This paper describes unusual properties of group II intron-encoded reverse transcriptases. They have higher processivity, fidelity, and thermostability than retroviral enzymes. Moreover, they have an unexpected proclivity for template switching that makes them very useful for a number of cloning applications.
Keywords: miRNA, next-generation sequencing, qRT-PCR, retrovirus, transcriptome
Abstract
Mobile group II introns encode reverse transcriptases (RTs) that function in intron mobility (“retrohoming”) by a process that requires reverse transcription of a highly structured, 2–2.5-kb intron RNA with high processivity and fidelity. Although the latter properties are potentially useful for applications in cDNA synthesis and next-generation RNA sequencing (RNA-seq), group II intron RTs have been difficult to purify free of the intron RNA, and their utility as research tools has not been investigated systematically. Here, we developed general methods for the high-level expression and purification of group II intron-encoded RTs as fusion proteins with a rigidly linked, noncleavable solubility tag, and we applied them to group II intron RTs from bacterial thermophiles. We thus obtained thermostable group II intron RT fusion proteins that have higher processivity, fidelity, and thermostability than retroviral RTs, synthesize cDNAs at temperatures up to 81°C, and have significant advantages for qRT-PCR, capillary electrophoresis for RNA-structure mapping, and next-generation RNA sequencing. Further, we find that group II intron RTs differ from the retroviral enzymes in template switching with minimal base-pairing to the 3′ ends of new RNA templates, making it possible to efficiently and seamlessly link adaptors containing PCR-primer binding sites to cDNA ends without an RNA ligase step. This novel template-switching activity enables facile and less biased cloning of nonpolyadenylated RNAs, such as miRNAs or protein-bound RNA fragments. Our findings demonstrate novel biochemical activities and inherent advantages of group II intron RTs for research, biotechnological, and diagnostic methods, with potentially wide applications.
INTRODUCTION
Reverse transcriptases (RTs), which synthesize cDNA copies of RNA substrates, are central to a variety of widely used methods in research and biotechnology including RT-PCR, transcriptome and miRNA profiling, next-generation RNA sequencing (RNA-seq), RNA structure mapping, and the analysis of protein- or ribosome-bound RNA fragments (Wang et al. 2009; Mayer et al. 2011; Ozsolak and Milos 2011). However, retroviral RTs, which have been the only ones available for use in these methods, have inherently low processivity and fidelity. Additionally, the synthesis of cDNAs from some RNA templates, including medically important diagnostic targets, is impeded by higher-order RNA structure, making it advantageous to carry out reverse transcription at elevated temperatures (Mayer et al. 2011). High temperatures can also improve the specificity of reverse transcription by discriminating against mispaired primers. Only a few RTs capable of functioning at high temperature have been available, and these have relatively high error rates. For example, SuperScript III, a widely used genetically engineered derivative of Moloney murine leukemia virus (M-MLV) RT, is active at temperatures up to 55°C and has an error rate of 4.5 × 10−5 (Potter et al. 2003). The Thermus thermophilus DNA polymerase, which has a half-life of 20 min at 95°C and is commonly used at 74°C, exhibits RT activity only in the presence of Mn2+, which greatly reduces its fidelity (error rate = 70 × 10−5) (Beckman et al. 1985). To address these problems, a number of derivatives of retroviral RTs have been developed that have increased thermostability and processivity, e.g., Affinityscript (Agilent) (Arezi and Hogrefe 2009), Maxima (ThermoScientific), Rocketscript (Bioneer), Thermoscript (Life Technologies), and Monsterscript (Illumina) or fidelity (AccuScript; Stratagene). An exceptionally improved derivative of M-MLV RT, which contains five mutations, is active at temperatures up to 70°C and has a processivity of 1000–1500 nt on a selected RNA template, but may have somewhat decreased fidelity (error rate reported as <10−4) (Baranauskas et al. 2012).
Retroviruses are only one of a number of different types of retroelements that are found in nature. As infectious viruses that must evade host responses, they benefit from encoding RTs with high error rates and low processivity, which favors RNA recombination, to introduce and propagate variations (Ji and Loeb 1992; Hu and Hughes 2012). Other families of retroelements, such as non-LTR-retrotransposons and mobile group II introns, have different lifestyles that require the synthesis of long continuous cDNAs with high fidelity, but remain untapped as a source of RTs for biotechnological applications. Mobile group II introns, the source of RTs used in this work, are retrotransposons that are found mainly in prokaryotes and fungal and plant organellar genomes and are thought to be evolutionary ancestors of spliceosomal introns and retrotransposons in higher organisms (Lambowitz and Zimmerly 2011). They consist of an autocatalytic intron RNA (“ribozyme”) and an intron-encoded RT, which act together in a ribonucleoprotein (RNP) particle to promote intron mobility by a mechanism (“retrohoming”) in which the excised intron RNA reverse splices directly into a DNA site and is reverse transcribed by the RT (Lambowitz and Zimmerly 2011).
Hundreds of group II intron RTs have been identified by genome sequencing (Candales et al. 2012). They typically contain four conserved domains: RT, with conserved sequence blocks (RT1–7) corresponding to the fingers and palm regions of retroviral RTs; X, a region corresponding to the RT thumb; D, a DNA target site-binding domain; and En, a DNA endonuclease domain that cleaves the DNA target site to generate a primer for reverse transcription of the intron RNA (Fig. 1A; Blocker et al. 2005). The En domain is missing in some group II intron RTs, which instead use nascent strands at DNA replication forks to prime reverse transcription (Lambowitz and Zimmerly 2011). The RT and X/thumb domains of group II intron RTs are larger than those of retroviral RTs due to an N-terminal extension (RT-0), and “insertions” (RT-2a, RT-3a, etc.) between the conserved RT sequence blocks, some of which are conserved in non-LTR-retrotransposon RTs (Malik et al. 1999; Blocker et al. 2005). It has been suggested that these larger RT and thumb domains enable more extensive interactions with RNA templates, leading to higher processivity during reverse transcription (Chen and Lambowitz 1997; Malik et al. 1999; Bibillo and Eickbush 2002a; Blocker et al. 2005). Unlike retroviral RTs, group II intron RTs lack an RNase H domain and have low DNA-dependent DNA polymerase activity in standard assays (Blocker et al. 2005; Smith et al. 2005; Lambowitz and Zimmerly 2011).
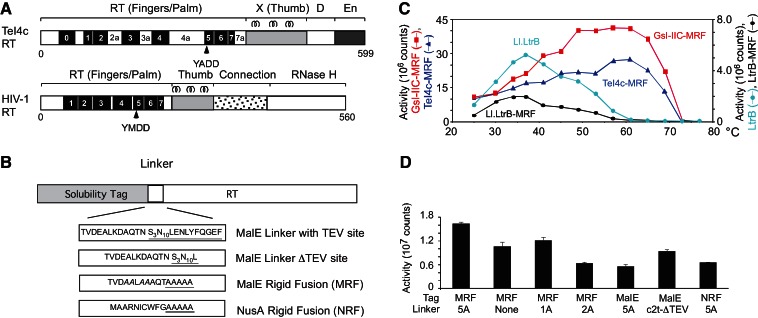
FIGURE 1.
Thermostable group II intron RT fusion proteins. (A) Comparison of group II intron TeI4c and retroviral HIV-1 RTs. Group II intron RT domains: RT with conserved sequence blocks RT-1 to RT-7, corresponding to the fingers and palm of retroviral RTs; X/thumb, with predicted α-helices (top) corresponding to those in the HIV-1 RT thumb; DNA-binding (D), and DNA endonuclease (En). Group II intron RTs have an N-terminal extension (RT-0) and “insertions” between the conserved RT sequence blocks (RT-2a, RT-3a, etc.) that are absent in retroviral RTs (Blocker et al. 2005; Lambowitz and Zimmerly 2011). Some group II intron RTs (e.g., GsI-IIC in this work) lack the En domain. (B) Group II intron RT fusion proteins. MalE-RT constructs have a MalE tag fused to their N terminus via a flexible linker with a TEV protease-cleavage site (underlined). MRF or NRF constructs have MalE or NusA solubility tags, respectively, fused to their N terminus via a rigid linker containing five alanines (underlined). For rigid fusions, the MalE tag has charged amino acid residues changed to alanines (italics), and the NusA tag is missing the two C-terminal amino acid residues. (C) Temperature profiles of RT activity. Poly(rA)/oligo(dT)42 and [32P]dTTP substrates were incubated with TeI4c-MRF (50 nM, 90 sec) or other indicated RTs (100 nM, 5 min), and polymerization of [32P]dTTP was quantified by binding to DE81 paper. Temperature profiles for additional group II intron RT fusion proteins in this assay are shown in Supplemental Figure S1. (D) RT activity of TeI4c-MRF RT constructs with different solubility tags and linkers. Assays were done as in C with 50 nM enzyme for 90 sec at 60°C. Bar graphs show the mean ± standard deviation (error bars) for three determinations.
During retrohoming, group II intron RTs must synthesize an accurate cDNA copy of the intron RNA, which is typically >2-kb long and folds into stable secondary and tertiary structures. Thus, group II intron RTs require high processivity and fidelity for their normal biological function. Indeed, retromobility of the Lactococcus lactis Ll.LtrB intron occurs in vivo with an error rate of ∼10−5, significantly lower than that of retroviral RTs (Conlan et al. 2005). Group II intron RTs from thermophiles are expected to combine these useful properties with thermostability. Thus far, however, only one mobile group II intron RT, the LtrA protein encoded by the Ll.LtrB intron, has been expressed in bacteria and purified with high yield and activity (Saldanha et al. 1999), while other group II intron RTs, including those from thermophiles, are poorly expressed and largely insoluble in the absence of bound RNAs (Vellore et al. 2004; Chee and Takami 2005; Ng et al. 2007). A further challenge for biotechnological development is that group II introns RTs often have mutations that decrease or abolish RT activity, reflecting that they are under selective pressure to suppress intron mobility, which is deleterious to their hosts (Mohr et al. 2010). Thus, it would be desirable to identify mobile group II introns that encode active RTs before investing the effort required for biochemical analysis and optimization. Recently, we identified group II introns in the thermophilic cyanobacterium Thermosynechococcus elongatus that are actively mobile and thermostable (Mohr et al. 2010), leading us to reinvestigate the expression and purification of thermostable group II intron RTs.
