The construction of 14 RNA nanoparticles resistant to RNase degradation, stable in serum for >36 h, and in vivo after systemic injection are described. Oligomers of various order (dimers to heptamers) were assembled using elementary modules isolated from the phi29 motor pRNA. These novel RNA nanostructures can harbor various functions for different applications in nanotechnology and medicine.
Keywords: nanobiotechnology, bacteriophage phi29, RNA nanotechnology, RNA nanoparticle, RNA therapeutics, target delivery
Abstract
Due to structural flexibility, RNase sensitivity, and serum instability, RNA nanoparticles with concrete shapes for in vivo application remain challenging to construct. Here we report the construction of 14 RNA nanoparticles with solid shapes for targeting cancers specifically. These RNA nanoparticles were resistant to RNase degradation, stable in serum for >36 h, and stable in vivo after systemic injection. By applying RNA nanotechnology and exemplifying with these 14 RNA nanoparticles, we have established the technology and developed “toolkits” utilizing a variety of principles to construct RNA architectures with diverse shapes and angles. The structure elements of phi29 motor pRNA were utilized for fabrication of dimers, twins, trimers, triplets, tetramers, quadruplets, pentamers, hexamers, heptamers, and other higher-order oligomers, as well as branched diverse architectures via hand-in-hand, foot-to-foot, and arm-on-arm interactions. These novel RNA nanostructures harbor resourceful functionalities for numerous applications in nanotechnology and medicine. It was found that all incorporated functional modules, such as siRNA, ribozymes, aptamers, and other functionalities, folded correctly and functioned independently within the nanoparticles. The incorporation of all functionalities was achieved prior, but not subsequent, to the assembly of the RNA nanoparticles, thus ensuring the production of homogeneous therapeutic nanoparticles. More importantly, upon systemic injection, these RNA nanoparticles targeted cancer exclusively in vivo without accumulation in normal organs and tissues. These findings open a new territory for cancer targeting and treatment. The versatility and diversity in structure and function derived from one biological RNA molecule implies immense potential concealed within the RNA nanotechnology field.
INTRODUCTION
One of the two approaches in nanotechnology is bottom-up assembly that uses modified or engineered building blocks (Schmidt and Eberl 2001; Balzani et al. 2002; Seeman and Belcher 2002; Shu et al. 2004; Xiao et al. 2009a,b, 2010). RNA is versatile in structure and function and even possesses enzymatic activity characteristic of some proteins (Nilsen 2007; Guo 2010; Shukla et al. 2011). It can also be designed, predicted, synthesized, and manipulated with a level of simplicity similar to that of DNA (Jaeger and Leontis 2000; Hansma et al. 2003; Grabow et al. 2011). Thus, RNA is a unique building block for the assembly of nanoparticles (Wilcox et al. 1992; Leontis et al. 2006; Guo 2010; Guo et al. 2012a,b). Several naturally or artificially functional RNAs, such as small interfering RNAs (siRNAs) (Brummelkamp et al. 2002; Carmichael 2002; Jacque et al. 2002; Li et al. 2002; Varambally et al. 2002; Ghildiyal and Zamore 2009; Guo et al. 2010; Wiedenheft et al. 2012), ribozymes (Sarver et al. 1990; Chowrira et al. 1991; Westhof 2012), microRNAs (He and Hannon 2004; Bartel 2009; Duchaine and Slack 2009; Fabian et al. 2010), riboswitches (Strobel and Cochrane 2007; Henkin 2008; Ogawa and Maeda 2008; Sudarsan et al. 2008), anti-sense RNAs (Coleman et al. 1985; Knecht and Loomis 1987), and RNA aptamers (Ellington and Szostak 1990; Tuerk and Gold 1990; Gold 1995; Srisawat and Engelke 2001) have been shown to play active roles at the cellular level by catalyzing biological reactions, regulating gene expression, sensing and communicating responses to cellular signals, or binding to specific targets for potential delivery of therapeutics. The incorporation of these functionalities into RNA nanoparticles could have immense applications in nanotechnology and medicine.
Although the concept of “RNA nanotechnology” was proven in 1998 (Guo et al. 1998), many scientists were adverse to RNA nanotechnology due to the susceptibility of RNA to RNase degradation. Due to structural flexibility, RNase sensitivity, and serum instability, the construction of RNA nanoparticles with concrete shapes for in vivo application remains challenging. The field of RNA nanotechnology has emerged only recently, and new methods for RNA nanoparticle constructions are in high demand presently. Here we report the construction of 14 different RNA nanoparticles with solid shapes for targeting cancers specifically.
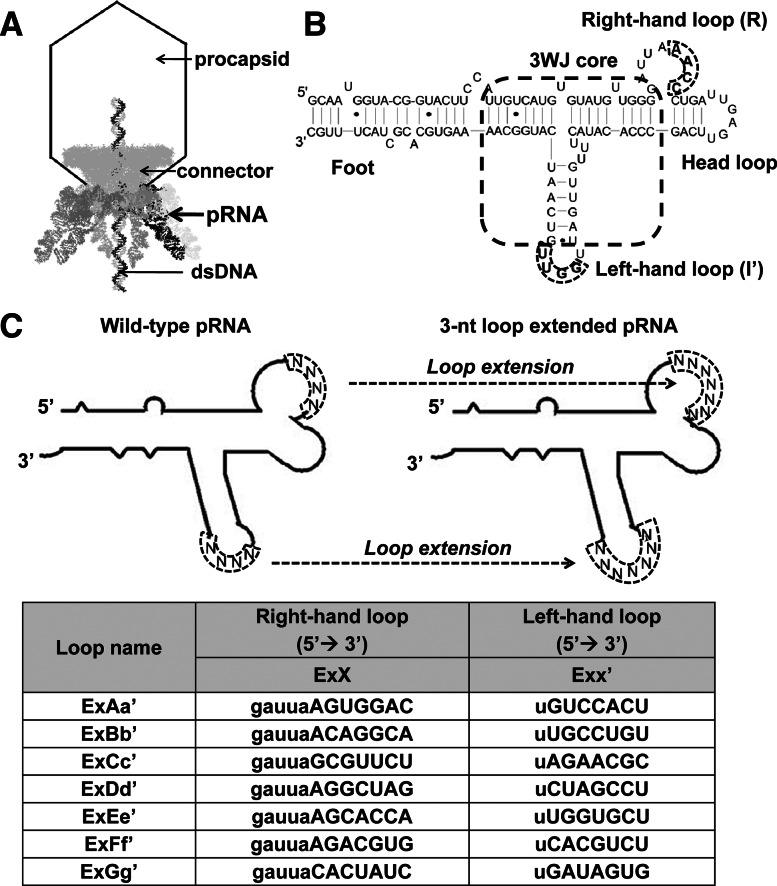
RNA is a polynucleic acid that possesses all the favorable attributes of conventional polymers. Several structural features of the bacteriophage phi29 packaging RNA (pRNA) are attractive for bottom-up assembly of RNA nanoparticles (Fig. 1A). Each pRNA contains a helical domain (5′/3′ paired ends denoted as the foot) (Fig. 1B; Cairns et al. 1988; Zhang et al. 1994; Reid et al. 1994a,b,c; Garver and Guo 1997), a central domain (bases 23–97) containing right- and left-hand loops for intermolecular interactions (Fig. 1B; Reid et al. 1994a; Garver and Guo 1997; Chen et al. 1999, 2000), and a three-way junction (3WJ) motif (see Figs. 1B, 5A below). Here we report the development of an array of techniques, or “toolkits,” derived from pRNA that utilize a variety of principles to construct RNA architectures with diverse shapes and angles. The resulting RNA nanoparticles are homogeneous with defined structure and stoichiometry and can be generated in a controllable and predictable manner. These toolkits will be very useful to construct higher-ordered, diverse RNA nanostructures with multiple functionalities as a polyvalent delivery system for nanotechnology and nanomedicinal applications.
FIGURE 1.
Sequences and principle for Toolkit I using the hand-in-hand interaction mechanism in the construction of RNA nanoparticles. (A) Bacteriophage phi29 DNA packaging motor. Six copies of packaging RNAs (pRNAs) assemble into a hexamer ring to gear the viral DNA packaging motor. (B) The primary sequence and secondary structure of wild-type pRNA. The two domains (helical foot and central R- and L-hand) are connected by a 3WJ core. (C) Extended pRNA interlocking loop sequences for Toolkit I.
FIGURE 5.
