AML1 (RUNX1) is a key transcription factor for hematopoiesis. RNA aptamers that bind specifically to the AML1 Runt domain were obtained. All the isolated aptamers contain the conserved sequence motif 5′-NNCCAC-3′ and 5′-GCGMGN′N′-3′ (M:A or C; N and N′ form Watson–Crick base pairs). Three guanines of the motif are directly recognized by three arginine residues of the Runt domain.
Keywords: RNA aptamer, AML1/RUNX1, NMR, SELEX
Abstract
AML1 (RUNX1) is a key transcription factor for hematopoiesis that binds to the Runt-binding double-stranded DNA element (RDE) of target genes through its N-terminal Runt domain. Aberrations in the AML1 gene are frequently found in human leukemia. To better understand AML1 and its potential utility for diagnosis and therapy, we obtained RNA aptamers that bind specifically to the AML1 Runt domain. Enzymatic probing and NMR analyses revealed that Apt1-S, which is a truncated variant of one of the aptamers, has a CACG tetraloop and two stem regions separated by an internal loop. All the isolated aptamers were found to contain the conserved sequence motif 5′-NNCCAC-3′ and 5′-GCGMGN′N′-3′ (M:A or C; N and N′ form Watson–Crick base pairs). The motif contains one AC mismatch and one base bulged out. Mutational analysis of Apt1-S showed that three guanines of the motif are important for Runt binding as are the three guanines of RDE, which are directly recognized by three arginine residues of the Runt domain. Mutational analyses of the Runt domain revealed that the amino acid residues used for Apt1-S binding were similar to those used for RDE binding. Furthermore, the aptamer competed with RDE for binding to the Runt domain in vitro. These results demonstrated that the Runt domain of the AML1 protein binds to the motif of the aptamer that mimics DNA. Our findings should provide new insights into RNA function and utility in both basic and applied sciences.
INTRODUCTION
AML1 (RUNX1) is an α subunit of the core binding factor (CBF), a heterodimeric transcription factor that binds to the core elements of many enhancers and promoters (de Bruijn and Speck 2004; Collins et al. 2009). Three α subunits (RUNX1–3; RUNX proteins) and one β subunit (CBFβ) are encoded in mammals. RUNX proteins contain a DNA-binding domain (DBD), known as the Runt domain, which recognizes a specific DNA element. CBFβ, which does not bind DNA itself, binds to the Runt domain and leads to increased DNA-binding affinity.
AML1 was originally isolated from a chromosomal break point in human acute leukemia (Miyoshi et al. 1991). Subsequently, AML1 was determined to be essential for definitive hematopoiesis during development and to play important roles in maintaining hematopoiesis and immune function in adults (Okuda et al. 1996; North et al. 2004). Several genetic aberrations that disrupt the function of AML1/CBFβ are frequently observed in human leukemia (Mitelman et al. 2007; Engel and Hiebert 2010). However, the precise function of AML1/CBFβ remains unknown. Tertiary structures of the Runt domain and the Runt/CBFβ heterodimeric complex have been investigated by X-ray crystallography and NMR (Nagata et al. 1999; Bravo et al. 2001; Tahirov et al. 2001; Backstrom et al. 2002). These studies revealed that the Runt domain folds primarily into a β-barrel structure classified as an s-type immunoglobulin (Ig) fold. This structure has homology to other DBDs such as those found in p53, NF-κB, NFAT, and other proteins. The Runt domain recognizes the DNA consensus sequence “YGYGGTY” (where Y = pyrimidine) (Melnikova et al. 1993) using two loop-containing regions and a C-terminal tail, which clamp around the sugar–phosphate backbone between the major and minor grooves. The C-terminal tail and one of the loop regions interact with the major groove. The three critical guanines are recognized by three arginine residues that form hydrogen bonds in the major groove. Another loop region interacts with the minor groove. Complex formation is further supported by several interactions between the side chains and backbone of the Runt domain and the sugar–phosphate backbone of DNA.
We obtained RNA aptamers that bind specifically to the AML1 protein to use as tools for better understanding AML1 and its potential utility for the diagnosis and treatment of AML1-related diseases. Aptamers are molecules that can bind with high affinity and specificity to a wide range of target molecules (Patel and Suri 2000; Oguro et al. 2003; Miyakawa et al. 2006, 2008; Ohuchi et al. 2006; Tanaka et al. 2007; Endo and Nakamura 2010). They can be generated by an in vitro selection process, known as systematic evolution of ligands by exponential enrichment or SELEX (Ellington and Szostak 1990; Tuerk and Gold 1990). We performed SELEX to obtain RNA aptamers against the Runt domain of AML1. We found that all the selected aptamers possessed a unique RNA motif and that one of these aptamers likely binds to the Runt domain in a manner similar to the DNA consensus binding sequence. The results of SELEX provided novel insight that the Runt domain could bind to RNA, with conserved sequence specificity, as well as to DNA.
RESULTS
Isolation of RNA aptamers that bind to the AML1 Runt domain
Two RNA selection experiments against the Runt domain (amino acid residues 1–188 of AML1), which contains the DBD, were performed using initial RNA pools of either 30 or 40 random nucleotide positions, referred to as the 30N and 40N RNA pools, respectively. Following nine rounds of selection, the 30N and 40N random RNA pools converged into eight and four independent sequences, respectively (Fig. 1A). Apt1 and Apt41 were the most frequently observed aptamers. All these aptamers contained the conserved sequence motif 5′-GCGMG-3′ and 5′-CCAC-3′ (M:A or C), although the order of the two sequences varied. The 5′-CCAC-3′ was located upstream of 5′-GCGMG-3′ in five sequences, including Apt1, among the 40 independent sequences analyzed. When 5′-CCAC-3′ was located upstream, the length of the spacer sequence was 4–6 nucleotides (nt), and when 5′-GCGMG-3′ was located upstream, the length of the spacer sequence was 3–5 nt. We also noted that 2 nt at the position 5′-adjacent to 5′-CCAC-3′ was always complementary to 2 nt at the position 3′-adjacent to 5′-GCGMG-3′, although the base sequences were not conserved (“NN” and “N′N′,” respectively). Therefore, the consensus sequences are 5′-NNCCAC-(4–6 nt)-GCGMGN′N′-3′ or 5′-GCGMGN′N′-(3–5 nt)-NNCCAC-3′ (Fig. 1B).
FIGURE 1.