RESULTS AND DISCUSSION
Expression and purification of group II intron RT fusion proteins
The expression and solubility of difficult proteins can sometimes be improved by fusion of a highly soluble protein, like maltose-binding protein (MalE) or N utilization substance A (NusA) (Nallamsetty and Waugh 2006). The MalE tag additionally enables facile protein purification via amylose-affinity chromatography. Thus, we tested whether group II intron RTs could be expressed and purified as MalE fusion proteins using a protocol that includes polyethyleneimine (PEI) precipitation, amylose-affinity chromatography, and a final heparin-Sepharose purification step (Materials and Methods). The PEI-precipitation step is used to remove tightly bound nucleic acids that would otherwise interfere with the use of exogenous RNA templates in biotechnological applications. Initial experiments in which a MalE tag was fused to the N terminus of the RT via a tobacco etch virus (TEV) protease-cleavable linker (Fig. 1B) gave proteins that have high thermostable RT activity, expressed well, and could be purified readily from Escherichia coli. However, when the MalE tag was removed by protease cleavage, the RTs precipitated immediately, whereas if the tag was not cleaved, the enzymes lost RT activity and were degraded within days, even when flash frozen in 50% glycerol. These findings were surprising because proteins that fold properly in the presence of a solubility tag ordinarily remain soluble after removal of the tag (Nallamsetty and Waugh 2006). The unusual behavior of the group II intron RTs may reflect that they are ordinarily coexpressed with and bind tightly to the intron RNA from which they are translated, forming an RNP complex in which both the protein and RNA are stabilized in an active conformation (Saldanha et al. 1999; Cui et al. 2004). Fortuitously, the solubility tag can substitute for the bound RNA, enabling group II intron RTs to remain soluble when endogenous RNAs are removed.
To overcome the difficulties with a cleavable MalE tag, we tested whether group II intron RTs could be stabilized by attaching the MalE tag via a noncleavable rigid linker of the type used to reduce conformational heterogeneity for protein crystallization (Smyth et al. 2003). Such MalE-rigid fusions typically have a linker region consisting of three to five alanine residues together with changes near the end of the MalE tag to replace charged amino acid residues with alanines. In initial experiments, we tested MalE rigid fusions of several group II intron RTs, including the mesophilic L. lactis Ll.LtrB intron RT (LtrA protein), several T. elongatus group II intron RTs that promote retrohoming at elevated temperatures in vivo (Mohr et al. 2010), and two Geobacillus stearothermophilus group II intron RTs, which were previously difficult to express and purify with high yield and activity (Vellore et al. 2004; Ng et al. 2007). We found that group II intron RTs expressed as MalE rigid fusions (denoted MRF) support retrohoming in an E. coli plasmid assay (LtrA-MRF RT, 35% wild-type efficiency at 30°C; TeI4h* RT, 87% wild-type efficiency at 48°C), indicating that they retain all required activities despite the presence of the MalE tag. Further, the group II intron fusion proteins have high thermostable RT activity (Fig. 1C; Supplemental Fig. S1) and could be expressed readily in E. coli with yields of up to 20 mg/L of >95% pure protein.
The two most active thermostable RTs identified in the initial experiments were GsI-IIC-MRF and TeI4c-MRF. In RT assays with the artificial substrate poly(rA)/oligo(dT)42, these RTs had temperature optima of 61°C, compared with 35°C for the mesophilic Ll.LtrB group II intron RT, and they retained activity up to at least 70°C, a temperature at which the assay may be limited by the stability of base-pairing between the oligo(dT)42 primer and poly(rA) template (calculated Tm = 62.3°C at 75 mM KCl) (Fig. 1C; Kibbe 2007). Adding maltose (10 μM to 1 mM), which can affect the conformation of the MalE tag, had no significant effect on TeI4c-MRF RT activity assayed with poly(rA)/oligo(dT)42 (data not shown). Additional RT assays with the TeI4c-MRF RT at 60°C showed that the optimal combination of tag and linker consists of a modified MalE tag fused to the N terminus of the RT via a linker of five alanine residues (Fig. 1D). Variants with a conventional MalE tag, a NusA tag fused by a rigid linker (denoted NRF), or shorter or no alanine linkers had lower RT activity, suggesting that the rigidity of the linker and optimal spacing of the solubility tag are important for maximal activity (Fig. 1D). Defining a unit of RT activity as the amount of enzyme required to polymerize 1 nmol of dNTP in 1 min at 60°C using poly(rA)/oligo(dT)42 as template, the TeI4c-MRF and GsI-IIc-MRF RTs have specific activities of 183 ± 71 units/μg and 1376 ± 421 units/μg, compared with 144 ± 76 and 222 ± 33 units/μg for SuperScript III in the same assay at 37°C and 55°C, respectively. In further experiments shown below, all assays used comparable RT activity units for all three enzymes.
Group II intron RT fusion proteins have high processivity and fidelity
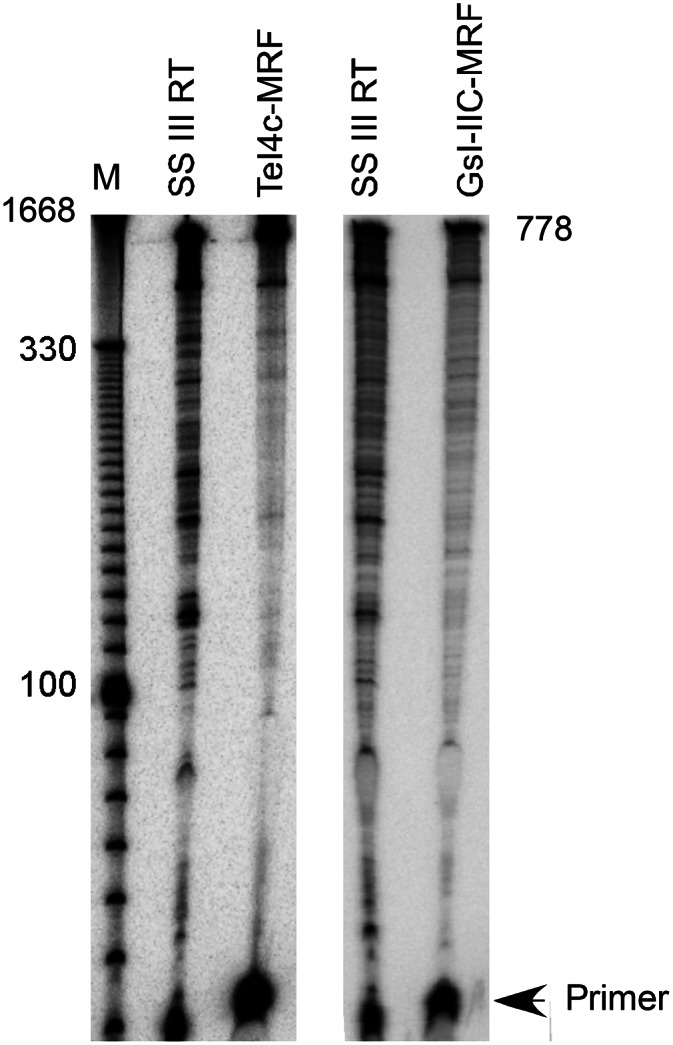
To test their suitability for cDNA synthesis applications, we compared the performance of the TeI4c- and GsI-IIC-MRF RTs with that of the commercially available thermostable RT, SuperScript III, in several standard assays of thermostability, processivity, and fidelity. In gel assays using a 509-nt in vitro-transcribed RNA template with a DNA primer annealed near its 3′ end, the TeI4c- and GsI-IIC-MRF RTs synthesized full-length cDNAs at temperatures up to 81°C and 69°C, respectively, while SuperScript III RT had no activity above 57°C (Fig. 2A; Supplemental Fig. S2). This assay measures thermostability in the presence of a bound RT substrate, and the results for SuperScript III are in agreement with the manufacturer’s product literature for this enzyme (Invitrogen.com).
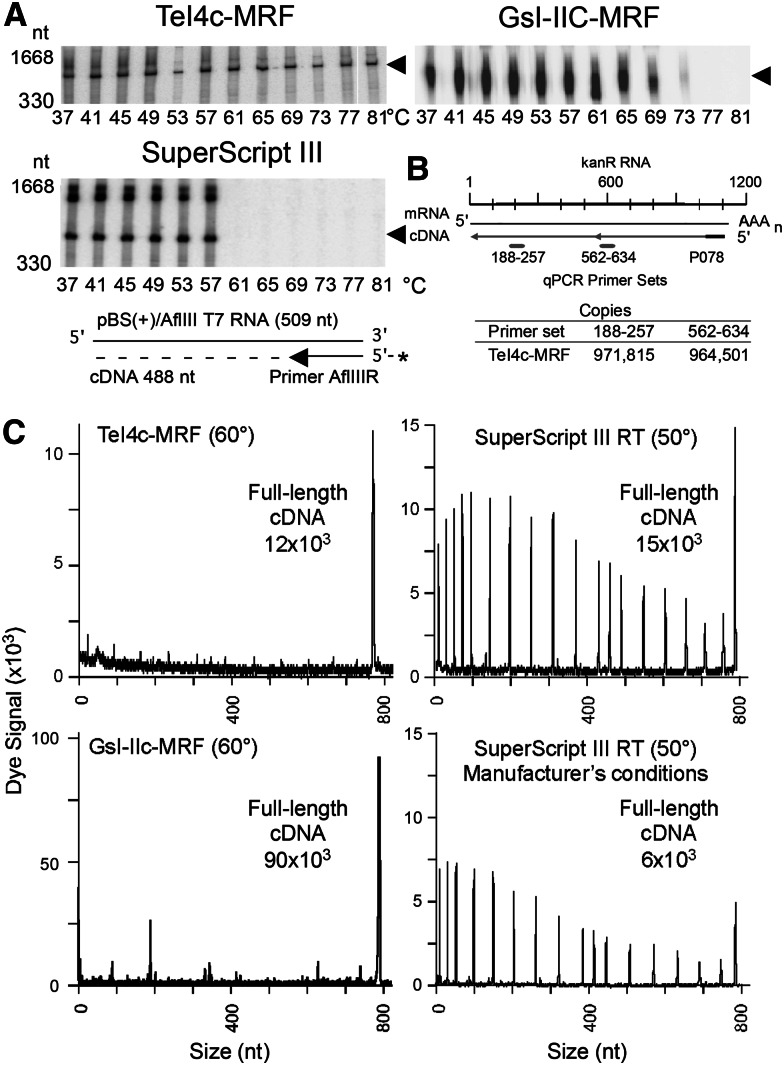
FIGURE 2.
Thermostability and processivity of group II intron RTs. (A) Gel assays of cDNA synthesis at different temperatures. A 509-nt in vitro–transcribed RNA (pBluescript KS(+)/AflIII) with a 5′-32P-labeled (star) primer (AflIIIR) annealed near its 3′ end was incubated for 30 min with TeI4c-MRF (2 μM), GsI-IIC-MRF (200 nM), or SuperScript III (10 units/μL) RTs, and the products were analyzed in a denaturing 6% polyacrylamide gel. Arrowheads to the right of the gel indicate the position of full-length cDNAs, and numbers to the left indicate positions of size markers (10-bp ladder). The regions of the gels containing the labeled DNA primer are shown in Supplemental Figure S2. (B) Taqman qRT-PCR. A 1.2-kb kanR RNA with primer P078 annealed near its 3′ end was reverse transcribed with TeI4c-MRF (200 nM) for 30 min at 60°C. The Table shows cDNA copies detected with primer sets 188–257 and 562–634, which detect cDNAs of 920 and 546 nt, respectively. (C) Capillary electrophoresis assays of cDNA synthesis. An 807-nt in vitro transcript containing an Ll.LtrB-ΔORF group II intron RNA with a fluorescently labeled DNA primer (5′ fluorophore WellRED) annealed near its 3′ end was reverse transcribed for 30 min with TeI4c-MRF RT (1 μM), GsI-IIC-MRF RT (200 nM), or SuperScript III (10 units/μL). cDNA lengths were determined relative to fluorescently labeled DNA markers (data not shown).
