Abstract
Sphingolipids are a diverse group of lipids that have essential cellular roles as structural components of membranes and as potent signaling molecules. In recent years, a detailed picture has emerged of the basic biochemistry of sphingolipids—from their initial synthesis in the endoplasmic reticulum (ER), to their elaboration into complex glycosphingolipids, to their turnover and degradation. However, our understanding of how sphingolipid metabolism is regulated in response to metabolic demand and physiologic cues remains incomplete. Here I discuss new insights into the mechanisms that ensure sphingolipid homeostasis, with an emphasis on the ER as a critical regulatory site in sphingolipid metabolism. In particular, Orm family proteins have recently emerged as key ER-localized mediators of sphingolipid homeostasis. A detailed understanding of how cells sense and control sphingolipid production promises to provide key insights into membrane function in health and disease.
Representing roughly 10%–20% of cellular lipids, sphingolipids are structural components of membranes and critical signaling molecules. Their homeostasis is mediated, at least in part, by ER-localized Orm proteins.
Eukaryotic cell membranes maintain a complex and tightly regulated complement of lipids and proteins that are essential for their function. These lipids can be divided into three broad classes—sterols, glycerolipids, and sphingolipids—on the basis of their distinct chemical structures and dedicated enzymatic machineries (Fig. 1A–C). Sphingolipids typically represent ∼10%–20% of cellular lipids and have essential functions arising both from their effects on the physical properties of membranes and from their roles as signaling molecules (van Meer et al. 2008). Additionally, the activities of many transmembrane and peripheral membrane proteins are dependent on their close association with sphingolipids (Lingwood and Simons 2010). Over recent years, sphingolipids have been shown to participate in an increasingly wide range of biological processes that includes secretion, endocytosis, chemotaxis, neurotransmission, angiogenesis, and inflammation (Hannun and Obeid 2008; Lingwood and Simons 2010; Lippincott-Schwartz and Phair 2010; Blaho and Hla 2011; Lingwood 2011).
Figure 1.
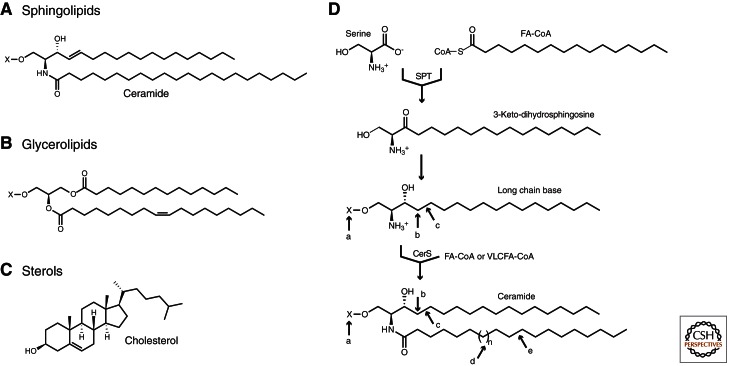
Structures of sphingolipids and other cellular lipids. (A–C) Representative structures of (A) sphingolipids, (B) glycerolipids, and (C) sterols. (D) Formation of sphingolipids from key building blocks, long chain bases (LCBs), and coenzyme A-linked fatty acids (FA-CoAs) that often have a very long acyl chain (VLCFA-CoA). Serine palmitoyltransferase (SPT) produces the LCB intermediate 3-keto-dihydrosphingosine, which is then reduced to yield LCBs that are used by ceramide synthase (CerS) to form ceramides. Sphingolipid structural diversity arises from (a) headgroup modifications including phosphorylation, glycosylation, or phosphocholine addition, (b) LCB hydroxylation, (c) LCB desaturation, (d) variability in the length of the N-linked acyl chain, and (e) desaturation of the N-linked acyl chain.
The focus of this article is the variety of regulatory mechanisms that cells use to ensure sphingolipid homeostasis. This task requires balancing sphingolipid levels in conjunction with sterols and glycerolipids and adapting sphingolipid metabolism in response to physiological cues and external stresses. A need for tight regulatory control is further highlighted by the potent signaling activities of many sphingolipid biosynthetic intermediates such as sphingosines and ceramides (Hannun and Obeid 2008; Fyrst and Saba 2010; Blaho and Hla 2011). Additionally, because most sphingolipids cannot move freely between different organelles, cells must regulate multiple intracellular pools of sphingolipids as well as lipid transport between these sites.
It is noteworthy that, despite great progress in defining the enzymes that carry out sphingolipid synthesis and degradation, how cells achieve sphingolipid homeostasis remains poorly understood. In this article, I will describe recent progress in the field and highlight outstanding questions. In particular, I will discuss the emergence of the endoplasmic reticulum (ER) as a key site for sphingolipid homeostasis. Several critical enzymes in sphingolipid metabolism are found in the ER, and recent studies have identified a mechanism for matching sphingolipid production to metabolic demand that depends on the ER-localized Orm family of proteins (Breslow et al. 2010). Although many details of Orm protein function remain to be discovered, Orm proteins provide a valuable model for understanding how cells sense sphingolipids and dynamically regulate sphingolipid metabolism.
SPHINGOLIPID FUNCTIONS AND METABOLISM
Sphingolipid Modes of Action
To provide a context for sphingolipid homeostasis, one must consider the mechanistic basis for sphingolipid functions as well as the enzymatic machinery that controls sphingolipid metabolism. Although a complete review of the functions of sphingolipids is beyond the scope of this article, their biological activities can be attributed to three primary modes of action. First, sphingolipids modulate the physical properties of membranes, including their fluidity, thickness, and curvature. These attributes underlie the key functions of sphingolipids in myelin (Chrast et al. 2011) and the epidermal barrier (Feingold 2009). These properties also enable sphingolipids to influence the activity and spatial organization of membrane proteins (Lingwood and Simons 2010). For example, the increase in sphingolipid content from the ER to the Golgi to the plasma membrane leads to a corresponding increase in membrane thickness, which is proposed to control vesicular trafficking and protein sorting through the secretory pathway (Lippincott-Schwartz and Phair 2010; Sharpe et al. 2010). Additionally, sphingolipids and sterols can self-associate to form dynamic, laterally segregated membrane microdomains or rafts. These microdomains act as dynamic platforms that regulate diverse membrane proteins including signaling receptors (Lingwood and Simons 2010).
A second major mode of sphingolipid function is as signaling molecules that serve as second messengers or as secreted ligands for cell-surface receptors. The primary signaling sphingolipids are long chain bases (LCBs, also known as sphingosines) and ceramides, as well as their phosphorylated derivatives. At steady state, levels of these biosynthetic intermediates are quite low, but they can be rapidly produced during signal transduction via de novo synthesis or breakdown of mature sphingolipids. Identifying the effectors of signaling sphingolipids is an area of ongoing research, with recent studies showing that intracellular targets include histone deacetylases (Hait et al. 2009), Bcl-2 family proteins (Chipuk et al. 2012), the ubiquitin ligase Traf2 (Alvarez et al. 2010), and kinases and phosphatases (for review, see Hannun and Obeid 2008). One of the best-characterized modes of sphingolipid signaling is seen for sphingosine-1-phosphate (S1P), which is an agonist for the G protein-coupled receptors (GPCRs) encoded by S1PR1-5 (for review, see Blaho and Hla 2011). These S1P receptors regulate a host of biological processes including immune cell trafficking, vascular permeability, and cardiac development; consequently, S1PRs are the focus of intensive drug development efforts that have recently yielded the multiple sclerosis drug fingolimod (Brinkmann et al. 2010).
Last, the exposure of diverse sphingolipid headgroups on the plasma membrane enables these lipids to mediate interactions between cells and their external environment. Binding between glycosphingolipids and lectins is important for the association of myelin with axons, the formation of nodes of Ranvier, and neutrophil adhesion to the endothelium (for review, see Lopez and Schnaar 2009; Lingwood 2011). Additionally, pathogens including SV40 virus, HIV, Vibrio cholerae, and Shigella dysenteriae co-opt glycosphingolipids as receptors to gain entry into host cells or to deliver virulence factors (Ewers and Helenius 2011; Lingwood 2011).