Runt-binding aptamer sequences. (A) RNA sequences selected from 30N and 40N randomized RNA libraries. Parental 30N or 40N sequences contained 5′- and 3′-constant sequences for primer annealing. The frequency of each sequence is shown in the right column as a percentage of the 60 sequences analyzed. (Red) Consensus sequences of aptamers. (B) The consensus sequences. M designates A or C; N designates any base; and N′ designates a base complementary to N. (Red) Conserved bases.
Enzymatic probing analysis of the secondary structure of Apt1
To determine sequence requirements for AML1–Runt binding, the secondary structure of Apt1 was predicted using RNAstructure 4.4 software (http://rna.chem.rochester.edu/). The predicted structure with the lowest free energy was a hairpin structure comprising one large apical loop, two small internal loops, three stem regions between the loops, and an extended 3′-single-stranded region (Fig. 2A). The consensus sequences are located in the apical loop structure.
FIGURE 2.

Enzymatic analysis of the secondary structure of Apt1. (A) The secondary structure of Apt1 predicted by RNAstructure 4.4. (Boldface) The 5′- and 3′-constant sequences of Apt1 for primer annealing; (red) the consensus sequences. The cleavage points of RNase V1 (open arrowhead), RNase T1 (solid arrowhead), and mung bean nuclease (striped arrowhead) and points uncleaved by RNase T1 (gray arrowhead) are indicated on the secondary structure of Apt1. The arrowhead length indicates the signal intensity of the digested fragments. (Dotted line) A predicted base pair. The signals >A55 were too strong to identify individual signals (lane 6 of panel B); therefore, the cleavage points by mung bean nuclease are not indicated from C56 to the 3′ terminus. (B) Autoradiogram of the RNase probing pattern of Apt1. The 5′-[32P]-labeled Apt1 (lane 2) was partially digested with alkali (lane 1), RNase T1 in the presence of 7 M urea (lane 3), RNase T1 (lane 4), RNase V1 (lane 5), and mung bean nuclease (lane 6). Digests were separated using denaturing polyacrylamide gel electrophoresis. (Left side of the autoradiogram) The positions of G-residues. The bar indicates the position of the full-length Apt1 (63-mer). (Right side) The estimated bases digested by RNase V1 and mung bean nuclease.
To analyze the secondary structure of Apt1, enzymatic probing was performed using single-strand-specific RNase T1 and mung bean nuclease as well as RNase V1, which preferentially cleaves double-stranded or otherwise structured regions. The results of RNase cleavage of 5′-[32P]-labeled Apt1 are shown in Figure 2B, and the summary of the results of enzymatic probing is shown in Figure 2A. Partial alkaline digests of 5′-labeled Apt1 were used as size markers (Fig. 2B, lane 1). Under denaturing conditions with 7 M urea, RNase T1 digested all the G residues of Apt1 (Fig. 2B, lane 3). However, G15, G28, G30, G31, G41, and (weakly) G11 and G12 were protected from digestion under native conditions (Fig. 2B, lane 4), suggesting that these G residues were base-pairing.
Fragments digested by RNase V1 and mung bean nuclease are known to retard migration in denaturing polyacrylamide gel electrophoresis (PAGE) by ∼1 nt because of their 3′-terminal hydroxyl compared with fragments digested by alkaline digestion and RNase T1 with a 3′-terminal phosphate (Fig. 2B, lanes 5,6; Favorova et al. 1981; Brown and Bevilacqua 2005). RNase V1 digested at least 16 sites in Apt1; however, it is known to recognize U residues in both single and double strands (Sobczak et al. 2010). Therefore, C7, A8, A9, A13, C18, C19, G31, C37, A38, C43, C44, and C45 were confirmed to be double stranded. Mung bean nuclease, which preferentially digests ApN in single strands, strongly digested at A23 and the 3′-terminal region beyond A55, and weakly digested at A8, C22, C24, G25, A47, G50, C53, and G54 in Apt1.
These results indicate that nucleotides from C22 to G26 are in a single strand, whereas C19 and G28 are in a double strand in the predicted apical loop. This finding suggests that the two conserved sequences of Apt1, 5′-ACCCAC-3′ and 5′-GCGAGGU-3′, are anti-parallel to each other and base pairs are formed between them, at least A16–U32, C17–G31, C18–G30, and C19–G28. Furthermore, G15 is in a double strand in the predicted internal loop. Thus, it is possible that G15 is stacked with the adjacent bases, which results in inefficient cleavage by RNase T1. As predicted, nucleotides from A47 to the 3′ terminus were shown to be single stranded (Fig. 2A). Enzymatic probing analysis revealed that Apt1 forms a stem–loop structure with a single-stranded tail, which is in agreement with the predicted secondary structure, and an additional base pair is likely formed involving the conserved motif within the apical loop structure.
Truncation of Apt1 to a 38-mer of Apt1-S
To characterize the aptamers, a truncated variant of Apt1, Apt1-S, was constructed using secondary structure information obtained from the enzymatic probing mentioned above. Apt1-S comprises the stem–loop from the 9th to the 39th nucleotide of Apt1, the 5′-GGG for effective transcription initiation by T7 RNA polymerase, and a 3′-CCCA for stabilizing the stem–loop structure, resulting in a length of 38 nt, which is 25 nt shorter than Apt1.
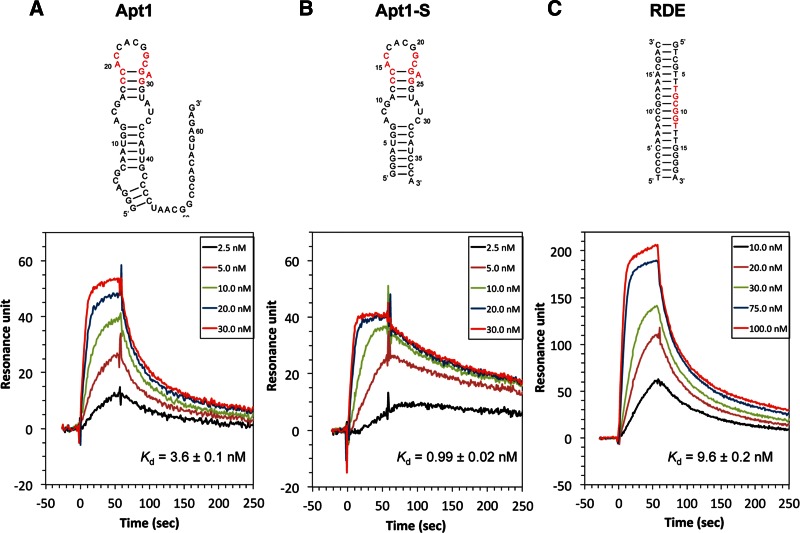
The apparent dissociation constants (Kd) of Apt1 and Apt1-S for Runt binding, calculated from their surface plasmon resonance (SPR) profiles, were 3.6 ± 0.1 nM and 0.99 ± 0.02 nM, respectively (Fig. 3A,B). Thus, it appears that the truncated 3′ and 5′ regions of Apt1 are dispensable for Runt binding, and the truncation leads to a stabilized structure that is more favorable for Runt binding. Apt1-S exhibited ∼10-fold higher affinity to the Runt domain compared with the Runt-binding double-stranded DNA element (RDE; Kd = 9.6 ± 0.2 nM) (Fig. 3C). Thus, the results indicated that the stem–loop region containing the consensus sequences is responsible for binding to the Runt domain with a higher affinity than RDE.