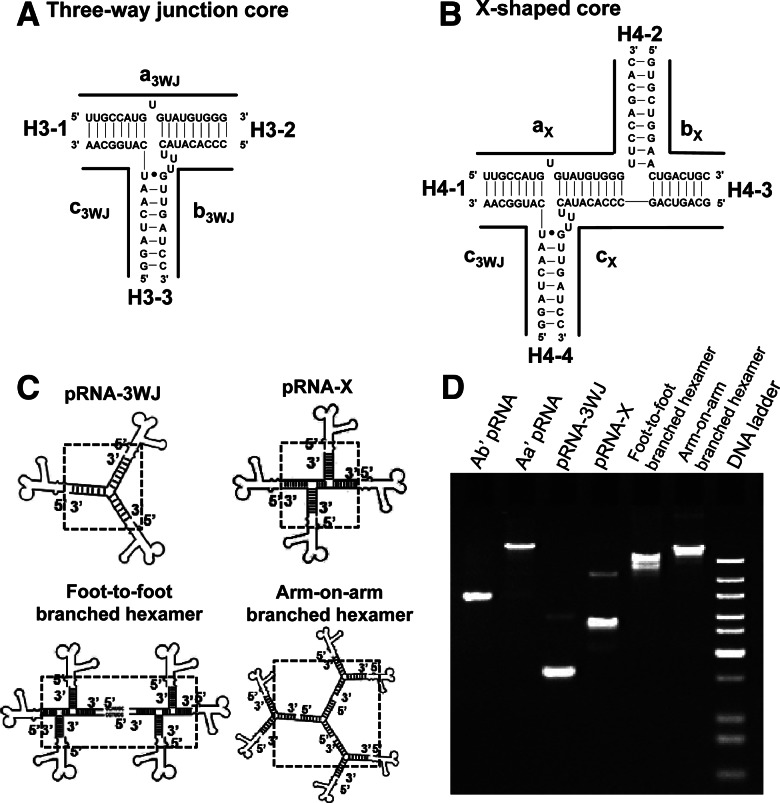
The application of Toolkit III based on pRNA 3WJ motif for the assembly of branched pRNA nanoparticles. (A) pRNA 3WJ motif for the construction of three-branch RNA nanoparticles in C. (B) pRNA 3WJ derived X-shaped pRNA motif for the construction of four-branch RNA nanoparticles in C. (C) Illustration of the assembled branched pRNA nanostructures based on pRNA 3WJ and X-shaped motif. (D) Twelve percent native PAGE gel demonstrating the efficient self-assembly of RNA nanoparticles using Toolkit III.
The incorporation of all functionalities was achieved prior to the assembly of the RNA nanoparticles, thus ensuring the production of homogeneous therapeutic nanoparticles with appropriate quality control for medical applications. The annealing of therapeutic RNA modules to the RNA (Grabow et al. 2011) or DNA (Lee et al. 2012a) nanoparticles after the assemblage of a scaffold has been reported. However, our novel bottom-up approach has demonstrated the feasibility of incorporation of all functionalities prior to the assemblage of RNA nanoparticles, without affecting the folding or function of both the core scaffold and the individual modules. This approach also ensures the high efficiency and homogeneity of the particles and avoids the dissociation of the RNA nanoparticles in vivo due to the presence of nicks in the component RNA functional modules of the RNA nanoparticles.
RESULTS
Toolkit I: Hand-in-hand toolkit for assemblage of pRNA nanoparticles by extending interlocking loops
Previously, we have reported that the interlocking loops of phi29 pRNA can be used for constructing RNA dimers, trimers, and hexamers through hand-in-hand interactions (Guo et al. 1998, 2005; Chen et al. 1999; Shu et al. 2003, 2011c; Khaled et al. 2005). However, the resulting particles were not found to be thermodynamically stable enough for nanotechnological applications. In this study, we tried to produce stable interlocking loops for nanoparticle construction by extending the loop sequences. But, random extension of the loop sequences failed due to interferences in RNA folding and 3D conformation. Extensive investigations have also resulted in a toolkit with a set of hand-in-hand loop sequences that can be used for the construction of stable RNA polygonal nanoparticles (Fig. 1C). The loop-extended pRNAs were constructed by replacing the complementary regions in the wild-type pRNA 4-nt R- and L-loops with a series of loop sequences (Fig. 1C). Since altering the nucleic acid sequences may have affected the global folding of pRNA molecules, the pRNA with re-engineered loop sequences were thoroughly analyzed with the online RNA folding program Mfold (Zuker 2003). A total of nine pairs of hand-in-hand loop sequences with predicted appropriate folding were selected out from the library and then used for constructing a variety of RNA polygonal particles via hand-in-hand interactions (Supplemental Fig. 1A).
The formation of pRNA homo-dimers is an excellent system by which to assay loop–loop interactions since the paired loop sequences are assigned to R- and L-loops within the same pRNA. The appropriately folded pRNA monomers will form self-dimers via the complementary R- and L-loops in the presence of Mg2+. After transcription, nine homo-dimers were refolded in TMS buffer for 1 h at 37°C and analyzed with 6% native PAGE to assay for dimer formation. The dimers were found to have migrated slower than the monomers, as shown in the native PAGE. Seven pairs of loop–loop sequences were able to facilitate homo-dimer formation (Supplemental Fig. 1B), and two pairs of loop–loop sequences showed weak dimer formation, indicating that the pRNA structure misfolded after the alteration of the loop sequences.
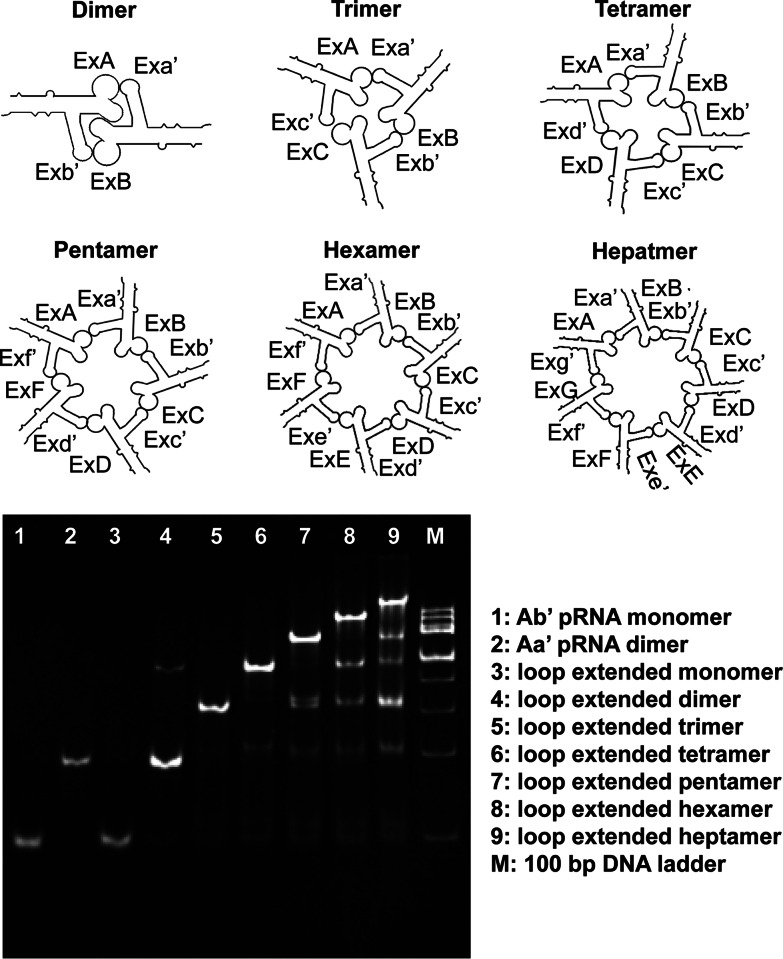
The seven pairs of loop–loop sequences that exhibited dimer formation were further investigated as potential candidates for building pRNA nanoscale assemblies. Using reengineered loop–loop interactions, pRNA dimer (ExAb′-ExBa′), trimer (ExBa′-ExCb′-ExAc′), tetramer (ExBa′-ExCb′-ExDc′-ExAd′), pentamer (ExBa′-ExCb′-ExDc′-ExFd′-ExAf′), hexamer (ExBa′-ExCb′-ExDc′-ExEd′-ExFe′-ExAf′), and heptamer (ExBa′-ExCb′-ExDc′-ExEd′-ExFe′-ExGf′-ExAg′) were assembled in the presence of Mg2+, as shown by native PAGE (Fig. 2; Supplemental Fig. 2). The formation of each nanoparticle was further confirmed by AFM imaging (see Fig. 5A–E, below).
FIGURE 2.
Native PAGE gel demonstrating the efficient self-assembly of RNA nanoparticles using Toolkit I based on the newly developed hand-in-hand interlocking loops from Figure 1.