Sphingolipid Biosynthesis
Sphingolipids differ from other lipids in their use of the amino acid serine as the backbone to which acyl chains are attached. Their synthesis therefore depends on dedicated enzymes that are essential for viability in organisms ranging from fungi to mammals (for a review, see Gault et al. 2010; Merrill 2011). The production of sphingolipids begins in the ER, where two key precursors, LCBs, and very-long-chain fatty acids (VLCFAs) are produced (Figs. 1D and 2). LCBs are generated by the condensation of serine with a coenzyme A-linked fatty acid (FA-CoA). This reaction is the rate-limiting step in de novo sphingolipid synthesis and is catalyzed by the heterodimeric, pyridoxal phosphate-dependent enzyme serine palmitoyltransferase (SPT). The SPT heterodimer is encoded by the homologous genes LCB1/2 in yeast and by SPTLC1-3 in mammals (Hanada 2003; Lowther et al. 2012), with Lcb2, Sptlc2, and Sptlc3 having the cofactor-binding motifs needed for catalysis. Consistent with the cytoplasmic localization of its substrates, the active site of this transmembrane enzyme is on the cytoplasmic face of the ER. SPT also requires small accessory subunits for maximal activity, although the precise role of these subunits is not yet known. The product of SPT is 3-keto-dihydrosphingosine, an intermediate that is reduced to yield the LCB dihydrosphingosine (Fig. 1D).
Figure 2.
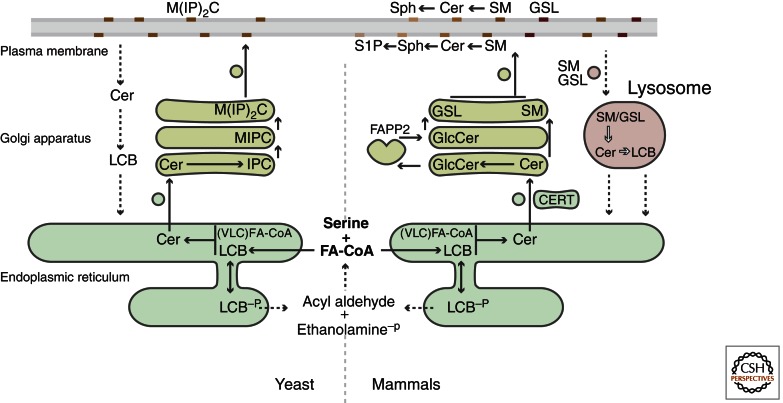
Overview of sphingolipid metabolism. Sphingolipid synthesis begins in the ER with the condensation of serine and coenzyme A-linked fatty acids (FA-CoAs) to form long chain bases (LCBs). LCBs may be phosphorylated to produce LCB-Ps or N-acylated to form ceramides (Cer). Ceramides are then transported to the Golgi by vesicular and nonvesicular means (e.g., the transport protein CERT). In mammalian cells (right), headgroup modifications in the Golgi yield sphingomyelin (SM) and glycosphingolipids (GSL) such as glucosylceramide (GlcCer). In yeast (left), ceramide is progressively modified to form inositolphosphorylceramide (IPC), mannosyl-inositolphosphorylceramide (MIPC), and mannosyl-diinositolphosphorylceramide (M[IP]2C). Sphingolipids then traffic to the plasma membrane, where they are most abundant. Most biosynthetic reactions are reversible, which enables turnover of mature sphingolipids and production of signaling molecules such as sphingosine (Sph) and sphingosine-1-phosphate (S1P). The lysosome is a key site of sphingolipid catabolism, generating breakdown products that can be recycled for sphingolipid synthesis. Alternatively, conversion to LCB-Ps enables terminal degradation of sphingolipids via a lyase enzyme that generates acyl aldehydes and ethanolamine phosphate. Filled arrows indicate biosynthetic steps; dashed arrows indicate degradative and recycling steps.
LCBs are subsequently N-acylated by ER-localized ceramide synthases to yield ceramides (Figs. 1D and 2). Ceramides represent the core structure of most cellular sphingolipids and are thus a key node in sphingolipid metabolism. Notably, the amide-linked acyl chains found in ceramides are often saturated and tend to be longer than those found in other glycerolipids (Lingwood and Simons 2010). These longer fatty acids, known as VLCFAs, are produced by ER-localized elongase complexes that catalyze cycles of two-carbon addition to shorter FA-CoAs produced by fatty acid synthase (Jakobsson et al. 2006). Each elongase isoform generates VLCFAs of a characteristic length that is determined by a molecular caliper mechanism (Denic and Weissman 2007). Similarly, several ceramide synthase isoforms are found in mammals, with each attaching specific acyl chains to LCBs (Mullen et al. 2012). Thus, the elongase enzymes and ceramide synthases provide a major source of structural diversity among sphingolipids. Importantly, these structural differences give rise to distinct functions, as exemplified by chain-length-specific roles for sphingolipids in the response to anoxia in Caenorhabditis elegans (Menuz et al. 2009) and in the barrier function of skin (Feingold 2009).
After ceramides are formed in the ER, they traffic to the Golgi, where they are further elaborated at the headgroup position (Fig. 2). This transport occurs by both vesicular and nonvesicular means and may be coupled to the movement of sphingolipids from the cytosolic leaflet of the membrane to the luminal/extracellular leaflet, where they are most abundant (Blom et al. 2011). Although the mechanisms controlling transbilayer flipping of sphingolipids remain poorly characterized, proteins that mediate sphingolipid transport have recently been identified. Specifically, CERT is a soluble protein that extracts ceramide from the ER and delivers it to the Golgi (Hanada et al. 2003), whereas FAPP2 performs a similar function for Golgi trafficking of glucosylceramide (D’Angelo et al. 2007).
In the Golgi, ceramides undergo species-specific modifications at the headgroup position (Fig. 2). The budding yeast Saccharomyces cerevisiae makes inositol- and mannose-containing glycosphingolipids, whereas mammalian cells produce more complex and diverse glycosphingolipids in addition to sphingomyelin (ceramide bearing a phosphocholine moiety) (Dickson et al. 2006; Gault et al. 2010). Sphingomyelin synthases and a range of glycosylation enzymes carry out these modifications before the ultimate delivery of sphingolipids to the plasma membrane.
Sphingolipid Turnover and Degradation
For virtually all of the biosynthetic steps described above, the reverse reaction is performed by enzymes dedicated to sphingolipid breakdown. For example, glycohydrolases and sphingomyelinases remove headgroup modifications, and ceramidases regenerate LCBs from ceramides (Fig. 2). These reactions take place within the plasma membrane (which can generate signaling sphingolipids) as well as in the lysosome, which is an important site of sphingolipid catabolism (Mao and Obeid 2008; Jenkins et al. 2009; Schulze and Sandhoff 2011). Lysosomal ceramides and LCBs generated from breakdown of mature sphingolipids exit this organelle by poorly characterized means and return to the secretory pathway, where they can be recycled to form new sphingolipids. Not all LCBs are recycled, however, as the ER also provides a pathway for terminal degradation of LCBs.
This ER-localized degradation mechanism arises from a key branchpoint in sphingolipid synthesis: Although most LCBs are acylated to form ceramides, LCBs can also be phosphorylated to generate long-chain base phosphates (LCB-Ps). LCB-Ps such as S1P are potent signaling molecules (Fyrst and Saba 2010; Blaho and Hla 2011), and they are also the sole substrates for a lyase enzyme that cleaves LCB-Ps to generate acyl aldehydes and phosphoethanolamine (Fig. 2). This reaction represents the only mechanism for the irreversible degradation of sphingolipids, and thus lyase activity is likely to be tightly regulated (Fyrst and Saba 2008). The ER enzymes that convert lyase-generated acyl aldehydes back into FA-CoAs for use in sphingolipid or glycerolipid synthesis have recently been identified, thus revealing the final steps of the sphingolipid metabolic pathway (Nakahara et al. 2012).
MECHANISMS OF SPHINGOLIPID HOMEOSTASIS
Given the structural diversity of sphingolipids and the wide range of biological processes in which they participate, it is not surprising that disruptions to sphingolipid homeostasis have deleterious effects. Genetic or pharmacologic inhibition of sphingolipid synthesis blocks cell proliferation in organisms ranging from mammals to fungi (Buede et al. 1991; Miyake et al. 1995; Hanada et al. 1998; Yamashita et al. 1999; Hojjati et al. 2005). Conversely, an excess of sphingolipids is also toxic, leading to ER stress and disruption of calcium homeostasis (Lloyd-Evans et al. 2008; Han et al. 2010). Increased levels of ceramides or LCB-Ps cause lethality in yeast (Schorling et al. 2001; Zhang et al. 2001), and a number of human lysosomal storage diseases, including Tay-Sachs disease, Niemann-Pick disease, and Gaucher disease, are attributable to mutations that block sphingolipid breakdown (Schulze and Sandhoff 2011). Additionally, a heredity sensory neuropathy has recently been shown to be caused by accumulation of nondegradable sphingolipid metabolites (Gable et al. 2010; Penno et al. 2010). Thus, sphingolipid metabolism must be finely balanced to avoid the harmful effects of insufficient or excess sphingolipids.
