Deep-sequencing of the small RNAs in two life stages of parasitic protozoan Trypanosoma brucei shows that the small RNAs can derive from various sources. Those generated from rRNAs and tRNAs are differentially expressed in the two life cycles studied.
Keywords: Trypanosoma brucei, high-throughput data analysis, small noncoding RNA, comparative transcriptome
Abstract
Trypanosoma brucei, a pathogen of human and domestic animals, is an early evolved parasitic protozoan with a complex life cycle. Most genes of this parasite are post-transcriptionally regulated. However, the mechanisms and the molecules involved remain largely unknown. We have deep-sequenced the small RNAs of two life stages of this parasite—the bloodstream form and the procyclic form. Our results show that the small RNAs of T. brucei could derive from multiple sources, including NATs (natural antisense transcripts), tRNAs, and rRNAs. Most of these small RNAs in the two stages were found to share uniform characteristics. However, our results demonstrate that their variety and expression show significant differences between different stages, indicating possible functional differentiation. Dicer-knockdown evidence further proved that some of the small interfering RNAs (siRNAs) could regulate the expression of genes. Based on the genome-wide analysis of the small RNAs in the two stages of T. brucei, our results not only provide evidence to study their differentiation but also shed light on questions regarding the origins and evolution of small RNA-based mechanisms in early eukaryotes.
INTRODUCTION
Trypanosoma brucei is an evolutionarily ancient protozoan that causes African sleeping sickness in humans and Nagana disease in animals (Pays et al. 2006; Brun et al. 2010). It has a complex life cycle with different stages in the blood-sucking insect vector (procyclic form [PF]) and in the mammalian hosts (slender form [SF]) (Siegel et al. 2010). During its life cycle, T. brucei needs to undergo substantial changes to adapt to different hosts (environments). These changes are orchestrated by specific gene expression-regulated mechanisms. Most genes of T. brucei are organized in long polycistronic units but lack the control of RNA polymerase II promoters (Borst 2002; Landfear et al. 2004). Therefore, post-transcriptional gene expression regulation plays important roles in this parasite’s development and differentiation (Clayton 2002; Berriman et al. 2005).
Noncoding RNAs, especially small RNAs, have been demonstrated to be important molecules in post-transcriptional regulation (Eddy 2001). MicroRNAs (miRNAs) and small interfering RNAs (siRNAs) have been considered the two prominent ones. miRNAs are 18–22 nt in length and originate from single-stranded precursors that form ∼70-nt hairpin structures with a few bulged nucleotides (Carrington and Ambros 2003; He and Hannon 2004). siRNAs are 21 to 26 nt in length (Hamilton et al. 2002) and are processed by the Dicer protein from long double-stranded RNAs (dsRNAs) (Golden et al. 2008). Both miRNAs and siRNAs can regulate gene expression by cleaving the corresponding mRNAs (perfect matching) or blocking their translation (nonperfect matching) by the enzyme Argonaute (Moazed 2009). miRNAs are prevalent in higher eukaryotes and have crucial impacts on development and differentiation (Nilsen 2008). In protozoa, however, the presence of miRNAs is still uncertain. It has been reported that miRNA-like molecules could be identified in some species of protozoa (Saraiya and Wang 2008; Braun et al. 2010; Huang et al. 2012), but many studies reported that the conserved miRNA or miRNA-like molecules could not be found in the protozoa examined (Grimson et al. 2008; Xue et al. 2008; Atayde et al. 2011). In contrast, siRNAs are ubiquitous and abundant in many protozoa (Robinson and Beverley 2003; Ullu et al. 2005; Zhang et al. 2008; Patrick et al. 2009; Lye et al. 2010; Tschudi et al. 2012), suggesting that siRNAs, but not miRNAs, may be the key molecules in the RNA interference (RNAi) pathway in protozoan species.
Trypanosoma brucei is one of the most ancient eukaryotes in which RNAi has been discovered and extensively studied (Patrick et al. 2009). It has been reported that endo-siRNAs in T. brucei could be derived from retroposons and repeat sequences to control retroposon transcript abundance (Djikeng et al. 2001). Our previous study revealed that in T. brucei, siRNAs derived from dsRNAs by pseudogenes and their homologous genes could regulate the expression of the parental gene (Wen et al. 2011). Recently, Tschudi et al. systematically investigated siRNAs from procyclic form and demonstrated that they were processed by two individual Dicer proteins (Tschudi et al. 2012). These results suggested that siRNAs in T. brucei might play important roles in post-transcriptional regulation.
In addition to miRNAs and siRNAs, ribosomal RNA-derived small RNAs also have been reported to play important roles. For example, in a collection of ∼1300 RITS (RNA-induced transcriptional silencing)-associated small RNAs, ∼30% of them could be mapped to rDNA (Cam et al. 2005). However, fragments of the abundant rRNAs are present in nearly all small RNA sequence libraries (Buhler et al. 2008). Early investigations suggested that these small RNAs were randomly degraded products, but more and more studies have revealed that they are functional molecules processed by special enzymes (Lee et al. 2009a; Li et al. 2012).
In recent years, a new kind of small RNA derived from tRNA (tRNA-derived fragments [tRFs]) has been identified in multiple species (Buhler et al. 2008; Li et al. 2008b; Cole et al. 2009; Lee et al. 2009b; Garcia-Silva et al. 2010; Peng et al. 2012). Increasing experimental evidence revealed that tRFs were usually synthesized when the cells or organisms were under stress, particularly in Trypanosoma cruzi (Garcia-Silva et al. 2010; Franzen et al. 2011). However, these tRFs could not be demosntrated in T. brucei, and the small RNAs derived from T. brucei tRNAs were considered as degradation products (Tschudi et al. 2012). The main difference between the two species is that RNAi is absent in T. cruzi but exists in T. brucei. These results raise the following question: Are tRFs the alternate mechanism for the RNAi pathway in T. cruzi?
To get a comprehensive knowledge of these small noncodingRNAs (ncRNAs) in T. brucei, we sequenced the small RNAs (≤30 nt) of the two stages (SF and PF) of T. brucei with high-throughput technology and analyzed various small ncRNAs at the genome level, then compared these small RNAs between the two stages.
RESULTS
Small RNAs in the slender form and procyclic form
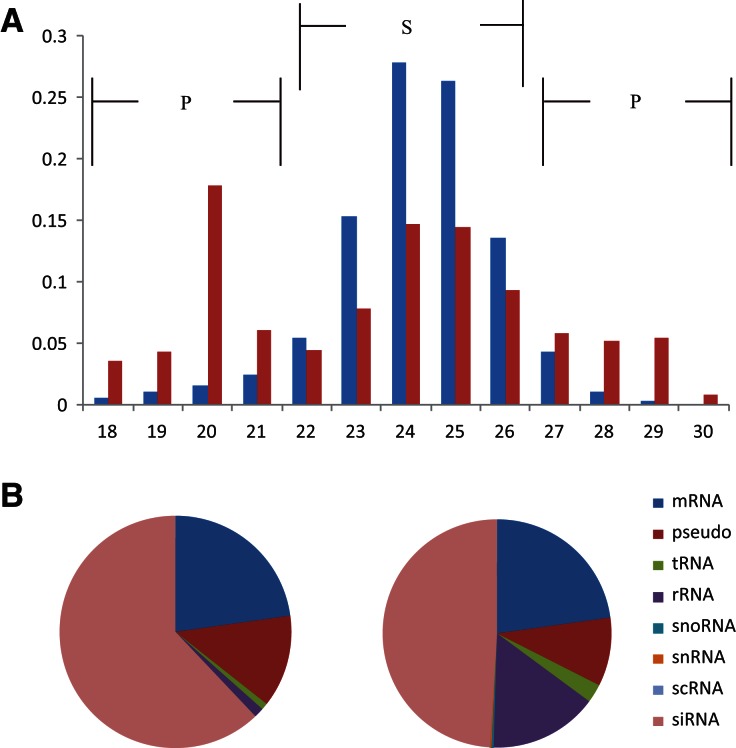
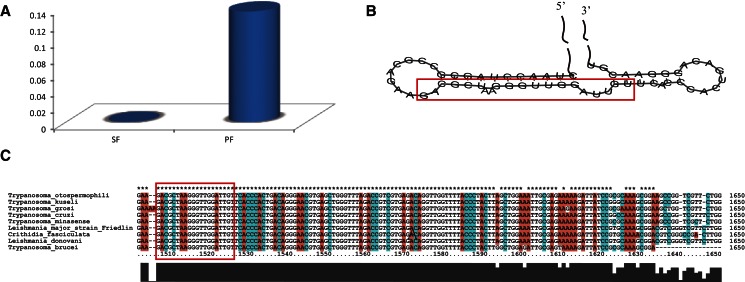
In total, 1,695,819 and 615,415 unique small RNAs were obtained from the slender and the procyclic forms, respectively (Table 1). Further analysis revealed that 1,751,956 unique small RNAs in these two forms could be mapped to the genome of T. brucei with two mismatches allowed. However, 67.6% of them (1,184,292/1,751,956) in the slender form, but only 24.1% (422,178/1,751,956) in the procyclic form, are stage-specific. A list of small RNAs that were demonstrated in both stages and showing differential expression (fold-change >2 and P value <0.01) is given in Supplemental Table S1. Also, the lengths of small RNAs are strikingly different between the two stages. Most small RNAs in the slender form are ∼23–26 nt, while in the procyclic form, the length of small RNAs is clustered in two ranges: 18–21 nt and 28–30 nt (Fig. 1A). Furthermore, interestingly, we found that the small RNAs could be derived from multiple sources and that the expression of siRNAs, rRNA-derived small RNAs, and tRNA-derived small RNAs are dramatically different in the two stages (Fig. 1B).
