Abstract
Despite the importance of arbuscular mycorrhizal fungi in the majority of terrestrial ecosystems, their ecology, genetics, and evolution are poorly understood, partly due to difficulties associated with detecting and identifying species. We explored the inter- and intraspecies variations of the 18S rRNA genes of the genus Gigaspora to assess the use of this marker for the discrimination of Gigaspora isolates and of Gigasporaceae populations from environmental samples. Screening of 48 Gigaspora isolates by PCR-denaturing gradient gel electrophoresis (DGGE) revealed that the V3-V4 region of the 18S rRNA gene contained insufficient variation to discriminate between different Gigaspora species. In contrast, the patterns of 18S ribosomal DNA (rDNA) heterogeneity within the V9 region of this marker could be used for reliable identification of all recognized species within this genus. PCR-DGGE patterns provided insight into some putative misidentifications and could be used to differentiate geographic isolates of G. albida, G. gigantea, and G. margarita but not G. rosea. Two major clusters were apparent based upon PCR-DGGE ribotype patterns, one containing G. albida, G. candida, G. ramisporophora, and G. rosea and the other containing G. decipiens and G. margarita. Dissection of the DGGE patterns by cloning, DGGE screening, and sequencing confirmed these groupings and revealed that some ribotypes were shared across species boundaries. Of the 48 isolates examined, only two displayed any spore-to-spore variation, and these exceptions may be indicative of coisolation of more than one species or subspecies within these cultures. Two Brazilian agricultural soils were also analyzed with a Gigasporaceae-specific nested PCR approach, revealing a dominance of G. margarita within this family.
Arbuscular mycorrhizal fungi (AMF) form one of the most common symbioses with plants (50), and their importance to natural and man-made ecosystems is well established (48, 50, 55). The AMF form a monophyletic group of obligate plant-symbiotic fungi belonging to the phylum Glomeromycota (44). Unfortunately, their ecology, genetics, and evolution are as yet poorly understood (20, 41). The main hurdles to AMF research are the inability to obtain axenic cultures and the difficulties associated with identifying AMF, especially in planta (13, 20). However, the recent application of molecular biological techniques for characterization of AMF has led to important advances in our understanding of the phylogeny (44, 45), ecology (23, 24, 26, 27, 30), genetics (20, 22), and evolution (19, 41) of this group of obligatory symbiotic fungi.
rRNA genes have become the most widely used targets for detection of AMF in environmental samples (13). Several PCR-based strategies targeting rRNA genes have recently been developed to detect AMF in DNA extracted from roots, soil, or spores (23, 29, 30, 56). Such strategies have provided new insights into AMF diversity by circumventing the need for trap cultures and morphological identifications, which can be highly biased, time-consuming, and inaccurate. Despite these advances, the operational taxonomic units obtained in most of these works can only be identified precisely to genus level or above. Thus, little progress has been made in species characterization and identification per se, which are still strongly dependent on morphological analysis and the investigator's level of expertise. Few studies have actually used the rRNA genes to identify species of AMF (38), with most analyses being limited to the detection of defined species of interest (32, 56).
Molecular analyses have revealed that a single AMF isolate or even individual spores may contain substantial heterogeneity among rRNA gene copies (2, 12, 31, 33, 34, 40; for a recent review, see reference 41), which may be unevenly distributed in the heterokaryotic nuclei of AMF spores (31, 54). Intraspecific rRNA heterogeneity seems to be a common phenomenon in AMF as well as in other groups of organisms, such as bacteria (1, 37), plants (11), insects (52), and crustaceans (18). However, little progress has been made in the interpretation of this heterogeneity. Such heterogeneity may lead to overestimations of the number of species when interpreting clone libraries of rRNA recovered from the environment (14). However, if the heterogeneity is consistent within a species, intraisolate heterogeneity might be used as an advantage to generate species-specific rDNA fingerprints for AMF detection and identification.
The genus Gigaspora represents an ecologically and economically (4, 42) important group within the Glomeromycota, and numerous studies have been dedicated to identify species within this genus and to study their ecology. The taxonomy of the genus Gigaspora has recently been revised by morphological (5), fatty acid methyl ester (6), molecular (3), and combined (32) approaches. Among the eight Gigaspora species described to date, Bentivenga and Morton (5) considered five to be valid species based on spore morphology: G. albida, G. decipiens, G. gigantea, G. margarita, and G. rosea. Two species were considered synonymous with previously described Gigaspora species (G. candida = G. rosea, and G. ramisporophora = G. margarita), and one species, G. tuberculata, was considered synonymous with Scutellospora persica. However, there are few useful morphological characters for Gigaspora species determination, and character ranges such as spore size and color often overlap between species (5). Bago et al. (3) used molecular signatures within the V9 region of the 18S rRNA gene as diagnostic characters for Gigaspora spp. identification. These authors were able to identify three distinct groups among the currently recognized species of Gigaspora: the Gigaspora rosea group (G. rosea and G. albida), Gigaspora margarita group (G. margarita and G. decipiens), and Gigaspora gigantea. Their analysis also revealed intraspore heterogeneity, which caused ambiguities in the signatures they found. Lanfranco et al. (32) continued the molecular characterization of selected Gigaspora species and proposed a number of species-specific primer sets, and Yokoyama et al. (58) developed primers based on satellite fragments to identify specific isolates of G. margarita. Thus, while important strides have been made recently, rapid and reliable methods to assess Gigaspora diversity are still lacking. Furthermore, little is known about the establishment, distribution, diversity, and competitiveness of Gigaspora spp. in the field (4, 42).
PCR-denaturing gradient gel electrophoresis (DGGE) was initially developed to study mutations. Nowadays, it has become one of the most applied culture-independent techniques to study the community structure of microorganisms (35). Separation in DGGE is based on differences in sequence composition that affect the melting behavior of the amplicons, causing a decrease in the electrophoretic mobility of a partially melted DNA molecule in a polyacrylamide gel containing a linearly increasing gradient of DNA denaturants (for more information, see reference 35). Recently, Kowalchuk et al. (30) successfully applied PCR-DGGE to study the community structure of AMF associated with Ammophila arenaria.
This study had three main goals: (i) to develop a rapid and reliable method to characterize and identify Gigaspora species, based upon PCR-DGGE analysis of variable regions of the 18S rRNA gene; (ii) to assess the level of intraspore and intraisolate 18S rDNA heterogeneity within the genus Gigaspora and evaluate PCR-DGGE as a method for studying this phenomenon; and (iii) to test the application of a Gigasporaceae-specific PCR-DGGE strategy for the assessment of Gigaspora diversity in environmental samples.
MATERIALS AND METHODS
AMF strains.
The AMF strains used as controls for standardization of PCR-DGGE protocols and to evaluate the level of discrimination between and within species are listed in Table 1. All Gigaspora species were represented by a reference isolate and as many additional isolates as we could collect from various sources (Table 1). The accession (catalogued) strain G. gigantea MN453A-7 was obtained from C. Walker, who received the material from the International Culture Collection of Arbuscular and Vesicular-Arbuscular Mycorrhizal Fungi (INVAM) in the form of spores dispersed in quartz sand since 9 October 1997. All Gigaspora strains used were characterized as pure cultures by morphological analysis.
TABLE 1.