It has long been appreciated that cells adjust sphingolipid production in response to metabolic need. Indeed it was reported more than 20 years ago that supplementing growth media with sphingosine leads to reduced de novo synthesis of LCBs in cultured cerebellar neurons (van Echten et al. 1990). However, only recently have the mechanisms that ensure sphingolipid homeostasis begun to be identified. Here a sharp contrast can be seen with the many regulatory pathways identified for sterols and glycerolipids (Nohturfft and Zhang 2009; Ye and DeBose-Boyd 2011). For instance, elegant studies have shown that cholesterol metabolism is controlled by multiple feedback mechanisms, including transcriptional regulation by SREBP, degradation of HMG-CoA reductase, and turnover of the LDL receptor responsible for cholesterol uptake (Chang et al. 2006; Brown and Goldstein 2009). Indeed, virtually all steps in glycerolipid and sterol metabolism are tightly regulated, and it would not be surprising if a comparable array of feedback pathways is ultimately found for sphingolipid homeostasis.
Below, I outline several regulatory pathways that enable cells to sense sphingolipid levels and tune their synthesis, degradation, and transport accordingly. Although the mechanisms that regulate sphingolipid headgroup modifications are the subject of ongoing investigation, I will focus here on the mechanisms that regulate levels of the core ceramide backbone and of other conserved metabolites. Additionally, the mechanisms used to acutely generate signaling sphingolipids are likely to be quite different from those that control steady-state levels (for reviews of sphingolipid signaling, see Hannun and Obeid 2008; Fyrst and Saba 2010; Blaho and Hla 2011).
Regulation of Sphingolipid Synthesis
The SPT-mediated condensation of serine with FA-CoAs to generate LCBs is the first and rate-limiting step in sphingolipid synthesis and is therefore a key point of regulation. In fact, one of the first studies of sphingolipid homeostasis showed that providing exogenous LCBs to cultured cells leads to a compensatory reduction in SPT activity (van Echten et al. 1990; Mandon et al. 1991). More recently, insights into SPT regulation have emerged from characterization of the biochemical properties of this enzyme. Specifically, it was found that SPT’s affinity for serine is similar to the intracellular concentration of this amino acid, suggesting that changes in serine levels may modulate LCB production. Indeed, in yeast, increases in serine during the heat shock response or when serine degradation is disrupted lead to elevated sphingolipid production (Cowart and Hannun 2007). Interestingly, sphingolipids may in turn down-regulate serine levels via induction of the serine catabolic enzyme Cha1, thus forming a potential feedback mechanism in which sphingolipids limit serine availability (Montefusco et al. 2012). These studies highlight substrate supply as a regulatory mechanism, and it will be important to determine whether sphingolipid metabolism is also regulated by the levels of FA-CoAs or by the extent to which these precursors are shunted to SPT versus other lipid metabolic processes.
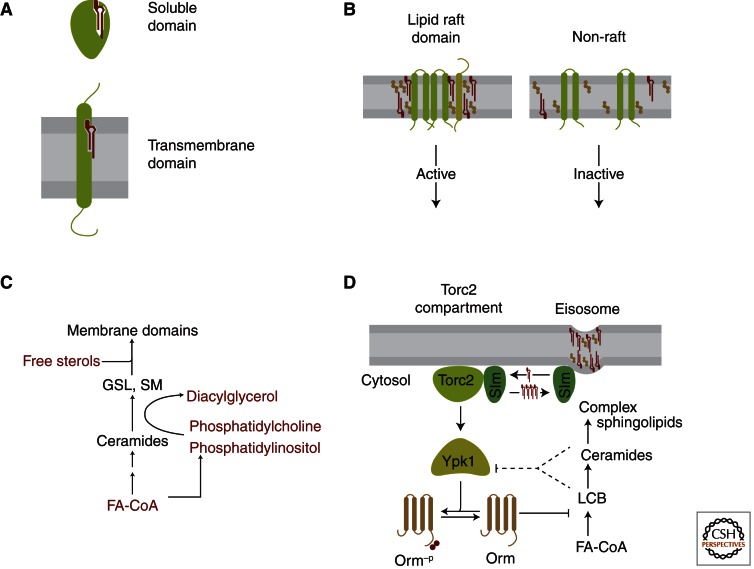
A second major mode of SPT regulation is mediated by Orm family proteins (Fig. 3). The ORM gene family encodes ER-resident membrane proteins and is conserved from yeast to human (Hjelmqvist et al. 2002), with the human ORMDL3 gene the subject of particular interest for its potential role in asthma development (Moffatt et al. 2007). A link between ORM genes and sphingolipid metabolism was first established for yeast ORM1 and ORM2 by systematic measurement of genetic interactions (Breslow et al. 2010). These functional genomic data suggested that Orm1/2 negatively regulate sphingolipid production, and consistent with this prediction, deletion of ORM1/2 leads to toxic accumulation of sphingolipids, whereas ORM1/2 overexpression reduces sphingolipid levels (Breslow et al. 2010; Han et al. 2010). Orm proteins regulate sphingolipid metabolism by forming a multiprotein complex with SPT termed the SPOTS complex (named for its components serine palmitoyltransferase, Orm1/2, the SPT accessory subunit Tsc3, and the phosphoinositide phosphatase Sac1) (Breslow et al. 2010). Thus, it is likely that Orm proteins directly inhibit SPT activity, although the precise mechanism for this inhibition remains unknown. The roughly stoichiometric amount of Sac1 in the SPOTS complex is also noteworthy and suggests an exciting new link between phosphoinositide and sphingolipid metabolism (see below and Breslow and Weissman 2010).
Figure 3.
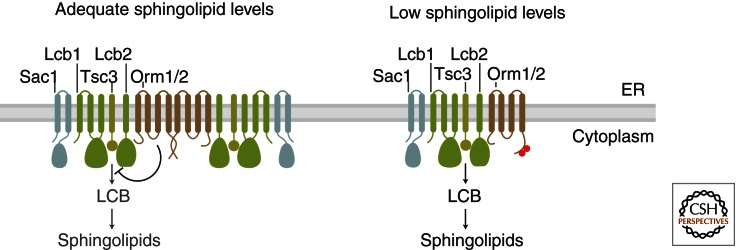
Sphingolipid homeostasis by Orm family proteins. The SPOTS complex contains serine palmitoyltransferase (formed in yeast by Lcb1, Lcb2, and the Tsc3 accessory subunit), Orm proteins, and the phosphoinositide phosphatase Sac1. Serine palmitoyltransferase carries out the first and rate-limiting step in sphingolipid synthesis: condensation of coenzyme A-linked fatty acids (FA-CoAs) with serine to yield long chain bases (LCBs). When the supply of sphingolipids is adequate (left), SPT activity is inhibited by Orm proteins, and the SPOTS complex shows an increased degree of dimerization/oligomerization. When sphingolipid levels become too low (right), Orm proteins are inactivated by phosphorylation, relieving their inhibition of SPT and enabling a compensatory increase in de novo sphingolipid synthesis. Phosphorylation also triggers a shift to monomeric SPOTS complex organization. Note that the membrane topology of SPT and Orm proteins is not definitely known; one potential topology is shown for simplicity.
A central feature of Orm proteins is that their SPT-inhibitory activity is subject to feedback regulation by sphingolipid levels (Breslow et al. 2010). Specifically, disruption of sphingolipid synthesis leads to inactivation of Orm1/2 via phosphorylation of their amino termini, thus enabling a compensatory increase in SPT activity (Fig. 3). This phosphorylation occurs in a graded fashion that may finely tune Orm activity in response to sphingolipid levels. Similarly, Orm1 activity is adjusted in response to manipulation of Orm2 expression levels, with increased Orm2 expression causing a corresponding increase in Orm1 phosphorylation and vice versa (Liu et al. 2012; Sun et al. 2012). The mechanism by which phosphorylation blocks Orm1/2-mediated inhibition of SPT requires further investigation, but one important feature is that phosphorylation triggers a change in the higher order assembly of the SPOTS complex from a more oligomeric state to a more monomeric state (Fig. 3) (Breslow et al. 2010).
The important role of Orm proteins in sphingolipid homeostasis is illustrated by mutations that impair their phosphoregulation. Specifically, mutations that block Orm protein phosphorylation reduce steady-state sphingolipid levels and prevent up-regulation of SPT activity when sphingolipid synthesis is disrupted. These mutations therefore cause a pronounced hypersensitivity to inhibitors of sphingolipid synthesis (Breslow et al. 2010); conversely, phosphomimetic mutations confer resistance to these drugs (Sun et al. 2012). The physiologic importance of Orm proteins in sphingolipid metabolism is further underscored by the finding that the acute increase in sphingolipid synthesis in response to heat stress is also mediated by Orm1/2 phosphorylation (Sun et al. 2012). Lastly, several groups have recently examined how changes in yeast sphingolipid levels are translated into changes in Orm activity (see section below on mechanisms for sensing sphingolipids).
