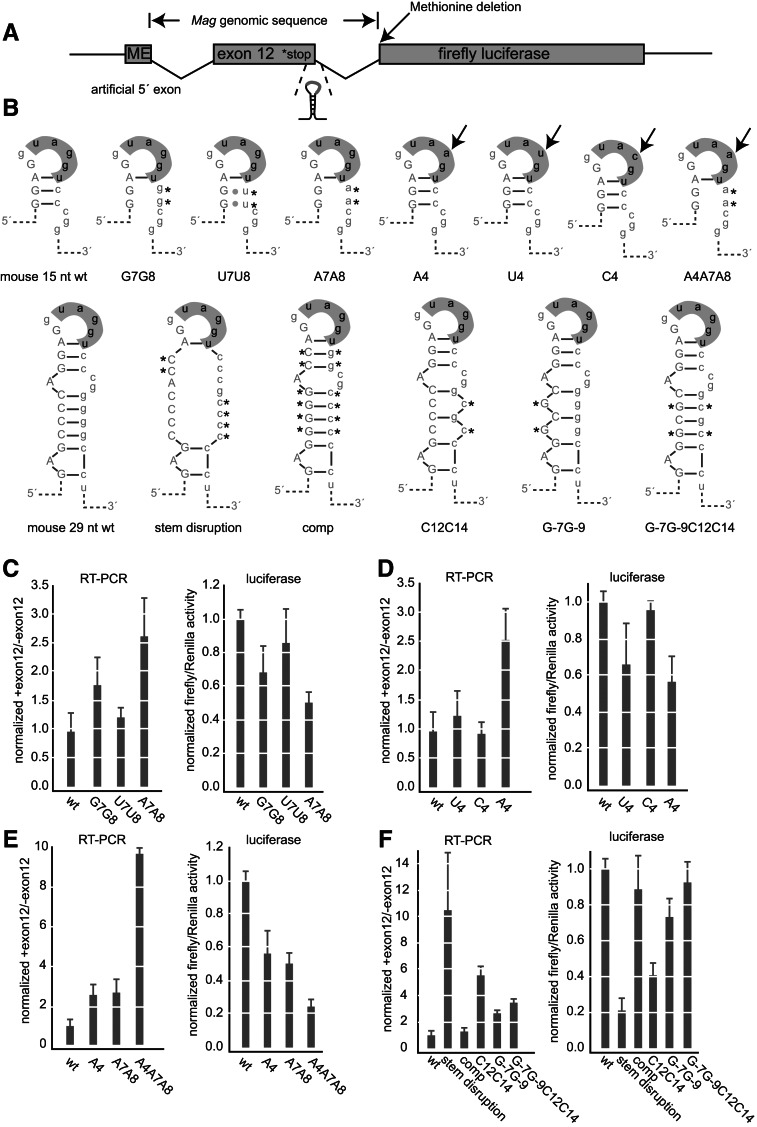
FIGURE 4.
The hnRNP A1-binding element and secondary structure control inclusion of Mag exon 12. (A) Diagram of the reporter construct. Mag exon 12 and its flanking introns were cloned upstream of firefly luciferase. The positions of the genomic insertion, stop codon, secondary structure, methionine deletion, and 5′ artificial exon are shown. (B) Diagram of the mutant sequences used in the reporter assay. The positions of the mutations are indicated. (C) Luciferase and RT-PCR analysis of upper right stem mutations. For the luciferase activity assay, firefly activity was divided by Renilla to control for transfection efficiency. Data were normalized to the expression of the wild-type construct. Data are the average and standard deviation of at least three independent transfections. For the RT-PCR assays, the ratio of the upper to the lower isoform after gel electrophoresis is shown. Error bars indicate the standard deviation of at least three replicates per construct. Data were normalized to the ratio of the wild-type construct. (D) Luciferase and RT-PCR analysis of loop mutations. Mutations are as indicated. Data were processed as in C. (E) Luciferase and RT-PCR analysis of combined upper stem and loop mutations. Data were processed as in C. (F) Luiferase and RT-PCR analysis of structural mutations. Data were processed as in C. (C–F) Data from all experimental replicates were aggregated for each construct. Expression data for constructs appearing on multiple graphs are the same from panel to panel, represented multiple times for clarity.