Species, strains, contributors or sources, origins, and germ plasm collections of the Gigaspora, Scutellospora, and Glomus isolates used in this studya
| No. | Species | Strain no. | Contributor or source | Origin | Germ plasm collectionb |
|---|---|---|---|---|---|
| 1 | Gigaspora albida* | BR607A | J. Morton | Brazil | INVAM |
| 2 | Gigaspora albida | BR601 | J. Morton | Brazil | INVAM |
| 3 | Gigaspora albida | UFLA24 | J.O. Siqueira | Brazil | UFLA |
| 4 | Gigaspora albida | CL151 | J. Morton | USA | INVAM |
| 5 | Gigaspora albida | FL713 | J. Morton | USA | INVAM |
| 6 | Gigaspora albida | INVAM927 | L. C. Maia | USA | CNPAB |
| 7 | Gigaspora candida* | BEG17 | V. Gianninazzi-Person | Taiwan | BEG |
| 8 | Gigaspora decipiens* | AU102 | J. Morton | Australia | INVAM |
| 9 | Gigaspora decipiens | W3516 | L. Abbott/C. Walker | Australia | Walker |
| 10 | Gigaspora gigantea | VA105C | J. Morton | USA | INVAM |
| 11 | Gigaspora gigantea | UFLA872 | J. O. Siqueira | Brazil | UFLA |
| 12 | Gigaspora gigantea* | MN453A-7 | C. Walker | USA | INVAM |
| 13 | Gigaspora gigantea | MA453A | J. Morton | USA | INVAM |
| 14 | Gigaspora gigantea | MN414D | J. Morton | USA | INVAM |
| 15 | Gigaspora gigantea | MN922A | J. Morton | USA | INVAM |
| 16 | Gigaspora gigantea | NC110A | J. Morton | USA | INVAM |
| 17 | Gigaspora gigantea | NC150 | J. Morton | USA | INVAM |
| 18 | Gigaspora gigantea | CUT | D. D. Douds | USA | USDA-ARS |
| 19 | Gigaspora gigantea | CUT | G. Bécard | USA | CNRS |
| 20 | Gigaspora margarita* | WV205A | INVAM | USA | INVAM |
| 21 | Gigaspora margarita | CNPAB1 | F. A. de Souza | Brazil | CNPAB |
| 22 | Gigaspora margarita | CNPAB16 | F. A. de Souza | Brazil | CNPAB |
| 23 | Gigaspora margarita | BEG34 Fr | V. Gianninazzi-Person | New Zealand | BEG |
| 24 | Gigaspora margarita | BEG34 It | V. Bianciotto | New Zealand | Torino |
| 25 | Gigaspora margarita | IES32 | R. HerreraPeraza | Cuba | IES |
| 26 | Gigaspora margarita | UFLA36 | J. O. Siqueira | Brazil | UFLA |
| 27 | Gigaspora margarita | TARLSM 478 | M. Saito | Taiwan | MAFF |
| 28 | Gigaspora margarita | K-1-520052 | M. Saito | Japan | MAFF |
| 29 | Gigaspora margarita | C-520054 | M. Saito | Japan | MAFF |
| 30 | Gigaspora margarita | Ni A | M. Saito | Nepal | MAFF |
| 31 | Gigaspora ramisporophora* | CNPAB22 | F. A. de Souza | Brazil | CNPAB |
| 32 | Gigaspora rosea* | FL105 | J. Morton | USA | INVAM |
| 33 | Gigaspora rosea | BR151A | J. Morton | Brazil | INVAM |
| 34 | Gigaspora rosea | BR227B | J. Morton | Brazil | INVAM |
| 35 | Gigaspora rosea | BR235 | J. Morton | Brazil | INVAM |
| 36 | Gigaspora rosea | FL219A | J. Morton | USA | INVAM |
| 37 | Gigaspora rosea | FL676 | J. Morton | USA | INVAM |
| 38 | Gigaspora rosea | KS885 | J. Morton | USA | INVAM |
| 39 | Gigaspora rosea | MA457C | J. Morton | USA | INVAM |
| 40 | Gigaspora rosea | NB103D | J. Morton | USA | INVAM |
| 41 | Gigaspora rosea | NC178 | J. Morton | USA | INVAM |
| 42 | Gigaspora rosea | NY328A | J. Morton | USA | INVAM |
| 43 | Gigaspora rosea | UT102 | J. Morton | USA | INVAM |
| 44 | Gigaspora rosea | WV187 | J. Morton | USA | INVAM |
| 45 | Gigaspora rosea | BEG9 | V. Gianninazzi-Person | Unknown | BEG |
| 46 | Gigaspora rosea | IES19 | R. HerreraPeraza | Brazil | IES |
| 47 | Gigaspora rosea | CI-520062 | M. Saito | Japan | MAFF |
| 48 | Gigaspora rosea | INVAM185 | D. D. Douds | USA | USDA-ARS |
| 49 | Gigaspora rosea | DAOM194757 | D. D. Douds | Canada | USDA-ARS |
| 50 | Gigaspora rosea | DAOM194757 | G. Bécard | Canada | CNRS |
| 51 | Gigaspora sp. | TW1-1 | M. Saito | Taiwan | MAFF |
| 52 | Scutellospora gregaria | CNPAB7 | F. A. de Souza | USA | CNPAB |
| 53 | Scutellospora heterogama | CNPAB2 | F. A. de Souza | Brazil | CNPAB |
| 54 | Scutellospora reticulata | CNPAB11 | F. A. de Souza | Brazil | CNPAB |
| 55 | Glomus clarum | CNPAB21 | F. A. de Souza | Brazil | CNPAB |
*, accession strains considered type or ex-type materials.
BEG, European Bank of Glomales, Dijon, France; CNPAB, Embrapa Agrobiologia, Rio de Janeiro, Brazil; CNRS, Centre National de la Recherche Scientifique, Toulouse, France; IES, Instituto Ecologia y Sistematica, Havana, Cuba; INVAM, International Culture Collection of Arbuscular and Vesicular-Arbuscular Mycorrhizal Fungi, Morgantown, W.V.; MAFF, Ministry of Agriculture, Forest and Fisheries, Ibaraki, Japan; Torino, Dipartimento Biologia Vegetale, Universita di Torino, Tonna, Italy; UFLA, Universidade Federal de Lavras, Minas Gerais, Brazil; USDA-ARS, U.S. Department of Agriculture Agricultural Research Service; Walker, C. Walker, personal collection, New Milton, England.
The curator of the INVAM collection sent us a blind test among the G. rosea strains we bought to test the capacity of our technique to differentiate between species.
Soil samples.
Samples were taken from an 8-year-old grassland field in a cattle farm in Brazil. The farm, Agropecuária Lopes, was located in Santo Antônio, Goiás State (16°28′00"S, 49°17′00"W, at 823 m above sea level), Brazil. The grassland was dominated by Brachiaria decumbens, which had replaced the native vegetation in the Cerrado (savannah) biome.
Intact soil cores (7.5 cm in diameter; 8.0 cm deep) were collected with polyvinyl chloride cylinders. The soil, a clayey dark red oxissol, had a pH of 5.5 (soil/water ratio, 1:2.5 [vol/vol]). It contained 0, 2.4, and 0.7 cmol of charge per kg (dry weight) of soil (cmolc) of Al, Ca, and Mg, respectively, dm−3 in a 1 N KCl extraction and 0.16 and 1 mg of P and K, respectively, dm−3 in a Mehlich I extraction. The samples were transported to Embrapa Agrobiologia, Seropédica, Rio de Janeiro, Brazil, and used to establish 10 trap cultures to assess the diversity of AMF. Brachiaria decumbens was used as the host plant. The trap cultures were sent to the Netherlands for further analysis. Two of these trap cultures were selected: one contained large numbers (sample A) and the other contained small numbers (sample B) of Gigaspora spores (Table 2). Spore identification and counting as well as DNA extraction were performed with three replicates of 30 g of soil inoculum each. The procedure for morphological identification of spores is described in de Souza et al. (16).
TABLE 2.
AMF spore number in trap cultures obtained from soil samples collected in an 8-year-old Brachiaria decumbens grassland field used for cattle in Goiás State, Brazil
| Species | No. of spores/30 g of dry soil
|
|
|---|---|---|
| Sample A | Sample B | |
| Acaulospora mellea | —a | 8.3 |
| Acaulospora morrowiae | 23.3 | — |
| Acaulospora rehmii | 8.3 | 19.0 |
| Acaulospora tuberculata | 1.7 | 36.7 |
| Acaulospora scrobiculata | 4.3 | — |
| Archaeospora gerdemannii | 95.3 | — |
| Gigaspora decipiens and Gigaspora margarita | 63.0 | 5.0 |
| Glomus macrocarpum | 57.3 | 2.3 |
| Glomus sp. strain N.1. | 17.0 | — |
| Glomus sp. strain N.2. | 128.0 | 28.0 |
| Glomus sp. strain N.3. | — | 4.7 |
| Scutellospora coralloidea | 0.7 | — |
| Scutellospora heterogama | 1.3 | — |
| Total | 400.2 | 104.0 |
—, not detected.
Spore extraction and preparation for DNA extraction.
Spores were extracted directly from the material received from the collections with the standard wet sieving technique, followed by centrifugation in water and subsequently in 50% sucrose solution (for details of spore extraction, see reference 15). After extraction, the spores were carefully selected under a binocular microscope, further cleaned by ultrasonication for 15 s, and rinsed in autoclaved ultrapure water (Millipore B.V., Etten-Leur, The Netherlands). This procedure was repeated four times. Individual clean and healthy-looking spores were selected and transferred to 1.5-ml microcentrifuge tubes at either 1 or 60 spores per tube and stored at −80°C until required. Individual spores were used to ensure purity and to compare the results with DNA extracted from multiple spores.
Control experiments with multiple target and nontarget AMF species.
In order to evaluate the effect of different target and nontarget species combinations on the detection limits and reproducibility of PCR-DGGE banding patterns, we combined DNA from strains Scutellospora heterogama CNPAB2 and G. margarita CNPAB16 at different ratios. The ratios used were Scutellospora heterogama CNPAB2 to G. margarita CNPAB16 at 1:1; 1:5; 1:10; 1:25; 1:50, and 1:100 and G. margarita CNPAB16 to S. heterogama CNPAB2 at the same ratios. In addition, we also combined nontarget DNA obtained from Glomus clarum CNPAB5 in ratios ranging up to 100:1 with G. margarita CNPAB16. Three replicas were performed for each combination.
Greenhouse experiment.
To ensure that the PCR-DGGE approach was sensitive enough to detect multiple species of Gigaspora and Scutellospora in root samples, we performed a greenhouse experiment. Clover plants (Trifolium pratense) were inoculated or not with a mixture of soil inoculum containing G. margarita (CNPAB16), S. gregaria (CNPAB7), S. heterogama (CNPAB2), and S. reticulata (CNPAB11). To ensure nodulation, the clover plants were also inoculated with soil filtrate containing rhizobia collected from clover field plots. The plants were grown in plastic cone pots containing 250 ml of a mixture of clay soil and sand (1:1, vol/vol) at pH 6.2 (soil/water ratio, 1:2.5). The pots were fertilized intermittently with 1/10th-strength nutrient solution (25) without N and P. After 2 months of growth, the pot contents were harvested. The soil was carefully removed from the roots with tap water, and the root system was cleaned by ultrasonication (60 W; B-2200 E1; Bransonic) twice for 3 min each in autoclaved water, followed by a final wash with autoclaved water. The roots from each pot were divided into subsamples for either DNA extraction or assessment of colonization rate by the method of Giovannetti and Mosse (21).