Toolkit II: Foot-to-foot toolkit for assemblage of higher-ordered pRNA nanoparticles based on palindrome sequences
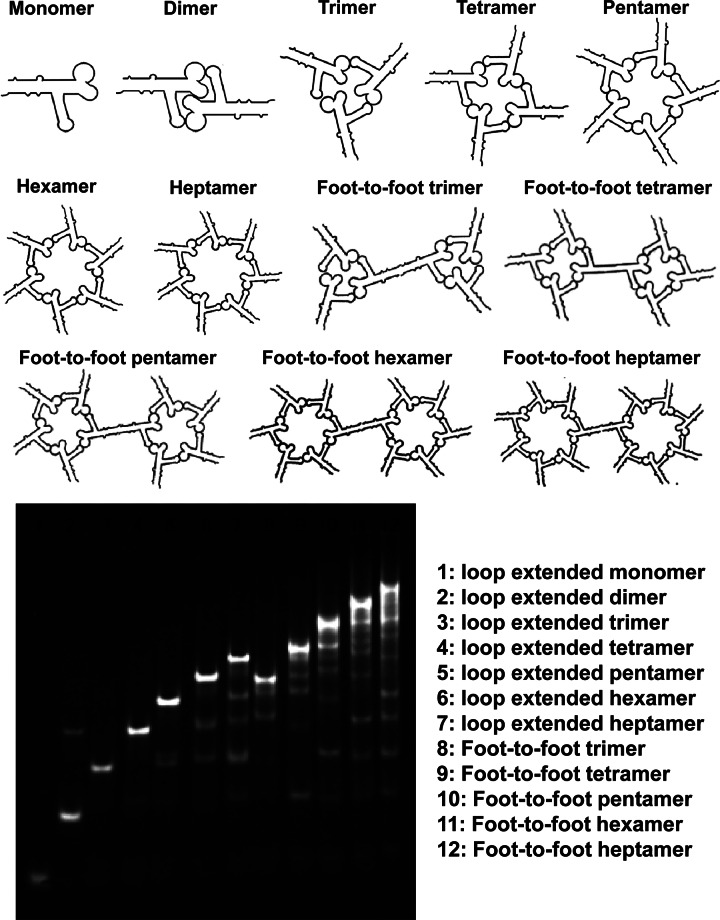
A palindrome sequence is one that reads the same in the 5′-to-3′ direction on one strand or from the 5′-to-3′ direction on the complementary strand. Six-nucleotide palindrome sequences were designed following the chart in Figure 3 and introduced at the 3′-end of the RNA building block. These RNA building blocks were able to form foot-to-foot self-dimers via intermolecular interactions mediated by the palindrome sequences. In a similar fashion, palindrome sequences were utilized to bridge RNA nanostructures, motifs, and scaffolds for the self-assembly of RNA hexamers, octamers, decamers, dodecamers, or any other duplex with an even number of subunits and were assembled into foot-to-foot structures, as revealed by native PAGE (Fig. 4) and AFM imaging (see Fig. 6F–J, below).
FIGURE 3.
Principle for the design of Toolkit II using foot-to-foot 6-nt palindrome sequences.
FIGURE 4.
Native PAGE gel demonstrating the efficient self-assembly of RNA nanoparticles using Toolkit II based on foot-to-foot palindrome sequence.
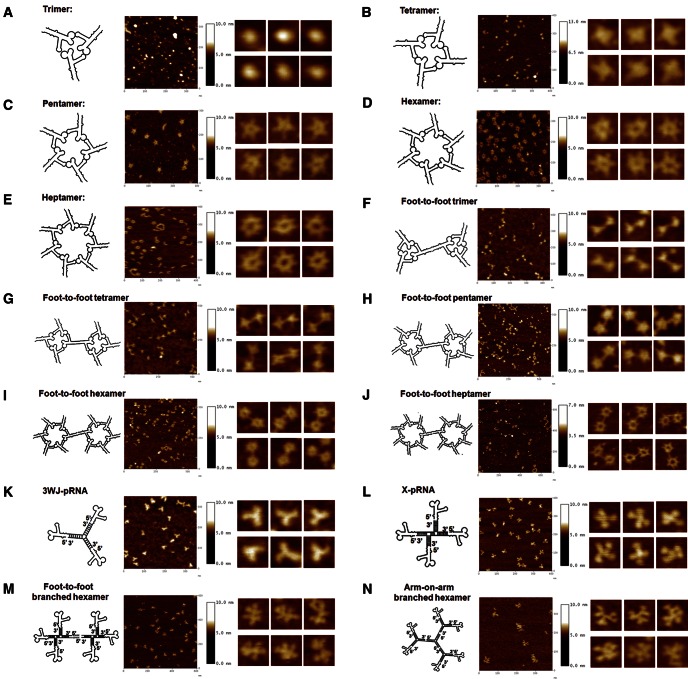
FIGURE 6.
AFM images of diverse pRNA nanoparticles (A–N) constructed using Toolkits I, II, and III. The second column is the magnified images of individual nanoparticles.
Toolkit III: Design of branched pRNA nanoparticles based on the 3WJ motif
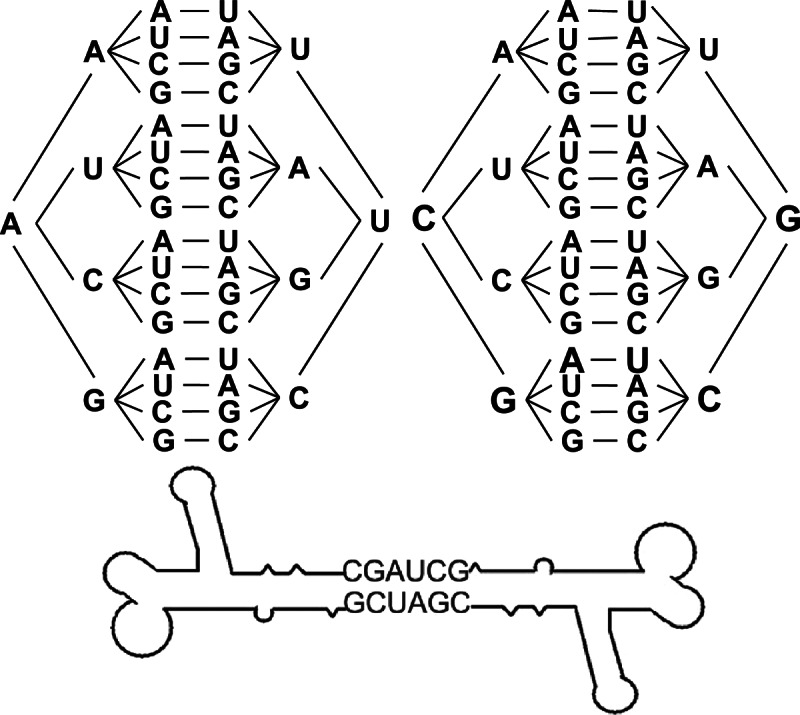
Large macromolecular assemblies of RNA nanoparticles were constructed via intramolecular interactions utilizing junction motifs. The pRNA-3WJ core was assembled with an unusually high efficiency from three RNA oligos mixed in the proper stoichiometric molar ratio in the absence of any metal ions (Shu et al. 2011a), as shown by native PAGE (Fig. 5A,C). The lengths of the helices H3-1, H3-2, and H3-3 were 8 bp, 9 bp, and 8 bp, respectively, and the resulting nanoparticles constructed were stable even under strong denaturing conditions.
The pRNA-X motif was constructed by opening the R-loop of the pRNA subunit and inserting a 9-bp sequence, thereby forming a double helical segment (H4-2) and extending the H4-3 helix by 4 bp (Fig. 5B; Haque et al. 2012). The length of the helices H4-1, H4-3, and H4-4 were each 8 bp long, while H4-2 was 9 bp long. The X-shaped motif could then be assembled from four RNA fragments mixed in a 1:1:1:1 ratio with a high affinity in absence of metal salts (Fig. 5C). Self-complementary palindrome sequences could then be added to either the 5′- or 3′-end of one of the strands of the core motifs to bridge two X-motifs harboring a total of six functionalities via intermolecular interactions, thereby generating a foot-to-foot branched hexamer (Fig. 5C).
Arm-on-arm branched hexavalent RNA nanoparticles were constructed from two 3WJ cores. A major consideration was to minimize the misfolding of the resulting complex after fusion of the sequences. One of the approaches involved the utilization of the thermodynamic properties of the strong-folding RNA cores, exemplified by the forward sequences and reverse sequences of the pRNA-3WJ core that generated two self-folded, stable scaffolds. Each of the three arms of the 3WJ harbored an overhanging, reverse 3WJ fragment that served as a sticky end to carry one of the monomer constructs, and produced either a trimeric (three identical molecules or structures) complex or a triplet (three varied molecules or structures) (Fig. 5C). Similarly, hexameric complexes were assembled efficiently in one step from RNA fragments with the following stoichiometric ratio: (a3WJ-FWD + a3WJ-REV):(a3WJ-FWD + a3WJ-REV):(a3WJ-FWD + a3WJ-REV):b3WJ:c3WJ = 1:1:1:3:3.
Each of the four aforementioned constructs could harbor functional modules at each of the branches, without affecting the folding of the core scaffolds. To test this, monomeric pRNA subunits were placed at each of the branches, and AFM images were produced that strongly indicated the formation of homogenous RNA nanoparticles with the desired structure and stoichiometry (Fig. 6K–N).