TABLE 1.
General view of small RNAs
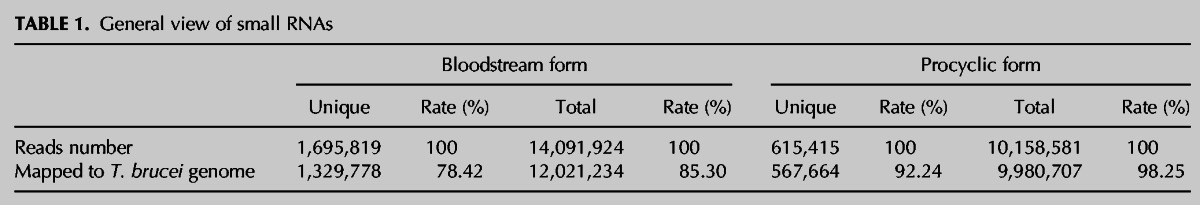
FIGURE 1.
Length and sources of total small RNA sequences in two stages. (A) The length distribution of total small RNAs in two stages. Blue bars represent the SF stage and red bars represent the PF stage. Region S lines show the size of small RNAs expressed more in the SF stage; P is for the PF stage. (B) The sources of small RNAs in two stages; left is the SF stage and right is the PF stage.
Small interfering RNA
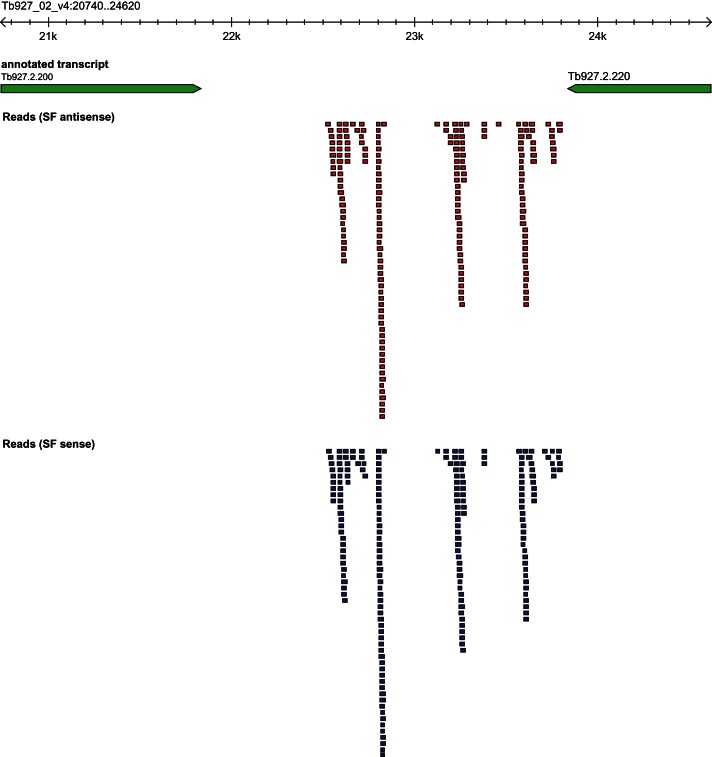
It has been known that siRNAs in T. brucei can be generated from transposable elements (TEs), including INGI, SLACS, and CIR147 (Djikeng et al. 2001; Patrick et al. 2009). In addition, results from a recent interesting study on siRNAs in the procyclic form of T. brucei revealed that siRNAs could originate from inverted repeats (IRs) and convergent transcription units (CTUs) (Tschudi et al. 2012). In the current study, we performed systematic inspection of siRNAs from these sources in the two stages of T. brucei and found that the siRNAs in the slender form and the procyclic form were derived from consistent sources, but the expressions were different between the two stages (Table 2). In addition, we found that siRNAs could also be derived from other sources. For example, siRNAs could be generated from the intergenic region in T. brucei (Fig. 2), similarly as reported in Drosophila (Okamura et al. 2008) and Arabidopsis (Borsani et al. 2005). This result indicates that the sources of siRNAs in T. brucei are more diverse than previously thought and may be similar to those found in higher organisms. Accumulating evidence from animals and plants has shown that natural antisense transcripts (NATs) can produce siRNAs (Tam et al. 2008; Watanabe et al. 2008; Guo et al. 2009; Zhou et al. 2009). Therefore, we have also investigated NAT-derived siRNAs in T. brucei.
TABLE 2.
Summary of siRNAs in SF and PF of T. brucei
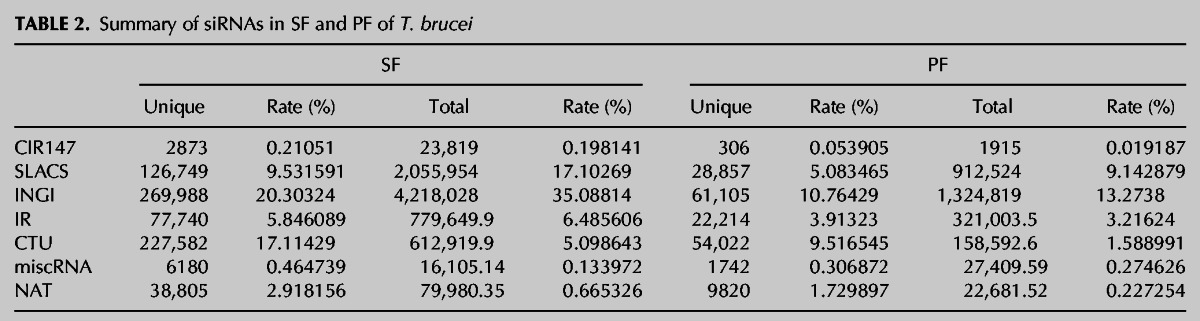
FIGURE 2.
An example of small RNAs derived from an intergenic region. The region on Tb927_02_v4 from 20,522 to 24,620 contains two protein coding genes (green arrows), Tb927.2.200 and Tb927.2.220, mapped by inverted overlapping small RNAs expressed in the intergenic region. The short segments indicate the siRNA pairs, and the colors represent their transcription direction (blue for sense and red for antisense).
NATs are a source of siRNAs in T. brucei
Once two RNAs are coexpressed in the same cell under the same conditions, they can form sense-antisense dsRNAs by complementary bases and generate siRNAs (NAT-siRNAs) (Zhou et al. 2009). In a recent study, we found that in T. brucei pseudogenes and their parental genes could form NATs and generate siRNAs (Wen et al. 2011). Herein, based on annotated transcripts and sequencing data, we identified all potential NAT pairs and their siRNA products in T. brucei (see Materials and Methods).