FIGURE 3.
Sequences and binding properties of aptamers and RDE. Predicted secondary structures of Apt1 (A), Apt1-S (B), and RDE (C) and their SPR sensorgrams for AML1–Runt binding. (Red) The consensus DNA sequence for binding to the Runt domain in RDE. The indicated concentrations of the Runt domain were added to the aptamers or RDE immobilized on the sensor chip. The dissociation constant Kd was determined as the ratio of the association rate constant kon and the dissociation rate constant koff as follows: Kd = koff/kon. The mean Kd value ± SE (n = 3) is shown. The kon and koff values are shown in Tables 1 and 2. Kinetic parameters were determined using BIAevaluation software (BIAcore AB).
Analysis of the secondary structure of Apt1-S by NMR
To investigate the precise secondary structure of Apt1-S, a NOESY spectrum was measured and imino proton resonances were assigned (Fig. 4). NOE connectivities for imino proton resonances of G1–G2–G3–U34–U5–G6–G7 and U27–G26–G25 revealed the formation of two stems. NOEs between NH2(C14)–NH (G23) and NH2(C16)–NH(G21) revealed the formation of C14–G23 and C16–G21 base pairs (Fig. 4). Because G23 and G25 are base-paired with C14 and C13, respectively, A24 should bulge out between these two G residues. For further analysis, we assigned H2, H5, H6, H8, H1′, and H2′ resonances of the conserved sequence motif region of Apt1-S using the standard method involving heteronuclear experiments (Varani et al. 1996). The NOE signal between H1′(G23) and H8(G25) was observed in the NOESY spectrum in D2O, suggesting that G23 and G25 are very close to each other and A24 is bulged out (Fig. 5). These NMR data suggested that Apt1-S has a CACG tetraloop and two stem regions separated by one internal loop (Fig. 6B) and that the conserved sequence motif region of Apt1-S has a stem comprising C13–G25, C14–G23, and C16–G21 base pairs containing an AC mismatch and a bulging out at A24.
FIGURE 4.
1D imino proton spectrum, 2D TOCSY, and 2D NOESY spectra of Apt1-S in H2O. 1D imino proton spectrum and 2D NOESY spectra of Apt1-S (mixing time = 150 msec). NOE connectivities are indicated by lines in the lower NOESY spectrum. Assignments were determined by NOE connectivities for imino proton resonances of G1–G2–G3–U34–U5–G6–G7 and U27–G26–G25 and are shown on top of the 1D imino proton spectrum. NOE connectivities are indicated by lines. Cross peaks between the NH2 (H41 and H42) –NH (H1) of C14–G23 and C16–G21 base pairs are shown. Cross peaks of intraresidue NOEs between H41–H42, NH2–H5, and H5–H6 of C14 and C16 are shown. (Red) The TOCSY spectrum superimposed on the NOESY spectrum. Assignments of NOE signals between NH2–NH were confirmed by these NOE connectivities. Assignments of NOE signals between H5–H6 were confirmed by sequential connectivities, as shown in Figure 5.
FIGURE 5.
2D NOESY spectrum of Apt1-S in D2O. The NOESY spectrum (mixing time = 400 msec) was recorded at 10°C, and cross peaks between aromatic H6 and H8 protons and ribose H1′ protons are shown. Sequential connectivities of the apical stem–loop (from C12 to G26) of Apt1-S are indicated by lines, and intraresidue NOEs are labeled by residues. The H1′ (G23) –H8 (G25) interresidue NOE is indicated.
FIGURE 6.
Amino acid residues required for the Runt domain to recognize DNA or RNA. (A) The secondary structure of RDE and amino acid residues that recognize bases, as determined by crystallography. (Black squares) Highly required amino acid residues; (dashed boxes) partially required amino acids. (Arrows) Direct interactions between an amino acid and a base. (B) The secondary structure of Apt1-S deduced by NMR analysis. (C) The secondary structure of the consensus motif. (Gray boxes) The conserved base pairs and a base.
The secondary structures of the other aptamers were predicted by sequence comparisons with Apt1. All the aptamers possessed the 5′-GCGMGNN-3′ and 5′-N′N′CCAC-3′ conserved motif (M:A or C; N and N′ form Watson–Crick base pairs), containing an AC mismatch and a single A/C bulge (Fig. 6C). This suggested that all these aptamers have the same secondary structure at the motif, although other regions are highly variable.
Base specificity of Apt1 for Runt binding
To confirm the importance of the motif for Runt binding, we performed mutation analysis of Apt1-S. Dissociation constants of mutants of Apt1-S containing base substitutions, deletions, or insertions were studied by SPR (Table 1). Mutations at the three CG base pairs (C14A, C16A, G21C, G23C, C16G–G21C, and C13G–G25C) markedly diminished the binding activity. Thus, it was suggested that the three consensus GC base pairs directly interact with the Runt domain. However, consistent with the results of SELEX, the C12G–G26C mutation had little effect on binding affinity, although the binding affinities of the C12G and G26C mutations, which disrupted base-pairing between the 12th and 26th bases, were markedly diminished. This finding indicates that the position of the C12–G26 base pair partakes in an NN′ base pair in the conserved sequence motif. Therefore, although the bases participating in NN′ base pairs are not involved in direct interactions with the Runt domain, they probably stabilize the structure of the motif.
TABLE 1.
Binding affinities of Apt1-S mutants to the Runt domain

Next, the effects of mutations at the bulged A24 position were investigated. A24C and A24U mutations had limited effects on binding affinity, suggesting that the base at A24 is not involved in direct interaction with the Runt domain and that the A bulge is not essential for binding. In particular, the A24C mutation had almost no effect on binding affinity, which is consistent with the SELEX results that the base equivalent to the A24 position is either an adenosine or a cytosine. In contrast, the A24G and A24Δ mutants showed a >103-fold reduction in binding affinity. When evaluating the effects of mutations at A15 and C22 in the AC mismatch, the A15G, A15C, A15U, C22A, C22G, C22U, and A15G–C22U mutations markedly diminished the binding activity, suggesting that the AC mismatch is important for the Runt-domain binding.
