
- Orci L, Ravazzola M, Le Coadic M, Shen WW, Demaurex N, Cosson P 2009. From the cover: STIM1-induced precortical and cortical subdomains of the endoplasmic reticulum. Proc Natl Acad Sci 106: 19358–19362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orso G, Pendin D, Liu S, Tosetto J, Moss TJ, Faust JE, Micaroni M, Egorova A, Martinuzzi A, McNew JA, et al. 2009. Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin. Nature 460: 978–983 [DOI] [PubMed] [Google Scholar]
- Osman C, Voelker DR, Langer T 2011. Making heads or tails of phospholipids in mitochondria. J Cell Biol 192: 7–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Roberts P, Chen Y, Kvam E, Shulga N, Huang K, Lemmon S, Goldfarb DS 2000. Nucleus-vacuole junctions in Saccharomyces cerevisiae are formed through the direct interaction of Vac8p with Nvj1p. Mol Biol Cell 11: 2445–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CY, Shcheglovitov A, Dolmetsch R 2010a. The CRAC channel activator STIM1 binds and inhibits L-type voltage-gated calcium channels. Science 330: 101–105 [DOI] [PubMed] [Google Scholar]
- Park SH, Zhu PP, Parker RL, Blackstone C 2010b. Hereditary spastic paraplegia proteins REEP1, spastin, and atlastin-1 coordinate microtubule interactions with the tubular ER network. J Clin Invest 120: 1097–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SK, Indig FE, Olivieri N, Levine ND, Latterich M 1998. Organelle membrane fusion: A novel function for the syntaxin homolog Ufe1p in ER membrane fusion. Cell 92: 611–620 [DOI] [PubMed] [Google Scholar]
- Peretti D, Dahan N, Shimoni E, Hirschberg K, Lev S 2008. Coordinated lipid transfer between the endoplasmic reticulum and the Golgi complex requires the VAP proteins and is essential for Golgi-mediated transport. Mol Biol Cell 19: 3871–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer SR 2001. Rab GTPases: Specifying and deciphering organelle identity and function. Trends Cell Biol 11: 487–491 [DOI] [PubMed] [Google Scholar]
- Pichler H, Gaigg B, Hrastnik C, Achleitner G, Kohlwein SD, Zellnig G, Perktold A, Daum G 2001. A subfraction of the yeast endoplasmic reticulum associates with the plasma membrane and has a high capacity to synthesize lipids. Eur J Biochem 268: 2351–2361 [DOI] [PubMed] [Google Scholar]
- Puhka M, Vihinen H, Joensuu M, Jokitalo E 2007. Endoplasmic reticulum remains continuous and undergoes sheet-to-tubule transformation during cell division in mammalian cells. J Cell Biol 179: 895–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puhka M, Joensuu M, Vihinen H, Belevich I, Jokitalo E 2012. Progressive sheet-to-tubule transformation is a general mechanism for endoplasmic reticulum partitioning in dividing mammalian cells. Mol Biol Cell 23: 2424–2432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S, Prinz WA 2008. Nonvesicular phospholipid transfer between peroxisomes and the endoplasmic reticulum. Proc Natl Acad Sci 105: 15785–15790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S, Im YJ, Hurley JH, Prinz WA 2006. Nonvesicular sterol movement from plasma membrane to ER requires oxysterol-binding protein-related proteins and phosphoinositides. J Cell Biol 173: 107–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rismanchi N, Soderblom C, Stadler J, Zhu PP, Blackstone C 2008. Atlastin GTPases are required for Golgi apparatus and ER morphogenesis. Hum Mol Genet 17: 1591–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzuto R, Brini M, Murgia M, Pozzan T 1993. Microdomains with high Ca2+ close to IP3-sensitive channels that are sensed by neighboring mitochondria. Science 262: 744–747 [DOI] [PubMed] [Google Scholar]
- Rizzuto R, Pinton P, Carrington W, Fay FS, Fogarty KE, Lifshitz LM, Tuft RA, Pozzan T 1998. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science 280: 1763–1766 [DOI] [PubMed] [Google Scholar]
- Rocha N, Kuijl C, van der Kant R, Janssen L, Houben D, Janssen H, Zwart W, Neefjes J 2009. Cholesterol sensor ORP1L contacts the ER protein VAP to control Rab7-RILP-p150 Glued and late endosome positioning. J Cell Biol 185: 1209–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos J, DiGregorio PJ, Yeromin AV, Ohlsen K, Lioudyno M, Zhang S, Safrina O, Kozak JA, Wagner SL, Cahalan MD, et al. 2005. STIM1, an essential and conserved component of store-operated Ca2+ channel function. J Cell Biol 169: 435–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Levine TP 2004. Multiple pools of phosphatidylinositol 4-phosphate detected using the pleckstrin homology domain of Osh2p. J Biol Chem 279: 44683–44689 [DOI] [PubMed] [Google Scholar]
- Schuck S, Prinz WA, Thorn KS, Voss C, Walter P 2009. Membrane expansion alleviates endoplasmic reticulum stress independently of the unfolded protein response. J Cell Biol 187: 525–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz SL, Cao C, Pylypenko O, Rak A, Wandinger-Ness A 2007. Rab GTPases at a glance. J Cell Sci 120: 3905–3910 [DOI] [PubMed] [Google Scholar]
- Scorrano L, Oakes SA, Opferman JT, Cheng EH, Sorcinelli MD, Pozzan T, Korsmeyer SJ 2003. BAX and BAK regulation of endoplasmic reticulum Ca2+: A control point for apoptosis. Science 300: 135–139 [DOI] [PubMed] [Google Scholar]