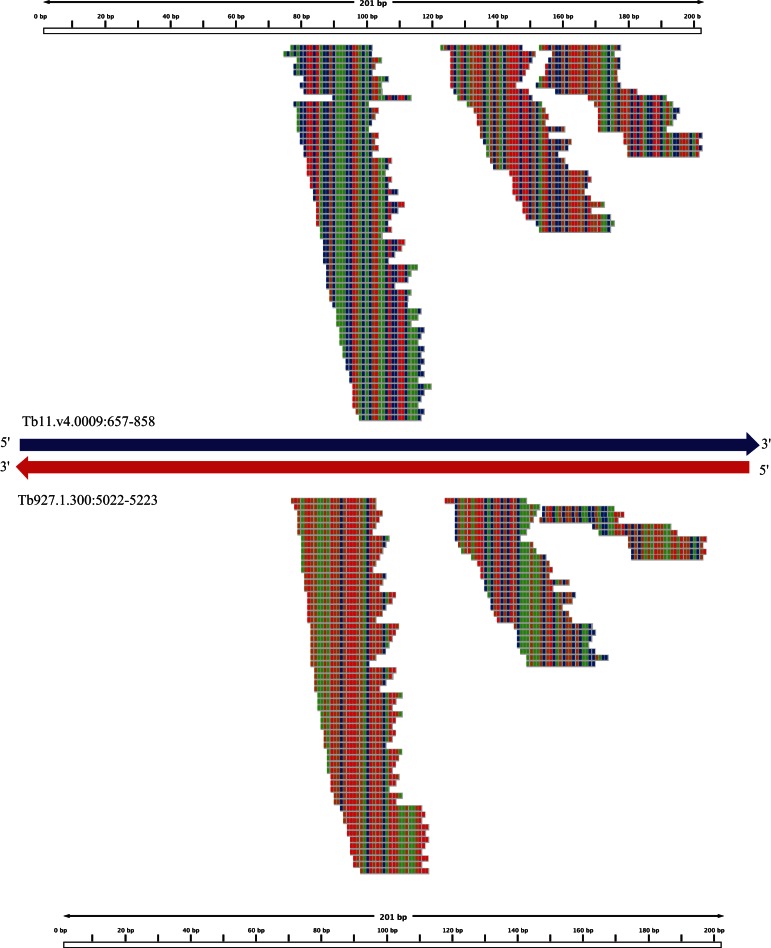
In total, 182 NAT pairs were identified based on the transcriptome of T. brucei (see Materials and Methods). Of these, 20.9% were composed of two coding genes, while 79.1% were composed of at least one noncoding gene, such as a pseudogene, snoRNA, snRNA, tRNA, and repeat elements. NATs can be divided into two types: cis-NATs, in which the two transcripts derive from the same genomic locus but opposite strands, and trans-NATs, in which the transcripts derive from different genomic loci (Zhou et al. 2009). Further analysis of 182 NATs reveals that 11 of them are cis-NATs and the others are trans- NATs (Supplemental Table S2). In the 182 pairs of NATs, most of them were found to generate small RNAs (136 in SF and 144 in PF) (Supplemental Table S2). For example, in the slender form, the transcripts of a VSG pseudogene (Tb11.v4.0009) and an RHS pseudogene (Tb927.1.300) were found to form a representative trans-NAT pair and to produce abundant small RNAs (Fig. 3). We then applied the raw data of TbAGO1-associated small RNAs in the procyclic form generated from a previous study (Tschudi et al. 2012). The results indicated that 147 NATs pairs were produced and that 134 of them are the same as those found from our procyclic data set (Supplemental Table S3). This result indicates that the methods to identify NAT-siRNAs are valid.
FIGURE 3.
Examples of small RNAs derived from a NAT pair. Long arrows indicate two NAT transcripts: Blue represents the sense transcript and red represents the antisense one; arrows point in the direction of transcription (5′ to 3′). The locations of small RNAs mapped to the overlap regions of the two transcripts were drawn by Integrative Genomics Viewer (IGV) (Thorvaldsdóttir et al. 2013). Thin lines above and below the arrows display the reads originated from sense and antisense, respectively. The colors indicate nucleotide type in the transcripts (green, A; blue, C; yellow, G; red, U).
The features of siRNAs in T. brucei
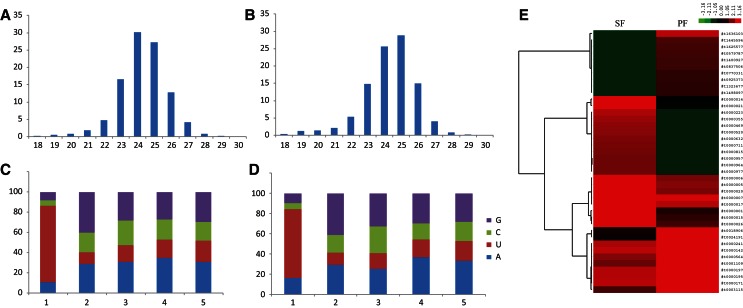
The majority of siRNAs found are ∼23–26 nt, and >60% preferred “U” as their 5′-terminal nucleotide (Fig. 4). The features of siRNAs in the two stages are similar; however, their expressions are different. Based on our analysis, 78.93% of the siRNAs were only identified in the slender form, whereas 13.8% were specifically expressed in the procyclic form. The other siRNAs that expressed in both stages also showed significant expression differences (Supplemental Table S4). In particular, 74.2% and 25.8% of these siRNAs had significantly higher expression in the slender form and in the procyclic form, respectively. Figure 4E shows the 10 most expressed and the 10 least expressed siRNAs in each stage, respectively. Results from the analysis of siRNA in the two stages of T. brucei demonstrated that the variety and quantity of the endogenous siRNAs were more abundant in the slender form than in the procyclic form.
FIGURE 4.
Features of all predicted siRNAs in the two life stages. (A,B) Length distribution of predicted siRNAs in SF (A) and PF (B). (C,D) 5′ end nucleotide of siRNAs in SF (C) and PF (D). (E) The 10 small RNAs expressed the most and the 10 small RNAs expressed the least in the two stages. A normalized method was used according to the following formula: normalized reads abundance = log10(C/N × 106), where C represents the number of siRNA reads and N represents the total number of mapped reads in the library.
siRNAs can regulate gene expression and target different genes in different stages
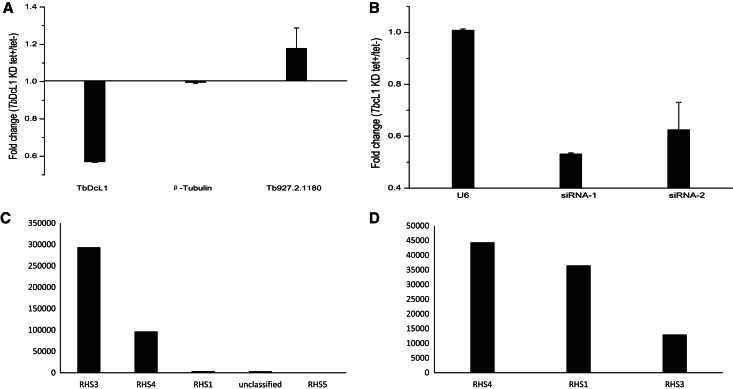
To test whether siRNAs could regulate gene expression, real-time PCR was performed on Dicer knock-down slender cells (Wen et al. 2011). For example, the transcript of Tb927.2.1180, which corresponds to the RHS3 gene and is targeted by most siRNAs, was up-regulated after Dicer knock-down (Fig. 5A). Correspondingly, the expression of siRNA-1, predicted to cleave Tb927.2.1180, was down-regulated (Fig. 5B). The results further show that siRNAs can regulate gene expression and are consistent with our previous study on pseudogene-derived siRNAs (Wen et al. 2011). Results from our analysis indicate that RHS genes are targeted by a great number of siRNAs. The RHS family includes six genes, RHS1 to RHS6, defined by their divergent C-terminal domains (Bringaud et al. 2002). In the slender form, most siRNAs were found to target RHS3, followed by RHS4, RHS1, and RHS5 (Fig. 5C). In the procyclic form, however, the RHS4 gene was targeted by most siRNAs, followed by RHS1 and RHS3 (Fig. 5D). Furthermore, 84 and 61 genes were targeted by stage-specific siRNAs found in the slender and procyclic form, respectively (Supplemental Tables S5, S6). These results indicate that the genes are not equally suppressed by siRNAs in different stages of T. brucei.
FIGURE 5.
