This study shows that debranching of lariat intermediates in splicing can be catalyzed by the spliceosome itself and that this reaction is in competition with reverse of the first step of splicing. New parallels between group II intron splicing and spliceosomal splicing are also revealed.
Keywords: spliceosome, debranching, reverse splicing, catalytic core
Abstract
Splicing of nuclear pre-mRNA occurs via two steps of the transesterification reaction, forming a lariat intermediate and product. The reactions are catalyzed by the spliceosome, a large ribonucleoprotein complex composed of five small nuclear RNAs and numerous protein factors. The spliceosome shares a similar catalytic core structure with that of fungal group II introns, which can self-splice using the same chemical mechanism. Like group II introns, both catalytic steps of pre-mRNA splicing can efficiently reverse on the affinity-purified spliceosome. The spliceosome also catalyzes a hydrolytic spliced-exon reopening reaction as observed in group II introns, indicating a strong link in their evolutionary relationship. We show here that, by arresting splicing after the first catalytic step, the purified spliceosome can catalyze debranching of lariat-intron-exon 2. The debranching reaction, although not observed in group II introns, has similar monovalent cation preferences as those for splicing catalysis of group II introns. The debranching reaction is in competition with the reverse Step 1 reaction influenced by the ionic environment and the structure of components binding near the catalytic center, suggesting that the catalytic center of the spliceosome can switch between different conformations to direct different chemical reactions.
INTRODUCTION
Splicing of pre-mRNA proceeds via two consecutive transesterification reactions, forming a lariat intermediate and product. In the first step of the reaction, the 2′-OH group of the branch adenosine residue attacks the phosphodiester bond of the 5′ splice site to free the 5′ exon and form lariat-intron-exon 2 through 2′-5′-phosphodiester linkage. In the second step, the 3′-OH group of the free 5′ exon attacks the phosphodiester bond of the 3′ splice site to join the two exons and excise the intron in the lariat form (Ruskin et al. 1984; Konarska et al. 1985). These reactions take place on the spliceosome, which is assembled by sequential binding of five small nuclear RNAs, U1, U2, U4, U5, and U6, and numerous protein factors to the pre-mRNA (for review, see Burge et al. 1999; Brow 2002; Will and Lührmann 2006; Wahl et al. 2009).
The spliceosome is a highly dynamic machine that undergoes multiple structural rearrangements throughout the assembly pathway. Structural changes of the spliceosome are mediated by DExD/H-box RNA helicases coupling with ATP hydrolysis (Staley and Guthrie 1998). Two DEAH-box ATPases are required for the catalytic steps. Prp2 is required for the first step to destabilize U2 snRNP components SF3a and SF3b (Warkocki et al. 2009; Lardelli et al. 2010; Liu and Cheng 2012). Subsequent binding of Yju2 and Cwc25 promotes the first reaction in an ATP-independent manner (Liu et al. 2007b; Chiu et al. 2009). After the reaction, Yju2 and Cwc25 need to be destabilized from the catalytic center, mediated by Prp16 (Tseng et al. 2011), which then allows the binding of Slu7, Prp18, and Prp22 to promote the second reaction independently of ATP (Frank and Guthrie 1992; Horowitz and Abelson 1993; Ansari and Schwer 1995; Schwer and Gross 1998). Prp22 is further required to catalyze the release of mRNA, dependent on its ATPase function (Company et al. 1991; Arenas and Abelson 1997; Wagner et al. 1998; Martin et al. 2002).
Despite the energy requirement for the spliceosome to remodel its structure during the catalytic steps, both transesterification reactions are readily reversible on affinity-purified spliceosomes when incubated under proper ionic conditions (Tseng and Cheng 2008). By blocking mRNA release with a Prp22 mutant protein defective in its helicase function, the purified spliceosome can catalyze reverse splicing of the second step (R2) when incubated in the absence of KCl, without requiring ATP or the addition of extra splicing factors. Upon addition of KCl, the reaction is reversed again to generate spliced products, suggesting that the presence of KCl favors the forward procession of the second reaction (F2). KCl is also required to promote the reverse reaction of the first transesterification (R1) on the spliceosome formed on 3′ splice site-mutated pre-mRNA ACAC, which is blocked for the second reaction. Furthermore, the presence of KCl also allows the occurrence of the spliced-exon reopening (SER) reaction (Tseng and Cheng 2008), which is hydrolytic cleavage of the mRNA precisely at the splice junction, and has been observed in some group II introns (Jarrel et al. 1988). The SER and R2 reactions appear to be in competition with each other, as incubation conditions favoring one reaction would disfavor the other (Tseng and Cheng 2008).
Here, we show that the spliceosome can also catalyze debranching of lariat intron-exon 2. Splicing intermediates accumulate on the spliceosome when Prp16 is depleted from the extract. When such spliceosomes were isolated and incubated, the splicing intermediates were converted to pre-mRNA by reverse splicing (R1), or debranched to form linear intron-exon 2 (DBR). The debranching reaction is independent of the debranching enzyme Dbr1 but requires KCl. R1 and DBR reactions also favor opposite ionic conditions, indicating competition between these two reactions. Intriguingly, adding tags to the amino terminus of Cwc25 or Yju2 biased the reaction preference toward R1 under all conditions. These results suggest that the reactions catalyzed by the spliceosome may be directed by the conformation of the spliceosome in the catalytic center, which can be modulated by changes of the ionic environment or the structure of the protein binding to the catalytic center.
RESULTS
The spliceosome catalyzes the debranching reaction
We have previously demonstrated reverse splicing of the first step by using a 3′ splice site-mutated pre-mRNA ACAC to block the second reaction and isolate the spliceosome containing splicing intermediates. Such R1 reactions require KCl. Reversal of the second step that occurs on the spliceosome blocked for message release, however, does not require KCl (Tseng and Cheng 2008). The spliceosome formed with ACAC pre-mRNA is arrested in the Step 2 conformation, after the replacement of Yju2/Cwc25 with Slu7/Prp18/Prp22. Conceivably, more drastic structural change of the spliceosome might be involved in this R1 reaction than in the R2 reaction, which occurs within the Step 2 conformation. We then examined ionic requirements for the R1 reaction that occurred within the Step 1 conformation by depletion of Prp16.
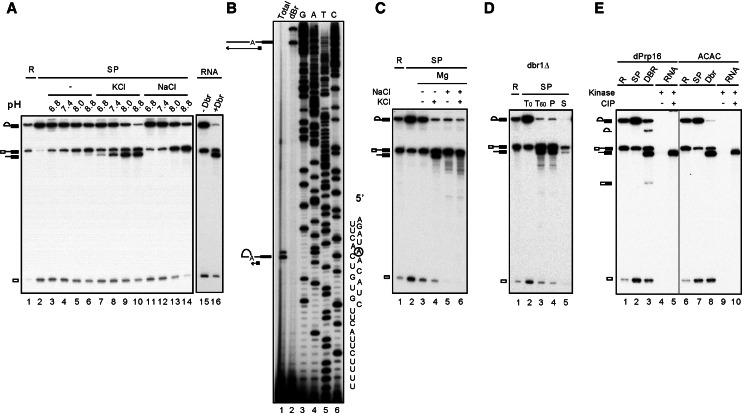
The spliceosome formed in Prp16-depleted extracts is arrested after the first reaction, accumulates splicing intermediates and Yju2/Cwc25, and is presumably retained in the Step 1 conformation. The spliceosome precipitated by anti-HA antibody, when Cwc25 is tagged with HA, contained primarily splicing intermediates (Fig. 1A, lane 2). When the purified spliceosome was incubated in buffers containing 4 mM MgCl2 of various pHs, splicing intermediates were found to be converted to pre-mRNA (lanes 3–6), indicating the occurrence of the R1 reaction. The efficiency of R1 increased with increasing pH. No errors were detected when 40 individual clones of RT-PCR products spanning the 5′ splice junction were sequenced (data not shown), indicating high precision of the R1 reaction. The results show that the R1 reaction that occurs within the Step 1 conformation does not require KCl. KCl is likely required for switching the Step 2 to Step 1 conformation without displacement of components. Interestingly, incubation in the presence of 150 mM KCl resulted in the generation of an RNA species that migrated slightly faster than pre-mRNA (lanes 7–10), corresponding in size to the linear intron-exon 2 generated by incubation of lariat-intron-exon 2 RNA with the debranching enzyme (lane 16). Primer extension analysis of gel-purified products confirmed its being linear intron-exon 2 (Fig. 1B). The presence of NaCl slightly increased the efficiency of the R1 reaction but did not promote formation of linear intron-exon 2 (Fig. 1A, lanes 11–14).