Several different assays were used to test the processivity of the enzymes. In Taqman qRT-PCR assays on a 1.2-kb kanR RNA template, primer sets near the middle (nt 562–634) and 5′ end (nt 188–257) of the RNA detected similar numbers of cDNAs for the TeI4c-MRF RT (971,815 and 964,501 copies, respectively), indicating high processivity (Fig. 2B). Similarly, in capillary electrophoresis assays in which each RT was tested at an optimal temperature, the TeI4c- and GsI-IIC-MRF RTs synthesized full-length cDNAs of a highly structured group II intron RNA with fewer premature stops than SuperScript III (Fig. 2C), a major problem in RNA-structure mapping and footprinting assays. In quantitative gel assays of processivity using a 5′-labeled group II intron RNA substrate with excess unlabeled substrate added after complex formation to trap dissociated RT, the average length of cDNAs synthesized by SuperScript III at 55°C was 176 ± 11 nt compared with 714 ± 16 nt for TeI4c-MRF RT and 708 ± 45 nt for GsI-IIC-MRF RT at 60°C (Fig. 3), mirroring the performance of the three enzymes in the capillary electrophoresis assays. Such processivity values are dependent upon the specific RNA template, and the group II intron RNA template used in this assay is a particularly challenging one due to its stable secondary and tertiary structures.
FIGURE 3.
Gel assay of processivity of cDNA synthesis. A 807-nt in vitro transcript containing an Ll.LtrB-ΔORF group II intron RNA with a 5′-32P-labeled primer annealed near its 3′ end was incubated for 30 min with TeI4c-MRF (2 μM) or GsI-IIc-MRF (1 μM) at 60°C or SuperScript III (10 units/μL) at 55°C in the presence of excess poly(rA)/oligo (dT)42 as a trap, and the products were analyzed in a denaturing 6% polyacrylamide gel alongside a 5′-labeled 10-bp ladder (M). The processivity (average length of template copied per initiation) was calculated by using the equation Σ(Ln.In)/Σ(In), where Ln is the length and In is the intensity of each analyzed cDNA fragment.
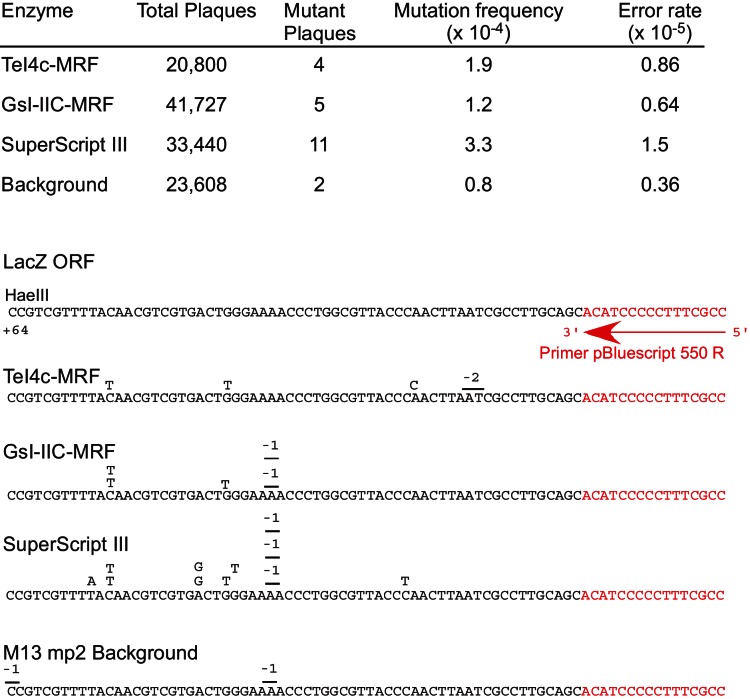
Finally, in tests of the fidelity of reverse transcription using an M13-based lacZ forward mutation assay, a standard assay for comparing the fidelity of different polymerases, the TeI4c and GsI-IIC-MRF RTs had two- to fourfold lower in vitro error rates than the retroviral RT (0.86 and 0.64 × 10−5 for the TeI4c- and GsI-IIC-MRF RTs, respectively, compared with 1.5 × 10−5 for SuperScript III and 0.36 × 10−5 for background) (Fig. 4). All three RTs gave a similar spectrum of mutations, including transitions, transversions, and frameshifts at runs of A-residues. Collectively, our results indicate that both of the thermostable group II intron RTs tested have higher thermostability, processivity, and fidelity than the retroviral RT.
FIGURE 4.
Error rates of different RTs determined by using an M13-based lacZ forward mutation assay. A 269-nt in vitro–transcribed RNA (pBluescript KS(+)/PvuI) encoding a segment of the LacZ α-fragment with annealed primer pBluescript 550R was reverse transcribed with TeI4c-MRF, GsI-IIC-MRF, or SuperScript III RTs, as described in the Materials and Methods. The resulting cDNAs were annealed to uracil-containing phage M13 single-stranded DNA, electroporated into E. coli MC 1061 F+ cells (Lucigen), and scored by plaque assays to determine the numbers of blue and white plaques. The mutation frequency was calculated as the ratio of white plaques to the total number of plaques. The error rate was calculated by dividing the mutation frequency by the number of nucleotide residues in the reverse-transcribed region at which changes would give a lacZ missense mutation. The background error rate was determined by electroporation of purified single-stranded M13 DNA. Sequence errors detected in cDNAs synthesized by different RTs are summarized below. “−1” and “−2” indicate −1 and −2 frameshifts, respectively; sequences complementary to the primer are shown in red.
Next-generation sequencing of human cDNA libraries
To globally compare the ability of the TeI4c-MRF and SuperScript III RTs to synthesize cDNAs of human mRNAs, we used these enzymes to reverse transcribe whole-cell RNAs from HeLa and MCF-7 cancer cells and analyzed the resulting cDNAs by next-generation sequencing. In these experiments, the RNA preparations were annealed with an oligo(dT)42 primer and reverse transcribed with the TeI4c-MRF RT at 60°C or SuperScript III at 50°C, a temperature recommended by the manufacturer for first-strand cDNA synthesis with this enzyme (http://tools.invitrogen.com/content/sfs/manuals/superscriptIIIfirststrand_pps.pdf). The second strand was then synthesized conventionally by using a commercial kit, and the resulting double-stranded DNAs were converted into RNA-seq libraries by using a transposome-based system for sequencing on an Illumina HiSeq instrument (Adey et al. 2010). The sequencing of the different samples produced between 18 and 58 million usable paired-end reads of 60 bases that mapped to human RefSeq transcripts.
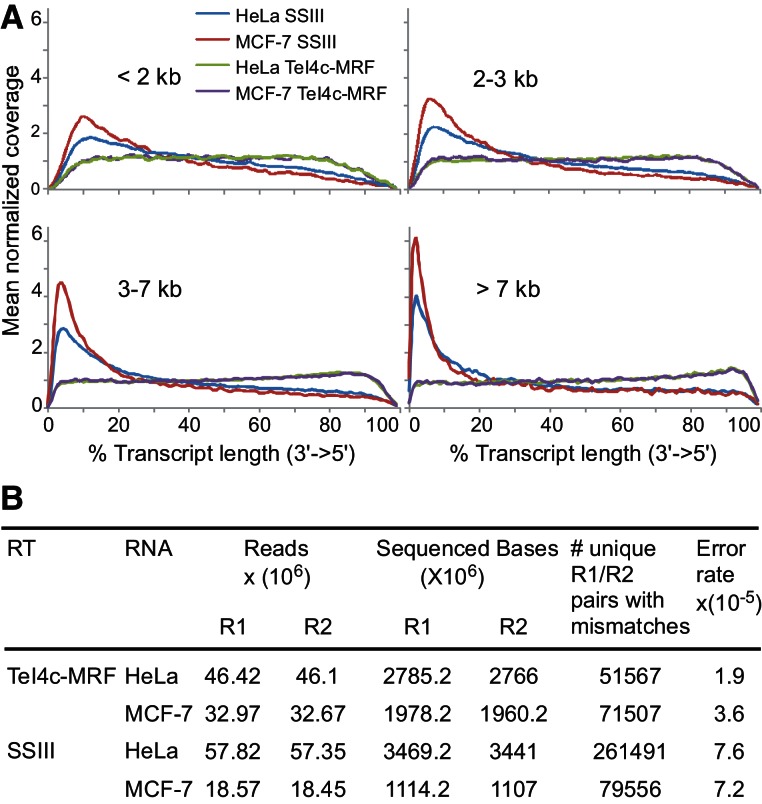
We compared the ability of the RTs to synthesize cDNAs by plotting the frequency of reads per unit length for 7203 curated human transcripts selected from the data sets (Fig. 5A). Because cDNA synthesis initiates from an oligo(dT)42 primer annealed to the poly(A) tail of mRNAs, the read density per unit length provides a measure of the processivity of the two enzymes on human mRNAs. The TeI4c-MRF samples had a fairly even distribution of read densities, even from transcripts >7 kb. In contrast, the SuperScript III samples displayed a pronounced 3′ bias in transcript coverage, even from transcripts of <2 kb. The slight 5′ bias seen for the longer transcripts in the group II intron RT samples may reflect internal initiations, which can occur for all RTs, but are either more frequent for the group II intron RT or not discernible for the retroviral RT due to its lower processivity. Similar data were obtained for the TeI4c-MRF libraries by SOLiD sequencing (Supplemental Fig. S3). The ability of the TeI4c-MRF RT to give a relatively uniform read distribution across transcript length utilizing an oligo(dT) primer enables RNA-seq of cellular mRNAs with minimal manipulation compared with standard RNA-seq methods, which require some combination of rRNA depletion/poly(A) selection, RNA fragmentation, or random priming to achieve uniformity (Wang et al. 2009; Ozsolak and Milos 2011).
FIGURE 5.