DNA extraction from spores, roots, and soil samples.
Spores were removed from the freezer (−80°C) and crushed with a micropestle (Treff AG, Degersheim, Switzerland) in 40 μl of 10 mM Tris-HCl buffer, pH 8.0, with 10 μl of 20% Chelex 100 (Bio-Rad Laboratories, Hercules, Calif.), for single spores. The same procedure was used with multiple spores except that the reagent concentrations were 80 and 40 μl for buffer and Chelex, respectively. The tubes were then incubated at 95°C for 10 min, chilled on ice, and centrifuged at 10,000 × g for 2 min. The supernatant was carefully transferred to a new tube and stored at −20°C until use. Samples from multiple spore isolations were treated with RNase before being stored. DNA extractions from trap plants with bulk soil and root material were performed with the UltraClean soil DNA isolation kit according to the manufacturer's instructions (MoBio Laboratories, Solana Beach, Calif.). Prior to DNA extraction, the samples (10 g of soil or 2 g of root) were homogenized and ground under liquid N2 with a mortar and pestle. A subsample of 0.5 g of bulk soil or root material was used for each DNA extraction. After extraction, the soil- and root-derived DNA was purified once more with the Wizard DNA purification kit (Promega, Madison, Wis.) as described by the manufacturer. For the greenhouse experiment, we extracted DNA from plant roots with the protocol described by Edwards et al. (17) with 50 mg of liquid nitrogen-powdered roots.
Nested PCR conditions for amplification from spore DNA.
The DNA from spores was first amplified with the forward primer NS1 in combination with reverse primer ITS4, covering the region from the beginning of the 18S rRNA gene through the 5′ end of the 25S rRNA gene (57). Primer positions are given in Fig. 1, and primer sequences, references, and PCR conditions are provided in Table 3. These reactions were performed in a final volume of 15 μl, with 5 μl of template DNA. The PCR mixture was composed of 200 μM each of the four deoxynucleoside triphosphates, 1.5 μM MgCl2, a 0.4 μM concentration of each primer, and 1 U of Expand high-fidelity DNA polymerase (Roche Diagnostics, Nederland B.V., Almere, The Netherlands) according to the manufacturer's recommended buffer conditions. All reactions were performed in a PTC200 thermal cycler (MJ Research; Waltham, Mass.).
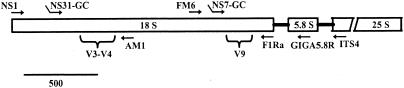
FIG. 1.
Cartoon focusing on the 18S rRNA gene. Approximate positions of primers (arrows not to scale) and variable regions targeted by PCR-DGGE analyses are shown. Bent tails on primers indicate the presence of a GC clamp.
TABLE 3.
rDNA primers, primer combinations, GC clamp, and PCR conditions used in this studya
| Primer | Sequence | Target group | Partner primer for PCR | PCR conditions | Product size (bp) |
|---|---|---|---|---|---|
| NS1 | 5′-GTAGTCATATGCTTGTCTC-3′ | Universal eukaryotes | ITS4 | 94°C for 60 s, 55°C for 240 s, 30 cycles | 2,300 |
| NS31-GC | 5′-TTGGAGGGCAAGTCTGGTGCC-3′ | Universal eukaryotes | AM1 | 94°C for 60 s, 61°C for 40 s, 30 cycles | 600 |
| FM6 | 5′-ACCTGCTAAATAGTCAGGCTA-3′ | Gigasporaceae | GIGA5.8R | 94°C for 60 s, 59°C for 45 s, 30 cycles | 700 |
| NS7-GC | 5′-GAGGCAATAACAGGTCTGTGATGC-3′ | Universal eukaryotes | F1Ra | 94°C for 60 s, 60°C for 28 s, 30 cycles | 400 |
| ITS4 | 5′-TCCTCCGCTTATTGATATGC-3′ | Universal eukaryotes | |||
| AM1 | 5′-GTTTCCCGTAAGGCGCCGAA-3′ | Fungi | |||
| GIGA5.8R | 5′-ACTGACCCTCAAGCAKGTG-3′ | Gigasporaceae | |||
| F1Ra | 5′-CTTTTACTTCCTCTAAATGACC-3′ | Fungi |
The product of this first PCR amplification was diluted 1:1,000, and 2 μl of this dilution was used as the template in a second round of PCR (reaction volume, 25 μl) designed to target either the V3-V4 or V9 region of the 18S rRNA gene (Fig. 1, Table 3). In each case, one of the primers contained a GC clamp to stabilize the amplicon's melting behavior for DGGE analysis (47).
Gigasporaceae-specific PCR conditions for the analysis of environmental samples.
To obtain Gigasporaceae-specific products from soil, roots, and spores from trap cultures and the greenhouse experiment, the first step of the nested PCR combined primers FM6 (this study) and GIGA5.8R (39) (see Fig. 1 and Table 3 for primer positions and sequences). PCR mix was prepared as described above, and the DNA extracted from soil and root samples was diluted 1:50 to 1:100 and used as template. The product of this first PCR amplification was diluted 500- to 1,000-fold, depending on product concentration, and used as the template for a second PCR with the primer pair NS7 (57) with GC clamp in combination with primer F1Ra (this study) as described above.
DGGE analysis.
All DGGE analyses were performed with the D-Gene system (Bio-Rad), with gradients of 25 to 40% and 32 to 42% denaturant and running conditions of 75 V for 16 h for the NS31 and GC/AM1 and 95 V for 17 h for the NS7 and GC/F1Ra primer combinations, respectively. Gels were run in 0.5× TAE (Tris-acetate-EDTA) buffer at a constant temperature of 60°C. Gels were stained for 20 min in MilliQ water containing 0.5 mg of ethidium bromide liter−1 and destained twice for 15 min in MilliQ water prior to UV transillumination. Gel images were digitally captured with the ImaGo system (B & L, Maarssen, The Netherlands).
DGGE banding patterns were assessed by cluster analysis with a Jaccard similarity coefficient, and similarities between profiles were depicted as a dendrogram constructed by the unweighted pair group method with arithmetic average (UPGMA) within the Bionumerics program, version 2 (Applied Maths, Kortrijk, Belgium). The banding pattern of G. rosea FL105 was used as a marker to standardize different gels.
Tests for reproducibility.
All the PCR amplifications and DGGE analyses were performed with three independent single-spore DNA isolates and compared with multiple-spore isolates. This was done to detect potential artifacts due to spore contamination (43) and to evaluate the reproducibility of the method. For soil and root DNA, three subsamples were compared for each trap culture.
Recovery of DNA from DGGE gels.
The most prominent bands obtained in the DGGE profiles of the trap culture spores were sequenced. The middle portion of selected DGGE band was excised, and approximately 60 mg of acrylamide gel material per band was transferred to a 0.5-ml microcentrifuge tube containing 40 μl of MilliQ water and frozen at −80°C for 1 h. Subsequently, the gel material was crushed with a plastic pellet mix (Treff AG), and the tubes were incubated at 37°C for 3 h. After centrifugation at 11,000 × g for 60 s, the supernatant was transferred to a new tube, and 1 μl of it was used as the template for subsequent PCR-DGGE analysis to check band position and purity. This procedure was repeated until a single sharp band was detected. After that, PCR was performed with the same primer pair used in the DGGE analysis without the GC clamp, and the product was prepared for sequencing analysis.
Cloning of AMF rDNA.
In order to obtain clones for different variants of the ribosomal genes present (ribotypes) in one species (spore), amplicons were obtained from DNA extracted from individual spores after PCR amplification with the primer pair NS1 and ITS4 (57) as described previously. PCR products were purified with the High Pure PCR product purification kit (Boehringer, Mannheim, Germany). Purified PCR products were then cloned into the pGEM-T easy vector, with Escherichia coli strain JM109 used for transformation, according to the procedure given by the manufacturer (Promega Benelux, Leiden, The Netherlands). The clones obtained were cultured and, after plasmid extraction by the Wizard Plus SV miniprep DNA purification system (Promega, Benelux), used as templates for PCR (see below).
Clone selection with DGGE and sequence analysis.
Plasmids containing an insert obtained from different Gigaspora isolates were used as the templates for reactions with the forward primer NS7-GC in combination with the reverse primer F1Ra as described above. DGGE screening was performed as described above, and the PCR products obtained from the original isolates were used as reference to select clones that corresponded to each of the ribotypes detected in each isolate examined.
Prior to sequencing, DNA templates were purified with Qiaquick purification columns. Sequencing reactions were performed with the Perkin Elmer Biosystems Big Dye terminator sequence reaction kit (Perkin Elmer, Foster City, Calif.) and run on a Perkin Elmer 3700 capillary sequencer at the National Institute for Public Health and the Environment (Bilthoven, The Netherlands).
Sequence alignments.
Sequences recovered from the GenBank/EMBL database or generated in this work were first aligned with Clustal-X (53), and then the alignment was improved by manual inspection. Phylogenetic analysis was conducted with the parsimony method in PAUP* version 4.0 Beta 10 (51).
Nucleotide sequence accession numbers.
The sequences and alignment generated in this study were deposited in the EMBL database under accession numbers AJ539236 to AJ539305 and alignment number ALIGN_000606.
RESULTS
DGGE profiles targeting the V3-V4 region of the 18S rRNA gene.