FIGURE 1.
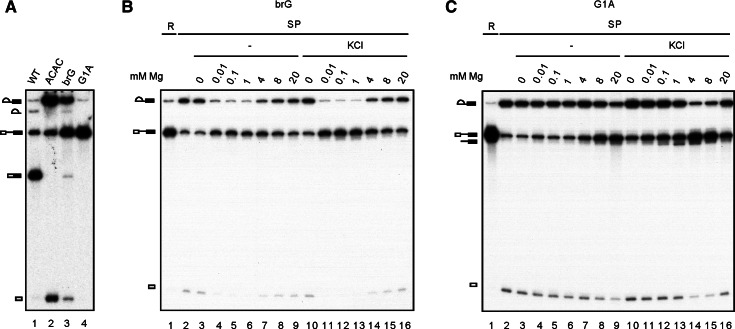
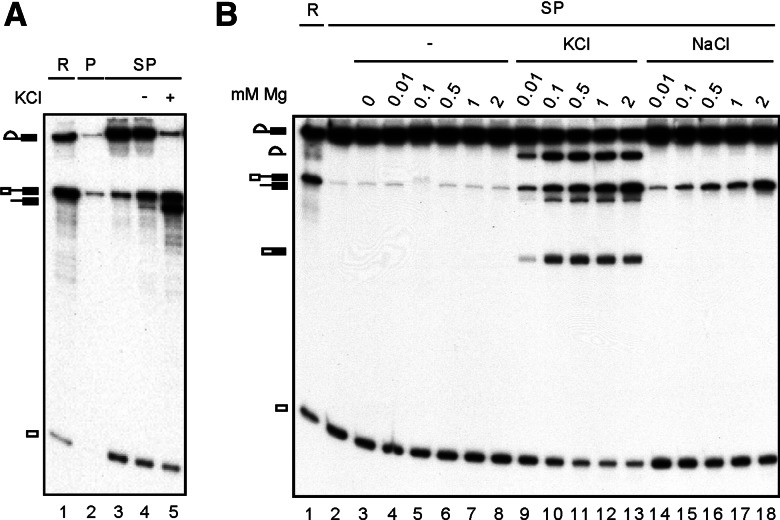
The spliceosome catalyzes the debranching reaction. (A) Splicing reaction was carried out in Prp16-depleted Cwc25-HA extracts, and the spliceosome was precipitated with anti-HA antibody. The purified spliceosome was incubated in 10 mM Tris-HCl, at pH 6.8, 7.4, 8.0, or 8.8, 4 mM MgCl2 without (lanes 3–6) or with 150 mM KCl (lanes 7–10) or 150 mM NaCl (lanes 11–14). RNA isolated as in lane 2 was also incubated in 10 mM Tris-HCl, pH 8.8, 4 mM MgCl2, and 150 mM of KCl without (lane 15) or with100 nM recombinant Dbr1 (lane 16). (B) The lariat (lane 1) and putative linear (lane 2) intron-exon 2 RNA were gel-purified and analyzed by primer extension with oligonucleotides A7. (C) The spliceosome precipitated with anti-Ntc20 antibody was incubated in 10 mM Tris-HCl, pH 8.8, 4 mM MgCl2 in the absence (lanes 3,4) or presence of 150 mM NaCl (lanes 5,6) for 1 h. The spliceosome was then washed with 1 mL 10 mM Tris-HCl, pH 8.8, and reincubated in 10 mM Tris-HCl, pH 8.8, 4 mM MgCl2 in the absence (lanes 3,5) or presence of 150 mM KCl (lanes 4,6) for 1 h. (D) The spliceosome formed in dbr1Δ extracts was incubated in 10 mM Tris-HCl, pH 8.8, 4 mM MgCl2, and 150 mM KCl for 1 h (lane 3), and the supernatant (lane 5) was separated from beads (lane 4). (E) A splicing reaction was carried out in Prp16-depleted extracts using wild-type (lanes 1–5) or ACAC (lanes 6–10) pre-mRNAs with high (lanes 1–3, 6–8) or low radioactivity (lanes 4,5,9,10), and the spliceosomes were precipitated with anti-Ntc20 antibody. Spliceosomes assembled on wild-type pre-mRNA (lane 2) were incubated in 10 mM Tris-HCl, pH 8.8, 4 mM MgCl2, and 150 mM KCl for 1 h (lanes 3–5), and RNA was isolated. RNA was also isolated from spliceosomes assembled on ACAC pre-mRNA and treated with Dbr1 (lanes 8–10). Purified RNA was subjected to phosphorylation using polynucleotide kinase and γ-32P-ATP without (lanes 4,9) or with (lanes 5,10) pretreatment with calf intestine alkaline phosphatase. (R) 1/10 of reaction mixture, (SP) spliceosome, (DBR) debranching reaction catalyzed by the spliceosome, (Dbr1) debranching reaction catalyzed by the debranchase.
Linear intron-exon 2 can be generated by either cleavage of pre-mRNA at the 5′ splice junction or by debranching of lariat-intron-exon 2. Group II introns have been shown to catalyze hydrolytic cleavage of the 5′ splice site to form linear intron-exon 2 (Podar et al. 1995; Daniels et al. 1996), but the debranching reaction has not been reported. We sought to determine whether linear intron-exon 2 was generated by debranching of lariat-intron-exon 2 or by cleavage of pre-mRNA. The spliceosome was formed in Prp16-depleted extracts to accumulate splicing intermediates and purified by precipitation with anti-Ntc20 antibody (Fig. 1C, lane 2). The purified spliceosome was incubated in the absence (lanes 3,4) or in the presence (lanes 5,6) of NaCl to drive the R1 reaction and accumulate pre-mRNA. The spliceosome in the pellet fraction was washed and reincubated with a buffer containing MgCl2 and KCl (lanes 4,6) to drive hydrolytic cleavage reactions. The result shows that linear intron-exon 2 was produced only from the spliceosome containing lariat-intron-exon 2 (lane 4) but not from that containing pre-mRNA (lane 6), suggesting that linear intron-exon 2 was the debranched product of lariat-intron-exon 2.