RNA-seq with a group II intron and retroviral RT. HeLa or MCF-7 RNAs were annealed with an oligo(dT)42 primer and incubated with TeI4c-MRF RT (1.24 μM) at 60°C or SuperScript III (10 units/μL) for 2 h at 50°C. The cDNAs were converted into RNA-seq libraries and paired-end sequenced on an Illumina HiSeq. (A) Distribution of reads per unit length for transcripts of different size classes. Reads were aligned using Eland-32 to a set of ∼7203 transcripts curated by selecting the longest isoform of each annotated gene from RefSeq (downloaded 11/2010), removing sequences containing ambiguous bases, and requiring that >95% of bases could be uniquely mapped to RefSeq and have mean base coverage >3X in standard brain and/or UHR mRNA data sets. (B) Error frequencies. Raw data were base-called using the Illumina Off-Line Basecaller (OLB version 1.9), and the resulting reads were aligned to human NCBI reference build 36 and splice junctions from UCSC refFlat (downloaded 02/2010) using Eland RNA (Casava 1.7) with default parameters. Potential RT errors were detected by looking for single-base mismatches relative to the reference sequence in overlapping sequence in both reads R1 and R2 of a paired-end cluster. Both R1 and R2 were required to have a base quality >25 and belong to a perfectly overlapping section of length ≥20 nt. Base mismatches common to both the TeI4c-MRF and SuperScript III libraries, which include sequence polymorphisms compared with the reference RNAs, were filtered out.
The paired-end sequencing data for the human cDNA libraries also provided an independent measure of RT error rate averaged for a large number of different transcripts. To calculate the error rate, we extracted only RefSeq mapped read pairs in which both reads have a mismatch to the reference in the same position. Unpaired mismatches or paired mismatches common to both enzymes were filtered out as either instrument error or sequence polymorphisms between the reference RNAs and experimental samples. The numbers of unique pairs containing a mismatch were then normalized to the total number of usable bases sequenced to obtain the error rate. Using this approach, we found that error rates for reverse transcription of the HeLa and MCF-7 RNAs by the TeI4c-MRF RT were 1.9 and 3.6 × 10−5, respectively, two- to fourfold lower than those for SuperScript III (7.6 and 7.2 × 10−5, respectively) (Fig. 5B). These results are in good agreement with the two- to fourfold lower error rates of the group II intron RTs in the M13-based lacZ forward mutation assay, where the fidelity of the enzymes was measured on a single RNA template (see above). In both assays, the error rates measured for the TeI4c and GsI-IIC group II intron RTs are lower than those reported in the literature for retroviral RTs (M-MLV RT; 3.6–6.7 × 10−5; HIV-1, >10−4) (Ji and Loeb 1992; Potter et al. 2003; Arezi and Hogrefe 2007).
Group II intron RT template switching enables attachment of adaptor sequences without RNA ligation
The cloning of cDNAs corresponding to nonpolyadenylated RNAs, such as miRNAs or protein-bound RNA fragments, requires the time-consuming and inefficient step of using an RNA ligase to attach oligonucleotide adaptors containing PCR primer-binding sites to the termini of the RNA or cDNA strand (Lau et al. 2001; Levin et al. 2010; Lamm et al. 2011). Moreover, RNA ligases commonly used for adaptor ligation have distinct preferences for the ends being ligated, thereby biasing representation of cDNAs in the resulting libraries (Linsen et al. 2009; Levin et al. 2010; Lamm et al. 2011). Some non-LTR-retroelements RTs differ from retroviral RTs in being able to template switch directly to the 3′ ends of new RNA templates that have little or no complementarity to the 3′ end of the cDNA (Kennell et al. 1994; Chen and Lambowitz 1997; Bibillo and Eickbush 2002b, 2004). We hypothesized that group II intron RTs might have a similar template-switching activity that could be used to synthesize a continuous cDNA that directly links an adaptor to a target RNA sequence without RNA ligation. The composite cDNA could then be circularized with CircLigase, an enzyme that efficiently circularizes single-stranded DNA (Polidoros et al. 2006) and PCR amplified with bidirectional primers that add barcodes for next-generation sequencing.
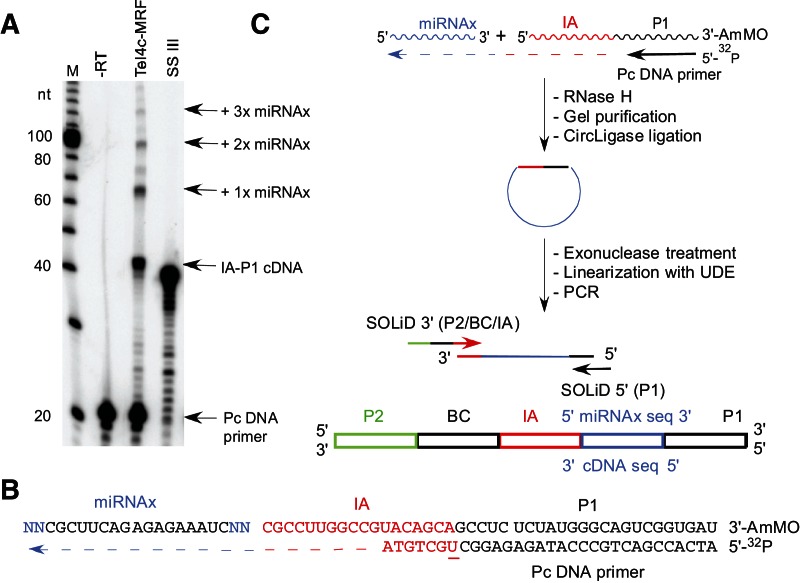
Figure 6A compares the ability of the TeI4c-MRF and SuperScript III RTs to template jump from a synthetic RNA template/DNA primer substrate containing the Internal Adaptor (IA) and P1 sequences for SOLiD next-generation sequencing to the 3′ end of a 21-nt RNA (denoted miRNAx), whose sequence is similar to that of a plant miRNA (Arabidopsis thaliana ath mir-173) (Fig. 6B). The miRNAx has two randomized nucleotide residues (N’s) at its 5′ and 3′ ends to assess biases during template switching, and the IA/P1 template RNA contains a 3′-aminomodifier (AmMO) to impede it from being recopied by template switching to its 3′ end (Fig. 6B). Whereas SuperScript III yields a single predominant product of ∼42 nt (IA–P1 cDNA) resulting from extension of the Pc primer to the 5′ end of the IA–P1 RNA, the TeI4c-MRF RT yields a similar but slightly larger product plus a series of bands of the sizes expected for template switching linking one, two, or three copies of the 21-nt miRNAx to the IA/P1 adaptor sequence.
FIGURE 6.
Template-switching activity of group II intron and retroviral RTs. (A) Gel assay. The initial 32P-labeled IA–P1 RNA/Pc DNA template-primer substrate (50 nM) and equimolar miRNAx were incubated with TeI4c-MRF RT (2 μM, 60°C) or SuperScript III (10 units/μL, 50°C; SSIII) for 15 min in the standard reaction medium for each enzyme (see Materials and Methods). The products were analyzed in a denaturing 20% polyacrylamide gel, which was scanned with a PhosphorImager. Lane “−RT” shows the IA–P1 RNA/Pc DNA substrate incubated under TeI4c-MRF RT conditions without RT. (Lane M) 32P-labeled 10-bp ladder size markers. (B) Template and primer sequences. The miRNAx target RNA has two randomized nucleotide residues (NN; blue) at each end to assess template-switching biases (Supplemental Fig. S4). The initial IA–P1 template RNA has a 3′ aminomodifier (AmMO) to impede template switching to that RNA end, and the Pc DNA primer is 5′ 32P-labeled and has an internal deoxyuridine (underlined) for relinearization of cDNAs after circularization with uracil–DNA excision mix (UDE; see below). (C) Protocol for the construction of cDNA libraries via group II intron RT template switching. In the first step, the group II intron RT template switches from the IA–P1 RNA/Pc DNA template/primer to miRNAx to generate a continuous cDNA that links the IA–P1 adaptor sequence to that of miRNAx. The products are then incubated with RNase H to digest the RNA template, gel-purified, and circularized with CircLigase. After digestion of unincorporated primers with exonuclease I, the cDNAs were relinearized with UDE at the deoxyuridine in the primer and amplified by PCR with primers that append adaptors and barcodes for next-generation sequencing.
The cDNA products synthesized by the TeI4c-MRF RT were excised from the gel, circularized with CircLigase, PCR amplified, and cloned and sequenced, as outlined in Figure 6C. The sequencing showed that the adaptor was linked seamlessly to the miRNA sequence by template switching and confirmed that the larger products resulted from single and multiple template switches linking one or more miRNAxs to the adaptor sequence (Supplemental Fig. S4). However, template switching under these conditions exhibited substantial bias, favoring miRNAs with a 3′ U-residue and disfavoring those with a 3′ A-residue. Related to this bias, the sequencing also showed that the TeI4c-MRF RT has a tendency to add extra nucleotide residues, mostly A-residues, to the 3′ ends of the cDNAs. Such “extra nucleotide addition,” sometimes referred to as terminal transferase activity, is a common property of RTs and DNA polymerases, a well-known application being TA cloning with Taq polymerase (Holton and Graham 1991). Although potentially useful for adding homopolymer tails to DNA ends, further analysis showed that the propensity of the TeI4c-MRF RT to add extra A residues to cDNA ends biases for template switching to a miRNA with a complementary 3′ U-residue and against template switching to a miRNA with a clashing 3′ A-residue.
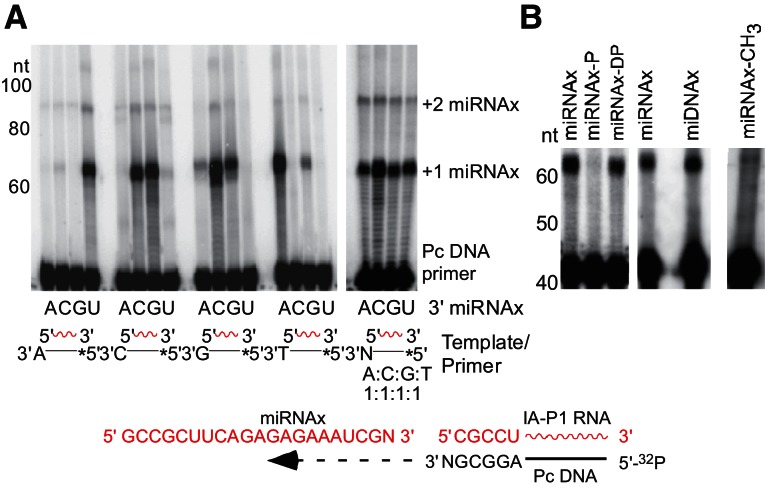
Although we developed methods for modulating this extra nucleotide-addition activity, resulting in a more uniform template switching (S Mohr and AM Lambowitz, unpubl.), we found a better approach to be that shown in Figure 7. In this approach, we circumvent biases resulting from uncontrolled extra nucleotide addition by using a mixture of initial template-primer substrates having different single-nucleotide 3′ overhangs of the priming strand, mimicking the structure expected for addition of a single extra nucleotide to the 3′ end of the cDNA. Figure 7 shows that such template-primer substrates favored template-switching to a miRNA having a complementary 3′-nucleotide residue, while an equimolar mixture of template-primer substrates with four different 3′ overhangs enabled more uniform template switching to miRNAs with different ends. Although retroviral RTs can template-switch by adding extra 3′-nucleotide residues to cDNAs, which then base pair to the new RNA template, at least two base pairs are required, one of which must be a relatively stable GC or CG pair (Oz-Gleenberg et al. 2011). The novel template-switching activity of the TeI4c-MRF RT can be used for the approach shown in Figure 7 because only a single base pair of any type is sufficient to promote template switching, even at 60°C, the operational temperature of this RT. Template-primers with different 3′ overhangs could be used separately to favor amplification of a specific RNA of known sequence (e.g., for qPCR quantitation) or together for cloning libraries of RNAs of unknown sequence. We note that although base-pairing appears to favor template switching, group II intron RTs may also be able to template switch without base-pairing, as appears to be the case for the Mauriceville retroplasmid RT (Kennell et al. 1994; Chen and Lambowitz 1997).