Substitution of the CACG tetraloop with GAAA or UUUU had little effect on binding affinity. The double-stranded mutant cleaved in the tetraloop, consisting of strands 1–18 and 19–37 of Apt1-S, retained binding activity, although it was reduced to one-twentieth. Thus, the loop structure likely contributes little to binding, which is consistent with the other aptamers containing the consensus sequences forming inverted stem–loop structures. These results suggest that having all the nucleotides comprising the consensus sequences is the most favorable for Runt binding. The three guanines—G21, G23, and G25—in the motif are indispensable for binding, similar to the three critical guanines in RDE, which are directly recognized by the three arginine residues of the Runt domain. Furthermore, the AC mismatch and A bulge-out are also important for its binding affinity.
Runt-domain amino acid residues required for Apt1 binding
Next, we performed mutational analysis of the Runt domain to determine whether Apt1-S binds to the same amino acid residues as during RDE binding. The crystal structure of the Runt domain–RDE complex showed that the amino acid residues R80, V170, D171, R174, and R177 were involved in the direct interaction with DNA bases and played important roles in defining the specificity of DNA recognition (Fig. 6A; Tahirov et al. 2001). R80 contacts the N7 and O6 atoms of G3, V170 contacts the C5 atom of C7′, D171 contacts the N4 atoms of C7′ and C6′, R174 contacts the N7 and O6 atoms of G5, and R177 contacts the N7 and O6 atoms of G6. Therefore, the aforementioned five residues were replaced with alanine, and binding affinities of RDE and Apt1-S were investigated by SPR (Table 2).
TABLE 2.
Binding affinities of Runt-domain mutants to RDE and Apt1-S

Alanine substitution at R80, D171, R174, or R177 reduced the binding to both RDE and Apt1-S. In contrast, the V170 mutant showed no marked change in binding to either Apt1-S or RDE. Tahirov et al. (2001) showed that V170A retained the DNA-binding activity, although a hydrophobic interaction was observed between V170 and the C5 position of C7′, which is similar to our results from mutational analysis of RDE.
Apt1 competes with DNA, but not CBFβ, for binding to the Runt domain
We further confirmed whether or not Apt1 actually competed with RDE for binding to the Runt domain by SPR. When the Runt domain was pre-incubated with equimolar concentrations of Apt1, RDE, or 30N RNA and then flowed to an RDE-bound sensor chip, 95% and 45% of the Runt domain with 30N RNA and RDE, respectively, bound to RDE on the sensor chip (Fig. 7A). However, almost none of the Runt domain with Apt1 bound to RDE on the sensor chip, suggesting that once Apt1 is bound to the Runt domain, it is hardly exchangeable with RDE because of its high binding affinity. Finally, we analyzed the effect of Apt1 on the heterodimerization of the Runt domain and the CBFβ subunit. The CBFβ subunit was added to the Runt domain bound to immobilized RDE or Apt1 on a sensor chip. The CBFβ subunit bound to the Runt domain and formed a complex with either RDE or Apt1 (Fig. 7B), suggesting that substituting RDE with Apt1 has little effect on the interaction between the Runt domain and CBFβ.
FIGURE 7.
Apt1 competes with RDE, but not CBFβ, for binding to the Runt domain. (A) SPR sensorgrams to analyze the competition between Apt1 and RDE. Following immobilization of the RDE to flow cell 1 on the sensor chip, the Runt domain (50 nM) mixed with 50 nM Apt1, RDE, 30N random RNA, or nothing was injected into flow cells 1 and 2 for 60 sec and dissociated for 180 sec at a flow rate of 20 μL/min. The signals of flow cell 1 were subtracted from that of flow cell 2 to eliminate nonspecific interactions. (B) Comparison of SPR sensorgrams for ternary complex formation between RDE–Runt–CBFβ (left) and Apt1–Runt–CBFβ (right). Following immobilization of the aptamer or RDE to flow cell 1 on the sensor chip, the Runt domain (50 nM) was injected into flow cells 1 and 2 for 60 sec at a flow rate of 20 μL/min. Then, 180 sec after injection of the Runt domain, CBFβ (500 nM) was injected into flow cells 1 and 2 for 60 sec and dissociated for 180 sec. (Arrows) Injection points. The signals of flow cell 1 were subtracted from that of flow cell 2 to eliminate nonspecific interactions, which results in the spikes of noise at the injection points due to the time lag between the flow cells 1 and 2.
DISCUSSION
In this study, we identified a conserved RNA sequence motif, 5′-GCGMGNN-3′ and 5′-N′N′CCAC-3′ (M:A or C; N and N′ form Watson–Crick base pairs), which binds to the DNA-binding site of the AML1 Runt domain with high affinity. Secondary structure analysis of one of these aptamers, Apt1, by enzyme probing and NMR showed that the two sequences form a stem structure containing an AC mismatch and a bulging out of A and that the spacer region forms a stable CACG tetraloop. Mutational analysis of this aptamer revealed that the three CG base pairs in the conserved sequence motif are important for Runt binding (Table 1). A comparison of the secondary structures of Apt1-S and RDE revealed that the nucleotides whose bases are recognized by the Runt domain in Apt1-S, C13, C14, G21, G23, and G25 correspond to the nucleotides C6′, C7′, G3, G5, and G6 in RDE (Fig. 6A,B). Therefore, it was suggested that the consensus sequences in Apt1-S bind to the Runt domain by mimicking the sequence and structure of RDE.
The binding affinities of some variants of Apt1-S with mutations in the bulged A24 (A24G and A24Δ) were markedly reduced. The A24G mutation may induce changes in base-pairing such as with C14–G24, which could result in disruption of the favorable topology of important nucleotides. Deletion of A24 could stabilize the A-form helix, which is unfavorable for Runt binding. Furthermore, all point mutations at A15 and C22, the AC mismatch, greatly reduced binding. The A15C, A15U, C22A, and C22G mutations may destabilize the tertiary structure of the aptamer because of disruption of the AC mismatch. In contrast, the A15G and C22U mutants could result in formation of a GC and an AU base pair, respectively, in lieu of an AC mismatch, which could stabilize the unfavorable A-form helix. The A15G–C22U mutant, which forms a GU wobble base pair, could also stabilize the unfavorable A-form helix. These results suggest that the distorted helix of the aptamer resulting from the A24 bulge and A15–C22 mismatch in the conserved sequence motif is favorable for Runt binding.