Functional assays for each module incorporated into pRNA nanoparticles
Similar to the principles of the fields of DNA and protein biotechnology and nanotechnology, one crucial factor to consider in the construction of RNA nanoparticles is the misfolding of the resulting complex during assembly. Using Toolkits I, II, and III, the resultant misfolding was significantly minimized. We constructed multivalent RNA nanoparticles harboring functional modules, such as siRNA, ribozymes, aptamers, and the folate (FA) ligand. It was revealed that (1) the incorporation of functional modules did not affect the folding of the central scaffolds and (2) the RNA modules within the nanoparticles folded into their respective authentic structures and retained their biological functions independently. Our assay revealed that the folding of functionalities was predictable and that each incorporated module folded and functioned independently within the nanoparticles.
Assessment of malachite green fluorescence
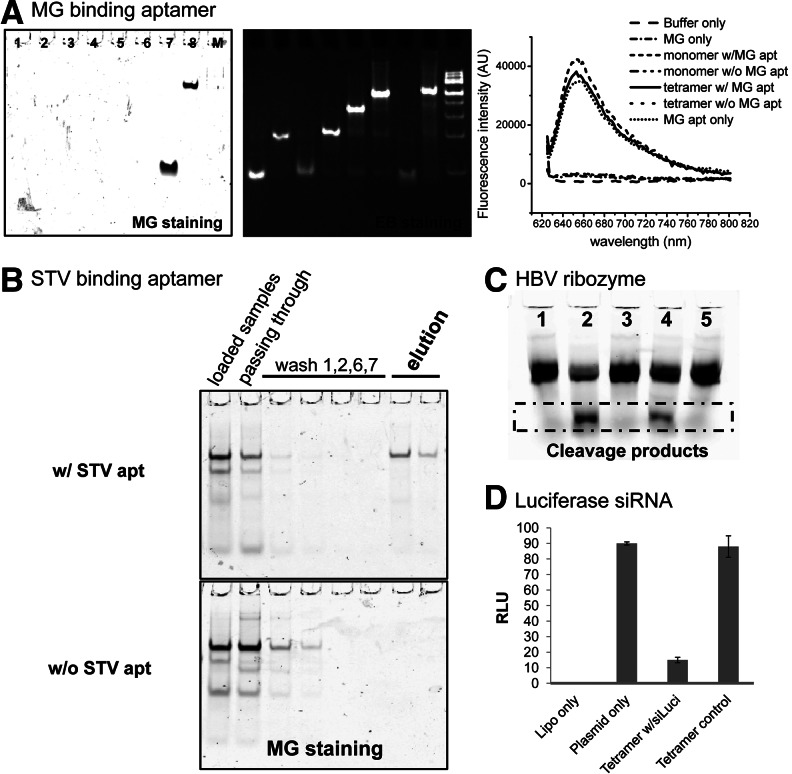
The fused malachite green (MG)–binding aptamer, in both the monomeric form and pRNA tetramer assemblies, retained its capacity to bind MG dye, as revealed by fluorescence emissions in native PAGE and by fluorometry (Fig. 7A; Baugh et al. 2000; Shu et al. 2011a; Reif et al. 2013). Free MG is not fluorescent by itself but emits fluorescent light only after binding to the RNA aptamer, a fact that supports our finding.
FIGURE 7.
Functional assay of each functional moiety incorporated in pRNA nanoparticles to demonstrate correct folding. (A) Fluorogenic assay demonstrating the emission of lights in the presence of the chemical MG binding to the MG aptamer incorporated in the pRNA vector using 6% native PAGE (lanes 1,2 are pRNA monomer and dimer control; lanes 3–6 are loop-extended pRNA monomer, dimer, trimer, and tetramer, respectively, without the MG aptamer; lanes 7,8 are loop-extended monomer and tetramers, respectively, harboring the MG binding aptamer), visualized by MG fluorescent imaging (left panel), ethidium bromide staining (center panel), and MG fluorescence emission (right panel). (B) Streptavidin (STV)-binding assay using STV affinity column to demonstrate the correct folding of the STV-aptamer incorporated into the RNA nanoparticles visualized by both MG staining in native PAGE gel. (C) Hepatitis B virus (HBV) ribozyme assay to demonstrate the correct folding and function of the HBV ribozyme incorporated in the RNA nanoparticles (lane 1, HBV polyA substrate; lane 2, HBV polyA substrate incubated with monomer harboring HBV ribozyme; lane 3, HBV polyA substrate incubated with monomer harboring disabled HBV ribozyme; lane 4, HBV polyA substrate incubated with tetramer harboring HBV ribozyme; and lane 5, HBV polyA substrate incubated with tetramer harboring disabled HBV ribozyme). (D) Dual-luciferase assay to demonstrate the correct annealing and function of the luciferase siRNA incorporated in the RNA nanoparticles.
Assay for streptavidin binding
Upon incubation with streptavidin (STV) resin, pRNA tetramers harboring the STV-binding aptamer (Srisawat and Engelke 2001) bound the STV resin and then were eluted out by biotin (Fig. 7B). Tetramers without the STV-binding aptamer did not appear in the elution fractions (Fig. 7B). The results demonstrated that the STV aptamer in the tetrameric complex maintained structural integrity and function after fusing with pRNA, as revealed by native PAGE. The tetramers that also contained the MG binding aptamer exhibited binding and staining, indicating that other functions were retained within tetramers after purification, as shown in Figure 7B. This STV-binding aptamer could potentially be utilized as the method of RNA nanoparticle purification.
Retention of catalytic activity of the hepatitis B virus ribozyme
Hepatitis B virus (HBV) ribozymes were able to cleave the 135-nt HBV RNA genome substrate into two fragments (60 nt and 75 nt) after being incorporated into the tetrameric pRNA nanoparticle (Fig. 7C), indicating that the ribozyme retained correct folding and function after it was incorporated into the pRNA nanoparticles.
Targeted gene silencing of luciferase
A dual-luciferase reporter assay system was used to measure the gene silencing effects of pRNA tetrameric constructs harboring four different siRNAs targeted toward the firefly luciferase gene (Shu et al. 2011b). The relative luciferase activity is proportional to the expression level of the firefly luciferase gene when normalized with the activity of the internal control, Renilla luciferase, which remains unaffected against siRNA. The results indicated that pRNA tetramers harboring four different firefly luciferase siRNAs displayed ∼70% decrease in firefly luciferase gene expression, compared with scrambled control siRNA (Fig. 7D).
Internalization of branched pRNA nanoparticles harboring FA
Many kinds of cancer cell lines, especially those from epithelial origins, overexpress FA receptors (FRs) on a plasma membrane by >1000-fold. FA has been extensively used as a targeting ligand, because it can be efficiently internalized into cancer cells via FR-mediated endocytosis (Lee and Low 1994; Lu and Low 2002). Accordingly, FA was incorporated in pRNA branched 3WJ nanoparticles to facilitate cancer cell targeting (Lu and Low 2002; Guo et al. 2006; Shu et al. 2011a). A Cy3 dye moiety was also incorporated into pRNA 3WJ strands by chemical conjugation, to serve as a fluorescent marker (Shu et al. 2011a). The pRNA nanoparticles harboring FA and Cy3 labels were compared with negative control nanoparticles harboring NH2 and Cy3 labels. Confocal imaging indicated binding of the pRNA nanoparticles harboring FA and their entry into the targeted cells, as demonstrated by the colocalization and overlap of the fluorescent 3WJ-pRNA nanoparticles (red) and cytoplasm (green) (Fig. 8A).
FIGURE 8.
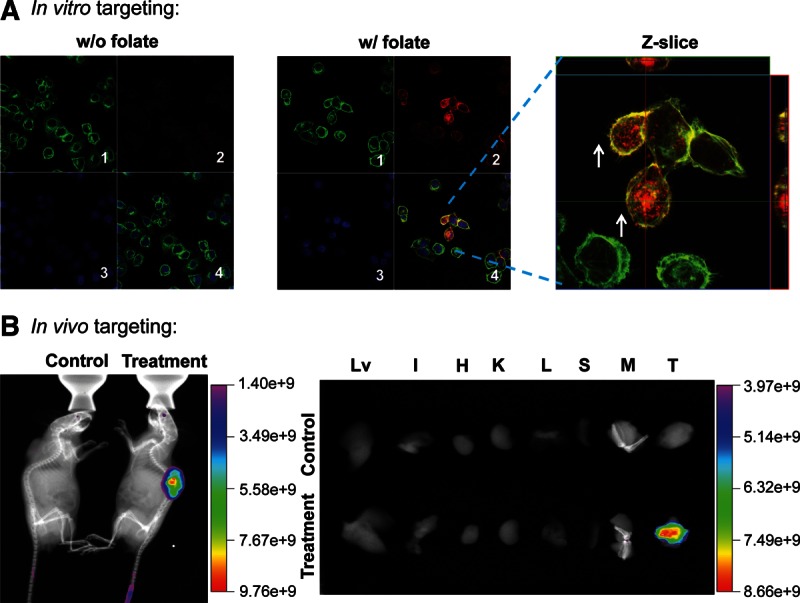
In vitro and in vivo binding and entry of pRNA nanoparticles into targeted cells. (A) Confocal images showing the targeting of RNA nanoparticles to folate receptor (FR)–positive KB cells by colocalization (overlap, 4; magnified, right panel) of cytoplasm (green, 1), RNA nanoparticles (red, 2), and nuclei (blue, 3). (B, left) Whole-body imaging revealed that pRNA nanoparticles targeted FR+ tumor xenografts upon systemic injection in nude mice. Treatment group was injected with pRNA nanoparticles containing folate and Alexa Fluor 647 label. Control group was injected with PBS. Images shown are X-ray plus fluorescence imaging. (Right) Organ imaging showing pRNA nanoparticles targeted to FR+ tumor (Lv indicates liver; K, kidney; H, heart; L, lung; S, spleen; I, intestine; M, muscle; and T, tumor). Scale bar, fluorescent intensity.
