Abstract
It is well established that the oxidation state of cysteine residues in proteins are critical to overall physical stability. The presence of disulfide bonds most often imparts thermodynamic stability, and as such, engineered disulfide bonds have become a means for improving the viability of protein therapeutics. In some cases, however, disulfide bonds can diminish stability. Because proteins are held together by numerous weak interactions, understanding the mechanisms by which stabilization is achieved is important to the design of new biotechnology products that better resist unfolding and aggregation. Mechanistic information describing how specific interactions influence stability is lacking, in part because the techniques typically used to study inherent stability do not provide sufficient detail. In the present study, a model protein system, phosphatase of regenerating liver (PRL-1), was used to investigate the role of cysteine residues on physical stability. A combination of chemical modulation and mutagenesis was employed to alter the redox state of the protein, and the effects were observed using a combination of low- and high-resolution methods. Specifically, solution NMR data revealed the stability of PRL-1 depends on cooperation between local interactions with the Cys side chains. This approach provides a means to better understand how protein stabilization is achieved.
INTRODUCTION
An understanding of the forces that contribute to protein stability is crucial to the development of new biotechnology products having improved half-lives and lower immunogenicity.1 Current methodologies to stabilize proteins include the addition of excipients, chemical modification of residues and/or the use of site-directed mutagenesis to produce a more stable protein species.2 More recently, engineering cysteine residues into the primary sequence to form a new or additional disulfide bonds has proved to be useful for the stabilization of antibodies.3-5 Disulfide bonds are valuable components of a protein's tertiary structure because they typically increase the half-life of the protein in terms of both cellular physiology and pharmacokinetics.6 Classical theory suggests that disulfide bonds stabilize proteins by reducing the entropy of the denatured state.7-9 More recent theories propose disulfide bonds stabilize the folded state enthalpically, presumably through favorable local interactions or by stabilizing the packing of hydrophobic residues.10,11 For example, they can restrict conformational motion and prevent changes in structure that can lead to unfolding and/or self-association/aggregation.12 Unfortunately, only a small amount of experimental data to support such a theory is available.13-15
Despite the aforementioned successes and many others, disulfide bonds are not always beneficial. Alterations to a protein's redox status in the form of disulfide exchange can promote aggregation by exposing previously buried, often hydrophobic, regions of a protein that can then associate.16,17 In the context of protein engineering, poorly placed disulfide bonds may alter the structure in an analogous manner, leading to aggregation.17,18. The currently accepted argument, however, is that the disulfide bond directly destabilizes the folded state by restricting energetically favorable conformational changes.6,17,19 Very few studies of such a phenomenon are reported in the current body of literature. In fact, the contribution of conformational dynamics to protein stability has only recently been evaluated,20-22and as such, very little is known about how the oxidation state of Cys residues contribute to the stability or instability of a protein with respect to conformational dynamics.19
Currently, a set of low-resolution experiments is commonly employed to assess structural stability (and more recently dynamics) and the approach has proven beneficial for rapidly identifying stabilizing conditions for individual proteins.20,23-25 The methods used provide general information about classes of bonds or structural features in the protein. Despite the obvious utility of these traditional methodologies, mechanistic understanding cannot be gleaned from these data sets to elucidate why stabilization is affected. Additional information about the contribution of individual residues can be obtained by comparing mutants to the wild-type protein using these methods and calculating changes in the free energy (ΔΔG) in an unfolding experiment.26,27 While this approach can determine whether a specific residue is required for folding, it does not reveal how cooperativity and conformational dynamics contribute to stabilization of the folded state. High-resolution or site-specific detail is required to explain why a protein is inherently more or less stable or is stabilized to a greater or lesser extent by specific solution conditions. Solution NMR studies of protein folding and unfolding are commonly employed to examine folding pathways,28-30 but this powerful technique has been greatly underutilized to examine the mechanisms of stabilization of the folded state from both the perspective of the packing arrangement in the protein as well as the effects of solution environment on the protein's conformational ensemble. Understanding how structure and dynamics affect the stability of the folded state is important for generating protein therapeutics that better resist unfolding and aggregation.
In the present study, the protein phosphatase of regenerating liver (PRL-1) was used as a model system to investigate the effects of disulfide bond formation on the protein's stability. PRL-1 has two discernable stable states, one of which contains a disulfide bond (oxidized, inactive) and one that does not (reduced, active). Here, the disulfide bond was disrupted in several ways and the protein carefully examined to assess the effects these two cysteines and their oxidation state have on the conformation of the protein. PRL-1 serves as an excellent model for this purpose for several reasons. First, the catalytic Cys (C104) is highly susceptible to inactivation by disulfide bond formation at the active site.31,32 Our group previously assigned the NMR resonances for the reduced and oxidized proteins and characterized formation of the disulfide bond between C104 and its partner C49.33,34 The reduction potential is approximately - 364 mV, which indicates that this protein strongly favors the oxidized state in vitro, but equilibrium control of the redox state is easily modulated using a redox buffer system to generate the reduced form.35 Second, a major conformational change occurs upon reduction of the protein, providing well-resolved, site-specific measurable differences between the two states using solution NMR spectroscopy.35 Additionally, the structures of both reduced and oxidized PRL-1 have been determined previously using X-ray crystallography, which facilitates interpretation of the changes observed in the NMR data with respect to the three-dimensional structure.32,36
Standard low-resolution techniques (circular dichroism and static light scattering) were used to show that the reduced form of the protein is slightly more stable than the oxidized. To understand how the reduced form compensates for the loss of the disulfide bond, a series of mutants were analyzed that disrupted disulfide bond formation in distinct ways. The physical stability of each variant is unique with respect to the others, indicating that the stability of the whole protein depends on a combination of local, synergistic interactions with the Cys side chains. High-resolution solution NMR provided site-specific and mechanistic information about how each interaction influences the protein's physical stability. This analysis reveals several key structural components that contribute to PRL-1's overall stability in the absence of disulfide bond formation. The results provide insight to explain how the reduced protein compensates for the loss of this structural feature and how local instability may lead to aggregation.