In cells, lariat introns are debranched by a debranching enzyme, Dbr1, after release from the spliceosome (Martin et al. 2002). The DBR1 gene can be disrupted with little effect on cellular growth (Chapman and Boeke 1991). To confirm that the debranching reaction was catalyzed by the spliceosome independent of Dbr1, we formed the spliceosome in Prp16-depleted extracts prepared from dbr1Δ yeast cells (Fig. 1D). The spliceosome was purified by precipitation with anti-Ntc20 antibody (lane 2). After incubation in the presence of KCl (lane 3), the supernatant (lane 5) and pellet (lane 4) fractions were separated. The results show that the spliceosome retained its ability to catalyze the R1 and debranching reactions. Furthermore, all RNA species remained associated with the spliceosome in the pellet fraction. These results confirm that the debranching reaction is catalyzed by the spliceosome independently of Dbr1.
Dbr1 catalyzes debranching of lariat-introns released after disassembly of the spliceosome, yielding linear introns with a phosphate at the 5′ end and a hydroxyl group at the 3′ end (Ruskin and Green 1985; Arenas and Hurwitz 1987). We examined whether the spliceosome-catalyzed debranching reaction also yielded 5′-phosphorylated RNA by testing whether the RNA can be phosphorylated without pretreatment with phosphatase (Fig. 1E). Spliceosomes were formed in Prp16-depleted extracts using pre-mRNA with low radioactivity and precipitated with anti-Ntc20 antibody. RNA was isolated after incubation to stimulate the debranching reaction and subjected to phosphorylation using polynucleotide kinase and γ-32P-ATP without (lane 4) or with (lane 5) pretreatment with calf intestine alkaline phosphatase. In parallel, the linear form of intron-exon 2 generated from the lariat form of ACAC pre-mRNA after treatment with Dbr1 was used as a control (lanes 9,10). In both cases, only pretreatment with phosphatase allowed phosphorylation of linear intron-exon 2 RNAs, indicating the presence of a phosphate group at their 5′ ends.
Characterization of the debranching reaction
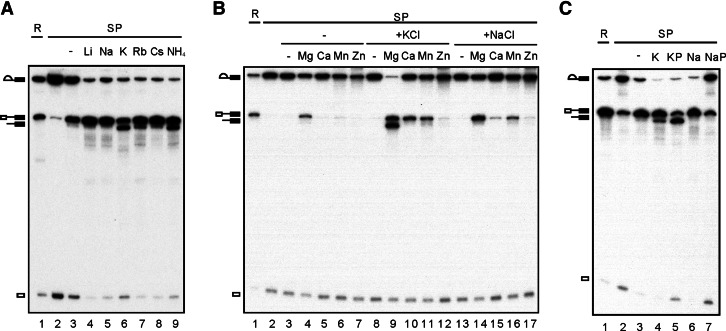
It is intriguing that KCl is required for the debranching reaction, whereas NaCl does not support the reaction. We, therefore, examined the ionic requirement for the debranching reaction in a systematic manner. Figure 2A shows that, while all monovalent cations efficiently stimulated the R1 reaction (cf. lane 3 for no addition of monovalent cation), K+ and NH4+ (lanes 6,9) promoted the debranching reaction most effectively. The efficiency of the debranching reaction decreased with increasing size of the monovalent cation (lanes 7,8), and no debranching reactions were detected with Na+ (lane 5) or Li+ (lane 4). Such ionic preference coincides with that of group II introns for supporting its structure and the splicing activity, as recently revealed from biochemical and crystallographic analyses (Marcia and Pyle 2012).
FIGURE 2.
Characterization of the debranching reaction. A splicing reaction was carried out in Prp16-depleted Cwc25-HA extracts (lane 1), and the spliceosome was precipitated with anti-HA antibody (lane 2). The spliceosome was incubated at 25°C for 1 h under various conditions: (A) 10 mM Tris-HCl, pH 8.8 and 4 mM MgCl2 alone (lane 3) or with 150 mM LiCl, NaCl, KCl, RbCl, CsCl, or NH4Cl (lanes 4–9); (B) 10 mM Tris-HCl, pH 8.8, alone (lanes 3,8,13) or with 4 mM MgCl2, CaCl2, MnCl2, and ZnCl2, in the absence (lanes 3–7) or presence of 150 mM KCl (lanes 8–12) or 150 mM NaCl (lanes 13–17); (C) 10 mM Tris-HCl, pH 8.8, 4 mM MgCl2, 150 mM KCl (lane 4), or 60 mM KPO4, pH 8.8, 4 mM MgCl2, and 30 mM KCl (lane 5), or 10 mM Tris-HCl, pH 8.8, 4 mM MgCl2, and 150 mM NaCl (lane 6), or 60 mM NaPO4, pH 8.8, 4 mM MgCl2, and 30 mM NaCl (lane 7). (R) 1/10 of reaction mixture, (SP) spliceosome, (KP) KPO4, (NaP) NaPO4.
Divalent cation Mg2+ is required for splicing of all types of introns and for the activity of many ribozymes. In many cases, other metal ions can replace the requirement of Mg2+ (Chen et al. 1997; Sontheimer et al. 1997; Shan et al. 1999; Frederiksen et al. 2012). Figure 2B shows that 4 mM Ca2+ or Mn2+ stimulated R1 in the presence of KCl (lanes 10,11) or NaCl (lanes 15,16). Zn2+ stimulated only a very low level of R1 (lanes 12,17). In the absence of a monovalent cation, significant R1 activity was observed only with Mg2+ (lanes 4–7). On the other hand, only Mg2+ could promote the debranching reaction at a 4 mM concentration (lane 9).
We also examined the effect of phosphate on the reverse and debranching reactions (Fig. 2C). Phosphate is required for high efficiency of yeast pre-mRNA splicing (Lin et al. 1985). In the presence of K+, phosphate also promoted debranching with high efficiency (lane 5), better than with KCl (lane 4). However, both R1 and debranching were inhibited in sodium phosphate (lane 7). Since phosphate promotes the splicing reaction, presumably it would inhibit reverse splicing. Conceivably, the combination of Na+ and phosphate would result in the inhibition of both debranching and reverse splicing. On the other hand, repression of reverse splicing by phosphate may give the spliceosome a greater chance to catalyze debranching in potassium phosphate. In this context, R1 and DBR may be in competition with one another, depending on the ionic environment of the spliceosome. In line with this, a previous study of the spliceosome, in which mRNA release was blocked, has suggested the reverse splicing reaction of the second step (R2) and the SER reaction being competitive with one another (Tseng and Cheng 2008).
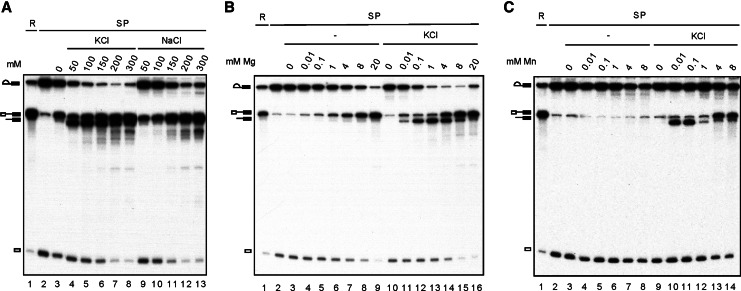
We, therefore, examined whether R1 and DBR also have different preferences for ionic environments. Titration with monovalent cations in the presence of 4 mM Mg2+ revealed that R1 and DBR have opposite preferences for KCl concentration (Fig. 3A). While R1 increased with increasing KCl or NaCl concentrations, DBR decreased with increasing KCl concentrations. Titration with Mg2+ revealed that the efficiency of R1 increased with increasing concentrations of Mg2+ up to 20 mM in the absence (Fig. 3B, lanes 3–7) or presence (lanes 8–12) of 150 mM KCl, although the efficiency was slightly higher in the presence than in the absence of KCl at the same concentration of Mg2+. In contrast, although the DBR could occur at low Mg2+ concentrations (0.01 and 0.1 mM), the efficiency decreased with increasing Mg2+ concentrations above 1 mM. These results demonstrated that above 1 mM Mg2+, increasing R1 results in decreasing DBR. Interestingly, titration with Mn2+ revealed that Mn2+ could also stimulate the debranching reaction but only at concentrations lower than 1 mM and also in a KCl-dependent manner (Fig. 3C, lanes 10,11). Above 1 mM Mn2+, DBR was severely weakened, and R1 became dominant. This was not observed for Ca2+ or Zn2+, neither of which stimulated DBR when examined across a wide range of concentrations (data not shown).