Amino acid residues in the Runt domain that are involved in direct interactions with DNA bases were also involved in binding to Apt1-S, based on the results of mutational analysis (Table 2). Only the R177A substitution resulted in a different response for RDE versus Apt1-S. Apt1-S binding was reduced more severely than RDE binding. R177 contacts the N7 and O6 atoms of G6 of RDE, which corresponds to G25 of Apt1-S. Therefore, R177 may play a more important role in Apt1-S binding than in RDE binding to stabilize the structure of the RNA motif. All these results indicate that the motif in Apt1 likely binds to the Runt domain by simply mimicking the sequence and structure of RDE, namely, three conserved G residues that interact directly with arginines in the Runt domain, an A24 bulge, and an AC mismatch, which position the G residues properly, similar to RDE. In a competition assay, Apt1 could bind to the Runt domain with higher affinity than that of RDE (Fig. 3), and Apt1 bound to the Runt domain could not be displaced by RDE (Fig. 7A). These results are consistent with our model that Apt1 binds to the Runt domain by mimicking RDE.
It is known that RDE binding to the Runt domain is promoted 10-fold by CBFβ, a partner of AML1. In this study, substituting RDE with Apt1 had little effect on the interaction between the Runt domain and CBFβ (Fig. 7B). Barton et al. (2009) recently reported on RNA aptamers against a Runt domain–CBFβ complex; however, their aptamers did not contain the consensus sequence found in ours. They showed that their aptamers could disrupt the Runt domain–DNA interaction in addition to the Runt domain–CBFβ interaction. A precise comparison of their aptamers with ours in terms of binding to the Runt domain may provide interesting information about the RNA–protein interaction.
The aptamer against the NF-κB p50 homodimer (p50)2 has been intensively analyzed (Huang et al. 2003; Ghosh et al. 2004; Reiter et al. 2008) and also mimics DNA and binds to the DNA-binding site of a transcription factor. NF-κB p50 is a transcription factor that regulates the expression of hundreds of genes by binding to target DNA sequences and comprises an N-terminal Ig-like domain that binds DNA and a C-terminal Ig-like domain. The (p50)2 homodimer shows preference for 11-bp targets comprising 5′-GGGRN A/T NYCCC. The 29-nt RNA aptamer against (p50)2 had no resemblance to the target DNA in terms of sequence and secondary structure. However, analysis of the crystal structure of the (p50)2–aptamer complex revealed that a DNA guanine base recognized by NF-κB p50 is replaced with two uracils in the NF-κB p50–aptamer complex. In the NF-κB p50–DNA complex, this guanine accepts two hydrogen bonds through its N7 and O6 atoms. In the NF-κB p50–aptamer complex, two successive uracil O4 atoms are present at homologous positions, allowing an arginine to mediate comparable contacts in both the complexes. Thus, the NF-κB p50–aptamer mimics the target DNA elements with its tertiary structure without conservation of bases.
In recent years, it has become evident that the vast majority of mammalian genomes are transcribed and several thousand noncoding transcripts that play regulatory roles in many physiological processes are produced (Amaral et al. 2008; Mattick 2009). Some of these noncoding RNAs are known to bind to transcriptional factors and regulate their activities, such as the growth arrest–specific 5 (Gas5) targeting glucocorticoid receptor (GR), steroid receptor RNA activator (SRA), and noncoding RNA repressor of NFAT (NRON) (Leygue 2007; Kino et al. 2010; Sharma et al. 2011). Conversely, some transcription factors such as Smads, p53, and ERα regulate the expression of small noncoding RNAs (microRNAs) during the processing of primary transcripts (Davis et al. 2008, 2010; Suzuki et al. 2009; Yamagata et al. 2009).
During these regulatory processes, several transcription factors, such as GR and Smads, bind to RNA as well as to DNA at related sites. However, the precise mechanisms of the RNA–protein interactions between noncoding RNAs and the DBD of transcription factors remain unknown. Our results from this study demonstrate that the DBD of the AML1 protein binds a sequence-conserved RNA motif that likely mimics DNA architecture by using an AC mismatch and bulge out. These results enhance our understanding of DNA-mimicking natural RNA motifs in regulatory noncoding RNAs. Furthermore, these results may be useful in the future for the rational design of aptamers against many other sequence-specific DNA-binding proteins.
MATERIALS AND METHODS
Expression and purification of proteins
The coding sequence of an AML1 N-terminal fragment (amino acids 1–188, referred to as the Runt domain) was cloned into the pDEST17 vector (Invitrogen). Recombinant His-tagged AML1–Runt was expressed in Escherichia coli strain BL21-AI (Invitrogen). The crude extract from a 1-L culture was applied to a 1-mL His trap HP column (GE Healthcare) equilibrated with buffer A (20 mM sodium phosphate at pH 7.0, 500 mM NaCl, 0.1% Triton X-100, and 2 mM DTT)/10 mM imidazole. A protease inhibitor cocktail (50 μL) for His-tagged proteins (Sigma-Aldrich) was added per gram of E. coli lysate. After washing with buffer A/10 mM imidazole, the protein was eluted using a 10–500 mM imidazole gradient, followed by dialysis against buffer B (10 mM sodium phosphate at pH 7.0, 100 mM NaCl, and 2 mM DTT). The sample was then applied to a 1-mL Resource S column (GE Healthcare) equilibrated with buffer C (30 mM sodium phosphate at pH 7.4, 30 mM sodium formate, and 60 mM sodium acetate) and eluted using a 0–1 M NaCl gradient. The purified protein was then dialyzed against buffer D (20 mM sodium phosphate at pH 7.4, 150 mM NaCl, 50% glycerol, and 1 mM DTT). The expression vectors containing Runt-domain mutants were obtained by PCR mutagenesis in the pDEST17–Runt vector.
The coding sequence of CBFβ with an N-terminal Flag-tag was cloned into the pDEST17 vector. Recombinant His-Flag-tagged CBFβ was expressed in E. coli BL21-AI, and the crude extract from a 1-L culture was applied to a 2.5-mL Ni–NTA agarose column (QIAGEN) equilibrated with buffer E (20 mM sodium phosphate at pH 8.0 and 300 mM NaCl)/10 mM imidazole. After washing with buffer E/20 mM imidazole, the protein was eluted with buffer A/250 mM imidazole, followed by dialysis against buffer F (10 mM Tris–HCl at pH 7.5 and 20 mM NaCl). The sample was then applied to a 1-mL Resource Q column (GE Healthcare) equilibrated with buffer G (50 mM 1-methyl piperazine, 50 mM Bis–Tris, and 25 mM Tris at pH 7.5) and eluted using a 0–800 mM NaCl gradient. The purified protein was then dialyzed against buffer H (10 mM Tris–HCl at pH 7.5, 100 mM NaCl, 50% glycerol, and 1 mM DTT).