Quantitative RT-PCR analysis of RNA levels in TbDCL1 knockdown cell lines. (A) mRNA level changes in conditional TbDCL1 knockdown cell lines (tet+) compared to noninduced cells (tet−). Housekeeping β-tubulin genes were used as an external control. Error bars (s.d.) are shown (n = 3). (B) Fold-change of selective NAT-siRNAs in conditional TbDCL1 knockdown cell lines (tet+) compared to noninduced cells (tet−); U6 small RNAs added to both induced and noninduced cell lines serve as an external control. Error bars (s.d.) are shown (n = 3). The sequences of siRNAs are listed in Supplemental Table S7. (C,D) Quantity of siRNA which targeted to the genes of the RHS family in SF (C) and PF (D).
rRNA-derived small RNAs
Data from our analysis in the procyclic form indicated that a great portion of differentially expressed small RNAs were derived from rRNA (Figs. 1B, 6A), and most of them are 20 nt long (Supplemental Fig. S1A). It was surprising to note that one of the small RNAs, which was found in both stages but its expression was dramatically increased in the procyclic form, accounted for 13.5% (1,370,060 of 10,158,581) of all sequencing small RNAs in this form (Table 3). Our results clearly demonstrated that it was derived from the hairpin structure at the 3′ end of the 28S rRNA (Fig. 6B). By comparison with the flanking sequences, we found that large numbers of small RNAs were mainly generated from this region (Supplemental Fig. S1B). After comparison with eight other species genetically related to T. brucei, we found that the sequences of this specific region in 28S rRNA are the same (Fig. 6C). The stage specificity and high expressivity, the restricted derivation of the region, and the high source conservation indicate that the rRNA-derived small RNAs may be functional.
TABLE 3.
Number of 20-nt small RNA derived from rRNA
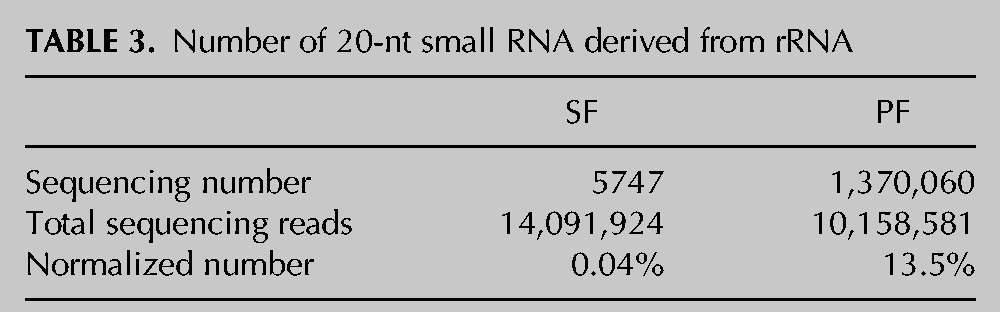
FIGURE 6.
rRNA-derived small RNA. (A) The expression difference of rRNA-derived small RNA in SF and PF. (B) The secondary structure of rRNA which generated small RNA. (C) Multiple alignment of rRNAs among nine species.
tRNA-derived small RNAs
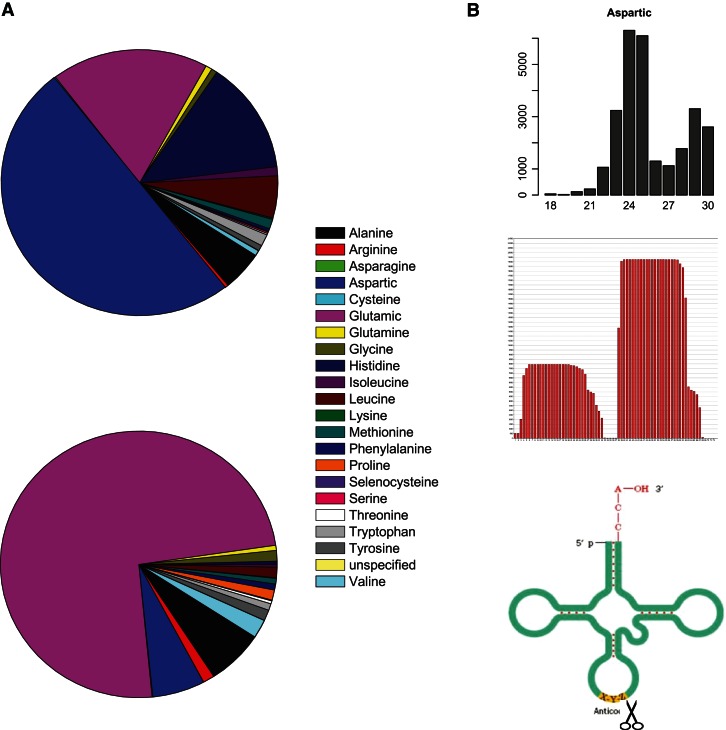
There are 66 tRNA genes in the genome of T. brucei which encode 22 tRNAs, including 20 conventional ones, one newly identified (selenocystein), and one nonspecific (Tb09.160.1075). After comparison with the small RNAs observed in the two stages, we found that the expression of tRNA-derived small RNAs was different. For instance, nearly half of the small RNAs were derived from tRNAAsp in the slender form, while in the procyclic form, the quantity was reduced to 6%. In contrast, tRNAGlu-derived small RNAs were mainly found in the procyclic form (74%), compared to 18% found in the slender form (Fig. 7A). Furthermore, we also found that small RNAs in the two stages were generated from specific locations of tRNAs repetitively (Supplemental Figs. S2, S3). For example, in the slender form, tRNAAsp could generate small RNAs in two clusters with different lengths (24 nt and 30 nt). The 30-nt fragments were mapped to the 5′ end of the tRNA, and the 24-nt fragments were derived from the 3′ end. However, no reads were mapped to the other parts of the tRNAAsp (Fig. 7B; Supplemental Fig. S2). These results indicate that the small RNAs derived from the tRNAs are unlikely to be the randomly degraded segments but are rather products carved by specific enzymes and may play biological functions in different stages of the life cycle.
FIGURE 7.
tRNA-derived small RNA. (A) Small RNAs derived from different types of tRNAs. The top is SF stage, and the bottom is PF stage. (B) An example of small RNAs in the slender form derived from tRNAAsp in relation to its secondary structure. The top is the length of tRNAAsp-derived small RNAs. The middle is the position of small RNA mapped to the sequence of tRNAAsp. The bottom is the sketch of tRNA structure and the cleavage site where small RNAs derived from.
DISCUSSION
It is well known that RNA interference is an important mechanism to control gene expression. However, the origin and evolution of RNAi remain an unsolved mystery (Okamura and Lai 2008). It has been considered that the origin of miRNAs might be independent in plants and animals (Grimson et al. 2008; Campo-Paysaa et al. 2011). However, data from the analysis of the features of siRNAs from various species (Hamilton et al. 2002; Ambros et al. 2003; Zilberman et al. 2003) have indicated that siRNAs in plants and animals share some similar characteristics (Ghildiyal and Zamore 2009; Smalheiser et al. 2011; Song et al. 2011). In order to understand whether siRNAs share similar features in eukaryotes, it is necessary to analyze the siRNAs in lower organisms. Thus, as a single cell, T. brucei is an ideal representative model for us to compare the categories, characteristics, and functions of siRNAs with those in other higher organisms.
It is well understood that endogenous siRNAs can derive from various sources. In animals, transposable elements, complementary annealed transcripts, and hairpin RNAs all can produce siRNAs (Okamura and Lai 2008). In plants, natural antisense siRNAs (NAT-siRNAs) (Borsani et al. 2005), trans-acting short interfering RNAs (tasiRNAs) (Axtell et al. 2006), heterochromatic siRNAs (Buhler et al. 2007), and long siRNAs (lsiRNAs) (Katiyar-Agarwal et al. 2007) have been reported to be sources of siRNAs. In the protozoan T. brucei, our current data demonstrate that siRNAs could originate from multiple sources (Table 2), similar to those found in higher eukaryotes. The length of most siRNAs in higher eukaryotes ranges from 21 nt to 26 nt (Hamilton et al. 2002), while in T. brucei, it ranges from 23 to 26 nt (Fig. 4A,B). Endo-siRNAs in animals and plants show various 5′-terminal nucleotides which are important for sorting into different AGO complexes (Mi et al. 2008). In T. brucei, the majority of endo-siRNAs prefer “U” as their 5′-terminal nucleotide (Fig. 4C,D), which might be corresponding to a single AGO (Shi et al. 2007). Results have shown that siRNAs in T. brucei play critical roles in genomic stability (Mi et al. 2008). Our results further demonstrate that siRNAs in T. brucei could also regulate gene expression at the post-transcriptional level (Fig. 5). Therefore, the critical roles of siRNAs should be considered to be conserved in all eukaryotes. As a matter of fact, based on previous results (Djikeng et al. 2001; Patrick et al. 2009; Tschudi et al. 2012) and our published data (Wen et al. 2011), the categories, characteristics, and functions of endo-siRNAs have been proved to be generally homologous in all eukaryotes.