FIGURE 3.
Characterization of the debranching reaction. A splicing reaction was carried out in Prp16-depleted Cwc25-HA extracts (lane 1), and the spliceosome was precipitated with anti-HA antibody (lane 2). The spliceosome was incubated at 25°C for 1 h under various conditions: (A) 10 mM Tris-HCl, pH 8.8 and 4 mM MgCl2 alone (lane 3) or with 50 mM, 100 mM, 150 mM, 200 mM, or 300 mM of KCl (lanes 4–8) or NaCl (lanes 9–13); (B) 10 mM Tris-HCl, pH 8.8, or with 0 mM, 0.01 mM, 0.1 mM, 1 mM, 4 mM, 8 mM, or 20 mM MgCl2 in the absence (lanes 3–9) or presence of 150 mM KCl (lanes 10–16); (C) 10 mM Tris-HCl, pH 8.8, with 0 mM, 0.001 mM, 0.01 mM, 0.1 mM, 1 mM, 2 mM, 4 mM, or 8 mM MnCl2 in the absence (lanes 3–8) or presence of 150 mM KCl (lanes 9–14). (R) 1/10 of reaction mixture, (SP) spliceosome.
Inhibition of the debranching reaction by N-terminally tagged Cwc25 and Yju2
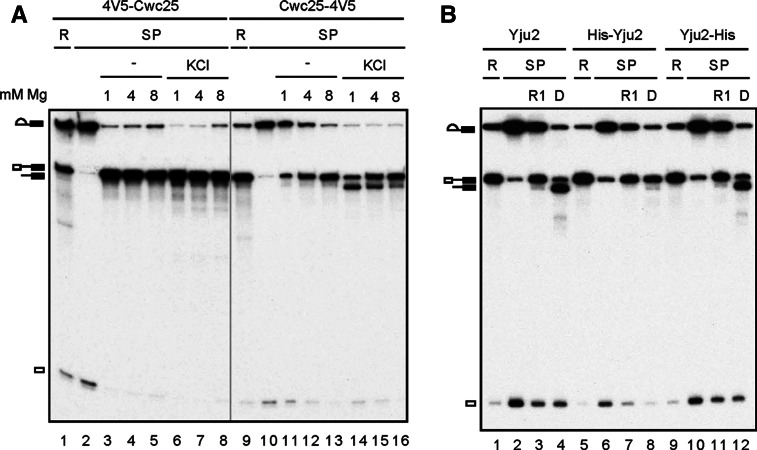
It is not surprising that the ionic environment of the pre-Prp16 spliceosome can direct the type of the reaction, presumably by influencing conformational dynamics of the spliceosome. Conditions that support the conformational state for R1 suppress the DBR, and vice versa. A conformational model has been proposed for the two catalytic steps of the spliceosome based on a wealth of genetic data from analysis of spliceosomal components, protein factors, and RNA elements, important for the catalytic reactions (Query and Konarska 2004; Liu et al. 2007a). It is reasonable to think that the conformation of the catalytic center can also be modulated by the structure of the proteins binding to the catalytic center of the spliceosome. Yju2 and Cwc25 are required for the first reaction. Cwc25, in particular, plays a key role in positioning the 5′ splice site and the branch site, as the reaction takes place immediately upon its binding to the spliceosome. We modified the structure of Cwc25 by adding four copies of the V5-epitope to either the N terminus (4V5-Cwc25) or C terminus (Cwc25-4V5) of Cwc25. Spliceosomes were assembled in Prp16-depleted 4V5-Cwc25 (Fig. 4A, lanes 1–8) or Cwc25-4V5 (lanes 9–16) extracts, and precipitated with anti-V5 antibody (lanes 2,10). The precipitated spliceosome was incubated with 1 mM, 4 mM, or 8 mM MgCl2 in the absence or presence of KCl to elicit R1 and DBR. When V5 was tagged at the C terminus of Cwc25, a similar pattern for R1 and DBR was observed as in untagged Cwc25, in which R1 favored higher Mg2+ concentrations and DBR favored lower Mg2+ concentrations, but only in the presence of KCl (lanes 11–16). Strikingly, when V5 was tagged at the N terminus of Cwc25, DBR was completely inhibited, and strong R1 was seen under all conditions with nearly all splicing intermediates converted to pre-mRNA (lanes 3–8). When the spliceosome was purified by precipitation with antibodies against Ntc20, the DBR inhibitory effect was not evident (data not shown), indicating that the binding of antibody to the V5-tag at the N terminus was essential to drive the spliceosome into an R1-permissive conformation.
FIGURE 4.
Inhibition of the debranching reaction by N-terminally tagged Cwc25 and Yju2. (A) Splicing was carried out in Prp16-depleted 4V5-Cwc25 or Cwc25-4V5 extracts. The spliceosome was precipitated with anti-V5 antibody, and then incubated at 25°C for 1 h in 10 mM Tris-HCl, pH 8.8, with 1 mM, 4 mM, or 8 mM MgCl2 in the absence (lanes 3–5, 11–13) or presence of 150 mM KCl (lanes 6–8, 14–16). (B) Splicing was carried out in Prp16-depleted (lanes 1–4) or in both Yju2- and Prp16-depleted Cwc25-HA extracts supplemented with recombinant His-Yju2 (lanes 5–8) or Yju2-His protein (lanes 9–12). The spliceosomes were immunoprecipitated with anti-HA antibody, and then incubated in 10 mM Tris-HCl, pH 8.8, 8 mM MgCl2 (lanes 3,7,11), or in 10 mM Tris-HCl, pH 8.8, 1 mM MgCl2, 150 mM KCl (lanes 4,8,12). (R) 1/10 of reaction mixture, (SP) spliceosome, (D) DBR.
We then examined the effect of the structure of Yju2 on directing R1 vs. DBR reactions and found that adding a His-tag to the N terminus of Yju2 was also inhibitory of the DBR (Fig. 4B). Splicing extracts prepared from the CWC25-HA strain were depleted of both Prp16 and Yju2 and supplemented with recombinant His-Yju2 or Yju2-His. The His-tag at the N terminus of Yju2 contains 14 extra amino acid residues in addition to six histidine residues, whereas at the C terminus, only six histidine residues are present in the tag. For the Yju2 untagged control, the extract was only depleted of Prp16 without the addition of recombinant Yju2. After the splicing reaction, spliceosomes were purified by precipitation with anti-HA antibody and incubated under conditions favoring R1 (8 mM MgCl2) (lanes 3,7,11) or DBR (1 mM MgCl2 and 150 mM KCl) (lanes 4,8,12). Like untagged Yju2, Yju2-His promoted R1 at 8 mM Mg2+ (lanes 3,11), and promoted DBR at 1 mM Mg2+ (lanes 4,12). With His-Yju2, DBR was nearly completely inhibited, but R1 was not further enhanced (lane 8). In this case, only 20 extra amino acid residues present at the N terminus of Yju2 were sufficient to prevent the spliceosome from switching to the DBR conformation, since the antibody did not bind Yju2. Together, these results show that changing the structure of either Cwc25 or Yju2 at the N terminus can influence conformational dynamics of the spliceosome, which may affect the geometry of the catalytic residues of the spliceosome to direct different reactions.