EXPERIMENTAL METHODS
Materials
Tris(hydroxymethyl)aminomethane hydrochloride (Tris-HCl), (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), anhydrous monobasic sodium phosphate, dibasic sodium phosphate heptahydrate, sodium chloride, and dithiothreitol (DTT) were all purchased from Fisher Scientific (Pittsburg, PA). All buffers were filtered through a 0.2 μm nylon filter before being applied to the protein samples. Ionic strength was controlled by the addition of an appropriate amount of sodium chloride. Because the exposure of HEPES to fluorescent light for extended periods of time can lead to the formation of hydrogen peroxide, HEPES buffer was stored at 4 °C in the dark and made fresh for each mutant studied.37,38 For NMR studies, 15N-ammonium chloride and D2O were purchased from Cambridge Isotopes (Andover, MA).
Protein Expression and Purification
Wild-type and C104S PRL-1 variants were expressed and purified as previously described.34,35 The primers used to generate the C49S, Y53E and Y53F mutants, 5’ to 3’, were ggagttaccacaatagtaagagtatGtgaagcaacttatgacac, gagtatgtgaagcaactGaAgacactactcttgtgg and gagtatgtgaagcaacttTtgacactactcttgtgg, respectively (Integrated DNA Technologies, Coralville, IA). Capital letters indicate the mutated base. All mutations were confirmed by bidirectional DNA dye terminator sequencing (Northwoods DNA Inc., Bemidji, MN). Purity of the final protein samples was assessed by SDS-PAGE and protein concentrations were determined by UV Absorbance at 280 nm using an extinction coefficient of 19420 M-1 cm-1. The extinction coefficient was calculated based on the reduced protein using the ProtParam program from ExPASy.39-41 Final samples were concentrated to 30 mg/mL and stored at 4 °C in 50 mM Tris-Cl, pH 7.4 with 100 mM NaCl until prior to analysis. These conditions are equivalent to purification conditions. All samples for NMR studies were grown on minimal media containing 15N-labeled ammonium chloride as the sole nitrogen source (Cambridge Isotopes, Andover, MA). Table 1 summarizes the purpose of studying each mutant. The activity of each mutant was tested using the standard generic substrate p-nitrophenyl phosphate (pNPP) in a colorimetric assay previously published.35
Table 1.
Purpose of PRL-1 Mutants
| PRL-1 Variant | Purpose |
|---|---|
| Wild Type (WT) | Permits chemical modification of the disulfide bond |
| C104S | Disrupts the disulfide bond directly; abolishes enzymatic activity |
| C49S | Disrupts the disulfide bond directly; retains activity |
| Y53E | Breaks hydrogen bond between Y53 and H103; increases solvent exposure of loop 3 (L3) |
| Y53F | Breaks hydrogen bond between Y53 and H103; retains apolar side chain |
Static Light Scattering
To assess the aggregation propensity of the various PRL-1 variants, the absorbance at 350 nm was monitored with increasing temperature. Protein samples were diluted to a final concentration of 1 mg/mL into 50 mM HEPES or 50 mM sodium phosphate, pH 7.5 (I=100 mM) with or without 10 mM dithiothreitol (DTT). All data were collected on a Cary 100 UV-VIS spectrophotometer equipped with 12-cell changer and temperature controller. Data points were collected every 0.5 °C at a rate of 5 °C per minute from 10-85 °C. Onset temperatures were determined from the plots of absorbance versus temperature. Each sample was analyzed in duplicate and the average of these is shown.
Circular Dichroism (CD)
CD spectra at 20 and 85 °C were acquired using a Jasco J-810 spectropolarimeter equipped with a 6-position Peltier temperature controller from 260-190 nm using a scanning speed of 50 nm/min and 0.5 nm resolution. Thermal melts were also performed by monitoring the CD signal at 222 nm every 1 °C from 20-85 °C. The temperature was gradually increased at a rate of 2 °C per minute, and the sample was equilibrated for five minutes at each temperature. The CD signal was converted to molar ellipticity using the Spectra Manager™ software (Jasco). Tms were determined by fitting the CD curve to a sigmoidal function with Prism 5 software (GraphPad, La Jolla, CA). Protein samples were diluted to a final concentration of 0.4 mg/mL in either 50 mM HEPES or 50 mM sodium phosphate, pH 7.5 (I=100 mM) with or without 1 mM DTT. Two independently prepared samples were analyzed in duplicate to generate error bars.