In vivo targeting to xenograft tumors
pRNA-X nanoparticles were constructed carrying FA to serve as a ligand for binding to cancer cells and Alexa Fluor 647 as a marker for imaging. The nanoparticles were systemically injected (via the tail vein) into athymic nude mice bearing KB cell xenografts in their subcutaneous flanks. Whole-body imaging revealed that fluorescence was located specifically in the tumor xenografts, expressing the FR 24 h after injection (Fig. 8B, left column). Interestingly, the fluorescent nanoparticles were not detected in any other organs in the body 3 h after injection, indicating that the particles were cleared from the normal organs quickly and were not trapped in the liver, lungs, or kidneys after systemic injection (Fig. 8B, right column). These results were very different from other studies that have shown nanoparticles that have accumulated in the liver, lungs, spleen, and kidneys and required 12–96 h to clear from the organs, as revealed by imaging (Altinoglu et al. 2008; Barth et al. 2010; Abdelmawla et al. 2011; Lee et al. 2012c; Ma et al. 2012). The ability of pRNA nanoparticles to effectively and selectively target cancer cells by this process would make this delivery system attractive for future clinical developments.
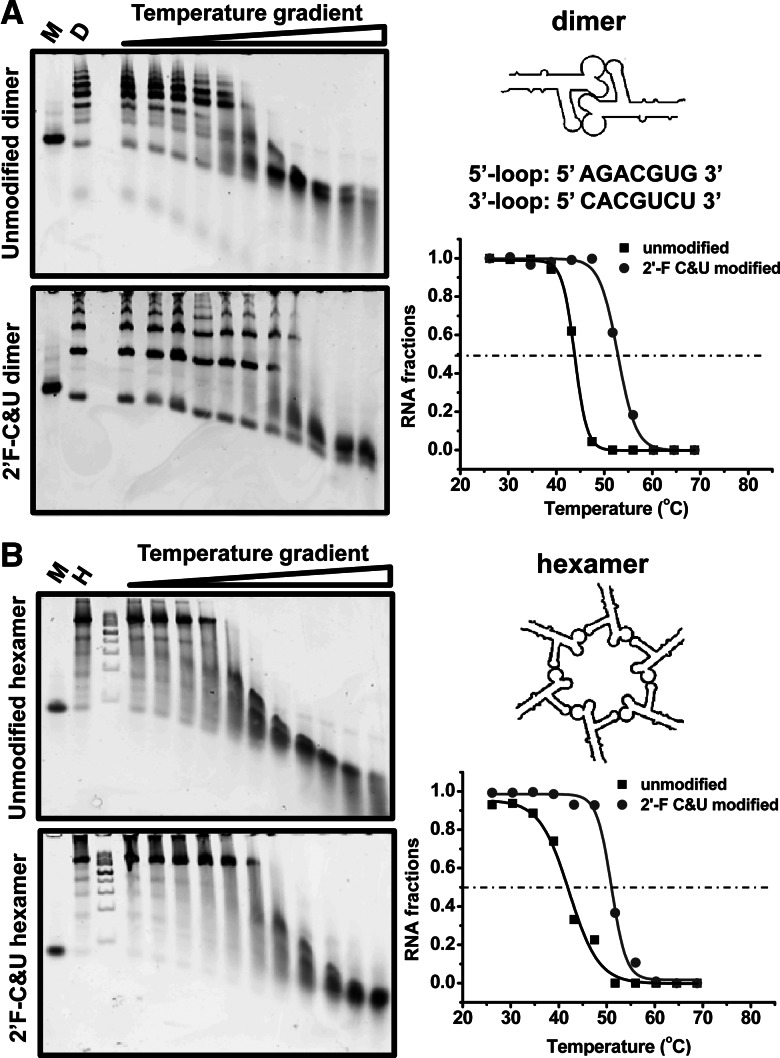
One concern of applying RNA nanoparticles for in vivo delivery, though, is the sensitivity of RNA to RNase degradation. To solve this, 2′-fluorine-deoxy-CTP (2′-F-dCTP) and 2′-fluorine-deoxy-UTP (2′-F-dUTP) were incorporated into the RNA strands (Liu et al. 2010). The 2′-F-dCTP & dUTP (2′-F C&U) modified RNA were found to be able to survive in 10% serum treatment for up to 36 h (Supplemental Fig. 3A), compared with unmodified RNA, which could only last for 2 h. The modified RNA could also survive in various concentrations of RNase A, compared with unmodified RNA, which only survived in very diluted RNase (Supplemental Fig. 3B). In order to determine whether 2′-F C&U modified RNA display a higher thermodynamic stability than unmodified RNA, the stability of unmodified and 2′-F C&U modified RNA assemblies isolated from native PAGE was compared by temperature gradient gel electrophoresis (TGGE) at 700 nM. Two constructs, a dimer and a hexamer, were assayed by TGGE, imaged, and then analyzed by ImageJ. The melting temperature (Tm) of each RNA nanoparticle was obtained by fitting the melting curves using nonlinear Sigmoidal fitting (Tm of unmodified dimer = 43.8°C; Tm of 2′-F C&U modified dimer = 52.8°C; Tm of unmodified hexamer = 42.2°C; Tm of 2′-F C&U modified hexamer = 50.9°C). For all the constructs tested (dimer and hexamer), the 2′-F C&U modified RNA assemblies (dimer or hexamer) were determined to be stable in temperatures ∼8°C–10°C higher than the conditions of the unmodified counterparts (Fig. 9). These results demonstrate the chemically and thermodynamically stable features of modified RNA nanoparticles for diverse applications in nanotechnology and nanomedicine.
FIGURE 9.
TGGE gel analysis of the thermodynamic stability of RNA nanoparticles. (A) pRNA dimer. (Left) TGGE gels of unmodified and 2′-F C&U modified pRNA dimer (M indicates pRNA monomer; D, pRNA dimer). (Right) Fitted melting curve of unmodified and 2′-F C&U modified pRNA dimer. (B) pRNA hexamer. (Left) TGGE gels of unmodified and 2′-F C&U modified pRNA hexamer (M indicates pRNA monomer; H, pRNA hexamer). (Right) Fitted melting curve of unmodified and 2′-F C&U modified pRNA hexamer.
DISCUSSION
Several mechanisms of RNA intermolecular interaction have been reported to play roles for the assembly of diverse RNA nanoparticles. These include (1) hand-in-hand (loop–loop interactions), as demonstrated in bacteriophage phi29 pRNA dimer, trimer, and hexamer formations (Guo et al. 1987, 1998; Shu et al. 2004, 2007); RNA nanorings based on RNAI/II inverse kissing complexes (Yingling and Shapiro 2007; Afonin et al. 2011; Grabow et al. 2011); kissing loops of HIV RNA (Chang and Tinoco 1994; Bindewald et al. 2008) and the hand-in-arm interactions of Drosophila bicoid mRNA (Wagner et al. 2004); (2) palindrome sequence-mediated formation of pRNA dimers, tetramers, and arrays (Shu et al. 2004); (3) RNA motifs as LEGO pieces to build quaternary structures via nontemplated assemblies including tecto-RNA, two-way junctions (2WJs), 3WJs, and four-way junctions (4WJs), and self-assembly by colE1 kissing loop interactions (Prats et al. 1990; Clever et al. 1996; Mujeeb et al. 1998; Jaeger and Leontis 2000; Lilley 2000; Shu et al. 2011a; Haque et al. 2012); (4) extension of arms of thermodynamically stable core to carry multiple therapeutic small RNAs (Shu et al. 2011a; Haque et al. 2012); (5) use of RNA binding proteins to serve as scaffolds for the formation of nanostructures, such as equilateral triangle constructs, where three proteins are bound to an RNA scaffold containing a kink-turn motif for protein binding (Schroeder et al. 2010; Ohno et al. 2011); and (6) combination of rolling circle transcription of RNA and self-assembly to produce giant spherical RNA particles (Lee et al. 2012b). Recently, we have reported the fabrication of ultrastable RNA nanoparticles using the phi29 3WJ core to carry siRNA, ribozymes, and aptamers for cancer targeting (Shu et al. 2011a; Haque et al. 2012), using the aforementioned approach 4. The thermodynamically stable pRNA-X nanoparticles are assembled from four small pieces of RNA fragment, which has proven to be stable in serum, remain intact at ultralow concentrations, and be resistant to denaturation in 8 M urea. The slope of the Tm curve is close to 90° (Shu et al. 2011a; Haque et al. 2012) suggesting thermostability. More importantly, various functionalities such as siRNA, ribozyme, or receptor-binding aptamer incorporated into the 3WJ core resulted in the formation of polyvalent particles displaying all the authentic functionalities in vitro and in vivo. Systemic injection of these RNA nanoparticles to mice showed that the RNA nanoparticles target specifically to the xenograft without accumulating in normal organs and tissues (Shu et al. 2011a; Haque et al. 2012). In this report, we extended our previous work in the aforementioned approach 1, in combination with approaches 2–5 to construct versatile RNA nanoparticles. Three “toolkits” were generated as hand-in-hand interaction (Toolkit I), palindrome sequences for foot-to-foot interaction (Toolkit II), and junction motifs from the phi29 motor pRNA to build larger macromolecular assemblies via intermolecular self-assembly (Toolkit III). Fourteen different homogenous RNA nanoparticles were fabricated in a controlled manner with defined size, structure, and stoichiometry. All the constructed RNA nanoparticles are thermodynamically and chemically stable and can harbor multiple functional modules (up to a maximum 12). We demonstrated incorporation of these functional modules by in vitro and in vivo functional assays. The results suggest that the toolkits can be broadly applied to construct versatile RNA nanoparticles for diverse applications in nanotechnology and nanomedicine.