The phenomenon of natural antisense transcripts is abundant in higher eukaryotes. In the human genome, 4%–9% of transcript pairs are overlapped, while in the murine genome, 1.7%–14% have been reported (Lehner et al. 2002; Shendure and Church 2002; Zhou et al. 2009). However, their prevalence in protozoa has not been well investigated. This study is the first systematic analysis of NAT pairs and NAT-siRNAs at a genome scale, using comprehensive deep-sequencing results in T. brucei. Hundreds of NAT pairs were identified. We found, in general, that when two transcripts in cis-NATs were from the same locus, each one of them was specifically complementary to the other. However, two complementary transcripts from different loci could also be involved in trans-NAT. Thus, it is suggested that one transcript is able to pair with several other transcripts to form a complex regulatory network (Zhou et al. 2009). Based on this consideration, we constructed the topology networks. A network was composed of several subnetworks, in which transcripts share similar functions. For instance, one subnetwork in a star structure is composed of 11 transcripts (Supplemental Fig. S4A), all of which are members of the variant surface glycoprotein (VSG) family, either a VSG gene or a VSG pseudogene. This indicates that VSG transcripts may form multiple sense-antisense RNA duplexes in T. brucei.
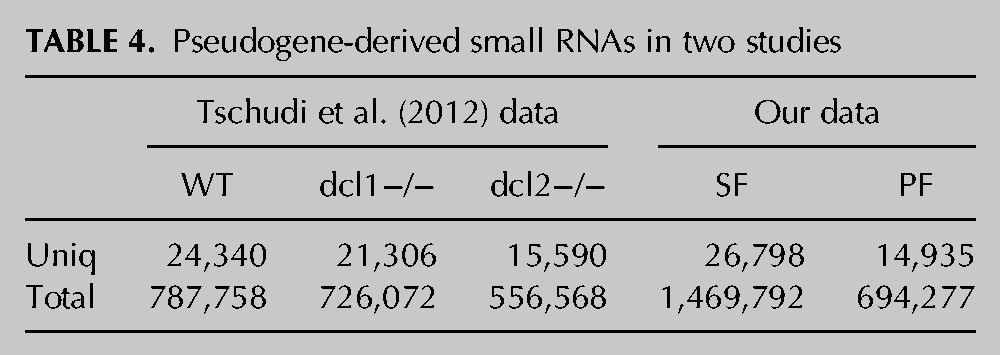
It is particularly interesting to know that there are many pseudogenes in the genome of T. brucei (10%) (Berriman et al. 2005). It has been reported that these pseudogenes are associated with the recombination of chromosomes (Thon et al. 1989). We have demonstrated that pseudogenes play important regulatory functions in T. brucei (Wen et al. 2011, 2012). In the present study, we have further demonstrated that pseudogenes could form a complex regulatory network with other transcripts in the NAT manner and could generate siRNAs, although this phenomenon was not observed by Tschudi and colleagues (Tschudi et al. 2012), who have suggested that the majority of pseudogenes do not generate small RNAs in the procyclic form of T. brucei. The reason for this difference might be the different mapping parameters used in the two studies. Because the sequences of pseudogenes and their parental genes are very similar, it is, thus, hard to identify a small RNA mapping to multiple loci. To avoid this problem, we used strict parameters on both our sequencing library and the other data sets (SRA057341) to identify the pseudogene-derived siRNAs (see Materials and Methods). As a result, tens of thousands of pseudogene-derived small RNAs were identified in all data sets of T. brucei from both studies (Table 4). These results indicate that siRNAs from pseudogenes could, indeed, derive in both stages of T. brucei.
TABLE 4.
Pseudogene-derived small RNAs in two studies
Based on the analysis for the targets of siRNA (Supplemental Tables S5, S6), our results further indicate that both RHS and VSG families are potentially regulated by siRNAs. Evidence suggests that the RHS family consists of at least six different RHS genes and a variable number of pseudogenes (Bringaud et al. 2002). In addition, it has been reported that the absence of AGO, a key enzyme in the RNAi pathway, led to an increase of steady-state transcripts of RHS genes and pseudogenes (Durand-Dubief et al. 2007). In our study, we found that a great number of siRNAs were targeted to the RHS family. This might explain why the RHS family was under the control of RNA interference (Wen et al. 2011). Furthermore, the siRNAs genes targeted to the RHS family were found to display obvious differences between the slender form and the procyclic form of T. brucei (Fig. 5). Given their genomic location on the subtelomeric regions, these results support the hypothesis that RHS proteins might be involved in antigenic variation (Pays 2005). Therefore, we suggest that the siRNAs may play critical roles in the antigenic variation in T. brucei.
VSG is an important antigen in the slender form of T. brucei. The extensive variety of this antigen allows the parasite to escape the immunological defense of the mammalian hosts. At any given time, only one VSG could be detected on the surface of the parasite (Vanhamme and Pays 1995). This indicates that, before novel immunological factors (antibodies) are produced in the mammalian hosts, the VSG in a few individuals of T. brucei has switched to a novel one, so that the parasites can escape the immunological reaction of the host in time. By comparison with the procyclic form, we found that some genes in the VSG family were specifically regulated by siRNAs in the slender form (Supplemental Table S5). Therefore, we propose that the siRNAs correspondingly evolve to eliminate the obsolete VSG transcripts as rapidly as possible, so that the novel VSG can be effective in a timely way. This might be similar to what is found in Giardia lamblia in which the expression of variant specific surface genes is under the control of RNA interference (Prucca et al. 2008).
RNAi has been shown to be functional in T. brucei but not found in two other trypanosomatids, i.e., Leishmania major and Trypanosoma cruzi (Robinson and Beverley 2003). However, the biological roles of RNAi in T. brucei had not been fully revealed (Owino et al. 2008). It was reported that RNAi in T. brucei is not essential for survival, either in the bloodstream or in the procyclic form (Janzen et al. 2006). Therefore, it has been considered that RNAi in this parasite might be of relatively minor significance. However, based on the analysis of the diverse categories, different expression in the two stages, gene regulatory function, and targeting to particular gene families, our current study has clearly demonstrated that the endo-siRNAs play biological roles in T. brucei. We suggest that the RNAi role in T. brucei might have been underestimated and should be reevaluated in light of new data.
It has been well known that miRNAs are important regulators in multicellular organisms. For example, in humans, up to 30% of the genes are regulated by miRNAs (John et al. 2004; Lewis et al. 2005). Interestingly, however, no miRNAs have been recorded in protozoan species, on the basis of the miRBase release 19.0 (Kozomara and Griffiths-Jones 2011). Although some reports have demonstrated miRNA-like molecules in protozoa, their types and abundance were much lower than those found in higher eukaryotes. Based on the analysis of our deep-sequenced data, we did not find typical miRNA molecules in the slender form or in the procyclic form of T. brucei, although bioinformatic analysis of the genome had predicted their existence (Mallick et al. 2008). We speculated that miRNAs in T. brucei might be too rare to be detected or even be absent in these stages. In contrast, however, a large number of siRNAs were found in the two stages of T. brucei, which have also been shown to play special regulatory roles in this parasite (Wen et al. 2011). Therefore, we suggest that siRNAs instead of miRNAs might significantly contribute to RNAi pathways in protozoan species.