NMR Experiments
For structural analysis of the various PRL-1 mutants, samples were concentrated to approximately 1 mM in 50 mM sodium phosphate buffer, 100 mM NaCl, pH 6.5, containing 5% D2O. 1H-15N Heteronuclear Single Quantum Coherence (HSQC) spectra were acquired on a Bruker Avance 800 MHz spectrometer using a cryogenic, triple resonance probe equipped with pulse field gradients. The spectra were acquired in 8 scans with 2048 points in 1H and 256* increments in 15N. All spectra were obtained at 37 °C. NMRpipe42 and SPARKY43 were used for spectral processing and data analysis. Chemical shift changes (Δδ) were calculated by subtracting the peak positions of the mutant protein (C104S or C49S) from the chemically reduced wild type. Chemical shift changes were expressed as the square root of the sum of the squares of chemical frequency differences in the 1H and 15N dimensions between two given proteins for each assigned residue as given in Equation 1.44
| (1) |
A change was deemed significant if Δf was greater than 33.81. This value represents the average line width of the reduced wild-type protein and was determined by summing the squares of the individual resonant line widths for 1H and 15N using Equation 1 and averaging. Line widths were measured in SPARKY.
Because the C49S mutant was significantly different in structure from the reduced wild-type protein, 2D 1H-15N HSQCs and 3D versions of the CBCA(CO)NH, HNCACB, C(CO)NH and HNCO experiments were collected to obtain sequential backbone assignments. The sample was concentrated to approximately 1.9 mM in 50 mM sodium phosphate, pH 6.5, with 100 mM NaCl and 5% D2O and analyzed in a Shigemi tube. Experiments were carried out on a Varian Inova 800 MHz NMR spectrometer using a cryogenic, triple-resonance probe equipped with pulse field gradients. All spectra were obtained at 37 °C. Spectra were processed using NMRpipe 42 and peak picked using Sparky.43 Peak lists were submitted to the PINE Server through NMRFAM, University of Wisconsin, Madison45 and cross-checked using the assignments for reduced wild type.34
For all NMR experiments described above, 1H chemical shifts were referenced with respect to an external DSS standard in D2O. Indirect referencing relative to 1H was determined for 13C and 15N, assuming ratios 13C/1H = 0.251449530 and 15N/1H = 0.101329118.46
RESULTS
Physical Stability of PRL-1
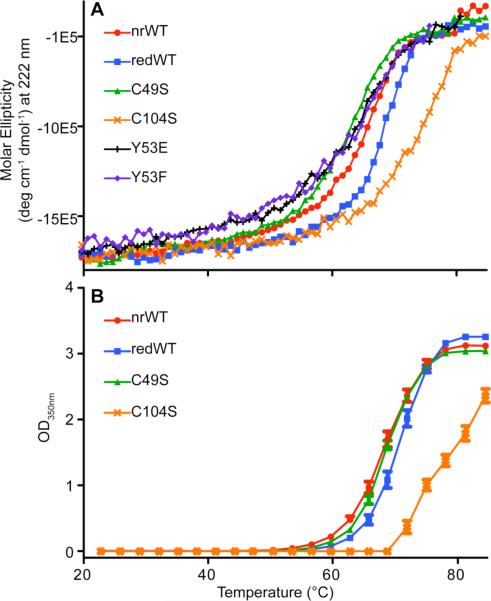
To determine the overall physical stability of the different forms of PRL-1, CD melts and SLS melts were performed on each variant. For CD, a Tm was calculated by fitting the data to a sigmoidal curve. For SLS, the onset of transition temperature is reported because saturation of the detector occurred in many cases. Table 2 summarizes this data. The CD and SLS plots for all PRL-1 variants in phosphate buffer are shown in Figure 1.
Table 2.
Physical Stability of PRL-1a
| Phosphate | HEPES | |||||||
|---|---|---|---|---|---|---|---|---|
| CD | SLS | CD | SLS | |||||
| Tm | ± SD | Onset T | ± SD | Tm | ± SD | Onset T | ± SD | |
| oxWT | 65.47 | 1.11 | 59.92 | 0.28 | 60.54 | 2.08 | 53.27 | 0.35 |
| redWTbc | 67.81 | 0.43 | 63.75 | 0.38 | 61.98 | 1.26 | 57.35 | 0.32 |
| redWTd | 79.01e | 0.66 | ndf | nd | nd | |||
| C49S | 62.79 | 2.00 | 59.68 | 0.22 | 56.25 | 0.32 | 49.95 | 0.32 |
| C104S | 76.31 | 1.07 | 72.1 | 0.32 | 64.85 | 2.06 | 59.28 | 0.22 |
| Y53E | 62.25 | 0.35 | nd | nd | nd | |||
| Y53F | 62.61 | 0.30 | nd | nd | nd | |||
All temperatures reported are in °C
For CD, reducing conditions were achieved by the addition of 1 mM DTT. For SLS, reducing conditions were achieved by the addition of 10 mM DTT.