MATERIALS AND METHODS
Nomenclature
Following standard pRNA nomenclature (Shu et al. 2011b), the right-hand (R-loop) sequence was assigned an uppercase letter (i.e., A, B, …); the left-hand (L-loop) sequence, a lowercase letter with a prime (i.e., a′, b′, …). A pair of the same uppercase and lowercase letters (e.g., Aa′) designated a complementary sequence in the R/L interlocking loop, while different letters indicate a lack of complementarity in the interlocking sequence. To distinguish from the wild-type pRNA, “Ex” was added before the letters (e.g., ExAa′) to indicate that the interlocking loop–loop interaction was extended.
The 3WJ domain of phi29 pRNA was constructed using three RNA oligos, denoted as a3WJ, b3WJ, and c3WJ. The three branches were named H3-1, H3-2, and H3-3, respectively (Fig. 5A). The 3WJ-derived, X-shaped pRNA was constructed using four RNA oligos and was denoted as aX, bX, cX, and c3WJ. The four branches were named H4-1, H4-2, H4-3, and H4-4, respectively (Fig. 5B).
In vitro synthesis, purification, and assay of RNA strands for nanoparticle assembly
Long RNA strands, with and without 2′-F-C&U modification, were synthesized by enzymatic methods, according to the method previously described (Zhang et al. 1994; Liu et al. 2010). Short DNA and RNA oligos, with and without 2′-F-C&U modification, were synthesized chemically by Integrated DNA Technologies and TriLink (Supplemental Tables 1–3).
Higher-order hand-in-hand pRNA polyvalent nanoparticles were formed by mixing the required subunits at an equal molar ratio in TMS buffer (5 mM MgCl2, 100 mM NaCl, 89 mM Tris-HCl at pH 7.6) for 1 h at 37°C and assayed with native 6% PAGE gel run in TBM buffer (5 mM MgCl2, 200 mM boric acid, 89 mM Tris-HCl at pH 7.6) for 3–4 h at 4°C. Foot-to-foot RNA complexes were constructed by extending the 3′-end of ExAb′ with a palindrome sequence (5′-CGAUCG-3′) and assayed by 4% native PAGE gel run in TBM buffer.
pRNA-3WJ core (a3WJ:b3WJ:c3WJ = 1:1:1), pRNA-X motif (aX-RNA:bX-RNA:cX-RNA:c3WJ = 1:1:1:1), foot-to-foot branched hexamer (aX-RNA:bX-RNA:cX-RNA:c3WJ+PS6 = 1:1:1:1), and arm-on-arm branched hexamer [(a3WJ-FWD + a3WJ-REV):(a3WJ-FWD + a3WJ-REV):(a3WJ-FWD + a3WJ-REV):b3WJ:c3WJ = 1:1:1:3:3)] complexes were assembled by mixing the individual component strands in a stoichiometric ratio (as shown in parenthesis) and assayed by 12% native PAGE gel run in TBM buffer.
AFM imaging
RNA nano-assemblies were imaged using specially modified mica surfaces (APS mica) (Lyubchenko and Shlyakhtenko 2009) with a MultiMode AFM NanoScope IV system (Veeco), operating in tapping mode, according to the method previously reported (Shu et al. 2011a).
Assay for each functional module incorporated within pRNA nanoparticles
Fluorescence assay for MG binding aptamer
A pRNA tetramer harboring an MG binding aptamer and a monomer also harboring an MG binding aptamer (100 nM) were mixed with MG dye (2 µM); the fluorescence was measured using a fluorospectrometer (Shu et al. 2011a). An pRNA tetramer without the MG binding aptamer served as a negative control, while an MG binding aptamer by itself served as the positive control. The same samples were also analyzed by 6% native PAGE run in TBM running buffer for 3–4 h at 4°C. The gel was stained with 5–10 µM MG in binding buffer and imaged by Typhoon FLA 7000 (GE Healthcare).
Binding assay for the STV-binding aptamer
pRNA tetramers harboring STV-binding aptamers were premixed and preassembled in binding buffer (PBS with 10 mM Mg2+) before incubation with STV Agarose resin (Thermo Scientific). Fifty microliters of STV resin was equilibrated at room temperature and washed with binding buffer. The tetramers harboring STV-binding aptamers (total, ∼4.8 μg) were added to each tube and incubated with the resin for 1 h at room temperature. Tetramers without an STV-binding aptamer served as a negative control. After incubation, the resin was spun at 500g for 1 min, and the supernatant (pass through) was removed. Then, 50 μL of binding buffer was added to the mixture and incubated for 15 min to wash the resin seven times (washes 1–7). RNA was eluted by 5 mM biotin. The samples were analyzed with 6% native PAGE run in TBM buffer and visualized by MG staining.
Activity assay for HBV ribozyme
HBV ribozyme activity was determined, according to the method reported previously (Hoeprich et al. 2003; Shu et al. 2011a). Briefly, HBV RNA substrate was fluorescently labeled with Cy3 (Mirus) and incubated with a pRNA tetramer harboring a HBV ribozyme (1:2 and 1:4 molar ratio). The pRNA/HBV ribozyme served as a positive control (Hoeprich et al. 2003), and a pRNA tetramer harboring a disabled HBV ribozyme was used as a negative control.
In vitro binding and entry of pRNA nanoparticles into targeted cells
KB cells (ATCC) were grown on glass coverslides in FA-free medium overnight. Cy3-labeled pRNA nanoparticles harboring a FA ligand were incubated with the cells for 2 h at 37°C, and imaged by confocal microscopy, according to the method previously reported (Shu et al. 2011a, b). A FA-free pRNA nanoparticle served as a negative control.
Assay for the silencing of luciferase genes in cancer cell model
Gene silencing assays were performed by cotransfecting 1.25 nM pRNA tetramer harboring firefly luciferase siRNA (Nakashima et al. 2011) with both pGL-3 and pRL-TK (Promega) plasmids coding for firefly and Renilla luciferase, respectively. The latter served as an internal control to normalize the luciferase data (Dual-Luciferase Reporter Assay System; Promega). pRNA tetramers harboring scrambled sequences served as a negative control.
Animal trial: in vivo targeting of tumor xenografts by systemic injection of pRNA nanoparticles
NOD/SCID IL2Rγ-null male mice (7 wk old) were maintained on a FA-free diet for 2 wk. The mice were then injected with 5 × 106 KB cells and maintained for 3 wk to allow tumor engraftment. Tumor xenograft was apparent ∼2 wk after injection of KB cells. The mice were then injected through the tail-vein with 250 µM (76 µL) pRNA-X nanoparticles harboring FA and Alexa Fluor 647 (Invitrogen) as a label. The mice were killed 24 h after injection and whole-body imaging was conducted using a MultiSpectral FX (Kodak Carestream) with a multi-angle 2D performance station. Following whole-body imaging, the mice were dissected and the major organs were isolated for imaging.
Temperature gradient gel electrophoresis
For TGGE analysis, the experimental setup was adjusted to have a linear temperature gradient perpendicular to the electric field. The temperature gradient was set from 26.1°C to 68.8°C. A 10 μL RNA sample (∼700 nM) was combined with 2 μL of gel loading buffer and run on 6% native PAGE at 20 W for 1 h. 10 mM MgCl2 was present in both gel and electrophoresis buffer. The gels were stained with ethidium bromide and imaged by Typhoon FLA 7000 (GE Healthcare). The dimer or hexamer fraction within the total RNA was analyzed by ImageJ, and the melting curve of each construct was fitted using nonlinear Sigmoidal fitting.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Emil Khisamutdinov for help with TGGE assays and Jeannie Haak for assistance in manuscript preparation. The research was supported by NIH grants EB003730 and CA151648 and the Arnold and Mabel Beckman Initiative for Macular Research External Grant 1108 to P.G. AFM images were obtained at Nanoimaging Core Facility supported by NIH SIG program and UNMC Program of ENRI to Y.L. P.G. is a cofounder of Kylin Therapeutics, Inc., and Biomotor and Nucleic Acid Nanotechnology Development Corp. Ltd.