All Gigaspora strains tested migrated to approximately the same position in the gel, with the G. margarita, G. decipiens, and Gigaspora sp. TW-1 strains showing a tight doublet and all other species showing only a single band (results not shown). The level of inter- and intraspecies heterogeneity within the V3-V4 region was not sufficient to discriminate among the different Gigaspora species tested. Nevertheless, two groups were distinct: the first group was formed by G. margarita, G. decipiens, and Gigaspora sp. strain TW-1 (double band), and the second group was composed of the other strains (single band).
DGGE profiles targeting the V9 region of the 18S rRNA gene.
PCR-DGGE analysis of the V9 region of the 18S rRNA gene could differentiate all Gigaspora species based on the type materials used, including those (G. candida and G. ramisporophora) declared to be invalid by morphological analysis (Fig. 2). Almost all isolates yielded multiple bands, indicating the presence of intraspore variation between ribotypes in this region in the strains examined. To test the consistency of the PCR-DGGE profile for a given isolate, we examined several single- and multiple-spore DNA isolations per isolate. No between-spore variations could be detected for any of the 48 isolates tested with the exception of G. albida CL151 and G. margarita UFLA36. The former produced two very similar patterns (CL151a and CL151b in Fig. 3), but in the CL151b type, one of bands were absent and the intensity of the lower band was higher than that in the CL151a type. The latter accession produced two very different banding patterns (UFLA36-T1 and UFLA36-T2 in Fig. 3). These two strains may each actually represent two coisolated populations (see the Discussion). No difference was observed between different cultures of the same accession strain maintained in different laboratories (data not shown).
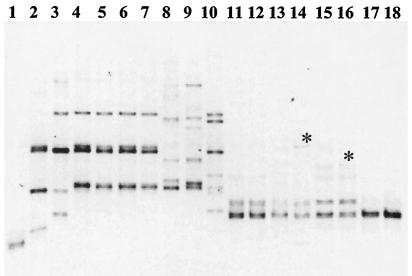
FIG. 2.
PCR-DGGE analysis of 18S rRNA gene fragments amplified from Gigaspora species and run for 15 h at 95 V for analysis of the V9 region. lanes: 1, G. gigantea UFLA872; 2, G. gigantea MN453A-7; 3, G. gigantea VA105C; 4, G. rosea BEG9; 5, G. rosea FL105; 6, G. rosea IES19; 7, G. albida INVAM927; 8, G. albida BR607A; 9, G. candida BEG17; 10, G. ramisporophora CNPAB22; 11, G. margarita CNPAB1; 12, G. margarita CNPAB16; 13, G. margarita IES32; 14, G. margarita WV205A; 15, G. margarita BEG34 France; 16, G. margarita BEG34 Italy; 17, G. decipiens AU102; 18, G. decipiens W3516. Asterisks show G. margarita strain-specific bands.
FIG. 3.
Dendrogram showing the distance tree and the PCR-DGGE banding patterns of 48 strains of Gigaspora and two divergent patterns found in strains G. albida CL151 and G. margarita UFLA36. Gels were run for 17 h at 95 V. Scale shows similarities of banding patterns; thicker lines indicate separation between major clades or subclades. Numbers indicate cophenetic correlations, which are estimates of the faithfulness of each subcluster of the dendrogram.
To determine the consistency of PCR-DGGE patterns within each species, we examined all the Gigaspora isolates that we could obtain (48 total). Dendrogram analysis of these banding patterns produced two major clades, the first containing the species G. albida, G. candida, G. ramisporophora, G. rosea and most of the G. gigantea strains and the second containing G. decipiens, G. margarita, and Gigaspora sp. strain TW-1 (Fig. 3). Within the first of these groups, the G. albida isolates formed a distinct subcluster. Of the 10 strains of G. gigantea analyzed, one, UFLA872, was unique and had no bands in common with any of the other 48 strains tested (Fig. 3).
The banding patterns for strains of the same species were generally highly similar and clustered together (Fig. 3). Isolate-specific bands could be identified for some strains of the species G. albida, G. gigantea, and G. margarita but generally not for G. decipiens and G. rosea, although accession G. rosea MA457C appeared to contain a number of unique ribotypes (Fig. 3).
PCR-DGGE detected potential misidentifications by revealing some patterns that did not cluster with their respective type material. For instance, within the species G. albida, accession UFLA24 clustered with the G. ramisporophora type material (CNPAB22) and accession INVAM927 produced a pattern identical to that of the large group of G. rosea isolates. Similarly, G. rosea BR235 grouped together with G. albida isolates; it was the blind sample sent by INVAM's curator. This result confirms the supposed misidentification of this accession based upon morphological characteristics (J. B. Morton, personal communication).
Although G. candida BEG17 and G. ramisporophora CNPAB22 are both considered to represent nonvalid species names based upon morphological evaluation (5), they could be clearly distinguished from the other species examined. Interestingly, the pattern of G. candida BEG17 was more similar to that of the G. albida isolates than to those of G. rosea, the species to which it was previously assigned (Fig. 3). G. ramisporophora CNPAB22 generated a banding pattern that was more similar to those of G. rosea (Fig. 3) than those of G. margarita, the species to which G. ramisporophora was assigned based upon morphological characteristics. A robust molecular characterization of the species G. candida and G. ramisporophora will require the analysis of additional isolates. Unfortunately, no other well-defined isolates of G. candida are available at this time, to the best of our knowledge, and very few well-defined isolates of G. ramisporophora are available.
Sequence analysis of PCR-DGGE banding patterns.
To gain further insight into the nature of the intra- and interspecies heterogeneity detected, sequence information was obtained for the region analyzed by PCR-DGGE for each of the ribotypes observed within representative isolates of each species (Table 4). Sequence analysis confirmed the identity of bands that displayed the same migratory behavior. Furthermore, sequence and phylogenetic analyses (Table 4) also confirmed a number of the relationships depicted in the dendrogram analysis of the PCR-DGGE patterns. For instance, G. margarita and G. decipiens are closely related and distinct from the group formed by G. albida, G. candida, G. ramisporophora, and G. rosea and that of G. gigantea. In addition, G. gigantea UFLA872 is quite distinct from all other accession strains and contains a DNA signature, CGCGTG, that has been reported to occur in Scutellospora species (3). With reference to the characterization of putative invalid species, sequence analysis confirmed ribotype overlap between G. candida BEG17 and G. albida BR607A, including a shared DNA signature, TAGGTT, which is distinct from that of G. rosea (3). The ribotype sequences obtained for G. ramisporophora were also more similar to those of G. albida, G. candida, and G. rosea than to those of G. margarita despite apparent similarity in the morphology of G. ramisporophora and G. margarita spores. Those results do not support the reclassification based on morphological analysis by which G. candida was reclassified as being synonymous with G. rosea and G. ramisporophora was reclassified as being synonymous with G. margarita (6).
TABLE 4.