CD and/or SLS were also used to study the physical stability of PRL-1 mutants under both reducing and nonreducing conditions, although no differences were detected. This data for this is not shown.
Reduced PRL-1-WT (redWT) was studied in 20 mM phosphate buffer at pH 6.5 with equivalent ionic strength (I=100 mM). All other values reported, including those for HEPES buffer, are at pH 7.5.
Melting was incomplete at the maximum temperature attainable and the data was extrapolated to determine the Tm.
Not determined
Figure 1. Physical Stability of PRL-1.
The physical stability of PRL-1 was monitored by either CD (A) or SLS (B). Tms determined from the data shown here are summarized in Table 2. All data presented in this figure were collected in phosphate buffer at pH 7.5. For CD analysis, data points represent the average of two replicates of two independently prepared samples. For SLS analysis, data points represent the average of two replicates.
Several trends in the CD data were observed with the PRL-1 variants. First, the reduced wild-type protein is slightly more stable than the oxidized protein, with melting temperatures at 67.81 ± 0.43 versus 65.47 ± 1.28, respectively. We anticipated the opposite result because, most often, disulfide bonds improve the ability of proteins to resist unfolding. To understand how the reduced protein compensates for the loss of the disulfide bond, the same analyses were also performed on each of the two cysteine mutants (C49S and C104S) that directly prevent formation of this bond under equivalent conditions (Table 1). Interestingly, the C49S and C104S mutants each have a unique melting profile that is different from each other, as well as the reduced wild type. Compared to the chemically reduced wild type, C49S has a significantly lower melting temperature (62.79 ± 2.00), while the C104S mutant has a significantly higher melting temperature (76.31 ± 1.07). Such large differences between the C49S, C104S and reduced wild-type proteins were quite unexpected because, with only a single atom changed, each should mimic the reduced conformation of PRL-1. The fact that these mutants have a 13.5 °C separation in their physical stability strongly indicates that the overall stability of PRL-1 is greatly impacted by local interactions with the side chain moieties of the C49 and C104 residues. Similar trends in the aggregation propensity of PRL-1 variants were also observed by SLS. In phosphate buffer, the reduced wild type was slightly less vulnerable to aggregation than the oxidized wild type (ΔTonset=3.83°C). The C104S protein is the least aggregation prone (Tonset=72.1 ± 0.32 °C), while the C49S protein self associates most readily (Tonset=59.68 ± 0.22). This data clearly shows that stability depends on more than just disulfide bond formation at the active site.
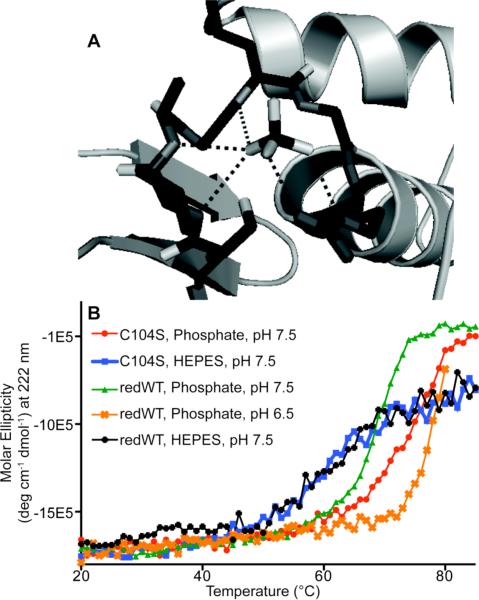
Table 2 shows that changing the buffer system from phosphate to HEPES reduces the physical stability of PRL-1 for all variants studied, decreasing Tm values by 5-6 °C. Similarly, the aggregation propensity measured by SLS also decreases consistently by approximately 6 °C with the same buffer switch. Because the biological role of PRL-1 is to facilitate phosphate hydrolysis from a phosphotyrosine substrate, this result suggests that the decrease in physical stability can be attributed to whether or not the protein can bind to the buffer species, albeit too weakly in this case to determine the KD even by NMR, placing it above the mid mM range (data not shown). The phosphate group is expected to bind at the active site of the protein, much like sulfate occupies the active site in the available crystal structure of C104S (Figure 2A).32,47 HEPES apparently does not participate in such an interaction, and each variant is destabilized to an equivalent extent by this buffer switch. Because both buffers have equivalent ionic strength, it is unlikely that the effect results from non-specific electrostatic influences, further suggesting that weak phosphate binding imparts stabilization to this protein.
Figure 2. Phosphate Stabilization of PRL-1.
A. The active site of the C104S crystal structure is shown occupied with a sulfate ligand, the oxygen atoms from which are coordinated to backbone amides and a conserved Arg side chain.47 Protein data bank (PDB) file 1XM2 was used to create this illustration.36 This figure was generated using Pymol. B. The physical stability of PRL-1-C104S was monitored by CD in either phosphate at pH 7.5 (red circle) or HEPES at pH 7.5 (blue square). Reduced wild type was monitored by CD in either phosphate at pH 7.5 (green triangle), phosphate at pH 6.5 (orange x) or HEPES at pH 7.5 (black diamond). In all cases, an equivalent ionic strength was maintained by the addition of sodium chloride. Calculated Tm values are reported in Table 2. For C104S, no differences in melting temperature were detected with the addition of reducing agent.