FIGURE 7.
Template-switching of group II intron RTs from 3′-overhang substrates. (A) Template-switching reactions were done with miRNAxs having different 3′-nucleotide residues (lanes A, C, G, U) and initial 32P-labeled RNA template/DNA primer substrates (IA–P1 RNA/Pc 3′-overhang DNA) having different single nucleotide 3′ overhangs (A, C, G, T, or an equimolar mixture of all four nucleotides [N]; shown schematically below gel). Reactions were with 2 µM TeI4c-MRF RT for 10 min at 60°C in a high-salt reaction medium (450 mM NaCl, 5 mM MgCl2, 20 mM Tris-HCl [pH 7.5], 1 mM DTT, 1 mM dNTPs), which reduces nontemplated nucleotide addition by the RT. The products were analyzed in a denaturing 20% polyacrylamide gel, which was scanned with a PhosphorImager. Numbers to left of the gel indicate positions of labeled size markers (10-bp ladder). (*) 32P-label at the 5′ end of primer. (B) Template switching from IA–P1 RNA/Pc DNA with equimolar single-nucleotide 3′ overhangs to an miRNAx with a 3′ phosphorylated C-residue before and after dephosphorylation with T4 polynucleotide kinase (P and DP, respectively); a DNA oligonucleotide of identical sequence (miDNAx); or an miRNAx with a 2′ O-methyl group (CH3) at its 3′ end.
Use of group II intron RT template-switching for miRNA cloning and sequencing
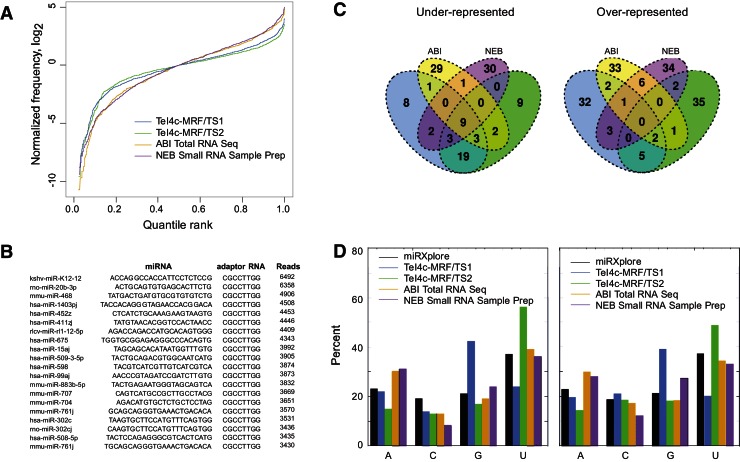
To assess its utility for library construction, we used group II intron RT template switching and two commercial kits utilizing conventional RNA-ligation methods to generate libraries for SOLiD sequencing of a reference set consisting of 963 equimolar miRNAs. We then compared the library abundance of 898 of these miRNAs with uniquely identifiable core sequences. The plots show that two libraries prepared by TeI4c-MRF RT template switching from initial template-primer substrates with different ratios of single-nucleotide 3′ overhangs (TS1 and TS2) have more uniform distributions of miRNA sequences (flatter lines) than those prepared by either commercial kit (Fig. 8A). Importantly, >97% of the miRNA sequences begin directly at the 3′ end of the miRNA and had seamless template-switching junctions with no extra nucleotide residues incorporated between the miRNA and the adaptor (Fig. 8B). Analysis of outliers identified nine miRNAs that were under-represented in all libraries, but otherwise little overlap between the miRNAs that were under- or over-represented by the different methods (Fig. 8C). Finally, we found that changes in the ratios of single-nucleotide 3′ overhangs of the initial template-primer substrates used for template switching affected the miRNA distribution in the libraries in the manner predicted for base-pairing of the 3′ overhang residue to the 3′-terminal residue of the miRNA (Fig. 8D). Thus, the ratio of 3′ overhangs could be adjusted to obtain either unbiased RNA profiles or to preferentially reverse transcribe specific target RNAs.
FIGURE 8.
Cloning and sequencing of miRNAs by using group II intron RT template switching. Template-switching reactions were done with TeI4c-MRF RT (2 µM) to a miRNA reference set (963 equimolar miRNAs, 110 nM; Miltenyi miRXplore) from an initial IA–P1 RNA template/Pc DNA primer substrate (100 nM). The latter had single A, C, G, or T 3′-overhangs mixed at an equimolar ratio (TS1) or at 2:0.5:1:1 (TS2) to adjust the representation of miRNAs with 3′ U- or G- residues. Reactions were done as in Figure 7 and cDNAs were cloned as in Figure 6C. Parallel RNA-seq libraries were prepared from equal aliquots of the miRNAs by using either a Total RNA-Seq kit (Applied Biosystems; ABI) or a small RNA sample prep set 3 kit (New England BioLabs; NEB). These kits ligate adaptors for SOLiD sequencing to the miRNA 3′ and 5′ ends simultaneously (ABI) or sequentially (NEB) and reverse transcribe with ArrayScript or SuperScript II using a DNA primer complementary to the 3′ adaptor. (A) Plots showing counts for a subset of 898 miRNA with uniquely identifiable 16-bp core sequences (nucleotides 4 through 20) ranked from the least to most abundant, median normalized, log2 transformed, and plotted to compare variance introduced by the library preparation method. To ensure no ambiguity of sequence mismatch across the miRNA reference panel while allowing for possible method-specific biases at the 3′ or 5′ ends, the distal sequencing adaptor sequence was concatenated to each mature miRNA sequence (the “concatenated reference”), and nucleotides 4 through 20 from each concatenated sequence were tested for occurrence within the concatenated reference anywhere in colorspace. Only concatenated sequences with no overlap to any other 16-bp core sequence were chosen for quantitation. (B) Template-switching junctions between the 3′ end of the miRNA and adaptor (IA) sequence of the 20 most frequent sequence reads from the TS1 library. (C) Venn diagrams showing overlap between under- and over-represented miRNAs in the RNA-Seq libraries prepared by the different methods. The 5% least and most abundant miRNAs in each library were identified using R and plotted using the VennDiagram R package (Chen and Boutros 2011). (D) Representation of miRNA 3′-terminal nucleotide residues in RNA-seq libraries. The bar graphs compare the percentage of miRNAs ending in each of the four bases in the miRXplore reference set (black) with the percentage of that base at the 3′ end of miRNAs in the RNA-seq libraries (TeI4c-MRF/TS1, blue; TeI4c-MRF/TS2, green; ABI Total RNA Seq, gold; NEB Small RNA Sample Prep, purple). (Left) The 3′-nucleotide residue of miRNAs in the RNA-seq libraries was identified as the base prior to the Internal Adaptor. To avoid primer-dimer, adaptor-only, and low-quality sequences, a perfect match to eight bases of the Internal Adaptor no closer than 15 bp from the start of each sequence was required when determining the terminal base in each sample. (Right) The distribution of 3′-nucleotide residues of the miRNAs in the RNA-seq libraries was inferred from the abundance-adjusted distribution of the set of 898 miRNAs identified by their unique core sequences. Similar trends were seen for both methods of identifying the 3′-terminal residue of the miRNA.
Further characterization showed that the group II intron RT template-switching reaction: (1) is inhibited by a 3′ phosphate, which would result from conventional RNase- or alkali-cleavage, but restored by 3′ phosphate removal; (2) occurs to DNA as well as RNA, indicating that a 2′ OH group on the 3′-terminal nucleotide is not required; and (3) occurs to a miRNA with a 2′ OMe at its 3′ end, albeit at reduced efficiency (∼10% that of the same oligonucleotide with a 2′ OH) (Fig. 7B). Thus, this reaction should be generally useful for cloning nonpolyadenylated RNAs, including protein-bound RNA fragments generated by RNase digestion in procedures such as HITS-CLIP/CRAC or ribosome profiling (Granneman et al. 2009; Ingolia et al. 2009; Zhang and Darnell 2011), and perhaps in the construction of DNA-seq libraries.
Collectively, our results demonstrate general methods for the high-level expression of group II intron RTs and advantages of these enzymes for cDNA synthesis, RT-PCR, and RNA-seq. In contrast to currently used methods utilizing retroviral RTs, the thermostable group II intron RT fusion proteins described here enable uniform transcriptome profiling of whole-cell RNAs without RNA fragmentation by using an oligo(dT) primer, preserving information, such as patterns of alternative splicing in long transcripts, that would otherwise be lost. The group II intron RT fusion proteins also enable less-biased profiling of miRNAs and other nonpolyadenylated RNAs and RNA fragments by template switching without the time-consuming and inefficient step of using RNA ligase for linker ligation. The high processivity of the enzymes should make them particularly useful for analysis of RNAs with stable secondary structures, and their high fidelity should be advantageous for the analysis of sequence variants in applications, such as tumor profiling. Finally, the methods developed here for the expression of highly active group II intron RTs with a rigidly linked solubility tag may be generally applicable to non-LTR retroelement RTs and other difficult to express enzymes.