The debranching reaction is inhibited by branchpoint A-to-G mutation
We further investigated whether the debranching reaction is affected by mutation of the nucleotides that form the branch structure. Pre-mRNA with an A to G change at the branchpoint (brG) splices less efficiently than wild-type pre-mRNA or the 3′ splice site mutant ACAC, whereas the 5′ splice site mutant, G1A, splices poorly. Like ACAC, splicing of brG or G1A pre-mRNA was severely impeded in the second step, resulting in the accumulation of lariat-intron-exon 2 (Fig. 5A). The G1A mutant spliced poorly, with only a very small amount of lariat-intron-exon 2 generated (lane 4), which could be enriched by precipitation of the spliceosome (Fig. 5C, lane 2). Spliceosomes formed in Prp16-depleted Cwc25-HA extracts were isolated by precipitation with anti-HA antibody and incubated with 0.01–20 mM MgCl2 in the presence or absence of KCl (Fig. 5B,C). As with wild-type pre-mRNA (Fig. 3B), the G1A spliceosome also catalyzed R1 and DBR (Fig. 5C), and the efficiency of R1 increased with increasing Mg2+ concentrations. Nevertheless, DBR is less efficient in the G1A mutant, indicating that changing G to A in the first position of the intron disfavors the conformation for DBR in the context of the lariat structure. The branchpoint nucleotide, however, has a much greater impact on DBR when changed to G, as no debranched product was detected (Fig. 5B). Interestingly, the brG mutant had an opposite preference for Mg2+ concentration for the R1 reaction. Efficient R1 reactions were observed at 0.01 mM Mg2+ both in the presence and absence of KCl, and the efficiency decreased with increasing Mg2+ concentrations. The mechanism for reverse preference is currently unknown.
FIGURE 5.
The debranching reaction is inhibited by branchpoint A-to-G mutation. (A) Splicing reactions were carried out with wild-type, ACAC, brG, or G1A pre-mRNA. (WT) wild type. (B,C) Spliceosomes assembled on brG (B) or G1A (C) pre-mRNA in Prp16-depleted Cwc25-HA extracts were individually precipitated with anti-HA antibody and incubated at 25°C for 1 h in 10 mM Tris-HCl, pH 8.8, or with 0 mM, 0.01 mM, 0.1 mM, 1 mM, 4 mM, 8 mM, or 20 mM MgCl2 in the absence of (lanes 3–9) or presence of 150 mM KCl (lanes 10–16). (R) 1/20 (B) or 1/100 (C) of reaction mixture. (SP) spliceosome.
The debranching reaction occurs on the Prp16-associated and post-Prp16 spliceosomes
The debranching reaction described above was observed on the spliceosome isolated at the pre-Prp16 stage with Cwc25 associated. To see whether the spliceosome can catalyze DBR only at this specific stage, we isolated Prp16-bound spliceosome and Prp22-bound spliceosome to see whether DBR can occur on these spliceosomes. While the Prp16-bound spliceosome is in the first-step conformational state, the Prp22-bound spliceosome is in the second-step conformation with Yju2/Cwc25 having been replaced by Slu7/Prp18/Prp22. For the Prp16-bound spliceosome, the splicing reaction was performed in Prp16-depleted extracts supplemented with HA-tagged Prp16 mutant protein D473A, and the spliceosome was isolated by precipitation with anti-HA antibody (Fig. 6A). The prp16-D473A protein, which carries a mutation in the ATPase motif, is able to bind to the spliceosome but unable to promote destabilization of Yju2/Cwc25 and itself (Tseng et al. 2011). When the purified spliceosome (lane 3) was incubated with Mg2+ in the absence (lane 4) or presence (lane 5) of KCl, efficient DBR was observed in the presence of KCl (lane 5), indicating no interference of DBR by the presence of Prp16.
FIGURE 6.
The debranching reaction occurs on the Prp16-associated and post-Prp16 spliceosomes. (A) A splicing reaction was carried out in Prp16-depleted extracts in the presence of recombinant HA-tagged Prp16 mutant D473A (lane 1), and the spliceosome was precipitated with unconjugated (lane 2) or anti-HA antibody-conjugated protein A-Sepharose (lanes 3–5). The precipitated spliceosome was incubated in 10 mM Tris-HCl, pH 8.8, 4 mM MgCl2, in the absence (lane 4) or presence of 150 mM KCl (lane 5). (B) A splicing reaction was carried out with ACAC pre-mRNA in Prp22-4V5 extracts (lane 1). The spliceosome was precipitated with anti-V5 antibody (lane 2), and the purified spliceosome was incubated at 25°C for 1 h in 10 mM Tris-HCl, pH 8.8, without (lane 3) or with 0.01 mM, 0.1 mM, 0.5 mM, 1 mM, or 2 mM MgCl2 in the absence (lanes 3–8) or presence of 150 mM KCl (lanes 9–13) or 150 mM NaCl (lanes 14–18). (R) 1/10 of reaction mixture, (P) protein A-Sepharose, (SP) spliceosome.
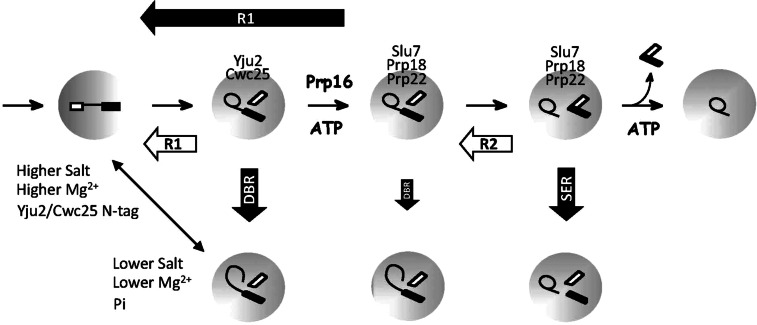
For Prp22-bound spliceosome, the spliceosome was assembled in Prp22-4V5 extracts with ACAC pre-mRNA to block the second reaction and isolated by precipitation with anti-V5 antibody. We have previously shown that, when the purified ACAC spliceosome was incubated in the presence of KCl, both splicing of the second step (F2) and reverse splicing of the first step (R1) could occur (Tseng and Cheng 2008). When the spliceosome was incubated with Mg2+ concentrations from 0.01 to 2 mM in the presence of KCl (Fig. 6B), linear intron-exon 2 was generated in addition to pre-mRNA and spliced products (lanes 9–13), indicating that DBR can also take place on the post-Prp16 spliceosome. Nevertheless, the reaction was less efficient but shows a similar Mg2+ dependency as the pre-Prp16 spliceosome. A summary for the reactions that the spliceosome can catalyze and conditions that favor R1 or DBR is shown in Figure 7.