Active site binding also explains the dramatic difference in physical stability between the C104S and reduced wild-type proteins. The catalytic Cys in the reduced wild type exists as a thiolate anion at physiological pH, which is typical for this family of enzymes because of the surrounding partially positive microenvironment and structural organization of the active site.31,32 With PRL-1-C104S, the thiolate side chain has been replaced with a hydoxyl group, which remains protonated, eliminating the negative charge at this position. Because less charge repulsion is present between phosphate and the –OH group from Ser than between phosphate and the negatively charged thiolate anion, binding by phosphate would occur to C104S more readily because of the differences in charge state of the active site. This conclusion is further supported by the fact that the melting temperatures measured by CD for the reduced wild type and C104S are statistically identical in HEPES buffer but differ significantly in phosphate buffer (ΔTm=2.87 and 8.5, respectively; Table 2). Although the onset temperatures for C104S and reduced wild type measured by SLS are statistically different in the same buffer system, the ΔTonset value for HEPES is considerably smaller than for phosphate buffer (1.93 versus 8.35 °C, respectively; Table 2), further indicating that phosphate binding imparts structural stability. As an alternative way of investigating the effect of charge repulsion on stability, the pH of the phosphate buffer was lowered to 6.5 from 7.5 while maintaining an equivalent ionic strength. This too enhanced the physical stability of the reduced wild type, as measured by CD (Tm=79.07). The primary phosphate species at pH 6.5 is in the -1 charge state whereas at 7.5, just over 50% of the phosphate is in the -2 charge state. The smaller degree of charge repulsion between the thiolate and singly charged phosphate ion allows the molecule to more readily occupy the active site and stabilize the protein. A comparison of the CD melts of these mutants in the different buffers is shown in Figure 2B.
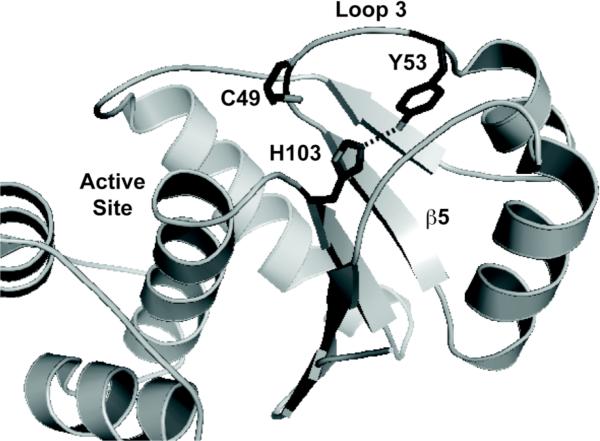
Phosphate binding does not fully explain the difference in structural stability of the reduced forms, as the Tm values measured by CD and onset temperatures measured by SLS remain consistently varied for the different forms across each buffer system (Table 2). When compared to the reduced wild-type protein in HEPES buffer, C49S consistently exhibits a lower melting temperature. This suggests that a specific local packing interaction involving C49 is perturbed in this protein that causes destabilization. The side chain of C49 is completely buried in the protein's hydrophobic core in the crystal structure of PRL-1-C104S. One notable feature in the crystal structure of the reduced state is the presence of a completely buried hydrogen bond between residues Y53 and H103 (Figure 3).32,36 In the reduced state, this hydrogen bond forms between the hydroxyl from the tyrosine side chain and the imidazolium Nε of the histidine and has a bond length of 2.85 Å. The C49 side chain is pointed inwards towards this interaction. In the oxidized state, the distance between these atoms lengthens to 3.07 Å, and the angle (107°) further deviates from ideality (120°)48 compared to the reduced form (113°). The C49 side chain is disulfide bound and points away from these residues. There is a concomitant rearrangement of residues in the region surrounding both the active site and the Y53-H103 hydrogen bond. Based on the small (2 °C) difference in Tm between the reduced and oxidized wild-type states, the optimization of the hydrogen bond and concomitant packing rearrangement around it more than compensate for the loss of the covalent disulfide bond. Based on this information, we hypothesized that the C49S mutant is destabilized because the introduction of the polar group favors greater solvent exposure and disrupts favorable packing interactions around the Y53-H103 hydrogen bond, despite being in the reduced conformation at the active site. To test this hypothesis, we created the Y53E and Y53F mutants and studied their physical stability by CD. As indicated by Figure 1 and Table 2, Y53E and Y53F have equivalent Tm values to the C49S mutant, suggesting a common mode of destabilization.
Figure 3. Y53-H103 Hydrogen Bonding in PRL-1.
PDB code 1XM2 was used to illustrate the proximity of the Y53 and H103 side chains.36 In the figure, the side chain of these residues, as well as C49 are shown as sticks. Various important regions of the protein, including loop 3 (L3) and the central β-strand (β5) are labeled. In the oxidized crystal structure (PDB 1ZCK),32 the interaction between Y53 and H103 is perturbed as these side chains move away from one another. This figure was generated using Pymol.