MATERIALS AND METHODS
Recombinant plasmids
pMalE-RT constructs (e.g., pMalE-TeI4c, pMalE-TeI4h*) contain the indicated RT ORF with an N-terminal MalE tag cloned behind the tac promoter in pMal-c2t (derived from pMal-c2x; New England BioLabs) (Kristelly et al. 2003). TeI4h* RT is a derivative of the native TeI4h RT, in which the YAGD motif in conserved sequence block RT-5 was changed to YADD (Mohr et al. 2010). pMalE-TeI4f, -TeI4c, and -TeI4h* were constructed by PCR amplifying the RT ORFs of Thermosynechococcus elongatus group II introns cloned in pET11 (TeI4f), pUC19 (TeI4c), or pACD2X (TeI4h*) (Mohr et al. 2010) with primers that append restriction sites, and then cloning the PCR products into the corresponding sites of pMal-c2t (TeI4c, EcoRI and PstI sites; TeI4f, BamHI site; TeI4h*, BamHI and PstI sites). pMalE-GsI-IIB and pMalE-GsI-IIC were constructed by PCR amplifying the RT ORFs from Geobacillus stearothermophilus strain 10 genomic DNA (obtained from Greg Davis, Sigma-Aldrich) with primers that add a C-terminal His6 tag and append BamHI and XbaI (GsI-IIB) or BamHI (GsI-IIC) sites and cloning the PCR products between the corresponding sites of pMal-c2t. GsI-IIB is a subgroup IIB2 intron that is inserted in the G. stearothermophilus recA gene and is related to previously described RT-encoding group II introns in the recA genes of Geobacillus kaustophilus (Chee and Takami 2005) and Bacillus caldolyticus (Ng et al. 2007). GsI-IIC is a group IIC intron found in multiple copies in the G. stearothermophilus genome (Moretz and Lampson 2010). The cloned GsI-IIC RT ORF corresponds to one of these genomic sequences and has three amino acid sequence changes compared with a related RT ORF cloned by Vellore et al. (2004). pMalE-LtrA was constructed by PCR amplifying the Ll.LtrB RT (LtrA protein) ORF of pImp-2 (Saldanha et al. 1999), using primers that append BamHI and HindIII sites and cloning the PCR product between the corresponding sites of pMal-c2t.
pMRF-RT constructs (e.g., pMRF-TeI4c) contain the indicated RT ORF with a MalE tag linked in frame to the N terminus of the ORF via a rigid fusion. They were derived from the corresponding pMalE-RT plasmids by replacing the TEV protease-cleavable linker (TVDEALKDAQTNS3N10LENLYFQG) with a rigid linker (TVDAALAAAQTAAAAA) by using QuikChange PCR mutagenesis with Accuprime polymerase (Life Technologies) (Makarova et al. 2000). Derivatives of pMRF-TeI4c with different linkers between the MalE tag and group II intron RT were also constructed by QuikChange. pNusA-RF-TeI4c-His expresses the TeI4c RT with a NusA tag fused to the N terminus of the protein via a rigid linker and a C-terminal His6 tag.
Protein purification
MalE and MRF group II intron RT fusion proteins were expressed in E. coli Rosetta 2 (EMD Chemicals) or ScarabXpress T7lac (Scarabgenomics). E. coli strains were transformed with the expression plasmid and grown at 37°C in 500-mL TB medium in 2.5-l Ultrayield flasks (Thompson Instrument Company) or 1-l LB medium in 4-l Erlenmeyer flasks. Expression was induced either by adding isopropyl β-D-1-thiogalactopyranoside (IPTG; 1 mM final) to mid-log phase cells (O.D.600 = 0.8; pMRF-TeI4c, -TeI4f, -TeI4h*, GsI-IIB, and GsI-IIC) or by growing cells in auto-induction medium (LB containing 0.2% lactose, 0.05% glucose, 0.5% glycerol, 24 mM (NH4)2SO4, 50 mM KH2PO4, 50 mM Na2HPO4; pMalE-LtrA and pMRF-LtrA). In either case, the cells were induced at 18°C–25°C for ∼48 h, pelleted by centrifugation, resuspended in buffer A (20 mM Tris-HCl [pH 7.5], 0.5 M KCl, 1 mM EDTA, 1 mM dithiothreitol [DTT]), and frozen at −80°C.
The TeI4c-, TeI4f-, TeI4h*-, GsI-IIC-, and LtrA-MRF RTs were purified by a procedure that involves cell disruption by freeze-thawing and sonication; polyethyleneimine (PEI) precipitation of nucleic acids; amylose-affinity chromatography; and heparin-Sepharose chromatography. The cell suspension was thawed, treated with lysozyme (1 mg/mL; Sigma) for 15 min on ice, then subjected to three cycles of freeze-thawing on dry ice, followed by sonication (Branson 450 Sonifier; amplitude 60% on ice; one 30-sec burst or three or four 10-sec bursts with 10 sec between bursts). After centrifuging to pellet cell debris, nucleic acids were precipitated by adding PEI to a final concentration of 0.1% and centrifuging at 15,000g for 15 min at 4°C (J16.25 rotor; Avanti J-E centrifuge; Beckman Coulter). The resulting supernatant was loaded onto an amylose column (Amylose High-Flow; New England BioLabs; 10-mL column equilibrated in buffer A), which was then washed with five column volumes each of buffer A containing 0.5, 1.5, or 0.5 M KCl, and eluted with buffer A containing 10 mM maltose. Protein fractions were pooled and purified further by heparin-Sepharose chromatography (three tandem 1-mL columns; GE Healthcare Biosciences). In initial experiments, the heparin-Sepharose column was equilibrated and the samples were loaded in 20 mM Tris-HCl (pH 7.5), 50–100 mM KCl, 1 mM EDTA, 1 mM DTT, 10% glycerol; but in later experiments, the KCl concentration in the loading buffer was increased to 500 mM, which improved solubility and yields. The proteins were applied to the column in the same buffer and eluted with a 40-column volume KCl gradient from the loading concentration to 2 M. Peak fractions of the RTs, which eluted at ∼800 mM KCl, were pooled and dialyzed against 20 mM Tris-HCl (pH 7.5), 0.5 M KCl, 1 mM EDTA, 1 mM DTT, and 50% glycerol, flash-frozen, and stored at −80°C. The TeI4c-NRF and GsI-IIB-MRF RTs were purified similarly, except that the amylose column was replaced (TeI4c-NRF) or followed (GsI-IIB-MRF) by a nickel column.
Protein concentrations were determined either by using the Bradford assay (Bradford 1976) with bovine serum albumin as a standard or by using the Qubit fluorescent assay according to the manufacturer’s instructions (Life Technologies). A unit of RT activity is defined as the amount of enzyme required to polymerize 1 nmol of dTTP in 1 min at 60°C, using poly(rA)/oligo(dT)42 as template, as described below. All protein preparations were >95% pure, and RT activity was unchanged after 6 mo of storage at −80°C. Very concentrated protein preparations (>15 mg/mL) tended to lose up to 20% of the protein due to precipitation over time, but the remaining soluble protein remained fully active, as determined by remeasuring activity before each use. The yields of TeI4c-MRF and GsI-IIC-MRF RTs grown in TB medium in Ultrayield flasks were 5–20 mg/L.
Retrohoming assays
The ability of group II intron RT fusion proteins to support retrohoming in vivo was tested by using an E. coli plasmid-based assay in which a group II intron with a phage T7 promoter sequence inserted near its 3′ end is expressed from a donor plasmid and retrohomes into a target site cloned in a recipient plasmid upstream of a promoterless tetR gene, thereby activating that gene (Guo et al. 2000; Karberg et al. 2001). The intron-donor plasmids, which carry a capR marker on the vector backbone, were derivatives of pACD2x (San Filippo and Lambowitz 2002) and use a T7lac promoter to express the group II intron RNA with the ORF deleted, followed in tandem by the RT being tested. The recipient plasmids, which carry an ampR marker on the vector backbone, were derivatives of pBRR3-ltrB (Guo et al. 2000; Karberg et al. 2001) and contain the target site for the intron being tested (positions −30 to +15 from the intron-insertion site). Retrohoming efficiencies were quantified in plating assays as the ratio of (TetR + AmpR)/AmpR colonies. The retrohoming efficiencies reported in Results were not normalized for protein expression levels.
Reverse transcription assays
Unless specified otherwise, reverse transcription reaction media were: TeI4c-MRF, 75 mM KCl, 10 mM MgCl2, 20 mM Tris-HCl (pH 7.5), 1 mM DTT; GsI-IIC-MRF, 10 mM NaCl, 10 mM MgCl2, 20 mM Tris-HCl (pH 7.5), 1 mM DTT; SuperScript III (Life Technologies; 75 mM KCl, 3 mM MgCl2, 50 mM Tris-HCl [pH 8.3], 5 mM DTT).
RT assays with poly(rA)/oligo(dT)42 were done by preincubating the RT (50 nM TeI4c-MRF RT or 100 nM of all other RTs) with poly(rA)/oligo(dT)42 (100 nM) in 75 mM KCl, 10 mM MgCl2, 20 mM Tris-HCl (pH 7.5), 1 mM DTT for 2 min at the desired temperature, and then initiating the reaction by adding 5 μCi [α-32P]dTTP (3000 Ci/mmol; PerkinElmer). The reactions were incubated for times that were within the linear range for each protein preparation and stopped by adding EDTA to a final concentration of 250 mM. Reaction products were spotted onto Whatman DE81 paper (10 × 7.5-cm sheets; GE Healthcare Biosciences), which was then washed three times with 0.3 M NaCl and 0.03 M sodium citrate, dried, and scanned with a PhosphorImager (Typhoon Trio Variable Mode Imager; GE Healthcare Biosciences) to quantify the bound radioactivity. For specific activity measurements, the assays were done at several different protein concentrations, with 1 mM unlabeled dTTP added to the reaction mixture.
For gel assays of cDNA synthesis at different temperatures (Fig. 2A), the RT (2 μM TeI4c-MRF; 200 nM GsI-IIC-MRF; or 10 units/µL SuperScript III [Life Technologies]) were preincubated with 100 nM RNA template annealed to a 5′-labeled DNA primer for 2 min at the desired temperature in RT reaction medium. The RNA template was a 509-nt RNA transcribed with phage T7 RNA polymerase from pBluescript KS(+) (Stratagene) digested with AflIII, and the annealed primer was AflIIIR (5′-CCGCCTTTGAGTGAGCTGATACCGCTCGCCGCAGCCG). The reactions were initiated by adding 1.25 mM dNTPs (1.25 mM each of dATP, dCTP, dGTP, and dTTP), incubated for 30 min, and terminated by adding 0.1% SDS/25 mM EDTA (final concentrations), followed by extraction with phenol-chloroform-isoamyl alcohol (25:24:1; phenol-CIA). The products were analyzed in a denaturing 6% polyacrylamide gel, which was dried and quantified with a PhosphorImager. A 5′-labeled 10-bp ladder (Life Technologies) was run in parallel to provide size markers. Gel assays for quantitative processivity measurements were done similarly with 50 nM substrate (an 807-nt in vitro transcript containing an Ll.LtrB-ΔORF + ΔA group II intron with the ORF and branch-point A-residue deleted [28-nt 5′ exon, 749-nt intron, 30-nt 3′ exon] with 5′-labeled primer Ll.LtrBΔA Rev [5′-GTGAAGAGGGAGGTACCGCCTTGTT] annealed near its 3′ end). For these assays, the enzyme was preincubated with the substrate for 30 min at room temperature prior to initiating the reaction by adding 1.25 mM dNTPs and 20–40 μM poly(rA)/oligo(dT)42 to trap dissociated RT, and the reaction was terminated by adding 0.1% SDS and 0.5 mg/mL proteinase K (final concentrations) and incubating at 37°C for 30 min. The processivity (average length of template copied per initiation) was calculated by using the equation Σ(Ln.In)/Σ(In), where Ln is the length and In is the intensity of each analyzed cDNA fragment.