DNA sequences of 70 PCR-DGGE-selected Gigaspora clones and two DGGE bands showing alignment of 24 parsimonious informative and two uninformative positions in the V9 18S rDNA region
| Species and clone code(s) (accession no.)a | Position in alignmentb
|
|||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 3 | |
| 5 | 7 | 7 | 7 | 1 | 4 | 7 | 9 | 3 | 6 | 6 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 8 | 8 | 8 | 8 | 9 | 9 | 9 | 0 | |
| 5 | 0 | 1 | 6 | 4 | 4 | 8 | 2 | 4 | 5 | 6 | 0 | 3 | 4 | 5 | 6 | 8 | 9 | 0 | 5 | 6 | 8 | 1 | 8 | 9 | 0 | |
| G. gigantea A1* UFLA872; B1* UFLA872 | C | C | G | C | C | C | T | A | T | C | G | C | C | G | C | g | t | G | G | C | C | C | G | T | A | C |
| G. decipiens 4; G. margarita M5, M6*, M7 (CNPAB1), T2 (CNPAB16), 7*, 8, 15* (BEG34), DGGE b2 | . | . | . | . | . | . | . | G | . | . | . | . | C | G | A | g | t | G | . | . | T | . | . | . | . | T |
| G. margarita F22, F44 (CNPAB1), T3 (CNPAB16) | T | . | . | . | . | . | . | G | . | . | . | . | C | G | A | g | t | G | . | . | T | . | . | . | . | T |
| G. margarita M9 (CNPAB1), T5 (CNPAB16), DGGE b1 | . | . | . | . | . | . | . | G | . | T | . | . | C | G | A | g | t | G | . | . | T | . | . | . | . | T |
| G. margarita 1, 21 (BEG34) | . | . | . | . | . | . | . | G | . | . | . | . | C | G | A | g | t | G | . | T | T | . | A | . | . | T |
| G. decipiens 2*; G. margarita 2, 10, 14 (BEG34) | . | . | . | . | . | . | . | G | . | . | . | . | C | G | A | g | t | G | . | . | . | . | . | . | . | T |
| G. decipiens 9; G. margarita M18 (CNPAB1), 5, 20 (BEG34) | . | . | . | . | . | . | . | G | . | . | . | . | T | G | A | g | t | G | . | . | T | . | . | . | . | T |
| G. margarita M8* (CNPAB1) | . | . | A | . | . | . | . | G | . | . | . | . | T | G | A | g | t | G | . | . | T | . | . | . | . | T |
| G. albida 13; G. candida C13, C17*; G. ramisporophora GP18 | . | . | . | . | . | T | . | G | . | . | . | . | T | A | A | g | t | G | . | . | T | . | . | C | T | T |
| G. albida 25* | . | . | . | . | . | T | . | G | C | . | . | . | T | A | A | g | t | G | . | . | T | . | . | C | T | T |
| G. ramisporophora GP16-GP11, GP22 | . | . | . | . | . | T | T | . | G | . | . | . | T | A | A | g | t | G | A | C | T | . | . | C | T | T |
| G. ramisporophora GP27, GP49 | . | T | . | . | . | T | . | G | . | . | . | T | T | A | A | g | t | G | . | . | T | . | A | C | T | T |
| G. rosea GenBank X58726 | . | . | . | . | . | T | . | G | . | . | . | . | T | A | A | g | t | G | . | . | T | . | . | C | T | . |
| G. albida 17; G. candida C4 | . | . | . | T | . | T | . | G | . | T | . | . | T | A | A | g | t | G | . | . | T | . | . | C | T | T |
| G. rosea R14, R16 | . | . | A | . | . | T | . | . | . | . | . | . | T | A | A | g | t | G | . | . | T | . | . | C | T | T |
| G. ramisporophora GP44 | . | . | . | . | . | T | . | G | . | . | . | . | T | A | A | g | t | G | . | . | T | . | . | C | C | T |
| G. albida Genbank Z14009 | . | . | . | . | . | T | . | G | . | . | . | . | T | A | A | g | t | G | . | . | T | . | . | C | . | . |
| G. ramisporophora GP14 | . | . | . | . | . | T | . | G | . | . | . | . | T | G | A | g | t | G | . | . | T | . | . | C | T | T |
| G. albida 14; G. candida b4, C3, C12, C14b, C16 | . | . | . | . | . | T | . | G | . | . | . | . | T | A | G | g | t | T | . | . | T | . | . | . | . | . |
| G. ramisporophora 16-1, 16-2 | . | . | . | . | T | T | . | G | . | . | . | . | T | A | T | g | t | T | . | . | T | . | . | . | . | . |
| G. ramisporophora GP33; G. rosea E2, R13, R15 | . | . | . | . | . | T | . | G | . | . | . | . | T | A | T | g | t | T | . | . | T | . | . | . | . | . |
| G. albida 19*, 31; G. candida C1, C14, C18, G1 | . | . | . | . | . | T | . | G | . | . | A | T | T | A | T | g | t | T | . | . | T | . | . | . | . | . |
| G. rosea R19 | . | . | . | . | . | T | . | G | C | . | . | . | T | A | T | g | t | T | . | . | T | . | . | . | . | . |
| G. gigantea 15, VA105C | . | . | . | . | . | T | . | G | . | . | . | . | T | A | A | g | t | T | . | . | T | T | . | . | . | . |
| G. gigantea 19, VA105C | . | . | . | . | . | T | . | G | . | . | . | . | T | A | A | g | t | T | . | . | T | . | . | . | . | . |
| G. gigantea 3*, 7*, 26*, VA105C | . | . | . | . | . | T | . | G | . | . | . | . | T | G | T | g | t | T | . | . | . | . | . | . | . | . |
| G. gigantea 10, VA105C | . | . | . | . | . | T | . | G | . | . | . | . | T | G | T | g | t | T | . | . | T | T | . | . | . | . |
| G. gigantea 6, VA105C | . | . | . | . | . | T | . | G | . | . | . | . | T | T | A | g | t | T | . | . | T | . | . | . | . | . |
| G. gigantea 20*, GenBank Z14010* | . | . | . | . | . | T | . | G | . | . | . | . | T | G | A | g | t | T | . | . | T | . | . | . | . | . |
Different clones in the same line have 100% DNA sequence similarity with one or more clones in the same line. Clones in the same line followed by * have less than 100% similarity with other clones in the same line. With those clones, the mutations found were not parsimoniously informative. Accession numbers not specified in the table are G. albida BR607A, G. candida BEG17, G. decipiens W3516, G. ramisporophora CNPAB22, and G. rosea BEG9.
Position 1 starts at the 5′ end of the primer NS7, according to the alignment provided at the European Bioinformatics Institute (EMBL-EBI) website (http://www.ebi.ac.uk/webin-align/webin_align_listali.html, alignment ALIGN_000606). Dots indicate bases identical to the corresponding base in the sequence on the first line, except within the DNA signatures of Bago et al. (3), in which all bases are shown, with divergent bases in bold type. The GT characters in lower case are not parsimoniously informative but are shown because they are part of the DNA signature proposed by Bago et al. (3) (underlined). The line spaces in the table separate the major clades obtained by phylogenetic analysis.
Detection of Gigasporaceae species in field samples. (i) Detection limit.
To test the specificity of the Gigasporaceae-specific primers used, nontarget DNA, in our case DNA from Glomus clarum CNPAB5 spores, was used. It did not interfere in the PCR amplification even when the nontarget species was provided in 100-fold-higher numbers. Multiple Gigaspora species could be detected in artificial spore mixtures when a given species represented 10% or more of the total, and the relative signal intensity roughly matched the spore volume (data not shown). When S. heterogama and G. margarita spores (both targeted by the primers used) were combined at various ratios, the larger spore size (i.e., more 18S rDNA targets per spore) of the latter species skewed the range within which both species could be detected. A single G. margarita spore could be detected in a background of up to 100 S. heterogama spores, whereas the S. heterogama signal was no longer detected when spores of this species were outnumbered fivefold or more by G. margarita spores. Thus, in the analysis of bulk samples containing large numbers of spores or DNA isolated directly from root or soil material, minor populations may not be detected. The analysis of individual spores, small groups of spores, or individual root pieces may therefore offer the best strategy for detecting the full breadth of Gigasporaceae diversity within a sample.
(ii) Detection of Gigasporaceae species in greenhouse experiments and environmental samples.
In a controlled greenhouse experiment, clover plants were inoculated with four AMF species, one species of Gigaspora and three of Scutellospora. Despite a colonization level of less than 20%, as determined by microscopic inspection, AMF-specific products could be easily detected with a nested PCR and DGGE approach. All four AMF species could be detected, although secondary bands had to be used to determine the presence of the two Scutellospora species, since the two species used presented a prominent DGGE band in the same position (Fig. 4).
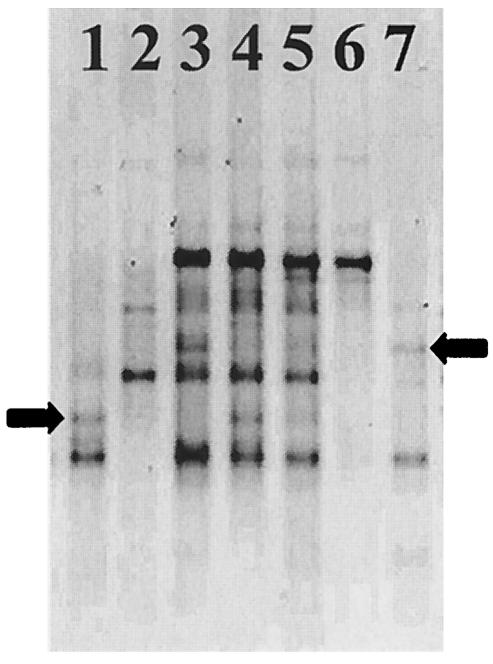
FIG. 4.
Detection and identification of Gigasporaceae from DNA extracted from 2-month-old clover roots by PCR-DGGE analysis of the V9 region of the 18S rRNA gene. DNA templates: lane 1, S. heterogama CNPAB2; lane 2, G. margarita CNPAB16; lane 3, Trifolium pratense replicate 1; lane 4, T. pratense replicate 2; lane 5, T. pratense replicate 3; lane 6, S. gregaria CNPAB7; lane 7, S. reticulata CNPAB11. Note the presence of secondary bands in lanes 4 and 5 (S. heterogama) and 3 (S. reticulata) (arrows). The gel was run for 17 h at 95 V.
Gigaspora spores were recovered from the trap cultures of the two Brazilian agricultural soil samples, as identified by morphological characteristics. These samples also contained spores belonging to the genera Archaeospora, Acaulospora, and Glomus (not targeted in our PCR-DGGE analysis), as well as small numbers of spores of the genus Scutellospora (targeted by the primers used in this study; Table 3). Nested PCR-DGGE analysis with DNA extracted from these Gigaspora spores as well as that isolated directly from soil and roots from the trap cultures generated banding profiles similar to those observed for the type material of G. margarita (Fig. 5), and band identity was confirmed by sequence analysis (Table 4). In addition, the band patterns of 30 individual spores recovered from sample A were identical and produced patterns similar to that of G. margarita strains CNPAB1, CNPAB16, and IES32. Although some Scutellospora spores were present in these samples, this genus was not detected via PCR-DGGE with DNA extracted directly from the soil and roots, even though the specificity of the PCR covered this genus. Recovered Scutellospora spores could be used as the template for PCR-DGGE analysis and could clearly distinguish them from the Gigaspora species detected (Fig. 5). The relative amount of Scutellospora material in the soil samples (0.5 g) used to extract DNA was apparently below the detection limit of our analysis.
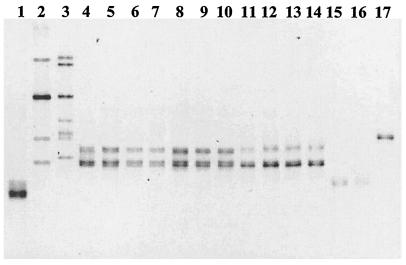
FIG. 5.