Aptamer selection procedures
The random RNA pools were prepared and in vitro selection was performed as described previously (Oguro et al. 2003) with some modifications. Transcription templates were prepared with a Klenow fragment (Takara) using the following synthetic oligonucleotides: 5′-TAATACGACTCACTATAGGGACACAATGGACG-3′ (primer 1) and 5′-CTCTCATGTCGGCCGTTA-(40N or 30N)-CGTCCATTGTGTCCCTATAGTGATCGTATTA-3′, where 40N and 30N represent 40 and 30 random nucleotide positions, respectively. An RNA pool of 40 or 30 random nucleotides was prepared by in vitro transcription using the AmpliScribe T7-Flash Transcription Kit (Epicentre Biotechnologies).
The RNA was heat-denatured for 3 min at 85°C and refolded by cooling on ice in binding buffer (20 mM sodium phosphate at pH 6.5, 2 mM magnesium acetate, 50–200 mM potassium acetate increased stepwise after every three cycles, 5% glycerol, 0.05% Triton X-100, and 5 mM β-mercaptoethanol). The RNA was incubated with the Runt domain fixed on affinity beads (Ni Sepharose HP [GE Healthcare], Talon metal affinity resin [Clontech], or Ni–NTA magnetic agarose [QIAGEN]) for 30 min in binding buffer at room temperature. The resins were washed five times with 400 µL of binding buffer, and bound RNA was eluted with 200 µL of binding buffer containing 500 mM imidazole, heat-denatured at 85°C, and precipitated with ethanol. The cDNA was synthesized using ReverTra Ace reverse transcriptase (TOYOBO) and amplified by PCR using primer 1 and primer 2 (5′-CTCTCATGTCGGCCGTTA-3′), followed by T7 transcription. After round 9, cDNA was collected for the selected RNA and cloned into the pGEM-T Easy vector (Promega) and sequenced.
Aptamer preparation
Apt1, Apt1-S, and mutants of Apt1-S were synthesized by in vitro transcription. The transcription template of Apt1 was amplified by PCR using primer 1 and primer 2 and a cloning vector containing Apt1. Primer 3 [5′-(T)16-CTCTCATGTCGGCCGTTA-3′] was used in lieu of primer 2 to attach an A16-tag for use in SPR assays. PCR products were purified using the QIAquick PCR Purification Kit (QIAGEN). The Apt1-S and related mutant templates for use in SPR assays were prepared by PCR using the T7 primer (5′-TAATACGACTCACTATAGGG-3′), A16-tagged primers, and the following synthetic oligonucleotide: 5′-GGGATGGGATACCTCGCCGTGGTGGGTCGTCCATCCCTATAGTGAGTCGTATTA-3′. The Apt1-S template for use in NMR studies was chemically synthesized. Transcription reactions were performed using the AmpliScribe T7-Flash Transcription Kit. For assignments of NMR signals, stable isotope-labeled [A, G-13C/15N] Apt1-S and [C, U-13C/15N] Apt1-S were synthesized using 13C- and 15N-labeled NTPs (Taiyo Nippon Sanso). The RNA samples were purified by denaturing PAGE.
SPR assays
SPR assays were performed as described previously (Mochizuki et al. 2005) using a BIAcore 2000 instrument (BIAcore AB). The 5′-biotinylated dT16 oligomer was immobilized to 100 Resonance Units (RU) in flow cells 2–4 on the surface of a streptavidin sensor chip (BIAcore AB). The sensor chip was washed and equilibrated in buffer I (20 mM sodium phosphate at pH 6.5, 2 mM magnesium acetate, 300 mM potassium acetate, 0.1% Tween 20, and 1 mM DTT) at 25°C. Following this, 4 μg/mL 3′-A16-tagged RNA or 3′-A16-tagged RDE (5′-GTCGTTTGCGGTTTGGGGAAAAAAAAAAAAAAAAA-3′ or 5′-TCCCCAAACCGCAAACGAC-3′) (Thornell et al. 1991; Crute et al. 1996; Wolf-Watz et al. 1999) dissolved in buffer I was immobilized in flow cells 2–4 for 60 sec at a flow rate of 50 μL/min. RDE was used as a positive control and the 30N random RNA pool as a negative control. The AML1–Runt domain in buffer I (2.5–1000 nM) was injected into flow cells 1–4 of the sensor chip for 60 sec and dissociated for 180 sec. Because the Runt protein was prone to precipitate at concentrations >1000 nM in buffer I, this concentration was the upper limit for stable measurements in SPR assays. To regenerate the sensor chip, bound material was completely removed by injecting 4 M urea. For monitoring the interaction using the BIAcore instrument, all procedures were automated to run repetitive cycles of sample injection. The signals of flow cells 2–4 were subtracted from that of flow cell 1 to eliminate nonspecific interactions. The sensorgrams were analyzed using BIA evaluation software (BIAcore AB). A Langmuir (1:1) binding model was used to analyze the association rate constant kon and the dissociation rate constant koff (M–1 sec–1 and sec–1, respectively). The dissociation constant Kd was also determined as the ratio of koff and kon as follows: Kd = koff/kon, and is represented by the mean value ± standard error from three independent measurements.
Enzymatic analysis of the secondary structure of RNA
The secondary structure of Apt1 was probed by RNase digestion as described previously (Sakamoto et al. 1997). Apt1 was first dephosphorylated with alkaline phosphatase from calf intestine (Roche) for 60 min at 50°C. The 5′ end of Apt1 was labeled with [γ-32P]ATP (Amersham) using T4 polynucleotide kinase (Takara). The labeled Apt1 was heated at 85°C, cooled on ice in buffer G (20 mM sodium phosphate at pH 6.5, 2 mM magnesium acetate, and 50 mM potassium acetate), and then incubated with 0.5 μg/μL yeast RNA (Ambion) and each RNase (RNase T1, RNase V1, or mung bean nuclease) for 10 min at 25°C. Apt1 was heated at 85°C and incubated in buffer H (20 mM citric acid at pH 5.0, 7 M urea, and 1 mM EDTA) and RNase T1 for 30 min at 50°C for RNase T1 digestion in the denatured condition. Alkaline digestion was performed in 50 mM sodium carbonate (pH 9.0) with 1 mM EDTA for 10 min at 90°C. The samples were analyzed by denaturing 15% PAGE.