In human cells, Ormdl proteins also negatively regulate sphingolipid levels and are found in a homologous SPOTS complex, indicating that their basic function is conserved (Breslow et al. 2010). However, the physiologic cues and regulatory mechanisms that control metazoan Orm protein activity are not yet known and may differ from what has been found in yeast. Notably, the phosphorylation sites found in yeast Orm1/2 are not conserved in higher organisms. There is, however, recent evidence for transcriptional regulation of mouse Ormdl genes. Specifically, up-regulation of Ormdl expression may provide a means to reduce SPT activity when sphingolipid degradation is comprised by deletion of the lyase enzyme (Hagen-Euteneuer et al. 2012). A detailed characterization of mammalian Orm protein regulation will be a key goal for future studies.
A second step in sphingolipid metabolism that is subject to tight regulation is the production of ceramides by ceramide synthase. In yeast, the Torc2 kinase complex is a positive regulator of ceramide synthase activity and therefore controls steady-state levels of LCBs and ceramides (Aronova et al. 2008). Torc2 activity is in turn induced by sphingolipid depletion (Berchtold et al. 2012), suggesting a potential feedback mechanism controlling ceramide production. It is not yet known how Torc2 regulates ceramide synthase. Interestingly, a recent phosphoproteomic study (Holt et al. 2009) identified phosphorylation sites in the yeast ceramide synthase Lag1 that share a similar sequence motif to the Orm1/2 sites, raising the possibility that these proteins are regulated by the same kinase pathway.
In mammalian cells, regulation of ceramide synthase activity is likely to involve a complex balance between the six enzyme isoforms (Mullen et al. 2011). In addition to transcriptional regulation, ceramide synthase activity was recently found to be modulated by reversible dimerization (Laviad et al. 2012). Different ceramide synthase isoforms physically interact, and the coexpression of a second ceramide synthase isoform can modulate the activity of another isoform. Furthermore, pharmacologic stimulation of ceramide synthesis leads to an increase in ceramide synthase dimerization, indicating that this process is dynamically regulated (Laviad et al. 2012). Additionally, the ceramide synthase isoform CerS2 has an S1P-binding motif, and S1P inhibits CerS2 activity in vitro (Laviad et al. 2008). In the future, it will be interesting to examine the in vivo significance of this regulation and to further characterize the physiologic signals that control ceramide synthase dimerization.
An additional regulator of sphingolipid synthesis was first identified on the basis of its homology with the sphingomyelin synthase enzyme. This ER-localized protein, SMSr is homologous to SM synthase but lacks SM synthase activity, having instead a weak ability to generate ceramide-phosphoethanolamine (CPE). Surprisingly, knockdown of SMSr causes a dramatic increase in ceramide levels that cannot be explained by a block in CPE production (Vacaru et al. 2009). Rather, SMSr may act as a sensor that detects ER ceramides and down-regulates ceramide production. How SMSr senses ceramide levels and inhibits ceramide production will be exciting topics for future study.
The studies described above have largely focused on the mechanisms that regulate individual enzymes, leaving open the possibility that the degree of coupling between different steps in sphingolipid synthesis serves as a point of regulation. For example, are the LCB products of SPT simply released into the ER membrane or could they be handed off to downstream enzymes to promote efficient ceramide production? If so, regulation of this “hand off” could provide a means to control sphingolipid production or direct excess sphingolipids for degradation. The possibility of coupling between enzymes in sphingolipid metabolism is suggested by work in yeast on the mechanism by which exogenous LCBs are used for ceramide synthesis. Specifically, it was shown that, to be efficiently incorporated into ceramides, LCBs must first be phosphorylated to form LCB-Ps and then dephosphorylated by the LCB phosphatase (Funato et al. 2003). This apparently futile cycle is required even when LCBs are added to microsomal membranes used in an in vitro assay. The mechanistic basis for this phenomenon remains unknown, but an appealing possibility is that LCBs are normally presented to ceramide synthase by SPT or the LCB phosphatase, and that exogenous LCBs are comparatively poor substrates. The spatial organization of different sphingolipid biosynthetic steps within the ER (and Golgi) remains poorly understood, but coordination between enzymes might provide a powerful means to regulate sphingolipid metabolism.
Regulation of Sphingolipid Transport and Turnover
The transport of sphingolipids is critical for their biosynthesis and also appears to be tightly regulated. In the case of CERT, the Hanada group has shown that phosphorylation inhibits CERT-dependent ceramide transport and that this phosphorylation is regulated by sphingolipid levels (Kumagai et al. 2007). Specifically, pharmacologic inhibition of SPT or enzymatic degradation of plasma membrane sphingomyelin induces CERT dephosphorylation and activation, which may enable a compensatory increase in sphingolipid production (Kumagai et al. 2007). An additional mechanism regulating sphingolipid transport is suggested from the fact that both CERT and FAPP2 contain pleckstrin homology (PH) domains that bind to phosphatidylinositol-4-phosphate (PI4P), a lipid that serves as a key Golgi marker (Di Paolo and De Camilli 2006). Thus, multiple pathways that regulate PI4P levels are also likely to impact sphingolipid transport. For example, nutrient or growth factor starvation induces relocalization of the PI4P phosphatase Sac1 to the Golgi, reducing Golgi PI4P levels and down-regulating secretion (Piao and Mayinger 2012). The dependence of both protein and lipid trafficking on PI4P may therefore provide a means to coordinately regulate their biogenesis. Lastly, because Sac1 is also part of the SPOTS complex, there may be as-yet-uncharacterized mechanisms by which PI4P links LCB production, sphingolipid transport, and protein trafficking (Breslow et al. 2010).
The turnover and degradation of sphingolipids is likely to play a key role in sphingolipid homeostasis, as evidenced by the accumulation of sphingolipids that occurs in lysosomal storage diseases or in mice lacking the lyase enzyme (Bektas et al. 2010; Schulze and Sandhoff 2011). Additionally, a hereditary sensory neuropathy was recently shown to be owing to mutations in SPT that cause the aberrant production of sphingolipids that cannot be degraded by the lyase. Specifically, these mutations allow SPT to use alanine or glycine in place of serine, yielding LCBs that lack the hydroxyl group necessary for lyase-mediated degradation; the subsequent accumulation of these sphingolipids leads to neurotoxicity (Gable et al. 2010; Penno et al. 2010). Degradation pathways may be especially important for sphingolipid homeostasis because, unlike glycerolipids and sterols, excess sphingolipids are not stored in lipid droplets (Walther and Farese 2012).
Notable progress has been made in characterizing several sphingolipid catabolic enzymes (for a review, see Mao and Obeid 2008; Jenkins et al. 2009). However, these studies have primarily focused on the roles of these enzymes in acute signaling responses rather than on their regulation in the context of sphingolipid homeostasis. Additionally, further studies are needed to understand how the lyase enzyme is regulated and how sphingolipid breakdown products are selected for recycling into new sphingolipids versus lyase-mediated degradation (for reviews of lyase function, see Fyrst and Saba 2008; Aguilar and Saba 2011). In yeast, there is evidence that the flux of LCBs to the lyase versus ceramide synthase is controlled by the activity of the LCB-P phosphatase Lcb3. Consistent with the fact that LCB-Ps are the sole substrates for lyase activity, LCB3 deletion increases LCB-P levels, promoting lyase-mediated degradation and inhibiting LCB incorporation into ceramides; conversely, overexpression of LCB3 has the opposite effect (Mao et al. 1997). It is not yet known if sphingolipid levels exert feedback control on Lcb3 or lyase activity; it is noteworthy, however, that Lcb3 may also be phosphorylated on a motif resembling that seen in Orm1/2 (Holt et al. 2009).
MECHANISMS FOR SENSING SPHINGOLIPIDS
How do cells sense sphingolipid levels to modulate sphingolipid metabolism accordingly? Several potential mechanisms for sensing sphingolipids can be envisioned (Fig. 4A–C), and there is evidence that a number of these strategies are used in vivo. First, cells could regulate sphingolipid metabolism by directly sensing a specific metabolite (Fig. 4A). Akin to the sterol-sensing domains found in regulators of sterol metabolism, a number of proteins possess domains that specifically recognize sphingolipids. These proteins include soluble transporters (e.g., CERT), the S1P GPCRs, and p24, a membrane protein that participates in COPI vesicle formation (Contreras et al. 2012). Recent crystallographic studies have illuminated the molecular interactions that enable specific recognition of sphingolipids, revealing hydrophobic binding pockets that can discriminate features such as headgroup structure and acyl chain length (Malinina et al. 2004, 2006; Kudo et al. 2008; Contreras et al. 2012; Hanson et al. 2012). Although the above proteins are not known to be homeostatic sensors, such sensors may use similar modes of molecular recognition.