Structural Analysis of PRL-1
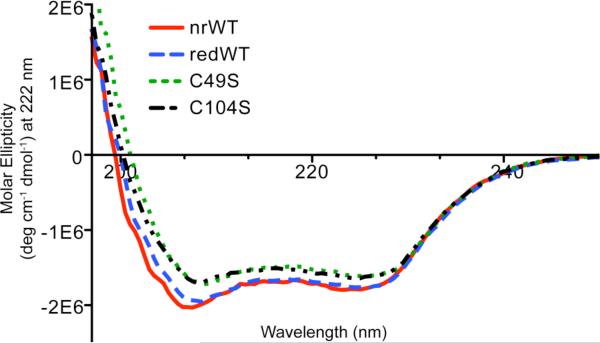
We first collected CD spectra of each PRL-1 variant to determine if substantial changes in the total secondary structure occur as the result of any of the mutations. The results are shown in Figure 4. Despite the significant deviations in physical stability between the reduced wild-type, C49S and C104S proteins, the total secondary structure of each mutant remains remarkably similar. Error bars were omitted in Figure 4 for presentation purposes. The values below 203 nm are statistically identical, whereas values in the α-helix and β-sheet regions are statistically different. These differences are disproportionately small compared to the differences in stability. Unfortunately, there is no systematic way to extract information from this data set to explain why the Tm values differ so substantially. High-resolution methods are needed to provide site-specific information capable of explaining the mechanism by which stabilization or destabilization is achieved with the introduction of various mutations or changes in solution conditions. We performed high-resolution solution NMR studies to obtain such information.
Figure 4. CD Spectra of PRL-1 Variants.
The secondary structure elements of the various mutants were analyzed by monitoring the CD signal from 190-250 nm. The data points represent the average of two replicates of two independently prepared samples. Error bars were omitted for figure quality purposes. All data points below 203 nm are statistically identical. At 208, 212 and 222 nm, where absorbance for α-helix and β-sheet are commonly observed, the values are statistically different.
The 1H-15N HSQC NMR experiment was used because this method selectively detects protons that are directly coupled to a nitrogen atom. This is particularly useful for the analysis of protein structure because of the repeating pattern of amides in the polypeptide backbone. A cross peak for each amide in the protein is observed, providing a fingerprint of the protein's unique structure. This experiment serves as the basis for assessing the overall fold and dynamic behavior of a protein. Because chemical shift is so sensitive to an atom's local environment, changes in a protein's tertiary structure, such as those introduced by point mutations, are manifest as changes in the position and/or shape of the peaks in the HSQC spectrum.49 For this analysis, backbone NMR assignments for reduced PRL-1, which we previously published, were used to assign the respective mutants and provide site-specific information.35 Because of the large number of changes between the reduced wild-type and C49S proteins (see below), independent backbone assignments for C49S were also made to identify additional peaks that could not be deduced using the reduced PRL-1 spectra.45
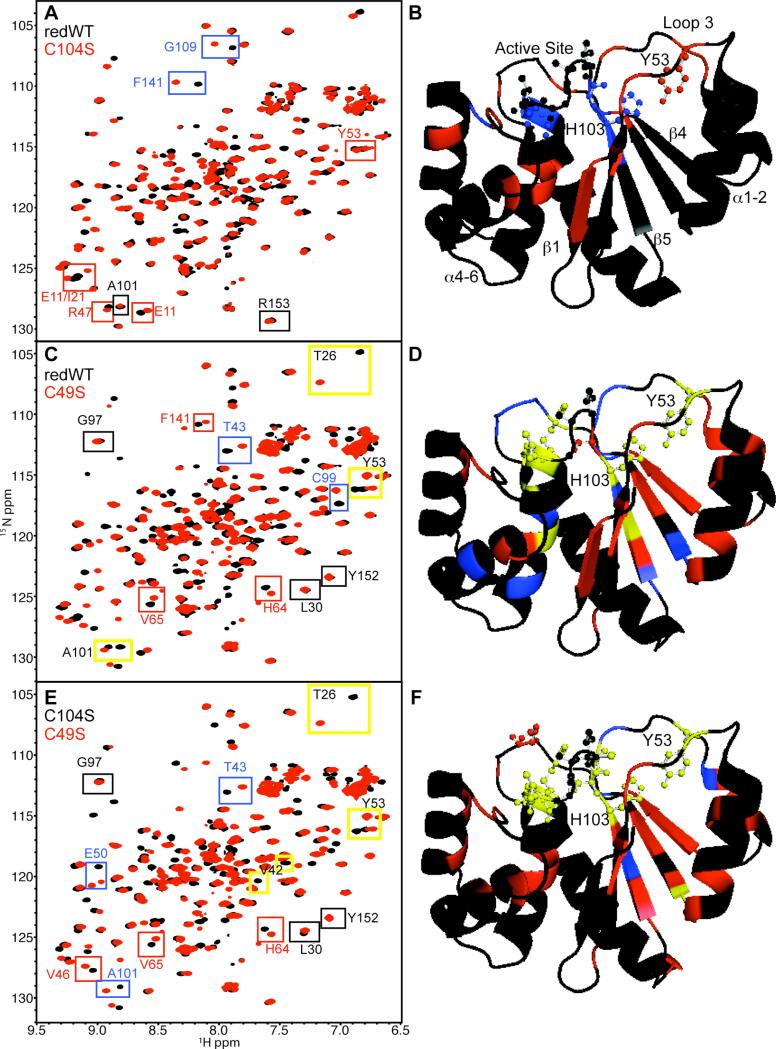
To determine which residues are perturbed by mutations that affect disulfide bond formation, we first compared the HSQC spectrum of the chemically reduced wild-type protein with that of C104S (Figure 5A). Although many of the peaks overlap, there are a number of significant changes observed between these two proteins, some of which are highlighted by boxes in the figure. To illustrate where these residues are located with respect to the mutation site, we mapped these changes to the available C104S crystal structure (Figure 5B).36 The largest chemical shift deviations (blue) are observed in or near the active site (V47, W68, V102-C104, G109, R110, F141), which is expected because the catalytic cysteine has been modified. The active site is illustrated in ball and stick format for reference. Other small changes (red) are observed near the active site and include H23, N24, R47, E50, F70, A74 and V81. Additional smaller changes (red) are present in α4-α6 (V113-V115, I133, G139-A140, L146) because a hydrogen bond between the active site and small loop containing F141 is perturbed. This change propagates, affecting