FIGURE 7.
Schematic of the spliceosome catalytic steps showing different chemical reactions that the spliceosome can catalyze. Filled thick arrows represent reactions that require monovalent cations, and open thick arrows represent those that do not require monovalent cations. The double-headed arrow represents competition between R1 and DBR reactions, with conditions favoring one or the other indicated.
DISCUSSION
In this study, we discovered and characterized a previously unidentified debranching reaction catalyzed by the spliceosome. Debranching of lariat-introns is the first step in degradation of excised introns after the splicing reaction (Martin et al. 2002). The debranching enzyme Dbr1 is not associated with the spliceosome and catalyzes the reaction on free lariat-intron RNA molecules after the spliceosome is disassembled. Although the debranched product, linear intron-exon 2, catalyzed by the spliceosome also contains a 5′ phosphate similar to that generated by the debranchase, the reaction is totally independent of the enzymatic activity of the debranchase, as the reaction takes place in dbr1Δ extracts. Notably, a putative debranched product of lariat-intron-exon 2 associated with the spliceosome has previously been observed but was not further characterized to clarify whether its formation was associated with the debranchase activity (Ansari and Schwer 1995). The DBR reaction represents a novel reaction that the spliceosome can catalyze in addition to the previously uncovered SER reaction.
The DBR reaction can occur efficiently on the spliceosome stalled in the first step by depletion of Prp16 or by using the ATPase mutant of Prp16 to prevent the release of Yju2 and Cwc25 from the spliceosome. At this conformational state, reverse of the first splicing reaction can also occur. The DBR reaction, being hydrolysis of the 2′-5′ phosphodiester bond of the lariat intermediate, can be viewed as the first half of the R1 reaction, in an analogy to SER as the first half of the R2 reaction. Reverse splicing of the first step via transesterification requires the 3′-OH of exon 1 to be positioned in a precise geometry. Conceivably, the lariat-intermediate may become more vulnerable to hydrolytic cleavage when the 3′-OH of exon 1 is out of place under certain conditions. Although DBR is not expected to occur during normal splicing reactions, as it proceeds more slowly than the forward or reverse transesterification reaction, it provides novel insights into chemical flexibility of the spliceosome, which is able to catalyze both hydrolytic and transesterification reactions in both steps. It also reveals that the spliceosome is designed in such a way that both catalytic steps are mechanistically analogous to each other.
The DBR reaction requires Mg2+, favors high pH, and has a preference for specific sizes of monovalent cations. K+ and NH4+ ions give the highest activity of the DBR, and the activity decreases with increasing size of ions. Monovalent cations of a smaller size, such as Li+ and Na+, hardly support the DBR reaction. Interestingly, a recent study of Oceanobacillus iheyensis (Oi) group IIC intron domains 1–5 shows a similar preference for the size of monovalent cations for the self-splicing activity of this ribozyme (Marcia and Pyle 2012). Crystal structural analysis of this intron has revealed binding of two K+ ions near the active site, together with two Mg2+ ions, forming a heteronuclear metal ion center (Marcia and Pyle 2012). The group II introns and spliceosomal introns share similar catalytic core structures and chemical mechanisms and are evolutionarily related. It is conceivable that spliceosomal introns also require binding of K+ ions to support the active structure for the debranching reaction. However, the reverse reaction does not require monovalent cations but is more efficient in their presence without a clear distinction between different sizes (Fig. 2A). Using purified precatalytic spliceosomes to chase the first reaction, we found that the forward splicing reaction is also independent of monovalent cations (data not shown), suggesting that binding of monovalent cations may influence the geometry of the catalytic core of the spliceosome to discriminate between transesterification and hydrolytic reactions. Coincidentally, splicing of the Oi group IIC intron proceeds through a hydrolytic pathway. It is worth noting that, while low K+ concentrations could promote DBR, the R1 reaction increases with increasing monovalent cation concentrations, tempting us to speculate on the existence of two K+ binding sites in the catalytic core, one with high affinity and the other with low affinity. In this scenario, occupancy of the high affinity, K+-specific site promotes DBR, whereas occupancy of the low affinity, nonspecific site facilitates R1. This is in contrast to the roles of K+ in binding to the Oi group IIC intron, in which proper sizes of the ion is necessary to rigidify the divalent ion binding sites and stabilize reaction intermediates (Marcia and Pyle 2012). How the catalytic residues are positioned in response to K+ binding on the spliceosome remains to be elucidated.
Although R1 can occur in the absence of monovalent cations, it proceeds more efficiently at higher Mg2+ concentrations, under which conditions DBR is greatly reduced. These results suggest that R1 and DBR may be in competition with one another, determined by the conformational state of the spliceosome in the catalytic center. Ionic environments can influence the equilibrium between the two conformational states so that conditions that promote the conformation for DBR would disfavor the R1 reaction, and vice versa.
Such a conformational model has been proposed for the two catalytic steps of the spliceosome (Query and Konarska 2004; Liu et al. 2007a). In this two-state conformational model, it has been hypothesized that the conformational states of Step 1 and Step 2 are in competition with one another. Mutations that affect the stability of one conformational state result in relative stabilization of the other and, consequently, stimulate the reaction of the other step (Liu et al. 2007a). Our results echo this conformational model and further extend it to multiple conformational states, which can be modulated by different ionic environments, as the basis of different chemical reactions. We have previously shown that both steps of transesterification are reversible (Tseng and Cheng 2008). Furthermore, the purified spliceosome arrested before mRNA release can catalyze the SER reaction when incubated in the presence of Mn2+. Like R1 and DBR, incubation conditions that promote the reverse reaction of the second step (R2) repress the SER reaction (Tseng and Cheng 2008), suggesting that the conformational states for R2 and SER are in competition with one another.
The debranching enzyme is a metallophosphoesterase that uses Mn2+ as a cofactor (Khalid et al. 2005). The optimal Mn2+ concentration for the debranching activity is 2–4 mM. Mg2+ can replace Mn2+ but is much less effective in stimulating its enzymatic activity (Khalid et al. 2005). The spliceosome-catalyzed debranching reaction was also stimulated by both Mg2+ and Mn2+. The optimal concentration for Mg2+ was 1–4 mM, and for Mn2+ was 0.01–0.1 mM, with no reaction detected at 0.001 mM (data not shown), indicating that Mn2+ is more effective in stimulating the debranching reaction. The spliceosome thus shares a similar property with the debranching enzyme in using Mn2+ for the debranching reaction, raising the question of the evolutionary relationship between the two catalytic systems. Interestingly, both DBR and R1 reactions occur at low frequency at 1 mM Mn2+, and R1 becomes more vigorous above 1 mM Mn2+ (Fig. 3C). It is conceivable that Mn2+ may have high affinity for the catalytic center of the spliceosome, and binding of a limiting amount of Mn2+ directs the spliceosome to the DBR conformational state. With increasing concentrations, more Mn2+ may bind to the spliceosome, which then directs the spliceosome to the R1 conformation. At 1 mM Mn2+, the spliceosome may be retained in an intermediate state, with low R1 and DBR activities.