REFERENCES
- Abdelmawla S, Guo S, Zhang L, Pulukuri S, Patankar P, Conley P, Trebley J, Guo P, Li QX 2011. Pharmacological characterization of chemically synthesized monomeric pRNA nanoparticles for systemic delivery. Mol Ther 19: 1312–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonin KA, Grabow WW, Walker FM, Bindewald E, Dobrovolskaia MA, Shapiro BA, Jaeger L 2011. Design and self-assembly of siRNA-functionalized RNA nanoparticles for use in automated nanomedicine. Nat Protoc 6: 2022–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altinoglu EI, Russin TJ, Kaiser JM, Barth BM, Eklund PC, Kester M, Adair JH 2008. Near-infrared emitting fluorophore-doped calcium phosphate nanoparticles for in vivo imaging of human breast cancer. ACS Nano 2: 2075–2084 [DOI] [PubMed] [Google Scholar]
- Balzani V, Credi A, Venturi M 2002. The bottom-up approach to molecular-level devices and machines. Chemistry 8: 5524–5532 [DOI] [PubMed] [Google Scholar]
- Bartel DP 2009. MicroRNAs: Target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth BM, Sharma R, Altinoglu EI, Morgan TT, Shanmugavelandy SS, Kaiser JM, McGovern C, Matters GL, Smith JP, Kester M, et al. 2010. Bioconjugation of calcium phosphosilicate composite nanoparticles for selective targeting of human breast and pancreatic cancers in vivo. ACS Nano 4: 1279–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh C, Grate D, Wilson C 2000. 2.8 Å crystal structure of the malachite green aptamer. J Mol Biol 301: 117–128 [DOI] [PubMed] [Google Scholar]
- Bindewald E, Hayes R, Yingling YG, Kasprzak W, Shapiro BA 2008. RNAJunction: A database of RNA junctions and kissing loops for three-dimensional structural analysis and nanodesign. Nucleic Acids Res 36: D392–D397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296: 550–553 [DOI] [PubMed] [Google Scholar]
- Cairns J, Overbaugh J, Miller S 1988. The origin of mutants. Nature 335: 142–145 [DOI] [PubMed] [Google Scholar]
- Carmichael GG 2002. Medicine: Silencing viruses with RNA. Nature 418: 379–380 [DOI] [PubMed] [Google Scholar]
- Chang KY, Tinoco I Jr 1994. Characterization of a “kissing” hairpin complex derived from the human immunodeficiency virus genome. Proc Natl Acad Sci 91: 8705–8709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Zhang C, Guo P 1999. Sequence requirement for hand-in-hand interaction in formation of pRNA dimers and hexamers to gear ϕ29 DNA translocation motor. RNA 5: 805–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Sheng S, Shao Z, Guo P 2000. A dimer as a building block in assembling RNA: A hexamer that gears bacterial virus phi29 DNA-translocating machinery. J Biol Chem 275: 17510–17516 [DOI] [PubMed] [Google Scholar]
- Chowrira BM, Berzal-Herranz A, Burke JM 1991. Novel guanosine requirement for catalysis by the hairpin ribozyme. Nature 354: 320–322 [DOI] [PubMed] [Google Scholar]
- Clever JL, Wong ML, Parslow TG 1996. Requirements for kissing-loop-mediated dimerization of human immunodeficency virus RNA. J Virol 70: 5902–5908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman J, Hirashima A, Inocuchi Y, Green PJ, Inouye M 1985. A novel immune system against bacteriophage infection using complementary RNA (micRNA). Nature 315: 601–603 [DOI] [PubMed] [Google Scholar]
- Duchaine TF, Slack FJ 2009. RNA interference and micro-RNA-oriented therapy in cancer: Rationales, promises, and challenges. Curr Oncol 16: 265–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellington AD, Szostak JW 1990. In vitro selection of RNA molecules that bind specific ligands. Nature 346: 818–822 [DOI] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N, Filipowicz W 2010. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem 79: 351–379 [DOI] [PubMed] [Google Scholar]
- Garver K, Guo P 1997. Boundary of pRNA functional domains and minimum pRNA sequence requirement for specific connector binding and DNA packaging of phage ϕ29. RNA 3: 1068–1079 [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD 2009. Small silencing RNAs: An expanding universe. Nat Rev Genet 10: 94–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold L 1995. The SELEX process: A surprising source of therapeutic and diagnostic compounds. Harvey Lect 91: 47–57 [PubMed] [Google Scholar]
- Grabow WW, Zakrevsky P, Afonin KA, Chworos A, Shapiro BA, Jaeger L 2011. Self-assembling RNA nanorings based on RNAI/II inverse kissing complexes. Nano Lett 11: 878–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P 2010. The emerging field of RNA nanotechnology. Nat Nanotechnol 5: 833–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P, Erickson S, Anderson D 1987. A small viral RNA is required for in vitro packaging of bacteriophage ϕ29 DNA. Science 236: 690–694 [DOI] [PubMed] [Google Scholar]
- Guo P, Zhang C, Chen C, Trottier M, Garver K 1998. Inter-RNA interaction of phage ϕ29 pRNA to form a hexameric complex for viral DNA transportation. Mol Cell 2: 149–155 [DOI] [PubMed] [Google Scholar]
- Guo S, Tschammer N, Mohammed S, Guo P 2005. Specific delivery of therapeutic RNAs to cancer cells via the dimerization mechanism of phi29 motor pRNA. Hum Gene Ther 16: 1097–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Huang F, Guo P 2006. Construction of folate-conjugated pRNA of bacteriophage phi29 DNA packaging motor for delivery of chimeric siRNA to nasopharyngeal carcinoma cells. Gene Ther 13: 814–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P, Coban O, Snead NM, Trebley J, Hoeprich S, Guo S, Shu Y 2010. Engineering RNA for targeted siRNA delivery and medical application. Adv Drug Deliv Rev 62: 650–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P, Haque F, Hallahan B, Reif R, Li H 2012a. Uniqueness, advantages, challenges, solutions, and perspectives in therapeutics applying RNA nanotechnology. Nucleic Acid Ther 22: 226–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P, Shu Y, Binzel D, Cinier M 2012b. Synthesis, conjugation, and labeling of multifunctional pRNA nanoparticles for specific delivery of siRNA, drugs and other therapeutics to target cells. Methods Mol Biol 928: 197–219 [DOI] [PubMed] [Google Scholar]
- Hansma HG, Oroudjev E, Baudrey S, Jaeger L 2003. TectoRNA and “kissing-loop” RNA: Atomic force microscopy of self-assembling RNA structures. J Microsc 212: 273–279 [DOI] [PubMed] [Google Scholar]
- Haque F, Shu D, Shu Y, Shlyakhtenko L, Rychahou P, Evers M, Guo P 2012. Ultrastable synergistic tetravalent RNA nanoparticles for targeting to cancers. Nano Today 7: 245–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Hannon GJ 2004. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet 5: 522–531 [DOI] [PubMed] [Google Scholar]
- Henkin TM 2008. Riboswitch RNAs: Using RNA to sense cellular metabolism. Genes Dev 22: 3383–3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeprich S, Zhou Q, Guo S, Qi G, Wang Y, Guo P 2003. Bacterial virus phi29 pRNA as a hammerhead ribozyme escort to destroy hepatitis B virus. Gene Ther 10: 1258–1267 [DOI] [PubMed] [Google Scholar]
- Jacque JM, Triques K, Stevenson M 2002. Modulation of HIV-1 replication by RNA interference. Nature 418: 435–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger L, Leontis NB 2000. Tecto-RNA: One-dimensional self-assembly through tertiary interactions. Angew Chem Int Ed Engl 39: 2521–2524 [DOI] [PubMed] [Google Scholar]
- Khaled A, Guo S, Li F, Guo P 2005. Controllable self-assembly of nanoparticles for specific delivery of multiple therapeutic molecules to cancer cells using RNA nanotechnology. Nano Letters 5: 1797–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knecht DA, Loomis WF 1987. Antisense RNA inactivation of myosin heavy chain gene expression in Dictyostelium discoideum. Science 236: 1081–1086 [DOI] [PubMed] [Google Scholar]
- Lee RJ, Low PS 1994. Delivery of liposomes into cultured KB cells via folate receptor-mediated endocytosis. J Biol Chem 269: 3198–3204 [PubMed] [Google Scholar]
- Lee H, Lytton-Jean AK, Chen Y, Love KT, Park AI, Karagiannis ED, Sehgal A, Querbes W, Zurenko CS, Jayaraman M, et al. 2012a. Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery. Nat Nanotechnol 7: 389–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JB, Hong J, Bonner DK, Poon Z, Hammond PT 2012b. Self-assembled RNA interference microsponges for efficient siRNA delivery. Nat Mater 11: 316–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Huh MS, Lee SY, Min S, Lee S, Koo H, Chu JU, Lee KE, Jeon H, Choi Y, et al. 2012c. Tumor-homing poly-siRNA/glycol chitosan self-cross-linked nanoparticles for systemic siRNA delivery in cancer treatment. Angew Chem Int Ed Engl 51: 7203–7207 [DOI] [PubMed] [Google Scholar]