the β1-containing residues V10-Y14 and β2-containing I21. On the opposite side of the active site, there are perturbations in two loops. Because the amides from these residues are already solvent exposed, the change in their chemical shift position is small. Residues Y53 and D54 in the flexible β2-α3 loop (loop three; L3) are slightly perturbed, as well as T26, which is located in loop two (L2) directly adjacent to L3. The perturbation to the Y53 backbone is likely a result of a change in conformational dynamics revolving around the orientation of H103, which results from active site stabilization upon phosphate binding. Overall, when comparing the NMR spectra of reduced wild type to that of C104S, greater than 97% of the changes observed originate from the active site. This strongly suggests that a major component of the stability difference between the chemically reduced wild type and C104S proteins is due to phosphate binding and the diminished charge repulsion at the active site in the hydroxyl-containing mutant. Moreover, increasing the concentration of phosphate in solution sharpens NMR line widths throughout the protein but more so for the resonances associated with active site. Because broadening of lines likely reflects conformational averaging, this data suggests that phosphate binding also shifts the dynamic equilibrium of the whole protein toward a more rigid state (data not shown).
Figure 5. Chemical Shift Mapping of PRL-1.
A. 1H-15N HSQC spectra of reduced of wild type (black) and C104S (red) are shown. Chemical shift changes between the two spectra were analyzed as described in the methods section. A representative example from each category described below is boxed in the spectra. Black boxes highlight residues with Δf < 33.8. Red boxes emphasize residues with 33.8 < Δf < 100 Hz. Blue boxes specify residues with 300 Hz < Δf > 100 Hz. B. Chemical shift changes (Δf) between reduced wild type and C104S were mapped to the available C104S (PDB 1XM2)36 crystal structure and color-coded as in A. The active site, Y53 and H103 residues are depicted as ball and sticks. Relevant regions of the protein are also labeled. C. 1H-15N HSQC spectra of reduced wild type (black) and C49S (red) with colored boxes highlighting chemical shift change from the corresponding categories. Additionally, residues with Δf > 300 Hz are specified by yellow boxes with black lettering. D. As in B except changes between the reduced wild type and C49S are mapped. The same chemical shift change categories used in panel C were used in panel D (no change=black; small change=red; medium change=blue; large change=yellow) E. 1H-15N HSQC spectra of C104S (black) and C49S (red) with colored boxes highlighting chemical shift changes from the corresponding categories. F. As in B and D except chemical shift changes between C104S and C49S were mapped. The same color-code applies. Panels B, E and F were generated using Pymol.
Despite the inability of the disulfide bound protein to bind phosphate at the active site, the choice of buffer still affects the stability of the oxidized species such that phosphate is more stabilizing. Based on the fact that PRL-1 precipitates at elevated phosphate concentrations (data not shown), it may be that the ion interacts with the oxidized form at another site or it may exert a non-specific salting-out effect. While studies to explain this phenomenon are beyond the scope of this work, a secondary phosphate interaction site has already been proposed. The C-terminal tail of PRL-1 contains a polybasic region, which has been shown to be important for binding to phospholipid membranes.50 The stabilization of oxidized wild type by phosphate may be a result of an interaction with this region of the protein. These residues are missing from the crystal structures but large perturbations corresponding to these residues in the NMR spectra occur upon active-site oxidation,35 suggesting they are organized differently in the two states.