Detection and identification of Gigasporaceae from DNA extracted from soil or single spores from trap cultures by PCR-DGGE analysis of the V9 region of the 18S rRNA gene. DNA templates: lane 1, G. gigantea UFLA872; lane 2, G. gigantea VA105C; lane 3, G. ramisporophora CNPAB22; lane 4, G. margarita CNPAB1; lanes 5 to 7, soil DNA extracted from trap culture A; lanes 8 to 10, soil DNA extracted from trap culture B; lanes 11 to 14, single-spore DNA from four different Gigaspora spores recovered from trap culture A; lanes 15 and 16, single-spore DNA from two different S. heterogama spores recovered from trap culture A; lane 17, single-spore DNA from S. coralloidea recovered from trap culture A. The gel was run for 15 h at 95 V.
DISCUSSION
PCR-DGGE as a tool to characterize, identify, and detect Gigaspora species.
By using PCR-DGGE targeting the V9 region of the 18S rRNA gene, we were able to generate highly reproducible profiles obtained from single-spore DNA isolations, which could be used to characterize and differentiate all Gigaspora species based on the type materials used. This included the discrimination of species previously thought to be invalid based upon morphological characteristics (G. ramisporophora and G. candida; see Fig. 2). While some intraspecific variation in PCR-DGGE banding patterns provided several markers that might be used to track specific isolates of a given species, species patterns were generally highly diagnostic (Fig. 3). This study provides the most complete molecular characterization available for the genus Gigaspora, and the specific PCR-DGGE method provided far better characterization and species identification than any other previously applied to this genus.
Due to the overlap of some ribotypes across species boundaries (e.g., G. albida, G. candida, G. ramisporophora, and G. rosea or G. decipiens and G. margarita; see Fig. 3 and Table 4), it became clear that the entire pattern of 18S rDNA types needed to be used for species characterization and identification, as opposed to just a single sequence. Thus, in contrast to other molecular approaches, such as PCR followed by cloning and sequencing, in which intraspore rDNA heterogeneity impairs interpretation, the PCR-DGGE approach uses this heterogeneity to advantage for the generation of highly reproducible isolate- or species-specific patterns. Furthermore, in comparison to some other molecular methods, PCR-DGGE is easy, rapid, and inexpensive to perform.
We used two different nested PCR strategies. The first nested PCR strategy uses family-specific primers in the first PCR. With that strategy, it was possible to analyze DNA extracted from environmental samples with small amounts of material extracted directly from spores, roots, or soil without the problem of nonspecific amplification. However, despite the family-specific nature of the nested PCR strategy and the presence of both Gigaspora and Scutellospora species in the mixed spores, roots, and soils analyzed, the latter genus was not detected. However, in a control experiment, we could detect the simultaneous presence of all four of the AMF species introduced into the root system (Fig. 4). PCR-DGGE strategies will generally fail to detect some minority populations (i.e., <1% of the total target) (10, 35), and our experiments showed that a total spore volume of 10% of the total was typically necessary to ensure the detection of a minority target species.
Furthermore, the DGGE signals detected are based on the relative number of template molecules of a given species in a sample. Thus, even assuming equal amplification efficiencies, it remains difficult to translate the levels of DGGE signal detected for certain AMF species, as the ratio of 18S rDNA target to fungal biomass and the ecological importance of this ratio are not known at present.
The second nested PCR strategy used universal primers that, in our case, amplified the whole 18S rRNA gene plus the internal transcribed spacer (ITS) region. That fragment was cloned, followed by PCR-DGGE selection targeting variable regions in the cloned fragment. That strategy proved to be an excellent approach to study inter- and intraspecies heterogeneity in the 18S rDNA. The principles of that strategy can be applied to study polymorphism in any other gene found in AMF. The application of a similar approach with faster-evolving genes might allow better discrimination between isolates and also might help to shed some light on evolutionary issues such as genetic drift and recombination in these ancient asexual organisms (19).
It is interesting that the DGGE system used was highly efficient in detecting sequence variation within the V9 region of the 18S rDNA gene. Within the 344 bp (without GC clamp) of sequence targeted, 303 bp were constant, 17 bp were parsimoniously uninformative, and 24 bp were parsimoniously informative (Table 4). The V9 18S rDNA fragments analyzed by PCR-DGGE are within the optimal size range for DGGE analysis, i.e., below 500 bp. Most of the mutations found were transitions (37 of 41), which allow discrimination by DGGE, as they always cause a change in the melting temperature. However, transversions were detected in four of the parsimoniously informative positions (positions 274, 275, 279, and 299; see Table 4), and some of them did not affect the melting temperature of the amplicons; consequently, they might not be detected if they were the only mutation present in the amplicon. Despite having the ability to discriminate most single-base-pair differences within 18S rDNA V9 region fragment analyzed here, any DGGE analysis will be limited by the amount of heterogeneity, type of mutation, and fragment size of the target region (36). This was exemplified by our analyses of the V3-V4 region, which contained less variation than the V9 region and also used a longer fragment (550 bp without the GC clamp). Interestingly, the V3-V4 region could be used to discriminate between Glomus species (30). Although the PCR-DGGE banding patterns were highly reproducible within a given isolate across various DNA isolations, exact banding patterns are dependent on the electrophoresis conditions used (data not shown), and analytical consistency and the use of type strains (isolates) as markers are critical when comparing samples.
The similarity between the sequences from the V9 region analyzed was high both between and within species (range, 98.6 to 100%; Table 4). Of the variable positions described, only 24 were phylogenetically informative, hampering robust phylogenetic analysis. As such, the comparison of V9 18S rDNA fingerprints (Fig. 3) provided a more reliable method of comparison than tree construction based on the sequence variations described in Table 4. With proper primer design, PCR-DGGE strategies as implemented here are also ideal for determining specific ribotypes of other AMF genera or other loci in experiments designed to address AMF reproduction and evolution (19, 41), as well as similar issues with respect to intraspecies heterogeneity among rDNA copies in bacteria (1, 14, 37). It should be noted that our analysis of single spores does not address the homokaryotic or heterokaryotic nature of AMF nuclei (20, 54), although the sensitivity of PCR should permit PCR-DGGE of single AMF nuclei.
Comparison between single- and multiple-spore DNA isolations and geographically diverse isolates.
The PCR-DGGE banding patterns of all single-spore DNA isolations tested for the same isolate were identical for 46 of 48 accession strains tested (Fig. 3). Thus, at least to the level of detection afforded by the system used here, a single spore appeared to contain the full range of variation of ribotypes present in an entire spore population. The two exceptions were both isolates recovered by trap cultures. Trap cultures are the most common way to isolate AMF from field samples. However, if more than one morphologically closely related species are coisolated in the same culture, further discrimination by spore morphology is difficult. One, G. margarita isolate, UFLA36, produced spores with two very different DGGE patterns that clustered apart from each other; one type (UFLA36-T1) clustered in the G. margarita-G. decipiens cluster, and the other type (UFLA36-T2) clustered with G. ramisporophora and G. rosea. In the case of G. albida CL151, the two spore types were similar, differing only by the absence of one band in one spore type and the relative intensity of one of the other bands (Fig. 3). In those cultures, further purification by single-spore pot culture followed by spore identification with PCR-DGGE can be used to purify and distinguish those populations.
Interestingly, some geographically distinct isolates were more similar than some isolates that were found at the same site. Within the species G. gigantea, for instance, strains NC110A and NC150 are rather different single-spore cultures recovered from the same site (7), whereas isolates CUT, MA453A, and MN453A-7 had identical rDNA patterns despite being isolated from disparate locations. Similarly, G. albida CL151 type a from the United States and BR601 from Brazil were identical (Fig. 3), while other sympatric populations showed more diversity. The presence of strain-specific bands should allow the tracking of specific AMF populations in studies dedicated to unraveling the ecological significance of such sympatric populations. The band intensities observed for different geographic isolates differed in some cases. This result suggests that the proportion of the different ribotypes may differ between isolates of the same species, but more quantitative methods, such as introduction of an internal standard in DGGE experiments (10), will be necessary to address this question.
PCR-DGGE characterization versus other schemes applied previously.
The lack of discrete and diagnostic characters for species identification within the genus Gigaspora has been a major obstacle to ecological studies of this genus. The use of PCR-DGGE as applied in this study clearly offers a higher level of discrimination and reliability than previous methods used to address this issue. For instance, based on spore morphology, G. candida was considered synonymous with G. rosea and G. ramisporophora was considered synonymous with G. margarita (5). However, our results not only do not support this reclassification, they show that the so-called invalid species are actually less related to their supposed synonymous species than to other species, based on the rDNA marker. We could also distinguish between species that were grouped together on the basis of the molecular signatures proposed by Bago et al. (3). Our detection of high degrees of heterogeneity within the V9 region of the 18S rRNA gene, which encompasses the sequence stretch examined by Bago et al. (3), also explains the ambiguities found within the signature sequences that they defined. The mixed PCR products recovered from an isolate could be resolved by PCR-DGGE (Table 4) but produced ambiguities at heterogeneous positions upon direct sequencing as preformed by Bago et al. (3).