Our results also showed that, when a His-tag was added to Yju2, or when four copies of V5-tag were added to the N terminus of Cwc25, the DBR reaction was completely inhibited, whereas the R1 reaction was not affected. Adding the tag to the C terminus of the protein did not alter the pattern of the reactions. It is not surprising that the structures of Yju2 and Cwc25 can also influence the conformation of the catalytic core of the spliceosome since these two proteins are required only for the first catalytic reaction (Liu et al. 2007b; Chiu et al. 2009). Cwc25, in particular, is recruited to the spliceosome immediately prior to the reaction and is destabilized after the reaction, mediated by Prp16 (Tseng et al. 2011). Cwc25 has also been shown to cross-link to the pre-mRNA in the 3′ tail a few bases downstream from the branchpoint, suggesting that it is situated at or near the catalytic center of the spliceosome during the first step (Chen et al. 2013). Conceivably, altering the structure of the proteins that bind at or near the catalytic center may impact the conformation in the catalytic core of the spliceosome. Thus, adding sequences to the N terminus of Yju2 or Cwc25 may distort the conformation of the spliceosome to prevent transition to the conformational state for DBR. In fact, when Cwc25 was tagged with V5 at the N terminus, the spliceosome precipitated with anti-V5 antibody strongly favored R1 under all conditions. This R1-prone effect is from the combination of the tag at the N terminus of Cwc25 and the binding of the antibody to the tag. Precipitation of the spliceosome with anti-Ntc20 antibody did not exhibit such an effect (data not shown), indicating that a large body of mass added to the N terminus of Cwc25 is needed for the R1-prone effect. Nevertheless, adding only 20 amino acid residues to the N terminus of Yju2 was sufficient to block the DBR. These results suggest that the N terminus of Yju2 may be in close contact with the other structural elements of the catalytic core so that even a small change in the structure can block the transition to the conformational state of DBR. On the other hand, the N terminus of Cwc25 may fit in the catalytic core more loosely and can tolerate more structural change without impacting the balance between different conformational states. However, further binding of the antibody greatly stabilizes the R1 conformation and completely inhibits the DBR. These results also suggest that the N termini of these two proteins may be organized spatially near each other in the catalytic core of the spliceosome and, therefore, exhibit similar effects on modulating the conformation, although to different extents.
Mutation at the branchpoint or at the 5′ splice site did not inhibit the R1 reaction. However, the DBR reaction was significantly suppressed in the G1A mutant and totally inhibited in the brG mutant. This indicates that, while the R1 conformation is insensitive to base changes at the branchpoint or the 5′ splice site, the DBR conformation is destabilized when these bases are changed. Unexpectedly, the brG mutant has an opposite Mg2+ preference for the R1 reaction, in favor of lower Mg2+ concentrations independent of KCl. The brU mutant behaves in a similar way to the brG mutant (data not shown). It is not clear how a change of the base at the branchpoint would reverse the Mg2+ preference for R1. Considering that Mg2+ concentrations as low as 10 µM are sufficient to efficiently stimulate R1 in the brG mutant, it is plausible that a high-affinity Mg2+ binding site may be created in brG to promote the R1 conformation. Increasing Mg2+ concentrations may result in binding of more Mg2+, which destabilizes the R1 conformation.
MATERIALS AND METHODS
Yeast strains
BJ2168: MATa prc1 prb1 pep4 leu2 trp1 ura3
YSCC22: MATa prc1 prb1 pep4 leu2 trp1 ura3 PRP22-4V5
YSCC25: MATa prc1 prb1 pep4 leu2 trp1 ura3 CWC25-HA
YSCC254: MATa prc1 prb1 pep4 leu2 trp1 ura3 4V5-CWC25
YSCC255: MATa prc1 prb1 pep4 leu2 trp1 ura3 CWC25-4V5
YSCC10: MATa prc1 prb1 pep4 leu2 trp1 ura3 dbr::LEU2
Oligonucleotides
The following oligonucleotides were used:
For sequencing
A1 CTTCATCACCAACGTAG
A2 TTTCACGCTTACTGCTT
A5 CGATTGCTTCATTCTT
For primer extension
A6 TCTTACAGTTAAATGGGATGG
A7 AAACATATAATATAGCAAC (primer extension of debranching product);
For construction of pRS406.CWC25.4V5
C25-1 GGCCGGATCCGACATATGGGGTCGGGCGATTT
C25-5 GGCCAAGCTTGTAGTCTAGGTCCGGAG
C25-6 GGCCTCTAGATAAACTTTCCTTTTCTTAGAGCTTGG
C25-7 CCGGGGTACCAGTCACCAAGGTGTTCC
Antibodies and reagents
The anti-V5 antibody was purchased from Serotec Inc. Protein A-Sepharose was from Sigma, AMV reverse transcriptase from Promega, SuperScript III reverse transcriptase from Invitrogen. and Ni-NTA Agarose from Qiagen.
Immunodepletion
Immunodepletion of Prp16 was performed by incubation of 100 μL of splicing extracts with 50 μL of the anti-Prp16 anti-serum coupled to 50 μL of protein A-Sepharose.
Immunoprecipitation
Splicing was carried out with Prp16-depleted extracts at 25°C for 30 min using wild-type or mutant actin pre-mRNA. The spliceosome was precipitated with anti-Ntc20 antibody, or with antibody against HA or V5 when protein was tagged with epitope, and washed three times with 1 mL NET-2 buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, and 0.05% NP-40) and once with 1 mL of 10 mM Tris-HCl, pH 8.8 (for pH titration, dH2O was used in the final wash instead). In experiments testing divalent cation effects, 50 μM of EDTA was included in NET-2, and EDTA was removed by one wash with 1 mL of NET-2.
Reactions on the precipitated spliceosome
The precipitated spliceosome was incubated with 30 μL of 10 mM Tris-HCl, pH 8.8 (or 6.8, or 7.4, or 8.0 in specific experiments), 4 mM MgCl2 (or MnCl2, or CaCl2, or ZnCl2), with or without 150 mM KCl or other monovalent ions at 25°C for 60 min.
Primer extension
The reaction mixtures for the debranching reaction were fractionated by electrophoresis on 8% polyacrylamide/8 M urea gels. Individual RNA species were excised from gels and eluted overnight in 450 μL of elution buffer containing 0.3 M NaOAc, pH 5, 2 mM EDTA, and 0.1% SDS. Following phenol-chloroform extraction and EtOH precipitation, RNA was dissolved in 7 μL of annealing buffer, containing 32P-labeled oligonucleotide A6 or A7, 50 mM Tris-HCl, pH 8.3, 75 mM KCl, and 3 mM MgCl2, and incubated at 100°C for 2 min, then 45°C for 5 min, and then mixed with 3 μL of a solution containing 1 unit RNasin, 50 units SuperScript III reverse transcriptase, and 33 mM DTT, and incubated for 1 h. Extension products were analyzed by electrophoresis on 12% polyacrylamide/8 M urea gels.
Sequencing of the splice junction
For sequence analysis of the splice junction, DNA fragments were generated by RT-PCR using primers A1 and A2 for the mRNA spliced junction and primers A1 and A5 for the 3′ splice site, and cloned into plasmid vector pGEM-T.
ACKNOWLEDGMENTS
We thank A. Pena for English editing and members of the Cheng laboratory for helpful discussions. This work was supported by a grant from the Academia Sinica and National Science Council (Taiwan), NSC100-2745-B-001-001-ASP.