Figure 4.
Mechanisms for sensing sphingolipids. (A) Sphingolipid sensing by direct recognition of a specific sphingolipid metabolite by a soluble or transmembrane sensor protein. (B) Sphingolipid sensing by detection of changes in membrane physical properties such as propensity to form sphingolipid-dependent membrane domains. (C) Sensing of non-sphingolipid metabolites that are functionally or metabolically coupled to sphingolipid biosynthesis. (D) Multiple sphingolipid-sensing mechanisms may control Orm-mediated sphingolipid homeostasis. Orm proteins are phosphorylated by Ypk1, and Ypk1 is activated by sphingolipid depletion via the Torc2 kinase complex. Torc2-mediated activation of Ypk1 is in turn regulated by Slm proteins. Slm proteins may be sphingolipid sensors, as their movement between two membrane domains—sphingolipid-containing eisosomes and a compartment containing Torc2—is regulated by sphingolipid levels. Disruption of sphingolipid synthesis causes relocalization of Slm proteins from eisosomes to Torc2 domains, leading to Ypk1 activation. In addition to Slm-mediated sensing of complex sphingolipids in the plasma membrane, upstream metabolites such as LCBs or ceramide may also regulate Orm1/2 phosphorylation (dotted lines).
A second means to sense sphingolipids is by detecting sphingolipid-dependent changes in the physical properties of membranes (Fig. 4B). This mechanism may be used by the yeast proteins Nce102, Slm1, and Slm2. These proteins dynamically localize to sphingolipid-dependent membrane domains, and their activity is likewise controlled by membrane domain association (Frohlich et al. 2009; Berchtold et al. 2012). For example, the transmembrane protein Nce102 localizes to sphingolipid-dependent domains known as eisosomes, where it inhibits the Pkh1 and Pkh2 kinases (Frohlich et al. 2009). Inhibition of sphingolipid synthesis causes Nce102 to rapidly exit eisosomes, thus relieving its inhibition of Pkh1/2 and providing a means to couple sphingolipid levels to kinase signaling. In the future it will be interesting to determine the molecular basis for the dynamic association of sensors such as Nce102 with sphingolipid-dependent domains.
Last, because sphingolipids are linked metabolically and functionally to other lipid types, changes in sphingolipid levels may be sensed indirectly via their impact on sterols and glycerolipids (Fig. 4C) (Breslow and Weissman 2010). Specifically, many sphingolipids physically associate with sterols, and thus changes in sphingolipid levels alter the amount and localization of free sterols (Gatt and Bierman 1980; Slotte and Bierman 1988). For example, a reduction in plasma membrane sphingolipids leads to flux of sterols from the plasma membrane to the ER, where they are detected by sterol sensors such as SREBP and OSBP (Chang et al. 2006; Brown and Goldstein 2009). These proteins initiate a variety of downstream responses, including OSBP-dependent activation of CERT (Ridgway et al. 1998; Perry and Ridgway 2006).
In contrast to the indirect effects of sphingolipids on sterols, sphingolipids have direct metabolic links to glycerolipids (Fig. 4C). Sphingolipids and glycerolipids are produced from the same pool of fatty acids. Additionally, they share headgroup modifications, with glycerolipids often providing the headgroup moieties for sphingolipids. For example, the production of inositolphosphorylceramide (IPC) in yeast relies on inositol derived from phosphatidylinositol (PI), with the latter being converted to diacylglycerol (DAG) (Brice et al. 2009). In mammalian cells, SM production is similarly coupled to conversion of phosphatidylcholine (PC) to DAG. Thus, glycerolipid sensors such as the DAG-binding domain in protein kinase C could provide a mechanism to monitor the rate of sphingolipid biosynthesis (Cerbon and del Carmen Lopez-Sanchez 2003; Cerbon et al. 2005).
With a range of potential sensing mechanisms, what is known about the actual strategies used to couple sphingolipid levels to sphingolipid synthesis? Here, the identification of a kinase pathway controlling phosphorylation of Orm proteins provides important insights (Fig. 4D). Specifically, Ypk1 was recently found to be the kinase that phosphorylates yeast Orm1/2 (Roelants et al. 2011). Furthermore, the increased phosphorylation of Orm1/2 caused by inhibition of sphingolipid synthesis is paralleled by a corresponding increase in Ypk1 activity (Roelants et al. 2011). This stimulation of Ypk1 activity is in turn the result of Torc2-mediated phosphorylation of Ypk1 at Thr662 (Roelants et al. 2011; Niles et al. 2012; Sun et al. 2012).
How then do sphingolipid levels control Torc2’s activation of Ypk1, and what sphingolipid species are sensed to regulate this key event? Here a prominent role for Slm proteins is suggested by reports that the PH-domain-containing proteins Slm1 and Slm2 are necessary and sufficient for recruitment of Ypk1 to Torc2 and for Ypk1 activation (Fig. 4D) (Berchtold et al. 2012; Niles et al. 2012). Furthermore, inhibition of sphingolipid synthesis causes a rapid relocalization of Slm proteins from eisosomes to membrane domains containing Torc2, and this relocalization is required for phosphorylation of Ypk1 at Thr662 (Berchtold et al. 2012). Thus, the dynamic localization of Slm1/2 links sphingolipid levels to Torc2-mediated activation of Ypk1.
Interestingly, redistribution of Slm proteins from eisosomes to Torc2-containing domains is triggered not only by pharmacologic inhibition of SPT, but also by inhibition of IPC synthase via the drug Aureobasidin A (AbA). Similarly, AbA also induces Ypk1 phosphorylation, suggesting that a complex sphingolipid (e.g., IPC or a downstream product) is sensed at the plasma membrane to control SPT activity (Berchtold et al. 2012). Furthermore, Slm proteins are also responsive to plasma membrane stretch, indicating that they may be more general sensors of plasma membrane stress (Berchtold et al. 2012). The molecular basis for dynamic changes in Slm protein localization promises to be an exciting topic for future studies.
Given the central role of SPT in sphingolipid homeostasis, Orm1/2 phosphorylation may also be regulated by sphingolipid metabolites upstream of plasma membrane IPC. Consistent with this possibility, an initial report found that SPT inhibition causes pronounced Orm phosphorylation, whereas repression of IPC synthase did not, suggesting that a metabolite upstream of IPC is sensed (e.g., LCBs or unmodified ceramides) (Breslow et al. 2010). A similarly mild degree of Orm protein phosphorylation is also seen in AbA-treated cells (Berchtold et al. 2012). Finally, it was recently proposed that LCBs directly regulate Orm phosphorylation, as exogenous LCBs induce dephosphorylation of Orm1/2 within 2 min, even in strains with reduced capacity to use LCBs to make ceramides (Sun et al. 2012). Thus, multiple metabolites throughout the sphingolipid pathway may be independently sensed to control sphingolipid synthesis (Fig. 4D).
CONCLUDING REMARKS
Although there has been notable recent progress in understanding sphingolipid homeostasis, there are many areas in which further studies are needed. First, it is likely that many of the mechanisms for sensing sphingolipids and regulating sphingolipid metabolism have yet to be identified. Similarly, how different regulatory pathways interact to provide an integrated homeostatic response remains poorly understood. Encouragingly, future studies that seek to answer these questions are likely to be aided by a number of new technologies. Mass spectrometry-based methods for sphingolipid analysis have emerged as powerful tools for quantitative measurement of sphingolipid levels (Ejsing et al. 2009; Shevchenko and Simons 2010; Wenk 2010) and for identification of protein–sphingolipid interactions (Alvarez et al. 2010; Gallego et al. 2010; Li et al. 2010). In particular, lipidomic analyses can provide a global view of how sphingolipid metabolism adapts to different perturbations or external cues (Guan et al. 2009; Breslow et al. 2010; Sampaio et al. 2011; Klose et al. 2012).