Our results suggest that G. albida INVAM927 should be reassigned as G. rosea (Fig. 3). G. albida INVAM927 was one of the strains used by Bentivenga and Morton (5) as type material to redescribe this species. In contrast, the material we used for this isolate came from the INVAM in Florida and not from West Virginia University. Unfortunately, a comparison between the material examined in this study and the original material used by Bentivenga and Morton (5) is no longer possible, as this accession strain has been lost from the INVAM collection (J. B. Morton, personal communication). The accession strain G. albida UFLA24 must also be reassigned as G. ramisporophora on similar grounds. One of the strains identified as G. rosea obtained from INVAM was actually G. albida, based on information sent by the curator of the collection. This isolate was correctly identified by our PCR-DGGE analysis (Fig. 3) as strain BR235, confirming the morphological analysis.
Despite being characterized as G. gigantea, accession UFLA872 presented a PCR-DGGE pattern and a DNA sequence that did not match those of any other isolate examined (Fig. 3). Spores of G. gigantea UFLA872 have the same size range as expected for G. gigantea and also exhibit a cytoplasm color typical of G. gigantea. This feature is considered a unique identifying characteristic of this species (46). Other authors have also described conflicts between morphological and molecular identifications of AMF. Bago et al. (3) suggested the reassignment of isolate DAOM194757, morphologically identified as G. margarita, to the G. rosea group. Lanfranco et al. (32) confirmed this result and also suggested reassignment of isolate E29 (G. margarita based on morphology) to the G. rosea group on similar grounds.
Implications of 18S rDNA heterogeneity for ecological and evolutionary studies.
Heterogeneity between rRNA markers within a species or single individual is a phenomenon that has been described for a wide range of organisms (1, 11, 14, 18, 31, 37, 41, 52). In our PCR-DGGE approach, we have not only used this heterogeneity to characterize and identify species, but have also combined this with a cloning and screening strategy to tease apart the relationships between the 18S rDNA variants detected in a single spore (species). Knowledge of this intraspecific variation is fundamental for proper assignment of operational taxonomic units for molecular sequence identification and phylogenetic analysis (14). It is possible that cloning and sequencing will underestimate the number of species if the clones chosen for analysis happen to harbor those ribotypes held in common between the different species present in a sample. It is more likely, however, that such studies will overestimate species diversity (14, 40). Our results show that some different morphospecies share certain ribotypes but also contain species-specific variants. Given the asexual nature of AMF (31), the mechanisms for the establishment and maintenance of such patterns of sequence diversity remain to be discovered.
Although rRNA genes have been highly useful markers for the phylogenetic study of microorganisms, including Gigaspora (44), data gained from this marker alone clearly cannot resolve all phylogenetic and evolutionary issues for these organisms. The analysis of other molecular markers is an urgent issue for reconstructing phylogenetic relationships within this genus and for other AMF. Thus, although we were able to detect and utilize interesting patterns of rDNA heterogeneity within the Gigasporaceae for detection and identification purposes, further studies are necessary to explain these patterns and understand how they fit into the scheme of AMF life history and evolution.
The ability to assess Gigaspora diversity directly in environmental samples opens new possibilities for studying the ecology of this group under field conditions without the need for trap cultures. Recently, G. margarita was found in Europe, and its occurrence seems to be affected by the tillage system used (28). Some Gigaspora species are known to harbor an endosymbiont of a proposed new bacterial genus, candidatus Glomeribacter gigasporarum (8, 9), and the characterization of fungal and bacterial partners might clarify the evolutionary and ecological aspects of that symbiosis. PCR-DGGE targeting the V9 region of the 18S rDNA provides a fast and reliable method to identify Gigaspora species, to assess Gigasporaceae diversity in field conditions, and to characterize inter- and intraspecies rDNA heterogeneity.
Acknowledgments
This work was funded by the Brazilian Council for Scientific and Technological Development (CNPq, grant 200850/98-9) and Embrapa Agrobiologia, Seropédica, Rio de Janeiro, Brazil (grant to F.A.D.S.).
We thank the following researchers for providing access to germ plasm: L. Abbott, G. Bécard, V. Bianciotto, D. D. Douds, V. Gianninazzi-Person (curator, European Bank of Glomeromycota), R. Herrera-Peraza, L. C. Maia, J. Morton (curator, INVAM), M. Saito, J. O. Siqueira, and C. Walker. We thank the anonymous reviewers for helpful comments.
Footnotes
Publication number 3268 of the NIOO-KNAW.
REFERENCES
- 1.Amann, G., K. O. Stetter, E. Llobet-Brossa, R. Amann, and J. Anton. 2000. Direct proof for the presence and expression of two 5% different 16S rRNA genes in individual cells of Haloarcula marismortui. Extremophiles 4:373-376. [DOI] [PubMed] [Google Scholar]
- 2.Antoniolli, Z. I., D. P. Schachtman, K. K. Ophel, and S. E. Smith. 2000. Variation in rDNA ITS sequences in Glomus mosseae and Gigaspora margarita spores from a permanent pasture. Mycol. Res. 104:708-715. [Google Scholar]
- 3.Bago, B., S. P. Bentivenga, V. Brenac, J. C. Dodd, Y. Piche, and L. Simon. 1998. Molecular analysis of Gigaspora (Glomales, Gigasporaceae). New Phytol. 139:581-588. [Google Scholar]
- 4.Balota, E. L., and E. S. Lopes. 1996. Introduction of arbuscular mycorrhizal fungi in coffee under field conditions: I. Persistence and interaction with native species. Rev. Bras. Ciência Solo 20:217-223. [Google Scholar]
- 5.Bentivenga, S. P., and J. B. Morton. 1995. A monograph of the genus Gigaspora, incorporating developmental patterns of morphological characters. Mycologia 87:719-731. [Google Scholar]
- 6.Bentivenga, S. P., and J. B. Morton. 1996. Congruence of fatty acid methyl ester profiles and morphological characters of arbuscular mycorrhizal fungi in Gigasporaceae. Proc. Natl. Acad. Sci. USA 93:5659-5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bever, J. D., P. A. Schultz, A. Pringle, and J. B. Morton. 2001. Arbuscular mycorrhizal fungi: More diverse than meets the eye, and the ecological tale of why. Bioscience 51:923-931. [Google Scholar]
- 8.Bianciotto, V., E. Lumini, L. Lanfranco, D. Minerdi, P. Bonfante, and S. Perotto. 2000. Detection and identification of bacterial endosymbionts in arbuscular mycorrhizal fungi belonging to the family Gigasporaceae. Appl. Environ. Microbiol. 66:4503-4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bianciotto, V., E. Lumini, P. Bonfante, and P. Vandamme. 2003. ‘Candidatus Glomeribacter gigasporarum’ gen. nov., sp nov., an endosymbiont of arbuscular mycorrhizal fungi. Int. J. Syst. Evol. Microbiol. 53:121-124. [DOI] [PubMed] [Google Scholar]
- 10.Brüggemann, J., J. R. Stephen, Y.-J. Chang, S. J. Macnaughton, G. A. Kowalchuk, E. Kline, and D. C. White. 2000. Competitive PCR-DGGE analysis of bacterial mixtures: an internal standard and an appraisal of template enumeration accuracy. J. Microbiol. Methods 40:111-123. [DOI] [PubMed] [Google Scholar]
- 11.Buckler, E. S. I., A. Ippolito, and T. P. Holtsford. 1997. The evolution of ribosomal DNA: divergent paralogues and phylogenetic implications. Genetics 145:821-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clapp, J. P., A. H. Fitter, and J. P. W. Young. 1999. Ribosomal small subunit sequence variation within spores of an arbuscular mycorrhizal fungus, Scutellospora sp. Mol. Ecol. 8:915-921. [DOI] [PubMed] [Google Scholar]
- 13.Clapp, J. P., T. Helgason, T. J. Daniell, and J. P. W. Young. 2002. Genetic studies of the structure and diversity of arbuscular mycorrhizal fungal communities, p. 201-224. In M. G. A. van der Heijden and I. R. Sanders (ed.), Ecological studies, vol. 157: mycorrhizal ecology. Springer-Verlag, Berlin, Germany.
- 14.Dahllof, I., H. Baillie, and S. Kjelleberg. 2000. rpoB-based microbial community analysis avoids limitations inherent in 16S rRNA gene intraspecies heterogeneity. Appl. Environ. Microbiol. 66:3376-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Souza, F. A., and R. L. L. Berbara. 1999. Ontogeny of Glomus clarum in Ri T-DNA transformed roots. Mycologia 91:343-350. [Google Scholar]
- 16.de Souza, F. A., S. F. B. Trufem, D. L. De Almeida, E. M. R. da Silva, and J. G. M. Guerra. 1999. Effect of pre-crops on the inoculum potential of arbuscular mycorrhizal fungi and cassava yield. Pesqui. Agropecu. Bras. 34:1913-1923. [Google Scholar]
- 17.Edwards, S. G., A. H. Fitter, and J. P. W. Young. 1997. Quantification of an arbuscular mycorrhizal fungus, Glomus mosseae, within plant roots by competitive polymerase chain reaction. Mycol. Res. 101:1440-1444. [Google Scholar]
- 18.Gandolfi, A., P. Bonilauri, V. Rossi, and P. Menozzi. 2001. Intraindividual and intraspecies variability of ITS1 sequences in ancient asexual Darwinula stevensoni (Crustacea: Ostracoda). Heredity 87:449-455. [DOI] [PubMed] [Google Scholar]
- 19.Gandolfi, A., I. R. Sanders, V. Rossi, and P. Menozzi. 2003. Evidence of recombination in putative ancient asexuals. Mol. Biol. Evol. 20:754-761. [DOI] [PubMed] [Google Scholar]
- 20.Gianinazzi-Pearson, V., D. van Tuinen, E. Dumas-Gaudot, and H. Dulieu. 2001. Exploring the genome of glomalean fungi, p. 3-17. In B. Hock (ed.), Fungal associations, vol. IX. The mycota. Springer-Verlag, Berlin, Germany.