- Leontis NB, Lescoute A, Westhof E 2006. The building blocks and motifs of RNA architecture. Curr Opin Struct Biol 16: 279–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Li WX, Ding SW 2002. Induction and suppression of RNA silencing by an animal virus. Science 296: 1319–1321 [DOI] [PubMed] [Google Scholar]
- Lilley DM 2000. Structures of helical junctions in nucleic acids. Q Rev Biophys 33: 109–159 [DOI] [PubMed] [Google Scholar]
- Liu J, Guo S, Cinier M, Shlyakhtenko L, Shu Y, Chen C, Shen G, Guo P 2010. Fabrication of stable and RNase-resistant RNA nanoparticles active in gearing the nanomotors for viral DNA packaging. ACS Nano 5: 237–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Low PS 2002. Folate-mediated delivery of macromolecular anticancer therapeutic agents. Adv Drug Deliv Rev 54: 675–693 [DOI] [PubMed] [Google Scholar]
- Lyubchenko YL, Shlyakhtenko LS 2009. AFM for analysis of structure and dynamics of DNA and protein-DNA complexes. Methods 47: 206–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Sadoqi M, Shao J 2012. Biodistribution of indocyanine green-loaded nanoparticles with surface modifications of PEG and folic acid. Int J Pharm 436: 25–31 [DOI] [PubMed] [Google Scholar]
- Mujeeb A, Clever JL, Billeci TM, James TL, Parslow TG 1998. Structure of the dimer initiation complex of HIV-1 genomic RNA. Nat Struct Biol 5: 432–436 [DOI] [PubMed] [Google Scholar]
- Nakashima Y, Abe H, Abe N, Aikawa K, Ito Y 2011. Branched RNA nanostructures for RNA interference. Chem Commun (Camb) 47: 8367–8369 [DOI] [PubMed] [Google Scholar]
- Nilsen TW 2007. RNA 1997–2007: A remarkable decade of discovery. Mol Cell 28: 715–720 [DOI] [PubMed] [Google Scholar]
- Ogawa A, Maeda M 2008. An artificial aptazyme-based riboswitch and its cascading system in E. coli. Chembiochem 9: 206–209 [DOI] [PubMed] [Google Scholar]
- Ohno H, Kobayashi T, Kabata R, Endo K, Iwasa T, Yoshimura SH, Takeyasu K, Inoue T, Saito H 2011. Synthetic RNA-protein complex shaped like an equilateral triangle. Nat Nanotechnol 6: 116–120 [DOI] [PubMed] [Google Scholar]
- Prats AC, Roy C, Wang PA, Erard M, Housset V, Gabus C, Paoletti C, Darlix JL 1990. cis elements and trans-acting factors involved in dimer formation of murine leukemia virus RNA. J Virol 64: 774–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid RJD, Bodley JW, Anderson D 1994a. Characterization of the prohead-pRNA interaction of bacteriophage phi29. J Biol Chem 269: 5157–5162 [PubMed] [Google Scholar]
- Reid RJD, Bodley JW, Anderson D 1994b. Identification of bacteriophage phi29 prohead RNA (pRNA) domains necessary for in vitro DNA-gp3 packaging. J Biol Chem 269: 9084–9089 [PubMed] [Google Scholar]
- Reid RJD, Zhang F, Benson S, Anderson D 1994c. Probing the structure of bacteriophage phi29 prohead RNA with specific mutations. J Biol Chem 269: 18656–18661 [PubMed] [Google Scholar]
- Reif R, Haque F, Guo P 2013. Fluorogenic RNA nanoparticles for monitoring RNA folding and degradation in real time in living cells. Nucleic Acid Ther 22: 428–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarver N, Cantin EM, Chang PS, Zaia JA, Ladne PA, Stephens DA, Rossi JJ 1990. Ribozymes as potential anti–HIV-1 therapeutic agents. Science 247: 1222–1225 [DOI] [PubMed] [Google Scholar]
- Schmidt OG, Eberl K 2001. Nanotechnology: Thin solid films roll up into nanotubes. Nature 410: 168. [DOI] [PubMed] [Google Scholar]
- Schroeder KT, McPhee SA, Ouellet J, Lilley DM 2010. A structural database for k-turn motifs in RNA. RNA 16: 1463–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman NC, Belcher AM 2002. Emulating biology: Building nanostructures from the bottom up. Proc Natl Acad Sci 99 (Suppl 2): 6451–6455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu D, Huang L, Hoeprich S, Guo P 2003. Construction of phi29 DNA-packaging RNA (pRNA) monomers, dimers, and trimers with variable sizes and shapes as potential parts for nanodevices. J Nanosci Nanotechnol 3: 295–302 [DOI] [PubMed] [Google Scholar]
- Shu D, Moll WD, Deng Z, Mao C, Guo P 2004. Bottom-up assembly of RNA arrays and superstructures as potential parts in nanotechnology. Nano Lett 4: 1717–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu D, Zhang H, Jin J, Guo P 2007. Counting of six pRNAs of phi29 DNA-packaging motor with customized single molecule dual-view system. EMBO J 26: 527–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu D, Shu Y, Haque F, Abdelmawla S, Guo P 2011a. Thermodynamically stable RNA three-way junctions for constructing multifuntional nanoparticles for delivery of therapeutics. Nat Nanotechnol 6: 658–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y, Cinier M, Fox SR, Ben-Johnathan N, Guo P 2011b. Assembly of therapeutic pRNA-siRNA nanoparticles using bipartite approach. Mol Ther 19: 1304–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y, Cinier M, Shu D, Guo P 2011c. Assembly of multifunctional phi29 pRNA nanoparticles for specific delivery of siRNA and other therapeutics to targeted cells. Methods 54: 204–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla GC, Haque F, Tor Y, Wilhelmsson LM, Toulme JJ, Isambert H, Guo P, Rossi JJ, Tenenbaum SA, Shapiro BA 2011. A boost for the emerging field of RNA nanotechnology. ACS Nano 5: 3405–3418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srisawat C, Engelke DR 2001. Streptavidin aptamers: Affinity tags for the study of RNAs and ribonucleoproteins. RNA 7: 632–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel SA, Cochrane JC 2007. RNA catalysis: Ribozymes, ribosomes, and riboswitches. Curr Opin Chem Biol 11: 636–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, Breaker RR 2008. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science 321: 411–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerk C, Gold L 1990. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249: 505–510 [DOI] [PubMed] [Google Scholar]
- Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, et al. 2002. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 419: 624–629 [DOI] [PubMed] [Google Scholar]
- Wagner C, Ehresmann C, Ehresmann B, Brunel C 2004. Mechanism of dimerization of bicoid mRNA: Initiation and stabilization. J Biol Chem 279: 4560–4569 [DOI] [PubMed] [Google Scholar]
- Westhof E 2012. Ribozymes, catalytically active RNA molecules. Introduction. Methods Mol Biol 848: 1–4 [DOI] [PubMed] [Google Scholar]
- Wiedenheft B, Sternberg SH, Doudna JA 2012. RNA-guided genetic silencing systems in bacteria and archaea. Nature 482: 331–338 [DOI] [PubMed] [Google Scholar]
- Wilcox CL, Crnic LS, Pizer LI 1992. Replication, latent infection and reactivation in neuronal culture with a herpes simplex virus thymidine kinase-negative mutant. Virology 187: 348–352 [DOI] [PubMed] [Google Scholar]
- Xiao F, Sun J, Coban O, Schoen P, Wang JC, Cheng RH, Guo P 2009a. Fabrication of massive sheets of single layer patterned arrays using lipid directed reengineered phi29 motor dodecamer. ACS Nano 3: 100–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F, Cai Y, Wang JC, Green D, Cheng RH, Demeler B, Guo P 2009b. Adjustable ellipsoid nanoparticles assembled from re-engineered connectors of the bacteriophage phi29 DNA packaging motor. ACS Nano 3: 2163–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F, Demeler B, Guo P 2010. Assembly mechanism of the sixty-subunit nanoparticles via interaction of RNA with the reengineered protein connector of phi29 DNA-packaging motor. ACS Nano 4: 3293–3301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yingling YG, Shapiro BA 2007. Computational design of an RNA hexagonal nanoring and an RNA nanotube. Nano Lett 7: 2328–2334 [DOI] [PubMed] [Google Scholar]
- Zhang CL, Lee C-S, Guo P 1994. The proximate 5′ and 3′ ends of the 120-base viral RNA (pRNA) are crucial for the packaging of bacteriophage f29 DNA. Virology 201: 77–85 [DOI] [PubMed] [Google Scholar]
- Zuker M 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.