Capillary electrophoresis assays of cDNA synthesis used the same 807-nt RNA group II intron RNA template described above for gel assays of processivity with a fluorescently labeled DNA primer (WellRED D4: 5′-/5D4/GTGAAGTAGGGAGGTACCGCCTTGTTC; IDT). The annealed template-primer substrate (100 nM) was incubated with TeI4c-MRF (1 μM) or GsI-IIC-MRF (200 nM) RTs for 2 min at reaction temperature in 75 mM KCl, 10 mM MgCl2, 20 mM Tris-HCl (pH 7.5), 5 mM DTT prior to initiating the reactions by adding 1 mM dNTPs. Reverse transcription with SuperScript III was done with 10 units enzyme/µL according to the manufacturer’s protocol either in the provided reaction medium or in the same reaction medium as the group II intron RTs. The reactions were incubated for 30 min at 60°C for the group II intron RTs or 50°C for SuperScript III and stopped by adding NaOH to a final concentration of 0.1 M, incubating at 95°C for 3 min, and neutralizing with HCl. After ethanol precipitation in the presence of 0.3 M NaOAc and glycogen carrier (5 μg; Fermentas), the cDNA pellets were washed with ice-cold 70% ethanol, dried, and dissolved in distilled water, and portions were analyzed by using a GenomeLab GeXP Genetic Analysis System (Beckman Coulter). Samples were denatured at 90°C for 180 sec, injected into the capillary array at 2.0 kV for 30 sec, and separated at 4.8 kV for 100 min. The temperature of the capillary array was maintained at 60°C throughout the separation. Peaks were discriminated from background by analyzing the raw data using MS Excel and Kaleidagraph, and cDNA lengths were assigned relative to WellRED dye D1-labeled DNA size standards (BioVentures), which were run together with the cDNAs.
Quantitative real-time reverse transcription-polymerase chain reaction (qRT-PCR)
cDNAs were synthesized at 60°C in 20-µL reactions containing 200 nM TeI4c-MRF RT, RT buffer (75 mM KCl, 10 mM MgCl2, 20 mM Tris-HCl at pH 7.5, 1 mM DTT), 1 mM dNTPs, and 5 × 108 copies of 1.2-kb KanR RNA (Promega) with annealed primer P078 (5′-GGTGGACCAGTTGGTGATTTTGAACTTTTGCTTTGCCACGGAAC). After a 2-min preincubation in reaction medium containing all other components at 60°C, reactions were initiated by adding 1 mM dNTPs, incubated at 60°C for 30 min, and terminated by freezing on dry ice.
To quantitate KanR cDNA, 25-µL reactions were done in triplicate in 96-well plates with optical caps with each well containing 5 µL of cDNA (corresponding to 1.25 × 107 copies of kanR RNA, 2X TaqMan Gene Expression Master Mix [Life Technologies], primer-probe mix [200 nM FAM-BFQ1 probe], and 300 nM forward and reverse primers) Primer set 188–257: Forward P09 kan-188F, 5′-GGGTATAAATGGGCTCGCG; Reverse P030 kan-257R, 5′-CGGGCTTCCCATACAATCG; Taqman probe P031 kan-213T, 5′-(6FAM, 6-carboxyfluorescein)-TCGGGCAATCAGGTGCGACAATC/3IABkFQ/(Iowa Black Fluorescence Quencher). Primer set 562–634: Forward P001 kan-562F 5′-CGCTCAGGCGCAATCAC; Reverse P002 kan-634R 5′-CCAGCCATTACGCTCGTCAT; Taqman probe P003 kan-581T 5′-(6-FAM)-ATGAATAACGGTTTGGTTGATGCGAGTGA (TAMRA, tetramethyl-6-carboxyrhodamine) (de Rozieres et al. 2004). Plasmid pET9a (EMD Chemicals) was used to generate a standard curve to quantitate KanR cDNA levels. qPCR was performed on the 7900HT Fast Real-Time PCR System (Applied Biosystems), using the 9600 emulation mode protocol (50°C for 2 min, 95°C for 10 min, then 45 cycles at 95°C for 15 sec, and 60°C for 60 sec). Data were collected and analyzed by using Life Technologies SDS Versions 2.3 software, and cycle thresholds for cDNA samples were plotted against the standard curve to determine copy number equivalents.
M13-based lacZ forward mutation assays of RT fidelity
M13-based lacZ forward mutations assays were as described (Ji and Loeb 1992) using a 269-nt RNA template corresponding to a segment of the LacZ α-fragment (positions +64 to +143) with a 5′-32P-labeled DNA primer annealed near its 3′ end. The 269-nt RNA was transcribed with T7 RNA polymerase from pBluescript KS(+) that had been digested with PvuI, and the annealed primer was pBluescript 550R (5′-CGCTATTACGCCAGCTGGCGAAAGGGGGATGT). Reverse transcription was done as for gel assays with the annealed template/primer substrate (100 nM), dNTPs (1 mM), and group II intron RT (2 μM) or SuperScript III (10 units/µL). The reactions were initiated by adding the RT, incubated for 15 min at 60°C (group II intron RTs) or 55°C (SuperScript III RT), and terminated by adding 125 mM EDTA. After hydrolyzing the RNA by incubating with 0.1 M NaOH at 95°C for 3 min and neutralizing with 0.1 M HCl, the cDNAs were purified in a denaturing 20% polyacrylamide gel, annealed to primer pBluescript HaeIII (5′ GTTGTAAAACGACGGCCAGTGAATTGTAATAC), and digested with HaeIII, then annealed to uracil-containing single-stranded M13 DNA (prepared as described at CSH protocols.org). The annealed cDNA was extended to synthesize the opposite M13 strand with phage T7 DNA polymerase (New England Biolabs) according to the manufacturer’s instructions. A portion of the extension reaction was electroporated into E. coli MC1061 F+ cells, which were plated at a density of 300–500 plaques per plate for blue/white screening. To identify mutations, the double-stranded M13 DNA was isolated from white plaques, as described (cshprotocols.org), and the lacZ α fragment was PCR amplified by using the M13 forward and reverse primers (http://cshprotocols.cshlp.org) and sequenced by the Sanger method using the M13 reverse primer. Background was determined by electroporating M13mp2 single-stranded DNA into MC1061 F+ and scoring for white plaques.
Next-generation sequencing of human cDNA libraries
RNA-seq of human mRNAs was done on whole-cell RNAs extracted from HeLa and MCF-7 cells using TRIzol (Life Technologies). A portion of the RNA preparation (500 ng) was mixed with oligo(dT)42 primer (3.3 μM final) and 1.6 mM of each of the four dNTPs in distilled water, heated to 65°C for 5 min, and cooled on ice for 5 min to anneal the primer before adding the remainder of the reaction medium. Reactions were initiated by adding TeI4c-MRF RT (1.24 μM) or SuperScript III (10 units/μL) and incubated for 2 h at 60°C and 50°C, respectively. The second DNA strand was synthesized with an NEBNext Second-Strand Synthesis kit (New England BioLabs), and the resulting double-stranded DNAs were either tagged by using a Nextera kit (Epicentre) and sequenced on an Illumina HiSeq instrument (Fig. 5) or fragmented by sonication (NEB Next protocol), tagged using an NEBNext kit, and sequenced on a SOLiD 4 instrument (Applied Biosystems; Supplemental Fig. S3).
Group II intron RT template switching
For group II intron RT template switching, we used an initial RNA template/DNA primer consisting of RNA oligonucleotide IA–P1 with a 3′ aminomodifier (AmMO, a primary amine attached via a linker of six to seven carbons; IDT) annealed at a 1:1.1 molar ratio to 5′-labeled primer Pc containing a deoxyuridine (sequences given in Fig. 6B). For reverse transcription reactions, the annealed template/primer substrate (50 or 100 nM) was incubated with eqimolar miRNAx and TeI4c-MRF RT (2–2.5 μM final) in 50–100 μL of its standard reaction medium (experiment in Fig. 6) or reaction medium containing 450 mM NaCl, 5 mM MgCl2, 20 mM Tris-HCl, (pH 7.5), 1 mM DTT, and 1 mM dNTPs (all other experiments) and incubated at 60°C for times indicated in the figure legends for individual experiments. The reactions were initiated by adding the RT and terminated by phenol–CIA extraction and ethanol precipitation. After incubating the products with thermostable RNase H (0.125 units/µL; Hybridase; Epicentre) for 5 min at 55°C, the cDNAs were size selected in a denaturing 20% polyacrylamide gel, extracted by soaking overnight in Tris-EDTA (10:1), followed by extraction with phenol-CIA and ethanol precipitation in the presence of 0.3 M sodium acetate and linear acrylamide carrier (0.005%). The cDNAs were circularized with CircLigase I (Epicentre; experiment in Supplemental Fig. S4) or CircLigase II (Epicentre; all other experiments) and treated with exonuclease I (Epicentre) to remove any remaining linear cDNA molecules, all according to the manufacturer’s instructions. The circularized cDNAs were then relinearized by using an Epicentre uracil DNA excision (UDE) kit according to the manufacturer’s instructions, and ethanol precipitated. The reaction products were amplified with Accuprime Pfx polymerase (Life Technologies) or Phusion Flash (New England BioLabs) using the SOLiD 5′ and 3′ primers (SOLiD 5′: 5′-CCACTACGCCTCCGCTTTCCTCTCTATGGGCAGTCGGTGAT; SOLiD 3′: 5′-CTGCCC CGGGTTCCTCATTCTCT/BARCODE/CTGCTGTACGGCCAAG GCG for 15 cycles of 95°C, 55°C, and 68°C for 5 sec each. The PCR products were band isolated from a 3% agarose gel (Wizard SV Gel and PCR Clean-Up Kit: Promega). They were then either TA cloned (Taq DNA polymerase, TOPO TA cloning kit; Life Technologies) and Sanger sequenced with the M13 F(-20) primer or sequenced on the 5500 XL (SOLiD) instrument (Applied Biosystems) to 35-bp of sequence.
The cloning and sequencing of the miRNA reference set (miRXplore; Miltenyi Biotech) was done similarly using the reference panel RNAs (110 nM) and initial IA–P1 RNA template/Pc DNA primer substrates (100 nM) with single nucleotides A, C, G, or T 3′-overhangs mixed at an equimolar ratio or at a ratio of 2:0.5:1:1 to adjust the representation of miRNAs with 3′ U or G residues.