- 21.Giovannetti, M., and B. Mosse. 1980. An evaluation of techniques for measuring vesicular arbuscular mycorrhizal infection in roots. New Phytol. 84:489-500. [Google Scholar]
- 22.Harrison, M. J. 1999. Molecular and cellular aspects of the arbuscular mycorrhizal symbiosis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50:361-389. [DOI] [PubMed] [Google Scholar]
- 23.Helgason, T., T. J. Daniell, R. Husband, A. H. Fitter, and J. P. W. Young. 1998. Ploughing up the wood-wide web? Nature 384:431. [DOI] [PubMed] [Google Scholar]
- 24.Helgason, T., J. W. Merryweather, J. Denison, P. Wilson, J. P. W. Young, and A. H. Fitter. 2002. Selectivity and functional diversity in arbuscular mycorrhizas of co-occurring fungi and plants from a temperate deciduous woodland. J. Ecol. 90:371-384. [Google Scholar]
- 25.Hoagland, D. R., and D. I. Arnon. 1950. The water-culture method for growing plants without soil. Circular 347. California Experimental Agricultural Station, Berkeley.
- 26.Husband, R., E. A. Herre, S. L. Turner, R. Gallery, and J. P. W. Young. 2002. Molecular diversity of arbuscular mycorrhizal fungi and patterns of host association over time and space in a tropical forest. Mol. Ecol. 11:2669-2678. [DOI] [PubMed] [Google Scholar]
- 27.Husband, R., E. A. Herre, and J. P. W. Young. 2002. Temporal variation in the arbuscular mycorrhizal communities colonising seedlings in a tropical forest. FEMS Microbiol. Ecol. 42:131-136. [DOI] [PubMed] [Google Scholar]
- 28.Jansa, J., A. Mozafar, T. Anken, R. Ruh, I. R. Sanders, and E. Frossard. 2002. Diversity and structure of AMF communities as affected by tillage in a temperate soil. Mycorrhiza 12:225-234. [DOI] [PubMed] [Google Scholar]
- 29.Kjoller, R., and S. Rosendahl. 2000. Detection of arbuscular mycorrhizal fungi (Glomales) in roots by nested PCR and SSCP (single stranded conformation polymorphism). Plant Soil 226:189-196. [Google Scholar]
- 30.Kowalchuk, G. A., F. A. de Souza, and J. A. van Veen. 2002. Community analysis of arbuscular mycorrhizal fungi associated with Ammophila arenaria in Dutch coastal sand dunes. Mol. Ecol. 11:571-581. [DOI] [PubMed] [Google Scholar]
- 31.Kuhn, G., M. Hijri, and I. R. Sanders. 2001. Evidence for the evolution of multiple genomes in arbuscular mycorrhizal fungi. Nature 414:745-748. [DOI] [PubMed] [Google Scholar]
- 32.Lanfranco, L., V. Bianciotto, E. Lumini, M. Souza, J. B. Morton, and P. Bonfante. 2001. A combined morphological and molecular approach to characterize isolates of arbuscular mycorrhizal fungi in Gigaspora (Glomales). New Phytol. 152:169-179. [DOI] [PubMed] [Google Scholar]
- 33.Lanfranco, L., M. Delpero, and P. Bonfante. 1999. Intrasporal variability of ribosomal sequences in the endomycorrhizal fungus Gigaspora margarita. Mol. Ecol. 8:37-45. [DOI] [PubMed] [Google Scholar]
- 34.Lloyd-MacGilp, S. A., S. M. Chambers, J. C. Dodd, A. H. Fitter, C. Walker, and J. P. W. Young. 1996. Diversity of the ribosomal internal transcribed spacers within and among isolates of Glomus mosseae and related mycorrhizal fungi. New Phytol. 133:103-111. [Google Scholar]
- 35.Muyzer, G., and K. Smalla. 1998. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Leeuwenhoek 73:127-141. [DOI] [PubMed] [Google Scholar]
- 36.Myers, R. M., T. Maniatis, and L. S. Lerman. 1987. Detection and localization of single base changes by denaturing gradient gel-electrophoresis. Methods Enzymol. 155:501-527. [DOI] [PubMed] [Google Scholar]
- 37.Nübel, U., B. Engelen, A. Felske, J. Snaidr, A. Wieshuber, R. I. Amann, W. Ludwig, and H. Backhaus. 1996. Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J. Bacteriol. 178:5636-5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Redecker, D., H. Thierfelder, C. Walker, and D. Werner. 1997. Restriction analysis of PCR-amplified internal transcribed spacers of ribosomal DNA as a tool for species identification in different genera of the order Glomales. Appl. Environ. Microbiol. 63:1756-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Redecker, D. 2000. Specific PCR primers to identify arbuscular mycorrhizal fungi within colonized roots. Mycorrhiza 10:73-80. [Google Scholar]
- 40.Sanders, I. R., M. Alt, K. Groppe, T. Boller, and A. Wiemken. 1995. Identification of ribosomal DNA polymorphisms among and within spores of the Glomales: application to studies on the genetic diversity of arbuscular mycorrhizal fungal communities. New Phytol. 130:419-427. [Google Scholar]
- 41.Sanders, I. R. 2002. Ecology and evolution of multigenomic arbuscular mycorrhizal fungi. Am. Nat. 160:S128-S141. [DOI] [PubMed] [Google Scholar]
- 42.Santos, A. L., F. A. de Souza, J. G. M. Guerra, R. L. L. Berbara. 2000. Estabelecimento e capacidade infectiva de Gigaspora margarita e Glomus clarum em solo sob erosão. Acta Bot. Bras. 14:127-139. [Google Scholar]
- 43.Schüssler, A. 1999. Glomales SSU rRNA gene diversity. New Phytol. 144:205-207. [Google Scholar]
- 44.Schüssler, A., D. Schwarzott, and C. Walker. 2001. A new fungal phylum, the Glomeromycota: phylogeny and evolution. Mycol. Res. 105:1413-1421. [Google Scholar]
- 45.Schwarzott, D., C. Walker, and A. Schussler. 2001. Glomus, the largest genus of the arbuscular mycorrhizal fungi (Glomales), is nonmonophyletic. Mol. Phylogenetics Evol. 21:190-197. [DOI] [PubMed] [Google Scholar]
- 46.Sejalon-Delmas, N., A. Magnier, D. D. Douds, and G. Bécard. 1998. Cytoplasmic autofluorescence of an arbuscular mycorrhizal fungus Gigaspora gigantea and nondestructive fungal observations in planta. Mycologia 90:921-926. [Google Scholar]
- 47.Sheffield, V. C., D. R. Cox, L. S. Lerman, and R. M. Myers. 1987. Attachment of a 40-base pair G+C-rich sequence (GC-clamp) to genomic DNA fragments by the polymerase chain reaction results in improved detection of single-base changes. Proc. Natl. Acad. Sci. USA 86:232-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sieverding, E. 1991. Vesicular-arbuscular mycorrhiza management in tropical agrosystems. Deutsche Gesellschaft für Technische Zusammenarbeit, Eschborn, Germany.
- 49.Simon, L., M. Lalonde, and T. D. Bruns. 1992. Specific amplification of 18S fungal ribosomal genes from vesicular-arbuscular mycorrhizal fungal communities. Appl. Environ. Microbiol. 58:291-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith, S. E., and D. J. Read. 1997. Mycorrhizal symbiosis, 2nd ed. Academic Press, London, England.
- 51.Swofford, D. L. 2003. PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4. Sinauer Associates, Sunderland, Mass.
- 52.Tang, J., L. Toe, C. Back, and T. R. Unnasch. 1996. Intraspecific heterogeneity of the rDNA internal transcribed spacer in the Simulium damnosum (Diptera: Simuliidae) complex. Mol. Biol. Evol. 13:244-252. [DOI] [PubMed] [Google Scholar]
- 53.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trouvelot, S., D. van Tuinen, M. Hijri, and V. Gianinazzi-Pearson. 1999. Visualization of ribosomal DNA loci in spore interphasic nuclei of glomalean fungi by fluorescence in situ hybridization. Mycorrhiza 8:203-206. [Google Scholar]
- 55.van der Heijden, M. G. A., J. N. Klironomos, M. Ursic, P. Moutoglis, E. R. Streitwolf, T. Boller, A. Wiemken, and I. R. Sanders. 1998. Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity. Nature 396:69-72. [Google Scholar]
- 56.van Tuinen, D., E. Jacquot, B. Zhao, A. Gollotte, and V. Gianinazzi-Pearson. 1998. Characterization of root colonization profiles by a microcosm community of arbuscular mycorrhizal fungi using 25S rDNA-targeted nested PCR. Mol. Ecol. 7:879-887. [DOI] [PubMed] [Google Scholar]
- 57.White, T. J., T. Bruns, S. Lee, and J. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-322. In M. A. Innis, D. H. Gelfand, J. J. Sminski, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, San Diego, Calif.
- 58.Yokoyama, K., T. Tateishi, T. Marumoto, and M. Saito. 2002. A molecular marker diagnostic of a specific isolate of an arbuscular mycorrhizal fungus, Gigaspora margarita. FEMS Microbiol. Lett. 212:171-175. [DOI] [PubMed] [Google Scholar]