In parallel with new techniques to measure sphingolipids, increasingly sophisticated approaches to functionally interrogate sphingolipid metabolism are now emerging. Here, the continued application of functional genomics (see Schuldiner and Weissman 2013) and transgenic mouse models are likely to have a key role. Inspiration may also be taken from the rich set of tools available for manipulating and monitoring phosphoinositide lipids (Várnai and Balla 2008; Chang-Ileto et al. 2012). In particular, sphingolipid sensors based on fluorescently tagged lipid-binding domains could serve as dynamic reporters of sphingolipid levels at different cellular sites. Such reagents would be especially powerful in combination with inducible enzymatic constructs that can be used to locally produce or degrade specific sphingolipid metabolites. Lastly, advances in human genetics may provide new insights into factors that regulate sphingolipid levels in health and disease (Hicks et al. 2009; Demirkan et al. 2012).
As an increasingly detailed picture emerges of the mechanisms that match sphingolipid supply to metabolic demand, a critical next step will be to understand the interrelationship between sphingolipid homeostasis and other cellular processes. Given the broad functions of sphingolipids, it is clear that many other cellular activities depend on sphingolipids; likewise, many other metabolic and secretory pathway processes influence and regulate sphingolipid metabolism. For instance, disruption of sphingolipid metabolism induces the unfolded protein response (UPR), whereas sphingolipid genes such as ORM2 are themselves UPR targets (Travers et al. 2000; Spassieva et al. 2009; Han et al. 2010; Liu et al. 2012). Similarly, heat shock induces LCB and ceramide production, and sphingolipids in turn promote thermotolerance (for a review, see Dickson et al. 2006). This interdependence illustrates that homeostasis at a cellular level arises from the coordination of sphingolipid metabolism with a range of pathways that control cell growth, stress responses, and differentiation. Thus, an integrated view of sphingolipid homeostasis promises to provide key insights into membrane and lipid function in cell biology and disease.
ACKNOWLEDGMENTS
I thank J. Weissman for helpful discussions and B. Toyama for excellent graphical assistance.
Footnotes
Editors: Susan Ferro-Novick, Tom A. Rapoport, and Randy Schekman
Additional Perspectives on The Endoplasmic Reticulum available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Aguilar A, Saba JD 2011. Truth and consequences of sphingosine-1-phosphate lyase. Adv Enzyme Regul 10.1016/j.advenzreg.2011.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez SE, Harikumar KB, Hait NC, Allegood J, Strub GM, Kim EY, Maceyka M, Jiang H, Luo C, Kordula T, et al. 2010. Sphingosine-1-phosphate is a missing cofactor for the E3 ubiquitin ligase TRAF2. Nature 465: 1084–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronova S, Wedaman K, Aronov P, Fontes K, Ramos K, Hammock B, Powers T 2008. Regulation of ceramide biosynthesis by TOR complex 2. Cell Metab 7: 148–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bektas M, Allende ML, Lee BG, Chen W, Amar MJ, Remaley AT, Saba JD, Proia RL 2010. Sphingosine 1-phosphate lyase deficiency disrupts lipid homeostasis in liver. J Biol Chem 285: 10880–10889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchtold D, Piccolis M, Chiaruttini N, Riezman I, Riezman H, Roux A, Walther TC, Loewith R 2012. Plasma membrane stress induces relocalization of Slm proteins and activation of TORC2 to promote sphingolipid synthesis. Nat Cell Biol 14: 542–547 [DOI] [PubMed] [Google Scholar]
- Blaho VA, Hla T 2011. Regulation of mammalian physiology, development, and disease by the sphingosine 1-phosphate and lysophosphatidic acid receptors. Chem Rev 111: 6299–6320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom T, Somerharju P, Ikonen E 2011. Synthesis and biosynthetic trafficking of membrane lipids. In The biology of lipids: Trafficking, regulation, and function (ed. Simons K), pp. 239–254 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow DK, Weissman JS 2010. Membranes in balance: Mechanisms of sphingolipid homeostasis. Mol Cell 40: 267–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow DK, Collins SR, Bodenmiller B, Aebersold R, Simons K, Shevchenko A, Ejsing CS, Weissman JS 2010. Orm family proteins mediate sphingolipid homeostasis. Nature 463: 1048–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brice S, Alford C, Cowart L 2009. Modulation of sphingolipid metabolism by the phosphatidylinositol-4-phosphate phosphatase Sac1p through regulation of phosphatidylinositol in Saccharomyces cerevisiae. J Biol Chem 284: 7588–7596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann V, Billich A, Baumruker T, Heining P, Schmouder R, Francis G, Aradhye S, Burtin P 2010. Fingolimod (FTY720): Discovery and development of an oral drug to treat multiple sclerosis. Nat Rev Drug Discov 9: 883–897 [DOI] [PubMed] [Google Scholar]
- Brown M, Goldstein J 2009. Cholesterol feedback: From Schoenheimer’s bottle to Scap’s MELADL. J Lipid Res 50: S15–SS27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buede R, Rinker-Schaffer C, Pinto W, Lester R, Dickson R 1991. Cloning and characterization of LCB1, a Saccharomyces gene required for biosynthesis of the long-chain base component of sphingolipids. J Bacteriol 173: 4325–4332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerbon J, del Carmen Lopez-Sanchez R 2003. Diacylglycerol generated during sphingomyelin synthesis is involved in protein kinase C activation and cell proliferation in Madin-Darby canine kidney cells. Biochem J 373: 917–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerbon J, Falcon A, Hernandez-Luna C, Segura-Cobos D 2005. Inositol phosphoceramide synthase is a regulator of intracellular levels of diacylglycerol and ceramide during the G1 to S transition in Saccharomyces cerevisiae. Biochem J 388: 169–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TY, Chang CC, Ohgami N, Yamauchi Y 2006. Cholesterol sensing, trafficking, and esterification. Annu Rev Cell Dev Biol 22: 129–157 [DOI] [PubMed] [Google Scholar]
- Chang-Ileto B, Frere SG, Di Paolo G 2012. Acute manipulation of phosphoinositide levels in cells. Methods Cell Biol 108: 187–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipuk JE, McStay GP, Bharti A, Kuwana T, Clarke CJ, Siskind LJ, Obeid LM, Green DR 2012. Sphingolipid metabolism cooperates with BAK and BAX to promote the mitochondrial pathway of apoptosis. Cell 148: 988–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrast R, Saher G, Nave KA, Verheijen MH 2011. Lipid metabolism in myelinating glial cells: Lessons from human inherited disorders and mouse models. J Lipid Res 52: 419–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras FX, Ernst AM, Haberkant P, Bjorkholm P, Lindahl E, Gonen B, Tischer C, Elofsson A, von Heijne G, Thiele C, et al. 2012. Molecular recognition of a single sphingolipid species by a protein’s transmembrane domain. Nature 481: 525–529 [DOI] [PubMed] [Google Scholar]
- Cowart L, Hannun Y 2007. Selective substrate supply in the regulation of yeast de novo sphingolipid synthesis. J Biol Chem 282: 12330–12340 [DOI] [PubMed] [Google Scholar]
- D’Angelo G, Polishchuk E, Di Tullio G, Santoro M, Di Campli A, Godi A, West G, Bielawski J, Chuang C, van der Spoel A, et al. 2007. Glycosphingolipid synthesis requires FAPP2 transfer of glucosylceramide. Nature 449: 62–67 [DOI] [PubMed] [Google Scholar]
- Demirkan A, van Duijn CM, Ugocsai P, Isaacs A, Pramstaller PP, Liebisch G, Wilson JF, Johansson A, Rudan I, Aulchenko YS, et al. 2012. Genome-wide association study identifies novel loci associated with circulating phospho- and sphingolipid concentrations. PLoS Genet 8: e1002490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denic V, Weissman J 2007. A molecular caliper mechanism for determining very long-chain fatty acid length. Cell 130: 663–677 [DOI] [PubMed] [Google Scholar]
- Dickson R, Sumanasekera C, Lester R 2006. Functions and metabolism of sphingolipids in Saccharomyces cerevisiae. Prog Lipid Res 45: 447–465 [DOI] [PubMed] [Google Scholar]
- Di Paolo G, De Camilli P 2006. Phosphoinositides in cell regulation and membrane dynamics. Nature 443: 651–657 [DOI] [PubMed] [Google Scholar]