- Shibata Y, Voeltz GK, Rapoport TA 2006. Rough sheets and smooth tubules. Cell 126: 435–439 [DOI] [PubMed] [Google Scholar]
- Shibata Y, Voss C, Rist JM, Hu J, Rapoport TA, Prinz WA, Voeltz GK 2008. The reticulon and DP1/Yop1p proteins form immobile oligomers in the tubular endoplasmic reticulum. J Biol Chem 283: 18892–18904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, Shemesh T, Prinz WA, Palazzo AF, Kozlov MM, Rapoport TA 2010. Mechanisms determining the morphology of the peripheral ER. Cell 143: 774–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth JT, Beg AM, Wu S, Putney JW Jr, Rusan NM 2012. Phosphoregulation of STIM1 leads to exclusion of the endoplasmic reticulum from the mitotic spindle. Curr Biol 10.1016/j.cub.2012.05.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa BA, Rothballer A, Kutay U, Schwartz TU 2012. LINC complexes form by binding of three KASH peptides to domain interfaces of trimeric SUN proteins. Cell 149: 1035–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefan CJ, Manford AG, Baird D, Yamada-Hanff J, Mao Y, Emr SD 2011. Osh proteins regulate phosphoinositide metabolism at ER-plasma membrane contact sites. Cell 144: 389–401 [DOI] [PubMed] [Google Scholar]
- Stone SJ, Vance JE 2000. Phosphatidylserine synthase-1 and -2 are localized to mitochondria-associated membranes. J Biol Chem 275: 34534–34540 [DOI] [PubMed] [Google Scholar]
- Suntharalingam M, Wente SR 2003. Peering through the pore: Nuclear pore complex structure, assembly, and function. Dev Cell 4: 775–789 [DOI] [PubMed] [Google Scholar]
- Szabadkai G, Bianchi K, Varnai P, De Stefani D, Wieckowski MR, Cavagna D, Nagy AI, Balla T, Rizzuto R 2006. Chaperone-mediated coupling of endoplasmic reticulum and mitochondrial Ca2+ channels. J Cell Biol 175: 901–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terasaki M, Chen LB, Fujiwara K 1986. Microtubules and the endoplasmic reticulum are highly interdependent structures. J Cell Biol 103: 1557–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolley N, Sparkes IA, Hunter PR, Craddock CP, Nuttall J, Roberts LM, Hawes C, Pedrazzini E, Frigerio L 2008. Overexpression of a plant reticulon remodels the lumen of the cortical endoplasmic reticulum but does not perturb protein transport. Traffic 9: 94–102 [DOI] [PubMed] [Google Scholar]
- Toulmay A, Prinz WA 2011. Lipid transfer and signaling at organelle contact sites: The tip of the iceberg. Curr Opin Cell Biol 23: 458–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner MD, Plutner H, Balch WE 1997. A Rab GTPase is required for homotypic assembly of the endoplasmic reticulum. J Biol Chem 272: 13479–13483 [DOI] [PubMed] [Google Scholar]
- Tzur YB, Wilson KL, Gruenbaum Y 2006. SUN-domain proteins: “Velcro” that links the nucleoskeleton to the cytoskeleton. Nat Rev Mol Cell Biol 7: 782–788 [DOI] [PubMed] [Google Scholar]
- Vance JE 1990. Phospholipid synthesis in a membrane fraction associated with mitochondria. J Biol Chem 265: 7248–7256 [PubMed] [Google Scholar]
- van der Zand A, Gent J, Braakman I, Tabak HF 2012. Biochemically distinct vesicles from the endoplasmic reticulum fuse to form peroxisomes. Cell 149: 397–409 [DOI] [PubMed] [Google Scholar]
- van Meer G, Voelker DR, Feigenson GW 2008. Membrane lipids: Where they are and how they behave. Nat Rev Mol Cell Biol 9: 112–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedrenne C, Hauri HP 2006. Morphogenesis of the endoplasmic reticulum: Beyond active membrane expansion. Traffic 7: 639–646 [DOI] [PubMed] [Google Scholar]
- Voelker DR 2000. Interorganelle transport of aminoglycerophospholipids. Biochim Biophys Acta 1486: 97–107 [DOI] [PubMed] [Google Scholar]
- Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA 2006. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell 124: 573–586 [DOI] [PubMed] [Google Scholar]
- Waterman-Storer CM, Salmon ED 1998. Endoplasmic reticulum membrane tubules are distributed by microtubules in living cells using three distinct mechanisms. Curr Biol 8: 798–806 [DOI] [PubMed] [Google Scholar]
- West M, Zurek N, Hoenger A, Voeltz GK 2011. A 3D analysis of yeast ER structure reveals how ER domains are organized by membrane curvature. J Cell Biol 193: 333–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak MJ, Bola B, Brownhill K, Yang YC, Levakova V, Allan VJ 2009. Role of kinesin-1 and cytoplasmic dynein in endoplasmic reticulum movement in VERO cells. J Cell Sci 122: 1979–1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Guan T, Gerace L 1997. Integral membrane proteins of the nuclear envelope are dispersed throughout the endoplasmic reticulum during mitosis. J Cell Biol 137: 1199–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Tsai MY 2006. The mitotic spindle matrix: A fibro-membranous lamin connection. Cell Cycle 5: 2345–2347 [DOI] [PubMed] [Google Scholar]
- Zurek N, Sparks L, Voeltz G 2011. Reticulon short hairpin transmembrane domains are used to shape ER tubules. Traffic 12: 28–41 [DOI] [PMC free article] [PubMed] [Google Scholar]