DATA DEPOSITION
RNA-seq data for the experiments in Figures 5, 8, and Supplemental Figure S3 have been deposited in NCBI’s SRA under the accession number SRP021468.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
COMPETING INTEREST STATEMENT
Thermostable group II intron RT fusion proteins and methods for their use are the subject of patent applications that have been licensed by the University of Texas to InGex, LLC, which sublicenses the technology for commercial use. A.M.L. and the University of Texas are minority equity holders in InGex, LLC, and S.M., E.G., S.K., and A.M.L. may receive royalty payments from the licensing of intellectual property. S.S. and S.K. are employed by companies that are potential licensees of the technology.
ACKNOWLEDGMENTS
We thank Gary Latham (Asuragen) for helpful discussions. This work was supported by NIH grants GM37949 and GM37951 and Welch Foundation grant F-1607 to A.M.L.
REFERENCES
- Adey A, Morrison HG, Asan, Xun X, Kitzman JO, Turner EH, Stackhouse B, MacKenzie AP, Caruccio NC, Zhang X, et al. 2010. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biol 11: R119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arezi B, Hogrefe HH 2007. Escherichia coli DNA polymerase III ε subunit increases Moloney murine leukemia virus reverse transcriptase fidelity and accuracy of RT–PCR procedures. Anal Biochem 360: 84–91 [DOI] [PubMed] [Google Scholar]
- Arezi B, Hogrefe H 2009. Novel mutations in Moloney Murine Leukemia Virus reverse transcriptase increase thermostability through tighter binding to template-primer. Nucleic Acids Res 37: 473–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranauskas A, Paliksa S, Alzbutas G, Vaitkevicius M, Lubiene J, Letukiene V, Burinskas S, Sasnauskas G, Skirgaila R 2012. Generation and characterization of new highly thermostable and processive M-MuLV reverse transcriptase variants. Protein Eng Des Sel 25: 657–668 [DOI] [PubMed] [Google Scholar]
- Beckman RA, Mildvan AS, Loeb LA 1985. On the fidelity of DNA replication: Manganese mutagenesis in vitro. Biochemistry 24: 5810–5817 [DOI] [PubMed] [Google Scholar]
- Bibillo A, Eickbush TH 2002a. High processivity of the reverse transcriptase from a non-long terminal repeat retrotransposon. J Biol Chem 277: 34836–34845 [DOI] [PubMed] [Google Scholar]
- Bibillo A, Eickbush TH 2002b. The reverse transcriptase of the R2 non-LTR retrotransposon: Continuous synthesis of cDNA on non-continuous RNA templates. J Mol Biol 316: 459–473 [DOI] [PubMed] [Google Scholar]
- Bibillo A, Eickbush TH 2004. End-to-end template jumping by the reverse transcriptase encoded by the R2 retrotransposon. J Biol Chem 279: 14945–14952 [DOI] [PubMed] [Google Scholar]
- Blocker FJ, Mohr G, Conlan LH, Qi L, Belfort M, Lambowitz AM 2005. Domain structure and three-dimensional model of a group II intron-encoded reverse transcriptase. RNA 11: 14–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Candales MA, Duong A, Hood KS, Li T, Neufeld RA, Sun R, McNeil BA, Wu L, Jarding AM, Zimmerly S 2012. Database for bacterial group II introns. Nucleic Acids Res 40: D187–D190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chee GJ, Takami H 2005. Housekeeping recA gene interrupted by group II intron in the thermophilic Geobacillus kaustophilus. Gene 363: 211–220 [DOI] [PubMed] [Google Scholar]
- Chen H, Boutros PC 2011. VennDiagram: A package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics 12: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Lambowitz AM 1997. De novo and DNA primer-mediated initiation of cDNA synthesis by the Mauriceville retroplasmid reverse transcriptase involve recognition of a 3′ CCA sequence. J Mol Biol 271: 311–332 [DOI] [PubMed] [Google Scholar]
- Conlan LH, Stanger MJ, Ichiyanagi K, Belfort M 2005. Localization, mobility and fidelity of retrotransposed group II introns in rRNA genes. Nucleic Acids Res 33: 5262–5270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Matsuura M, Wang Q, Ma H, Lambowitz AM 2004. A group II intron-encoded maturase functions preferentially in cis and requires both the reverse transcriptase and X domains to promote RNA splicing. J Mol Biol 340: 211–231 [DOI] [PubMed] [Google Scholar]
- de Rozieres S, Swan CH, Sheeter DA, Clingerman KJ, Lin YC, Huitron-Resendiz S, Henriksen S, Torbett BE, Elder JH 2004. Assessment of FIV-C infection of cats as a function of treatment with the protease inhibitor, TL-3. Retrovirology 1: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman S, Kudla G, Petfalski E, Tollervey D 2009. Identification of protein binding sites on U3 snoRNA and pre-rRNA by UV cross-linking and high-throughput analysis of cDNAs. Proc Natl Acad Sci 106: 9613–9618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Karberg M, Long M, Jones JP III, Sullenger B, Lambowitz AM 2000. Group II introns designed to insert into therapeutically relevant DNA target sites in human cells. Science 289: 452–457 [DOI] [PubMed] [Google Scholar]
- Holton TA, Graham MW 1991. A simple and efficient method for direct cloning of PCR products using ddT-tailed vectors. Nucleic Acids Res 19: 1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu WS, Hughes SH 2012. HIV-1 reverse transcription. Cold Spring Harb Perspect Med 2: a006882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji JP, Loeb LA 1992. Fidelity of HIV-1 reverse transcriptase copying RNA in vitro. Biochemistry 31: 954–958 [DOI] [PubMed] [Google Scholar]
- Karberg M, Guo H, Zhong J, Coon R, Perutka J, Lambowitz AM 2001. Group II introns as controllable gene targeting vectors for genetic manipulation of bacteria. Nat Biotechnol 19: 1162–1167 [DOI] [PubMed] [Google Scholar]
- Kennell JC, Wang H, Lambowitz AM 1994. The Mauriceville plasmid of Neurospora spp. uses novel mechanisms for initiating reverse transcription in vivo. Mol Cell Biol 14: 3094–3107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kibbe WA 2007. OligoCalc: An online oligonucleotide properties calculator. Nucleic Acids Res 35: W43–W46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristelly R, Earnest BT, Krishnamoorthy L, Tesmer JJ 2003. Preliminary structure analysis of the DH/PH domains of leukemia-associated RhoGEF. Acta Crystallogr D Biol Crystallogr 59: 1859–1862 [DOI] [PubMed] [Google Scholar]
- Lambowitz AM, Zimmerly S 2011. Group II introns: Mobile ribozymes that invade DNA. Cold Spring Harb Perspect Biol 3: a003616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamm AT, Stadler MR, Zhang H, Gent JI, Fire AZ 2011. Multimodal RNA-seq using single-strand, double-strand, and CircLigase-based capture yields a refined and extended description of the C. elegans transcriptome. Genome Res 21: 265–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858–862 [DOI] [PubMed] [Google Scholar]
- Levin JZ, Yassour M, Adiconis X, Nusbaum C, Thompson DA, Friedman N, Gnirke A, Regev A 2010. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat Methods 7: 709–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linsen SE, de Wit E, Janssens G, Heater S, Chapman L, Parkin RK, Fritz B, Wyman SK, de Bruijn E, Voest EE, et al. 2009. Limitations and possibilities of small RNA digital gene expression profiling. Nat Methods 6: 474–476 [DOI] [PubMed] [Google Scholar]
- Makarova O, Kamberov E, Margolis B 2000. Generation of deletion and point mutations with one primer in a single cloning step. Biotechniques 29: 970–972 [DOI] [PubMed] [Google Scholar]
- Malik HS, Burke WD, Eickbush TH 1999. The age and evolution of non-LTR retrotransposable elements. Mol Biol Evol 16: 793–805 [DOI] [PubMed] [Google Scholar]
- Mayer G, Muller J, Lunse CE 2011. RNA diagnostics: Real-time RT-PCR strategies and promising novel target RNAs. Wiley Interdiscip Rev RNA 2: 32–41 [DOI] [PubMed] [Google Scholar]
- Mohr G, Ghanem E, Lambowitz AM 2010. Mechanisms used for genomic proliferation by thermophilic group II introns. PLoS Biol 8: e1000391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretz SE, Lampson BC 2010. A group IIC-type intron interrupts the rRNA methylase gene of Geobacillus stearothermophilus strain 10. J Bacteriol 192: 5245–5248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallamsetty S, Waugh DS 2006. Solubility-enhancing proteins MBP and NusA play a passive role in the folding of their fusion partners. Protein Expr Purif 45: 175–182 [DOI] [PubMed] [Google Scholar]
- Ng B, Nayak S, Gibbs MD, Lee J, Bergquist PL 2007. Reverse transcriptases: Intron-encoded proteins found in thermophilic bacteria. Gene 393: 137–144 [DOI] [PubMed] [Google Scholar]
- Oz-Gleenberg I, Herschhorn A, Hizi A 2011. Reverse transcriptases can clamp together nucleic acids strands with two complementary bases at their 3′-termini for initiating DNA synthesis. Nucleic Acids Res 39: 1042–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F, Milos PM 2011. RNA sequencing: Advances, challenges and opportunities. Nat Rev Genet 12: 87–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polidoros AN, Pasentsis K, Tsaftaris AS 2006. Rolling circle amplification-RACE: A method for simultaneous isolation of 5′ and 3′ cDNA ends from amplified cDNA templates. Biotechniques 41: 35–36, 38, 40 passim [DOI] [PubMed] [Google Scholar]
- Potter J, Zheng W, Lee J 2003. Thermal stability and cDNA synthesis capability of SuperScript III reverse transcriptase. Focus (Invitrogen Newsletter) 25: 19–24 [Google Scholar]
- Saldanha R, Chen B, Wank H, Matsuura M, Edwards J, Lambowitz AM 1999. RNA and protein catalysis in group II intron splicing and mobility reactions using purified components. Biochemistry 38: 9069–9083 [DOI] [PubMed] [Google Scholar]
- San Filippo J, Lambowitz AM 2002. Characterization of the C-terminal DNA-binding/DNA endonuclease region of a group II intron-encoded protein. J Mol Biol 324: 933–951 [DOI] [PubMed] [Google Scholar]
- Smith D, Zhong J, Matsuura M, Lambowitz AM, Belfort M 2005. Recruitment of host functions suggests a repair pathway for late steps in group II intron retrohoming. Genes Dev 19: 2477–2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DR, Mrozkiewicz MK, McGrath WJ, Listwan P, Kobe B 2003. Crystal structures of fusion proteins with large-affinity tags. Protein Sci 12: 1313–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellore J, Moretz SE, Lampson BC 2004. A group II intron-type open reading frame from the thermophile Bacillus (Geobacillus) stearothermophilus encodes a heat-stable reverse transcriptase. Appl Environ Microbiol 70: 7140–7147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M 2009. RNA-Seq: A revolutionary tool for transcriptomics. Nat Rev Genet 10: 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Darnell RB 2011. Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data. Nat Biotechnol 29: 607–614 [DOI] [PMC free article] [PubMed] [Google Scholar]