- Ejsing C, Sampaio J, Surendranath V, Duchoslav E, Ekroos K, Klemm R, Simons K, Shevchenko A 2009. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc Natl Acad Sci 106: 2136–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewers H, Helenius A 2011. Lipid-mediated endocytosis. In The biology of lipids: Trafficking, regulation, and function (ed. Simons K), pp. 255–268 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feingold KR 2009. The outer frontier: The importance of lipid metabolism in the skin. J Lipid Res 50: S417–S422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohlich F, Moreira K, Aguilar P, Hubner N, Mann M, Walter P, Walther T 2009. A genome-wide screen for genes affecting eisosomes reveals Nce102 function in sphingolipid signaling. J Cell Biol 185: 1227–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funato K, Lombardi R, Vallee B, Riezman H 2003. Lcb4p is a key regulator of ceramide synthesis from exogenous long chain sphingoid base in Saccharomyces cerevisiae. J Biol Chem 278: 7325–7334 [DOI] [PubMed] [Google Scholar]
- Fyrst H, Saba JD 2008. Sphingosine-1-phosphate lyase in development and disease: Sphingolipid metabolism takes flight. Biochim Biophys Acta 1781: 448–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyrst H, Saba JD 2010. An update on sphingosine-1-phosphate and other sphingolipid mediators. Nat Chem Biol 6: 489–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gable K, Gupta SD, Han G, Niranjanakumari S, Harmon JM, Dunn TM 2010. A disease-causing mutation in the active site of serine palmitoyltransferase causes catalytic promiscuity. J Biol Chem 285: 22846–22852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego O, Betts MJ, Gvozdenovic-Jeremic J, Maeda K, Matetzki C, Aguilar-Gurrieri C, Beltran-Alvarez P, Bonn S, Fernandez-Tornero C, Jensen LJ, et al. 2010. A systematic screen for protein-lipid interactions in Saccharomyces cerevisiae. Mol Syst Biol 6: 430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatt S, Bierman EL 1980. Sphingomyelin suppresses the binding and utilization of low density lipoproteins by skin fibroblasts. J Biol Chem 255: 3371–3376 [PubMed] [Google Scholar]
- Gault CR, Obeid LM, Hannun YA 2010. An overview of sphingolipid metabolism: From synthesis to breakdown. In Sphingolipids as signaling and regulatory molecules (ed. Chalfant C, Del Poeta M), pp. 1–23 Landes Bioscience, Austin, TX: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan XL, Souza CM, Pichler H, Dewhurst G, Schaad O, Kajiwara K, Wakabayashi H, Ivanova T, Castillon GA, Piccolis M, et al. 2009. Functional interactions between sphingolipids and sterols in biological membranes regulating cell physiology. Mol Biol Cell 20: 2083–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen-Euteneuer N, Lutjohann D, Park H, Merrill AH Jr, van Echten-Deckert G 2012. Sphingosine 1-phosphate (S1P) lyase deficiency increases sphingolipid formation via recycling at the expense of de novo biosynthesis in neurons. J Biol Chem 287: 9128–9136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hait NC, Allegood J, Maceyka M, Strub GM, Harikumar KB, Singh SK, Luo C, Marmorstein R, Kordula T, Milstien S, et al. 2009. Regulation of histone acetylation in the nucleus by sphingosine-1-phosphate. Science 325: 1254–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Lone MA, Schneiter R, Chang A 2010. Orm1 and Orm2 are conserved endoplasmic reticulum membrane proteins regulating lipid homeostasis and protein quality control. Proc Natl Acad Sci 107: 5851–5856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K 2003. Serine palmitoyltransferase, a key enzyme of sphingolipid metabolism. Biochim Biophys Acta 1632: 16–30 [DOI] [PubMed] [Google Scholar]
- Hanada K, Hara T, Fukasawa M, Yamaji A, Umeda M, Nishijima M 1998. Mammalian cell mutants resistant to a sphingomyelin-directed cytolysin. Genetic and biochemical evidence for complex formation of the LCB1 protein with the LCB2 protein for serine palmitoyltransferase. J Biol Chem 273: 33787–33794 [DOI] [PubMed] [Google Scholar]
- Hanada K, Kumagai K, Yasuda S, Miura Y, Kawano M, Fukasawa M, Nishijima M 2003. Molecular machinery for non-vesicular trafficking of ceramide. Nature 426: 803–809 [DOI] [PubMed] [Google Scholar]
- Hannun Y, Obeid L 2008. Principles of bioactive lipid signalling: Lessons from sphingolipids. Nat Rev Mol Cell Biol 9: 139–150 [DOI] [PubMed] [Google Scholar]
- Hanson MA, Roth CB, Jo E, Griffith MT, Scott FL, Reinhart G, Desale H, Clemons B, Cahalan SM, Schuerer SC, et al. 2012. Crystal structure of a lipid G protein-coupled receptor. Science 335: 851–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks AA, Pramstaller PP, Johansson A, Vitart V, Rudan I, Ugocsai P, Aulchenko Y, Franklin CS, Liebisch G, Erdmann J, et al. 2009. Genetic determinants of circulating sphingolipid concentrations in European populations. PLoS Genet 5: e1000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjelmqvist L, Tuson M, Marfany G, Herrero E, Balcells S, Gonzalez-Duarte R 2002. ORMDL proteins are a conserved new family of endoplasmic reticulum membrane proteins. Genome Biol 3: RESEARCH0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hojjati MR, Li Z, Jiang XC 2005. Serine palmitoyl-CoA transferase (SPT) deficiency and sphingolipid levels in mice. Biochim Biophys Acta 1737: 44–51 [DOI] [PubMed] [Google Scholar]
- Holt LJ, Tuch BB, Villen J, Johnson AD, Gygi SP, Morgan DO 2009. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science 325: 1682–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson A, Westerberg R, Jacobsson A 2006. Fatty acid elongases in mammals: Their regulation and roles in metabolism. Prog Lipid Res 45: 237–249 [DOI] [PubMed] [Google Scholar]
- Jenkins RW, Canals D, Hannun YA 2009. Roles and regulation of secretory and lysosomal acid sphingomyelinase. Cell Signal 21: 836–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose C, Surma MA, Gerl MJ, Meyenhofer F, Shevchenko A, Simons K 2012. Flexibility of a eukaryotic lipidome—Insights from yeast lipidomics. PLoS ONE 7: e35063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo N, Kumagai K, Tomishige N, Yamaji T, Wakatsuki S, Nishijima M, Hanada K, Kato R 2008. Structural basis for specific lipid recognition by CERT responsible for nonvesicular trafficking of ceramide. Proc Natl Acad Sci 105: 488–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai K, Kawano M, Shinkai-Ouchi F, Nishijima M, Hanada K 2007. Interorganelle trafficking of ceramide is regulated by phosphorylation-dependent cooperativity between the PH and START domains of CERT. J Biol Chem 282: 17758–17766 [DOI] [PubMed] [Google Scholar]
- Laviad EL, Albee L, Pankova-Kholmyansky I, Epstein S, Park H, Merrill AH Jr, Futerman AH 2008. Characterization of ceramide synthase 2: Tissue distribution, substrate specificity, and inhibition by sphingosine 1-phosphate. J Biol Chem 283: 5677–5684 [DOI] [PubMed] [Google Scholar]
- Laviad EL, Kelly S, Merrill AH Jr, Futerman AH 2012. Modulation of ceramide synthase activity via dimerization. J Biol Chem 287: 21025–21033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Gianoulis TA, Yip KY, Gerstein M, Snyder M 2010. Extensive in vivo metabolite-protein interactions revealed by large-scale systematic analyses. Cell 143: 639–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingwood CA 2011. Glycosphingolipid functions. In The biology of lipids: Trafficking, regulation, and function (ed. Simons K), pp. 149–174 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingwood D, Simons K 2010. Lipid rafts as a membrane-organizing principle. Science 327: 46–50 [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Phair RD 2010. Lipids and cholesterol as regulators of traffic in the endomembrane system. Annu Rev Biophys 39: 559–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Huang C, Polu SR, Schneiter R, Chang A 2012. Regulation of sphingolipid synthesis through Orm1 and Orm2 in yeast. J Cell Sci 125: 2428–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd-Evans E, Morgan AJ, He X, Smith DA, Elliot-Smith E, Sillence DJ, Churchill GC, Schuchman EH, Galione A, Platt FM 2008. Niemann-Pick disease type C1 is a sphingosine storage disease that causes deregulation of lysosomal calcium. Nat Med 14: 1247–1255 [DOI] [PubMed] [Google Scholar]
- Lopez PHH, Schnaar RL 2009. Gangliosides in cell recognition and membrane protein regulation. Curr Opin Struct Biol 19: 549–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowther J, Naismith JH, Dunn TM, Campopiano DJ 2012. Structural, mechanistic and regulatory studies of serine palmitoyltransferase. Biochem Soc Trans 40: 547–554 [DOI] [PubMed] [Google Scholar]