NMR measurements
The RNA samples were annealed by heating for 5 min at 95°C followed by snap cooling on ice. Unlabeled Apt1-S and stable isotope-labeled [A, G-13C/15N] Apt1-S and [C, U-13C/15N] Apt1-S were dissolved in 20 mM sodium phosphate (pH 6.5) containing 50 mM NaCl. NMR spectra were measured using DRX-500 and DRX-600 spectrometers (Bruker BioSpin). Spectra were recorded at a 10°C probe temperature. The imino proton resonances of G and U residues within RNA in H2O were distinguished by the 1H–15N HSQC spectrum measured with [A, G-13C/15N] Apt1-S and [C, U-13C/15N] Apt1-S. Exchangeable proton resonances were assigned by NOESY in H2O with a mixing time of 150 msec using the jump-and-return scheme for water suppression (Plateau and Gueron 1982). Nonexchangeable proton resonances were assigned by NOESY (mixing time of 400 msec), TOCSY (mixing time of 50 msec), 2D HSQC, HCCH-COSY, and HCCH-TOCSY in D2O using a standard method (Varani et al. 1996).
Competition experiments between the Apt1 aptamer and RDE
The assays were performed at 25°C using a BIAcore X instrument. The 5′-biotinylated dT16 oligomer was immobilized to 100 RU in flow cell 1 on the surface of the streptavidin sensor chip (BIAcore AB). Then 4 μg/mL 3′-A16-tagged RDE dissolved in buffer I was immobilized in flow cell 1 for 60 sec at a flow rate of 20 μL/min. The AML–Runt domain (50 nM) mixed with 50 nM Apt1, RDE, or 30N random RNA in buffer I was injected into flow cells 1 and 2 for 60 sec and dissociated for 180 sec at a flow rate of 20 μL/min. The signals of flow cell 1 were subtracted from that of flow cell 2 to eliminate nonspecific interactions.
SPR analysis of the aptamer–Runt–CBFβ ternary complex formation
Assays were performed at 25°C using a BIAcore X instrument. The 5′-biotinylated dT16 oligomer was immobilized to 100 RU in flow cell 1 on the surface of the streptavidin sensor chip (BIAcore AB). Following immobilization of the aptamer or RDE in flow cell 1 of the sensor chip, the AML1–Runt domain (50 nM) in buffer I was injected into flow cells 1 and 2 of the sensor chip for 60 sec at a flow rate of 20 μL/min. Then, 180 sec after injection of the AML1–Runt domain, CBFβ (500 nM) in buffer I was injected into flow cells 1 and 2 of the sensor chip for 60 sec and dissociated for 180 sec. The signals of flow cell 1 were subtracted from that of flow cell 2 to eliminate nonspecific interactions.
ACKNOWLEDGMENTS
We thank Akira Ishiguro, Eri Sakota, and Miho Saito for their technical assistance. This study was supported by the Core Research of Evolutional Science & Technology (CREST) of the Japan Science and Technology Agency (JST) and in part by the Program for Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation (NIBIO) and research grants from The Ministry of Education, Sports, Culture, Science, and Technology of Japan (MEXT).
REFERENCES
- Amaral PP, Dinger ME, Mercer TR, Mattick JS 2008. The eukaryotic genome as an RNA machine. Science 319: 1787–1789 [DOI] [PubMed] [Google Scholar]
- Backstrom S, Wolf-Watz M, Grundstrom C, Hard T, Grundstrom T, Sauer UH 2002. The RUNX1 Runt domain at 1.25 Å resolution: A structural switch and specifically bound chloride ions modulate DNA binding. J Mol Biol 322: 259–272 [DOI] [PubMed] [Google Scholar]
- Barton JL, Bunka DH, Knowling SE, Lefevre P, Warren AJ, Bonifer C, Stockley PG 2009. Characterization of RNA aptamers that disrupt the RUNX1–CBFβ/DNA complex. Nucleic Acids Res 37: 6818–6830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo J, Li Z, Speck NA, Warren AJ 2001. The leukemia-associated AML1 (Runx1)–CBFβ complex functions as a DNA-induced molecular clamp. Nat Struct Biol 8: 371–378 [DOI] [PubMed] [Google Scholar]
- Brown TS, Bevilacqua PC 2005. Method for assigning double-stranded RNA structures. BioTechniques 38: 368–372 [DOI] [PubMed] [Google Scholar]
- Collins A, Littman DR, Taniuchi I 2009. RUNX proteins in transcription factor networks that regulate T-cell lineage choice. Nat Rev Immunol 9: 106–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crute BE, Lewis AF, Wu Z, Bushweller JH, Speck NA 1996. Biochemical and biophysical properties of the core-binding factor α2 (AML1) DNA-binding domain. J Biol Chem 271: 26251–26260 [DOI] [PubMed] [Google Scholar]
- Davis BN, Hilyard AC, Lagna G, Hata A 2008. SMAD proteins control DROSHA-mediated microRNA maturation. Nature 454: 56–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BN, Hilyard AC, Nguyen PH, Lagna G, Hata A 2010. Smad proteins bind a conserved RNA sequence to promote microRNA maturation by Drosha. Mol Cell 39: 373–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruijn MF, Speck NA 2004. Core-binding factors in hematopoiesis and immune function. Oncogene 23: 4238–4248 [DOI] [PubMed] [Google Scholar]
- Ellington AD, Szostak JW 1990. In vitro selection of RNA molecules that bind specific ligands. Nature 346: 818–822 [DOI] [PubMed] [Google Scholar]
- Endo K, Nakamura Y 2010. A binary Cy3 aptamer probe composed of folded modules. Anal Biochem 400: 103–109 [DOI] [PubMed] [Google Scholar]
- Engel ME, Hiebert SW 2010. Proleukemic RUNX1 and CBFβ mutations in the pathogenesis of acute leukemia. Cancer Treat Res 145: 127–147 [DOI] [PubMed] [Google Scholar]
- Favorova OO, Fasiolo F, Keith G, Vassilenko SK, Ebel JP 1981. Partial digestion of tRNA–aminoacyl-tRNA synthetase complexes with cobra venom ribonuclease. Biochemistry 20: 1006–1011 [DOI] [PubMed] [Google Scholar]
- Ghosh G, Huang DB, Huxford T 2004. Molecular mimicry of the NF-κB DNA target site by a selected RNA aptamer. Curr Opin Struct Biol 14: 21–27 [DOI] [PubMed] [Google Scholar]