REFERENCES
- Ansari A, Schwer B 1995. SLU7 and a novel activity, SSF1, act during the PRP16-dependent step of yeast pre-mRNA splicing. EMBO J 14: 4001–4009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenas JE, Abelson JN 1997. Prp43: An RNA helicase-like factor involved in spliceosome disassembly. Proc Natl Acad Sci 94: 11798–11802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenas J, Hurwitz J 1987. Purification of a RNA debranching activity from HeLa cells. J Biol Chem 262: 4274–4279 [PubMed] [Google Scholar]
- Brow DA 2002. Allosteric cascade of spliceosome activation. Annu Rev Genet 36: 333–360 [DOI] [PubMed] [Google Scholar]
- Burge CB, Tuschl TH, Sharp PA 1999. Splicing of precursors to mRNAs by the spliceosome. In RNA world II (ed. Gesteland RF, et al. ), pp. 525–560 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Chapman KB, Boeke JD 1991. Isolation and characterization of the gene encoding yeast debranching enzyme. Cell 65: 483–492 [DOI] [PubMed] [Google Scholar]
- Chen Y, Li X, Gegenheimer P 1997. Ribonuclease P catalysis requires Mg2+ coordinated to the pro-Rp oxygen of the scissile bond. Biochemistry 36: 2425–2438 [DOI] [PubMed] [Google Scholar]
- Chen H-C, Tseng C-K, Tsai R-T, Chung C-S, Cheng S-C 2013. Link of NTR-mediated spliceosome disassembly with DEAH-box ATPases Prp2, Prp16 and Prp22. Mol Cell Biol 33: 514–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu Y-F, Liu Y-C, Chiang T-W, Yeh T-C, Tseng C-K, Wu NY, Cheng S-C 2009. Cwc25 is a novel splicing factor required after Prp2 and Yju2 to facilitate the first catalytic reaction. Mol Cell Biol 29: 5671–5678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Company M, Arenas J, Abelson J 1991. Requirement of the RNA helicase-like protein PRP22 for release of messenger RNA from spliceosomes. Nature 349: 487–493 [DOI] [PubMed] [Google Scholar]
- Daniels DL, Michels WJ Jr, Pyle AM 1996. Two competing pathways for self-splicing by group II introns: A quantitative analysis of in vitro reaction rates and products. J Mol Biol 256: 31–49 [DOI] [PubMed] [Google Scholar]
- Frank D, Guthrie C 1992. An essential splicing factor, SLU7, mediates 3′ splice site choice in yeast. Genes Dev 6: 2112–2124 [DOI] [PubMed] [Google Scholar]
- Frederiksen JK, Li N-S, Das R, Herschlag D, Piccirilli JA 2012. Metal-ion rescue revisited: Biochemical detection of site-bound metal ions important for RNA folding. RNA 18: 1123–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz DS, Abelson J 1993. Stages in the second reaction of pre-mRNA splicing: The final step is ATP independent. Genes Dev 7: 320–329 [DOI] [PubMed] [Google Scholar]
- Jarrel KA, Peebles CL, Dierich RC, Romiti SL, Perlman PS 1988. Group II intron self-splicing. Alternative reaction conditions yield novel products. J Biol Chem 263: 3432–3439 [PubMed] [Google Scholar]
- Khalid MF, Damha MJ, Shuman S, Schwer B 2005. Structure–function analysis of yeast RNA debranching enzyme (Dbr1), a manganese-dependent phosphodiesterase. Nucleic Acids Res 33: 6349–6360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarska MM, Grabowski PJ, Padgett RA, Sharp PA 1985. Characterization of the branch site in lariat RNAs produced by splicing of mRNA precursors. Nature 313: 552–557 [DOI] [PubMed] [Google Scholar]
- Lardelli RM, Thompson JX, Yates JR III, Stevens SW 2010. Release of SF3 from the intron branchpoint activates the first step of pre-mRNA splicing. RNA 16: 516–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin RJ, Newman AJ, Cheng SC, Abelson J 1985. Yeast mRNA splicing in vitro. J Biol Chem 260: 14780–14792 [PubMed] [Google Scholar]
- Liu H-L, Cheng S-C 2012. The interaction of Prp2 with a defined region of the intron is required for the first splicing reaction. Mol Cell Biol 32: 5056–5066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Query CC, Konarska MM 2007a. Opposing classes of prp8 alleles modulate the transition between the catalytic steps of pre-mRNA splicing. Nat Struct Mol Biol 14: 519–526 [DOI] [PubMed] [Google Scholar]
- Liu Y-C, Chen H-C, Wu N-Y, Cheng S-C 2007b. A novel splicing factor Yju2 is associated with NTC and acts after Prp2 in promoting the first catalytic reaction of pre-mRNA splicing. Mol Cell Biol 27: 5403–5413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcia M, Pyle AM 2012. Visualizing group II intron catalysis through the stages of splicing. Cell 151: 497–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Schneider S, Schwer B 2002. Prp43 is an essential RNA-dependent ATPase required for release of lariat-intron from the spliceosome. J Biol Chem 277: 17743–17750 [DOI] [PubMed] [Google Scholar]
- Podar M, Perlman PS, Padgett RA 1995. Stereochemical selectivity of group II intron splicing reverse splicing, and hydrolysis reactions. Mol Cell Biol 15: 4466–4478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Query CC, Konarska MM 2004. Suppression of multiple substrate mutations by spliceosomal prp8 alleles suggests functional correlations with ribosomal ambiguity mutants. Mol Cell 14: 343–353 [DOI] [PubMed] [Google Scholar]
- Ruskin B, Green MR 1985. An RNA processing activity that debranches RNA lariats. Science 229: 135–140 [DOI] [PubMed] [Google Scholar]
- Ruskin B, Krainer AR, Maniatis T, Green MR 1984. Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro. Cell 38: 317–331 [DOI] [PubMed] [Google Scholar]
- Schwer B, Gross CH 1998. Prp22, a DExH-box RNA helicase, plays two distinct roles in yeast pre-mRNA splicing. EMBO J 17: 2086–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan SO, Yoshida A, Sun SG, Piccirilli JA, Herschlag D 1999. Three metal ions at the active site of the Tetrahymena group I ribozyme. Proc Natl Acad Sci 96: 12299–12304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontheimer EJ, Sun S, Piccirilli JA 1997. Metal ion catalysis during splicing of premessenger RNA. Nature 388: 801–805 [DOI] [PubMed] [Google Scholar]
- Staley JP, Guthrie C 1998. Mechanical devices of the spliceosome: Motors, clocks, springs, and things. Cell 92: 315–326 [DOI] [PubMed] [Google Scholar]
- Tseng C-K, Cheng S-C 2008. Both catalytic steps of nuclear pre-mRNA splicing are reversible. Science 320: 1782–1784 [DOI] [PubMed] [Google Scholar]
- Tseng C-K, Liu H-L, Cheng S-C 2011. DEAH-box ATPase Prp16 has dual roles in remodeling of the spliceosome in catalytic steps. RNA 17: 145–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner JDO, Jankowsky E, Company M, Pyle AM, Abelson JN 1998. The DEAH-box protein PRP22 is an ATPase that mediates ATP-dependent mRNA release from the spliceosome and unwinds RNA duplexes. EMBO J 17: 2926–2937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Lührmann RL 2009. The spliceosome: Design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]
- Warkocki Z, Odenwälder P, Schmitzová J, Platzmann F, Stark H, Urlaub H, Ficner R, Fabrizio P, Lührmann R 2009. Reconstitution of both steps of Saccharomyces cerevisiae splicing with purified spliceosomal components. Nat Struct Mol Biol 16: 1237–1243 [DOI] [PubMed] [Google Scholar]
- Will CL, Lührmann R 2006. Spliceosome structure and function. In The RNA world (ed. Gesteland RF, et al. ), pp. 369–400 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]