- Malinina L, Malakhova ML, Teplov A, Brown RE, Patel DJ 2004. Structural basis for glycosphingolipid transfer specificity. Nature 430: 1048–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinina L, Malakhova ML, Kanack AT, Lu M, Abagyan R, Brown RE, Patel DJ 2006. The liganding of glycolipid transfer protein is controlled by glycolipid acyl structure. PLoS Biol 4: e362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandon E, van Echten G, Birk R, Schmidt R, Sandhoff K 1991. Sphingolipid biosynthesis in cultured neurons. Down-regulation of serine palmitoyltransferase by sphingoid bases. Eur J Biochem 198: 667–674 [DOI] [PubMed] [Google Scholar]
- Mao C, Obeid LM 2008. Ceramidases: Regulators of cellular responses mediated by ceramide, sphingosine, and sphingosine-1-phosphate. Biochim Biophys Acta 1781: 424–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao C, Wadleigh M, Jenkins G, Hannun Y, Obeid L 1997. Identification and characterization of Saccharomyces cerevisiae dihydrosphingosine-1-phosphate phosphatase. J Biol Chem 272: 28690–28694 [DOI] [PubMed] [Google Scholar]
- Menuz V, Howell K, Gentina S, Epstein S, Riezman I, Fornallaz-Mulhauser M, Hengartner M, Gomez M, Riezman H, Martinou J 2009. Protection of C. elegans from anoxia by HYL-2 ceramide synthase. Science 324: 381–384 [DOI] [PubMed] [Google Scholar]
- Merrill AH Jr 2011. Sphingolipid and glycosphingolipid metabolic pathways in the era of sphingolipidomics. Chem Rev 111: 6387–6422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake Y, Kozutsumi Y, Nakamura S, Fujita T, Kawasaki T 1995. Serine palmitoyltransferase is the primary target of a sphingosine-like immunosuppressant, ISP-1/myriocin. Biochem Biophys Res Commun 211: 396–403 [DOI] [PubMed] [Google Scholar]
- Moffatt M, Kabesch M, Liang L, Dixon A, Strachan D, Heath S, Depner M, von Berg A, Bufe A, Rietschel E, et al. 2007. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature 448: 470–473 [DOI] [PubMed] [Google Scholar]
- Montefusco DJ, Newcomb B, Gandy JL, Brice SE, Matmati N, Cowart LA, Hannun YA 2012. Sphingoid bases and the serine catabolic enzyme CHA1 define a novel feedforward/feedback mechanism in the response to serine availability. J Biol Chem 287: 9280–9289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen TD, Spassieva S, Jenkins RW, Kitatani K, Bielawski J, Hannun YA, Obeid LM 2011. Selective knockdown of ceramide synthases reveals complex interregulation of sphingolipid metabolism. J Lipid Res 52: 68–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen TD, Hannun YA, Obeid LM 2012. Ceramide synthases at the centre of sphingolipid metabolism and biology. Biochem J 441: 789–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahara K, Ohkuni A, Kitamura T, Abe K, Naganuma T, Ohno Y, Zoeller RA, Kihara A 2012. The Sjögren-Larsson syndrome gene encodes a hexadecenal dehydrogenase of the sphingosine 1-phosphate degradation pathway. Mol Cell 46: 461–471 [DOI] [PubMed] [Google Scholar]
- Niles BJ, Mogri H, Hill A, Vlahakis A, Powers T 2012. Plasma membrane recruitment and activation of the AGC kinase Ypk1 is mediated by target of rapamycin complex 2 (TORC2) and its effector proteins Slm1 and Slm2. Proc Natl Acad Sci 109: 1536–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohturfft A, Zhang SC 2009. Coordination of lipid metabolism in membrane biogenesis. Annu Rev Cell Dev Biol 25: 539–566 [DOI] [PubMed] [Google Scholar]
- Penno A, Reilly MM, Houlden H, Laura M, Rentsch K, Niederkofler V, Stoeckli ET, Nicholson G, Eichler F, Brown RH Jr, et al. 2010. Hereditary sensory neuropathy type 1 is caused by the accumulation of two neurotoxic sphingolipids. J Biol Chem 285: 11178–11187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry RJ, Ridgway ND 2006. Oxysterol-binding protein and vesicle-associated membrane protein-associated protein are required for sterol-dependent activation of the ceramide transport protein. Mol Biol Cell 17: 2604–2616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao H, Mayinger P 2012. Growth and metabolic control of lipid signalling at the Golgi. Biochem Soc Trans 40: 205–209 [DOI] [PubMed] [Google Scholar]
- Ridgway ND, Lagace TA, Cook HW, Byers DM 1998. Differential effects of sphingomyelin hydrolysis and cholesterol transport on oxysterol-binding protein phosphorylation and Golgi localization. J Biol Chem 273: 31621–31628 [DOI] [PubMed] [Google Scholar]
- Roelants FM, Breslow DK, Muir A, Weissman JS, Thorner J 2011. Protein kinase Ypk1 phosphorylates regulatory proteins Orm1 and Orm2 to control sphingolipid homeostasis in Saccharomyces cerevisiae. Proc Natl Acad Sci 108: 19222–19227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampaio JL, Gerl MJ, Klose C, Ejsing CS, Beug H, Simons K, Shevchenko A 2011. Membrane lipidome of an epithelial cell line. Proc Natl Acad Sci 108: 1903–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorling S, Vallee B, Barz W, Riezman H, Oesterhelt D 2001. Lag1p and Lac1p are essential for the Acyl-CoA-dependent ceramide synthase reaction in Saccharomyces cerevisae. Mol Biol Cell 12: 3417–3427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Schuldiner M, Weissman JS 2013. The contribution of systematic approaches to characterizing the proteins and functions of the endoplasmic reticulum. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a013284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze H, Sandhoff K 2011. Lysosomal lipid storage diseases. In The biology of lipids: Trafficking, regulation, and function (ed. Simons K), pp. 287–306 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe HJ, Stevens TJ, Munro S 2010. A comprehensive comparison of transmembrane domains reveals organelle-specific properties. Cell 142: 158–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Simons K 2010. Lipidomics: Coming to grips with lipid diversity. Nat Rev Mol Cell Biol 11: 593–598 [DOI] [PubMed] [Google Scholar]
- Slotte JP, Bierman EL 1988. Depletion of plasma-membrane sphingomyelin rapidly alters the distribution of cholesterol between plasma membranes and intracellular cholesterol pools in cultured fibroblasts. Biochem J 250: 653–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spassieva SD, Mullen TD, Townsend DM, Obeid LM 2009. Disruption of ceramide synthesis by CerS2 down-regulation leads to autophagy and the unfolded protein response. Biochem J 424: 273–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Miao Y, Yamane Y, Zhang C, Shokat KM, Takematsu H, Kozutsumi Y, Drubin DG 2012. Orm protein phosphoregulation mediates transient sphingolipid biosynthesis response to heat stress via the Pkh-Ypk and Cdc55-PP2A pathways. Mol Biol Cell 23: 2388–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers K, Patil C, Wodicka L, Lockhart D, Weissman J, Walter P 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101: 249–258 [DOI] [PubMed] [Google Scholar]
- Vacaru A, Tafesse F, Ternes P, Kondylis V, Hermansson M, Brouwers J, Somerharju P, Rabouille C, Holthuis J 2009. Sphingomyelin synthase-related protein SMSr controls ceramide homeostasis in the ER. J Cell Biol 185: 1013–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Echten G, Birk R, Brenner-Weiss G, Schmidt R, Sandhoff K 1990. Modulation of sphingolipid biosynthesis in primary cultured neurons by long chain bases. J Biol Chem 265: 9333–9339 [PubMed] [Google Scholar]
- van Meer G, Voelker DR, Feigenson GW 2008. Membrane lipids: Where they are and how they behave. Nat Rev Mol Cell Biol 9: 112–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Várnai P, Balla T 2008. Live cell imaging of phosphoinositides with expressed inositide binding protein domains. Methods 46: 167–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TC, Farese RV Jr 2012. Lipid droplets and cellular lipid metabolism. Annu Rev Biochem 81: 687–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenk MR 2010. Lipidomics: New tools and applications. Cell 143: 888–895 [DOI] [PubMed] [Google Scholar]
- Yamashita T, Wada R, Sasaki T, Deng C, Bierfreund U, Sandhoff K, Proia RL 1999. A vital role for glycosphingolipid synthesis during development and differentiation. Proc Natl Acad Sci 96: 9142–9147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, DeBose-Boyd R 2011. Regulation of cholesterol and fatty acid synthesis. In The biology of lipids: Trafficking, regulation, and function (ed. Simons K), pp. 175–188 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Skrzypek M, Lester R, Dickson R 2001. Elevation of endogenous sphingolipid long-chain base phosphates kills Saccharomyces cerevisiae cells. Curr Genet 40: 221–233 [DOI] [PubMed] [Google Scholar]