In T. brucei, we found that rRNAs could also generate some small RNAs, ∼20 nt long, with “G” as the preferred nucleotide at the 5′-terminal end. Interestingly, these small RNAs were dramatically highly expressed in the procyclic form. This might indicate that they could be produced by some specific enzymes and are functional. rRNA-derived small RNAs have been reported in Neurospora crassa with damaged DNA. They are ∼20–21 nt long, with a preference for uridine at the 5′-terminal end, and mostly originate from 18S, 5.8S, and 26S rDNA. They are processed by an argonaute protein QDE-2 and called qiRNAs (Lee et al. 2009a). QDE is a protein of the AGO family that has been considered to be associated with post-transcriptional gene silencing (Fagard et al. 2000). In T. brucei, only TbAGO1 has been identified (Durand-Dubief and Bastin 2003). Analysis of the data for the procyclic form from another study (Tschudi et al. 2012) shows affinity of purified TbAGO1 from procyclic trypanosomes. The results show that the expression number of 20-nt rRNA-derived small RNA is very low and does not change greatly after knockout of the TbDCL1 or TbDCL2, two other key RNases in RNA interference in this parasite. This suggests that the synthesis of rRNA-derived small RNAs in T. brucei might be dependent on other mechanisms instead of on the RNA interference pathway. Moreover, these rRNA fragments are present in both stages of T. brucei but extremely highly expressed in the procyclic form; this might be closely related to the energy metabolism in this form or even be associated with the differentiation of T. brucei. More investigations will be required for revealing the important functions of rRNA-derived small RNAs.
tRNA-derived fragments are evolutionarily conserved molecules that have been reported in various species. Our results in T. brucei are consistent with that conclusion in this parasite and in other organisms and support the idea that tRNA-derived small RNAs are not degradation products but a novel class of small RNAs (Li et al. 2008b; Lee et al. 2009b; Garcia-Silva et al. 2010; Peng et al. 2012). The expression of tRFs was considered to be in response to physiological stress or nutritional deficiency (Li et al. 2008b; Lee et al. 2009b; Peng et al. 2012). tRFs have been discovered in Giardia (Li et al. 2008b) and T. cruzi (Garcia-Silva et al. 2010). In the two protozoan species, these small RNAs have been shown to be expressed more when the cells were under nutritional deficiency. In our study, we found that tRNAGlu produced more small RNAs in the procyclic form, while tRNAAsp-derived small RNAs were expressed more in the slender form. This indicates that the small RNA derived from tRNA might play roles in differentiation in the life cycle of this parasite. In contrast to T. cruzi, in which the RNAi pathway is absent, in T. brucei, although the RNAi pathway is functional, we found that the expression of tRNAAsp-derived small RNAs did not change after Dicer knockdown (data not shown). It seems that both in T. cruzi and T. brucei, tRFs are independent of the RNAi pathway, although the mechanisms and molecules involved in tRF regulation are far from being understood.
In conclusion, our results show that the siRNAs in T. brucei can derive from different sources and are able to regulate gene expression at the post-transcriptional level. Also, rRNA and tRNA can generate small RNAs with expression dramatically different in different stages of the life cycle. This clearly shows the presence of small RNAs in T. brucei and indicates their likely prevalence in other protozoan species.
MATERIALS AND METHODS
Trypanosome cultivation
Trypanosoma brucei brucei STIB 920 slender forms were isolated from infected mice blood collected at the peak parasitaemia by DEAE cellulose (DE-52, Whatman) (Lanham and Godfrey 1970). Some of these slender form cells were cultivated in HMI-11 medium at 37°C with 5% CO2 and 95% air. After subculture several times, the cells were transferred to TVM-1 medium and incubated at 27°C, where trypanosomes were differentiated into the procyclic form. The slender form and procyclic form cells were both abundantly collected and suspended in TRIZOL (Invitrogen). The TbDCL1 knockdown clones obtained have been described in a previous study (Wen et al. 2011).
Data sources
Eleven megabase-sized chromosomes and scaffold sequences of T. brucei were downloaded from the TriTrypDB (release-2.1, update time: 04-Mar-2010 15:23), one of EuPathDB (http://tritrypdb.org/tritrypdb/) (Aslett et al. 2010). Kinetoplast DNA maxi-circle sequences were downloaded from NCBI GenBank number M94286.1. Transcript sequences and annotation information were downloaded from TriTrypDB and rearranged by a Perl script. Ingi repeat elements were downloaded from NCBI (accession number X05710) and Spliced-Leader Associated Conserved Sequence (SLACS) was retrieved from NCBI (accession number X17078); the CIR147 sequence was obtained from previous reports (Kritikou 2008). The small RNAs used for comparison, sequenced by Dr. Elisabetta Ullu (Yale University), were downloaded from the NCBI Sequence Read Archive (SRA) at http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi, under accession no. SRA057341. Other sequences for comparison were obtained from the same article and rearranged by a Perl script (Tschudi et al. 2012).
Small RNA deep-sequencing and analysis
Total RNAs were extracted from trypanosome cells by TRIZOL, following the manufacturer’s protocol, and run on agarose gels for quality control. Small RNA library preparation and Solexa sequencing were performed by the Beijing Genomics Institute (BGI) in ShenZhen, Guangdong, China. sRNAs ranging from 18 to 30 nt were gel-purified and ligated to the 3′ adaptor (5′-pUCGUAUGCCGUCUUCUGCUUGidT-3′;p,phosphate;idT,inverted deoxythy midine) and 5′ adaptor (5′-GUUCAGAGUUCUACAGUCCGACGAUC-3′). Ligation products were gel-purified, reverse-transcribed, and amplified using Illumina’s sRNA primer set (5′-CAAGCAGAAGACGGCATACGA-3′; 5′-AATGATACGGCGACCACCGA-3′). Samples were sequenced on an Illumina 1G Genome Analyzer. The adaptor sequences were removed from the Illumina-generated reads at BGI Shenzhen using a dynamic programming algorithm that required at least a 5-nt overlap between 35-nt reads and the 3′ adaptor sequence. After removing the reads without the adaptor sequences, poly-A reads, and 5′ adaptor contaminants, the remaining 18- to 30-nt reads were mapped to the T. brucei genome assembly from TritrypDB (Aslett et al. 2010) using bowtie with two mismatches allowed (Langmead and Salzberg 2012). The mapped reads were then annotated against known RNA transcripts. All reads in the “mapped” data set were compiled into a set of unique sequences, with the number of reads for each sequence reflecting relative abundance (Liao et al. 2010). The read count of each unique sequence was normalized to reads per million (RPM), according to the following formula: RPM = C/N × 106, where C represents the number of sequencing small RNA reads and N represents the total number of mapped reads.
NATs and NAT-derived siRNAs identification
Cis-NAT pairs were identified by the location of transcripts in the genome. If two transcripts were transcribed from the opposite strands of the same locus and the overlap region was longer than 20 bp, a value also adopted in other studies (Werner and Berdal 2005), then they were considered to form a pair of cis-NATs.
Trans-NAT pairs were identified by pairwise alignment (Altschul et al. 1990). If a pair of transcripts from different genomic loci could form an RNA-RNA duplex with a BLAST (Li et al. 2008a) E-value less than 10−9, they were considered to form a candidate trans-NAT pair. For each NAT, we computed the density of small RNA loci in the overlapping region by the same method used in another study (Guo et al. 2009). In brief, small RNAs were respectively mapped to the sense and antisense transcripts of a NAT pair, and the number of small RNAs in the complementary region of the NAT were calculated. The number was then divided by the length of the complementary region to get the density. If the densities of both sense and antisense strands were more than 0.04, this region was considered to have the ability to generate NAT-siRNAs.
siRNA pairs identification
The features of siRNA biogenesis are different from the other small RNAs (Allen et al. 2005). Using these features as criteria, we developed an approach for strict siRNA pairs identification via high-throughput sequence results. The pairs of siRNAs should meet three conditions: (1) The reads are perfectly mapped to the T. brucei genome; (2) the two reads are of the same length; and (3) the two reads can form a duplex with 2-nt overhangs at the end of the 3′ terminus. Examples are shown in Figures 2 and 3 and Supplemental Fig. S4B.
Identification of the expression of small RNAs
The Bayesian test method was used to determine the expression difference between the two stages. The approach was based on the comparison of tag counts generated from digital expression analyses (Audic and Claverie 1997). The small RNAs, whose fold-change between two stages was more than 2 and the P-value <0.01, were considered to be expressed significantly differently. We drew a heat map of the small RNAs (Fig. 4E), which expressed the 10 most abundant and the 10 least abundant in each stage by using the following formula: normalized reads abundance = log10(C/N × 106), where C represented the number of siRNA reads and N represented the total number of mapped reads.
tRNA and rRNA-derived small RNA
tRNA sequences were retrieved from TritrypDB. Small RNAs were mapped to these tRNAs with up to two mismatches allowed. Each type of tRNA that generated small RNAs was normalized by the number of tRNA copies of this type. rRNA sequences were downloaded from the SILVA rRNA database (Pruesse et al. 2007). Clustalx (Thompson et al. 1997) was used for multiple alignments of rRNAs.