- Huang DB, Vu D, Cassiday LA, Zimmerman JM, Maher LJ III, Ghosh G 2003. Crystal structure of NF-κB (p50)2 complexed to a high-affinity RNA aptamer. Proc Natl Acad Sci 100: 9268–9273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP 2010. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal 3: ra8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leygue E 2007. Steroid receptor RNA activator (SRA1): Unusual bifaceted gene products with suspected relevance to breast cancer. Nucl Recept Signal 5: e006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS 2009. The genetic signatures of noncoding RNAs. PLoS Genetics 5: e1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikova IN, Crute BE, Wang S, Speck NA 1993. Sequence specificity of the core-binding factor. J Virol 67: 2408–2411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitelman F, Johansson B, Mertens F 2007. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer 7: 233–245 [DOI] [PubMed] [Google Scholar]
- Miyakawa S, Oguro A, Ohtsu T, Imataka H, Sonenberg N, Nakamura Y 2006. RNA aptamers to mammalian initiation factor 4G inhibit cap-dependent translation by blocking the formation of initiation factor complexes. RNA 12: 1825–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyakawa S, Nomura Y, Sakamoto T, Yamaguchi Y, Kato K, Yamazaki S, Nakamura Y 2008. Structural and molecular basis for hyperspecificity of RNA aptamer to human immunoglobulin G. RNA 14: 1154–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi H, Shimizu K, Kozu T, Maseki N, Kaneko Y, Ohki M 1991. t(8;21) breakpoints on chromosome 21 in acute myeloid leukemia are clustered within a limited region of a single gene, AML1. Proc Natl Acad Sci 88: 10431–10434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K, Oguro A, Ohtsu T, Sonenberg N, Nakamura Y 2005. High affinity RNA for mammalian initiation factor 4E interferes with mRNA-cap binding and inhibits translation. RNA 11: 77–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata T, Gupta V, Sorce D, Kim WY, Sali A, Chait BT, Shigesada K, Ito Y, Werner MH 1999. Immunoglobulin motif DNA recognition and heterodimerization of the PEBP2/CBF Runt domain. Nat Struct Biol 6: 615–619 [DOI] [PubMed] [Google Scholar]
- North TE, Stacy T, Matheny CJ, Speck NA, de Bruijn MF 2004. Runx1 is expressed in adult mouse hematopoietic stem cells and differentiating myeloid and lymphoid cells, but not in maturing erythroid cells. Stem Cells 22: 158–168 [DOI] [PubMed] [Google Scholar]
- Oguro A, Ohtsu T, Svitkin YV, Sonenberg N, Nakamura Y 2003. RNA aptamers to initiation factor 4A helicase hinder cap-dependent translation by blocking ATP hydrolysis. RNA 9: 394–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohuchi SP, Ohtsu T, Nakamura Y 2006. Selection of RNA aptamers against recombinant transforming growth factor-β type III receptor displayed on cell surface. Biochimie 88: 897–904 [DOI] [PubMed] [Google Scholar]
- Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR 1996. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell 84: 321–330 [DOI] [PubMed] [Google Scholar]
- Patel DJ, Suri AK 2000. Structure, recognition and discrimination in RNA aptamer complexes with cofactors, amino acids, drugs and aminoglycoside antibiotics. J Biotechnol 74: 39–60 [DOI] [PubMed] [Google Scholar]
- Plateau P, Gueron M 1982. Exchangeable proton NMR without base-line distortion, using new strong-pulse sequences. J Am Chem Soc 104: 7310–7311 [Google Scholar]
- Reiter NJ, Maher LJ III, Butcher SE 2008. DNA mimicry by a high-affinity anti-NF-κB RNA aptamer. Nucleic Acids Res 36: 1227–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, Kawai G, Katahira M, Kim MH, Tanaka Y, Kurihara Y, Kohno T, Watanabe S, Yokoyama S, Watanabe K, et al. 1997. Hairpin structure of an RNA 28-mer, which contains a sequence of the enzyme component of a hammerhead ribozyme system: Evidence for tandem G:A pairs that are not of side-by-side type. J Biochem (Tokyo) 122: 556–562 [DOI] [PubMed] [Google Scholar]
- Sharma S, Findlay GM, Bandukwala HS, Oberdoerffer S, Baust B, Li Z, Schmidt V, Hogan PG, Sacks DB, Rao A 2011. Dephosphorylation of the nuclear factor of activated T cells (NFAT) transcription factor is regulated by an RNA–protein scaffold complex. Proc Natl Acad Sci 108: 11381–11386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobczak K, Michlewski G, de Mezer M, Krol J, Krzyzosiak WJ 2010. Trinucleotide repeat system for sequence specificity analysis of RNA structure probing reagents. Anal Biochem 402: 40–46 [DOI] [PubMed] [Google Scholar]
- Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K 2009. Modulation of microRNA processing by p53. Nature 460: 529–533 [DOI] [PubMed] [Google Scholar]
- Tahirov TH, Inoue-Bungo T, Morii H, Fujikawa A, Sasaki M, Kimura K, Shiina M, Sato K, Kumasaka T, Yamamoto M, et al. 2001. Structural analyses of DNA recognition by the AML1/Runx-1 Runt domain and its allosteric control by CBFβ. Cell 104: 755–767 [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Akagi K, Nakamura Y, Kozu T 2007. RNA aptamers targeting the carboxyl terminus of KRAS oncoprotein generated by an improved SELEX with isothermal RNA amplification. Oligonucleotides 17: 12–21 [DOI] [PubMed] [Google Scholar]
- Thornell A, Hallberg B, Grundstrom T 1991. Binding of SL3-3 enhancer factor 1 transcriptional activators to viral and chromosomal enhancer sequences. J Virol 65: 42–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerk C, Gold L 1990. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249: 505–510 [DOI] [PubMed] [Google Scholar]
- Varani G, Aboul-ela F, Allain FHT 1996. NMR investigation of RNA structure. Prog Nucl Magn Reson Spectrosc 29: 51–127 [Google Scholar]
- Wolf-Watz M, Xie XQ, Holm M, Grundstrom T, Hard T 1999. Solution properties of the free and DNA-bound Runt domain of AML1. Eur J Biochem 261: 251–260 [DOI] [PubMed] [Google Scholar]
- Yamagata K, Fujiyama S, Ito S, Ueda T, Murata T, Naitou M, Takeyama K, Minami Y, O’Malley BW, Kato S 2009. Maturation of microRNA is hormonally regulated by a nuclear receptor. Mol Cell 36: 340–347 [DOI] [PubMed] [Google Scholar]