Pseudogene-derived small RNA
Sequenced small RNAs were mapped to all the annotated mRNAs in T. brucei with no mismatches. All these mapped reads were removed from the data set, and the remaining reads were mapped to the annotated pseudogenes; only the sense mapped reads were kept. These small RNAs were believed to be pseudogene-derived small RNAs.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
This work was supported by Grant (2011CB811300) from the National Basic Research program (“973” program) to L.-H.Q., by Grants from the National Natural Science Foundation of China (#31071995) and Sun Yat-Sen University (12lgjc11) to Z.-R.L., and China Postdoctoral Science Foundation (2012M510204) to Y.-Z.W. The authors thank Dr. E. Ullu of Yale University for providing the adaptor sequences of their sequencing experiments for our comparative analysis and the many people from our laboratories who provided valuable comments and suggestions.
REFERENCES
- Allen E, Xie Z, Gustafson AM, Carrington JC 2005. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ 1990. Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D 2003. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr Biol 13: 807–818 [DOI] [PubMed] [Google Scholar]
- Aslett M, Aurrecoechea C, Berriman M, Brestelli J, Brunk BP, Carrington M, Depledge DP, Fischer S, Gajria B, Gao X, et al. 2010. TriTrypDB: A functional genomic resource for the Trypanosomatidae. Nucleic Acids Res 38(Database issue): D457–D462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atayde VD, Tschudi C, Ullu E 2011. The emerging world of small silencing RNAs in protozoan parasites. Trends Parasitol 27: 321–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audic S, Claverie JM 1997. The significance of digital gene expression profiles. Genome Res 7: 986–995 [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Jan C, Rajagopalan R, Bartel DP 2006. A two-hit trigger for siRNA biogenesis in plants. Cell 127: 565–577 [DOI] [PubMed] [Google Scholar]
- Berriman M, Ghedin E, Hertz-Fowler C, Blandin G, Renauld H, Bartholomeu DC, Lennard NJ, Caler E, Hamlin NE, Haas B, et al. 2005. The genome of the African trypanosome Trypanosoma brucei. Science 309: 416–422 [DOI] [PubMed] [Google Scholar]
- Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK 2005. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123: 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borst P 2002. Antigenic variation and allelic exclusion. Cell 109: 5–8 [DOI] [PubMed] [Google Scholar]
- Braun L, Cannella D, Ortet P, Barakat M, Sautel CF, Kieffer S, Garin J, Bastien O, Voinnet O, Hakimi MA 2010. A complex small RNA repertoire is generated by a plant/fungal-like machinery and effected by a metazoan-like Argonaute in the single-cell human parasite Toxoplasma gondii. PLoS Pathog 6: e1000920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringaud F, Biteau N, Melville SE, Hez S, El-Sayed NM, Leech V, Berriman M, Hall N, Donelson JE, Baltz T 2002. A new, expressed multigene family containing a hot spot for insertion of retroelements is associated with polymorphic subtelomeric regions of Trypanosoma brucei. Eukaryot Cell 1: 137–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun R, Blum J, Chappuis F, Burri C 2010. Human African trypanosomiasis. Lancet 375: 148–159 [DOI] [PubMed] [Google Scholar]
- Buhler M, Haas W, Gygi SP, Moazed D 2007. RNAi-dependent and -independent RNA turnover mechanisms contribute to heterochromatic gene silencing. Cell 129: 707–721 [DOI] [PubMed] [Google Scholar]
- Buhler M, Spies N, Bartel DP, Moazed D 2008. TRAMP-mediated RNA surveillance prevents spurious entry of RNAs into the Schizosaccharomyces pombe siRNA pathway. Nat Struct Mol Biol 15: 1015–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cam HP, Sugiyama T, Chen ES, Chen X, FitzGerald PC, Grewal SI 2005. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat Genet 37: 809–819 [DOI] [PubMed] [Google Scholar]
- Campo-Paysaa F, Semon M, Cameron RA, Peterson KJ, Schubert M 2011. microRNA complements in deuterostomes: Origin and evolution of microRNAs. Evol Dev 13: 15–27 [DOI] [PubMed] [Google Scholar]
- Carrington JC, Ambros V 2003. Role of microRNAs in plant and animal development. Science 301: 336–338 [DOI] [PubMed] [Google Scholar]
- Clayton CE 2002. Life without transcriptional control? From fly to man and back again. EMBO J 21: 1881–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JW, Green PJ, Barton GJ, Hutvagner G 2009. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA 15: 2147–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djikeng A, Shi H, Tschudi C, Ullu E 2001. RNA interference in Trypanosoma brucei: Cloning of small interfering RNAs provides evidence for retroposon-derived 24–26-nucleotide RNAs. RNA 7: 1522–1530 [PMC free article] [PubMed] [Google Scholar]
- Durand-Dubief M, Bastin P 2003. TbAGO1, an argonaute protein required for RNA interference, is involved in mitosis and chromosome segregation in Trypanosoma brucei. BMC Biol 1: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand-Dubief M, Absalon S, Menzer L, Ngwabyt S, Ersfeld K, Bastin P 2007. The Argonaute protein TbAGO1 contributes to large and mini-chromosome segregation and is required for control of RIME retroposons and RHS pseudogene-associated transcripts. Mol Biochem Parasitol 156: 144–153 [DOI] [PubMed] [Google Scholar]
- Eddy SR 2001. Non-coding RNA genes and the modern RNA world. Nat Rev Genet 2: 919–929 [DOI] [PubMed] [Google Scholar]
- Fagard M, Boutet S, Morel JB, Bellini C, Vaucheret H 2000. AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc Natl Acad Sci 97: 11650–11654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzen O, Arner E, Ferella M, Nilsson D, Respuela P, Carninci P, Hayashizaki Y, Aslund L, Andersson B, Daub CO 2011. The short non-coding transcriptome of the protozoan parasite Trypanosoma cruzi. PLoS Negl Trop Dis 5: e1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Silva MR, Frugier M, Tosar JP, Correa-Dominguez A, Ronalte-Alves L, Parodi-Talice A, Rovira C, Robello C, Goldenberg S, Cayota A 2010. A population of tRNA-derived small RNAs is actively produced in Trypanosoma cruzi and recruited to specific cytoplasmic granules. Mol Biochem Parasitol 171: 64–73 [DOI] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD 2009. Small silencing RNAs: An expanding universe. Nat Rev Genet 10: 94–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden DE, Gerbasi VR, Sontheimer EJ 2008. An inside job for siRNAs. Mol Cell 31: 309–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, King N, Degnan BM, Rokhsar DS, Bartel DP 2008. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature 455: 1193–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Zhang Z, Gerstein MB, Zheng D 2009. Small RNAs originated from pseudogenes: cis- or trans-acting? PLoS Comput Biol 5: e1000449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton A, Voinnet O, Chappell L, Baulcombe D 2002. Two classes of short interfering RNA in RNA silencing. EMBO J 21: 4671–4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Hannon GJ 2004. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet 5: 522–531 [DOI] [PubMed] [Google Scholar]
- Huang PJ, Lin WC, Chen SC, Lin YH, Sun CH, Lyu PC, Tang P 2012. Identification of putative miRNAs from the deep-branching unicellular flagellates. Genomics 99: 101–107 [DOI] [PubMed] [Google Scholar]
- Janzen CJ, van Deursen F, Shi H, Cross GA, Matthews KR, Ullu E 2006. Expression site silencing and life-cycle progression appear normal in Argonaute1-deficient Trypanosoma brucei. Mol Biochem Parasitol 149: 102–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS 2004. Human microRNA targets. PLoS Biol 2: e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Gao S, Vivian-Smith A, Jin H 2007. A novel class of bacteria-induced small RNAs in Arabidopsis. Genes Dev 21: 3123–3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S 2011. miRBase: Integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res 39(Database issue): D152–D157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kritikou E 2008. Small RNAs: Endo-siRNAs truly endogenous. Nat Rev Mol Cell Biol 9: 426–427 [Google Scholar]
- Landfear SM, Ullman B, Carter NS, Sanchez MA 2004. Nucleoside and nucleobase transporters in parasitic protozoa. Eukaryot Cell 3: 245–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanham SM, Godfrey DG 1970. Isolation of salivarian trypanosomes from man and other mammals using DEAE-cellulose. Exp Parasitol 28: 521–534 [DOI] [PubMed] [Google Scholar]
- Lee H-C, Chang S-S, Choudhary S, Aalto AP, Maiti M, Bamford DH, Liu Y 2009a. qiRNA is a new type of small interfering RNA induced by DNA damage. Nature 459: 274–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Shibata Y, Malhotra A, Dutta A 2009b. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev 23: 2639–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner B, Williams G, Campbell RD, Sanderson CM 2002. Antisense transcripts in the human genome. Trends Genet 18: 63–65 [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20 [DOI] [PubMed] [Google Scholar]
- Li JT, Zhang Y, Kong L, Liu QR, Wei L 2008a. Trans-natural antisense transcripts including noncoding RNAs in 10 species: Implications for expression regulation. Nucleic Acids Res 36: 4833–4844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Luo J, Zhou H, Liao JY, Ma LM, Chen YQ, Qu LH 2008b. Stress-induced tRNA-derived RNAs: A novel class of small RNAs in the primitive eukaryote Giardia lamblia. Nucleic Acids Res 36: 6048–6055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Ender C, Meister G, Moore PS, Chang Y, John B 2012. Extensive terminal and asymmetric processing of small RNAs from rRNAs, snoRNAs, snRNAs, and tRNAs. Nucleic Acids Res 40: 6787–6799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao JY, Ma LM, Guo YH, Zhang YC, Zhou H, Shao P, Chen YQ, Qu LH 2010. Deep sequencing of human nuclear and cytoplasmic small RNAs reveals an unexpectedly complex subcellular distribution of miRNAs and tRNA 3′ trailers. PLoS One 5: e10563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lye LF, Owens K, Shi H, Murta SM, Vieira AC, Turco SJ, Tschudi C, Ullu E, Beverley SM 2010. Retention and loss of RNA interference pathways in trypanosomatid protozoans. PLoS Pathog 6: e1001161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallick B, Ghosh Z, Chakrabarti J 2008. MicroRNA switches in Trypanosoma brucei. Biochem Biophys Res Commun 372: 459–463 [DOI] [PubMed] [Google Scholar]
- Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, Wu L, Li S, Zhou H, Long C, et al. 2008. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133: 116–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D 2009. Small RNAs in transcriptional gene silencing and genome defence. Nature 457: 413–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen TW 2008. Endo-siRNAs: Yet another layer of complexity in RNA silencing. Nat Struct Mol Biol 15: 546–548 [DOI] [PubMed] [Google Scholar]
- Okamura K, Lai EC 2008. Endogenous small interfering RNAs in animals. Nat Rev Mol Cell Biol 9: 673–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Balla S, Martin R, Liu N, Lai EC 2008. Two distinct mechanisms generate endogenous siRNAs from bidirectional transcription in Drosophila melanogaster. Nat Struct Mol Biol 15: 581–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owino VA, Masiga DK, Limo MK 2008. RNA interference: A pathway to drug target identification and validation in trypanosomes. Afr J Biochem Res 2: 66–73 [Google Scholar]
- Patrick KL, Shi H, Kolev NG, Ersfeld K, Tschudi C, Ullu E 2009. Distinct and overlapping roles for two Dicer-like proteins in the RNA interference pathways of the ancient eukaryote Trypanosoma brucei. Proc Natl Acad Sci 106: 17933–17938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pays E 2005. Regulation of antigen gene expression in Trypanosoma brucei. Trends Parasitol 21: 517–520 [DOI] [PubMed] [Google Scholar]
- Pays E, Vanhollebeke B, Vanhamme L, Paturiaux-Hanocq F, Nolan DP, Perez-Morga D 2006. The trypanolytic factor of human serum. Nat Rev Microbiol 4: 477–486 [DOI] [PubMed] [Google Scholar]
- Peng H, Shi J, Zhang Y, Zhang H, Liao S, Li W, Lei L, Han C, Ning L, Cao Y, et al. 2012. A novel class of tRNA-derived small RNAs extremely enriched in mature mouse sperm. Cell Res 22: 1609–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prucca CG, Slavin I, Quiroga R, Elias EV, Rivero FD, Saura A, Carranza PG, Lujan HD 2008. Antigenic variation in Giardia lamblia is regulated by RNA interference. Nature 456: 750–754 [DOI] [PubMed] [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glockner FO 2007. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35: 7188–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson KA, Beverley SM 2003. Improvements in transfection efficiency and tests of RNA interference (RNAi) approaches in the protozoan parasite Leishmania. Mol Biochem Parasitol 128: 217–228 [DOI] [PubMed] [Google Scholar]
- Saraiya AA, Wang CC 2008. snoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS Pathog 4: e1000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shendure J, Church GM 2002. Computational discovery of sense-antisense transcription in the human and mouse genomes. Genome Biol 3: RESEARCH0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Tschudi C, Ullu E 2007. Depletion of newly synthesized Argonaute1 impairs the RNAi response in Trypanosoma brucei. RNA 13: 1132–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel TN, Hekstra DR, Wang X, Dewell S, Cross GA 2010. Genome-wide analysis of mRNA abundance in two life-cycle stages of Trypanosoma brucei and identification of splicing and polyadenylation sites. Nucleic Acids Res 38: 4946–4957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalheiser NR, Lugli G, Thimmapuram J, Cook EH, Larson J 2011. Endogenous siRNAs and noncoding RNA-derived small RNAs are expressed in adult mouse hippocampus and are up-regulated in olfactory discrimination training. RNA 17: 166–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song R, Hennig GW, Wu Q, Jose C, Zheng H, Yan W 2011. Male germ cells express abundant endogenous siRNAs. Proc Natl Acad Sci 108: 13159–13164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, et al. 2008. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 453: 534–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG 1997. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thon G, Baltz T, Eisen H 1989. Antigenic diversity by the recombination of pseudogenes. Genes Dev 3: 1247–1254 [DOI] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, Mesirov JP 2013. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief Bioinform 14: 178–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschudi C, Shi H, Franklin JB, Ullu E 2012. Small interfering RNA-producing loci in the ancient parasitic eukaryote Trypanosoma brucei. BMC Genomics 13: 427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullu E, Lujan HD, Tschudi C 2005. Small sense and antisense RNAs derived from a telomeric retroposon family in Giardia intestinalis. Eukaryot Cell 4: 1155–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhamme L, Pays E 1995. Control of gene expression in trypanosomes. Microbiol Rev 59: 223–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al. 2008. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 453: 539–543 [DOI] [PubMed] [Google Scholar]
- Wen YZ, Zheng LL, Liao JY, Wang MH, Wei Y, Guo XM, Qu LH, Ayala FJ, Lun ZR 2011. Pseudogene-derived small interference RNAs regulate gene expression in African Trypanosoma brucei. Proc Natl Acad Sci 108: 8345–8350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen YZ, Zheng LL, Qu LH, Ayala FJ, Lun ZR 2012. Pseudogenes are not pseudo any more. RNA Biol 9: 27–32 [DOI] [PubMed] [Google Scholar]
- Werner A, Berdal A 2005. Natural antisense transcripts: Sound or silence? Physiol Genomics 23: 125–131 [DOI] [PubMed] [Google Scholar]
- Xue X, Zhang Q, Huang Y, Feng L, Pan W 2008. No miRNA were found in Plasmodium and the ones identified in erythrocytes could not be correlated with infection. Malar J 7: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Ehrenkaufer GM, Pompey JM, Hackney JA, Singh U 2008. Small RNAs with 5′-polyphosphate termini associate with a Piwi-related protein and regulate gene expression in the single-celled eukaryote Entamoeba histolytica. PLoS Pathog 4: e1000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Sunkar R, Jin H, Zhu JK, Zhang W 2009. Genome-wide identification and analysis of small RNAs originated from natural antisense transcripts in Oryza sativa. Genome Res 19: 70–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Cao X, Jacobsen SE 2003. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299: 716–719 [DOI] [PubMed] [Google Scholar]