We next compared the NMR spectrum of the chemically reduced wild-type protein to that of C49S (Figure 5C). Surprisingly, there are far more changes between these two protein forms than observed between the chemically reduced wild type and C104S. Representative large (yellow), medium (blue) and small (red) changes are highlighted with corresponding colored boxes in the figure. Again for visualization, the changes were mapped to the available C104S crystal structure with each residue colored according to the appropriate chemical shift change category (Figure 5D).36 Many of the largest chemical shift changes are located in or near the active site (V48-E50, W68, F70-A74, H103, C104, L108-R110). Similar to the C104S-reduced wild type comparison, several additional perturbations to α4-α6 are observed (L114-L117, L119, Q131, Q135, F141 and Q145). These changes are again propagated to β1 and β2 (V10-V12). Because changes in α4-α6, β1 and β2 are confined to the internal face and are relatively small across variants with differing stabilities, we conclude that this region of the protein does not correlate with aggregation propensity. Loop three containing Y53 is also altered. Interestingly, in each region of the protein described above, the number and magnitude of chemical shift changes are larger in C49S, and the specific residues involved in some cases differ. For example, Y53 and nearby residues, especially T26, are perturbed to a much greater extent with the C49S mutation than with the C104S. The 1H line width of the Y53 peak increases from 22.7 to 53.9 Hz (137% increase), despite the fact that the average line width for all C49S peaks is approximately the same as that for the reduced wild type (33.05 and 31.8 Hz, respectively). This data supports our hypothesis that burial of the C49 side chain stabilizes the Y53-H103 interaction and incorporation of a polar moiety at this position increases conformational flexibility in this region.
To further probe the role of the hydrogen bond between Y53 and H103, we eliminated the interaction by substituting Phe for Tyr. An NMR spectrum of Y53F was collected, and the broad line widths of the peaks indicate that this protein exists in a molten globule-like state. With Y53F, the Phe side chain is likely exchanging between a buried and exposed conformation of loop three, and consequently, the conformation of this residue and the surrounding residues is not restricted to a specific organization. Because the entire set of peaks is broadened to a similar extent in this spectrum, it appears that the Y53-H103 hydrogen bond organizes a particular conformation of the protein, which has increased physical stability. Y53E was made because it was expected that mutation to Glu would favor a more exposed state for this region, due to the introduction of a charged moiety. The NMR spectrum of Y53E was collected and compared with that of C49S (Supporting information, Figure 1). Overall, the NMR spectrum of Y53E has many more peaks than would be predicted for a single stable conformation, and many of the peaks in the central portion of the spectrum are overlapped and unresolved. Based on the line widths, it appears that a mixture of two or more distinct conformations exists, possibly a set of more open conformers that undergo slow conformational exchange on the NMR timescale. Despite this complexity, critical portions of the Y53E spectrum are remarkably similar to C49S. Of importance is the fact that the peaks for Y53 and E53 are virtually identical. This information supports our hypothesis that the C49S mutation also alters the status of the Y53-H103 hydrogen bond. To eliminate the possibility that mutation of Y53 might alter the redox state of the protein, resulting in the increased number of peaks, reducing agent was added to the sample. The addition of a reducing agent does not alter the appearance of the Y53E (or Y53F) spectrum in any way (data not shown), indicating that the protein exists as multiple conformers independent of disulfide bond formation. As such, the formation of the disulfide bond appears to depend on the presence of the Y53-H103 bond, and the local packing organization imparted to the active site and L3 are a result of this interaction.
Based on the above data and the assumption that the charge state of the active site for C49S and chemically reduced wild type are approximately the same, it seems that the active site is coupled to the rest of the protein through H103 via its hydrogen bond to Y53 in L3. Although more extensive structural analysis is needed to fully explain the cooperativity, the beta sheet also packs against the large hydrophobic surface generated by the Y53-H103 hydrogen bond. An unusually large number of substantial differences appear in the central β-strand (β5) located in the hydrophobic core of the protein variants. A101 is the central residue of this structural feature, and the C49S mutation causes a considerably larger change in frequency than does C104S for this residue when compared to the chemically reduced wild type (Δδ1H and Δδ15N for C49S are 0.116 and 2.54 ppm, respectively; Δδ1H and Δδ15N for C104S are 0.000 and 0.073 ppm, respectively). As can be observed in Figure 5D, the largest changes are observed in the central strand (β5) with residues C99, A101, V102 and H103. The remaining blue change is V42 in β2, and smaller changes are observed for all residues in β4. These changes may be a reflection of the altered packing arrangement of surrounding residues and/or dynamics in L3 where both C49 and Y53, which hydrogen bonds to H103 positioned at the edge of the active site in β5, are located. To better illustrate the differences between C104S and C49S, the chemical shift changes observed between the NMR spectra of these two proteins (Figure 5E) were again mapped to the structure. The results are shown in Figure 5F.
C49S, which lacks both the Y53-H103 hydrogen bond and C49-C104 disulfide bond, is consistently less stable than all other forms studied, indicated by its dramatically lower melting temperature in both phosphate and HEPES and increased propensity to aggregate. As such, increased protein aggregation is correlated with increased flexibility in this region of the protein. This is supported by the fact that the oxidized wild type, which contains the disulfide bond but lacks the hydrogen bond, is more stable than C49S (Table 3). When comparing the reduced and oxidized wild type proteins,35 similar changes to the central β-sheet are observed, as well as large changes in α-helix two (α2). Similar changes to α2 are detected when comparing C49S to reduced wild type and/or C104S and can be seen in Figures 5D and F. A close up of α2 is provided in the supporting information (Figure S2) to better illustrate the location and magnitude of these changes. Because β4 is composed of entirely hydrophobic residues and such sequences are prone to aggregation,51 we hypothesize that stable packing of L3 and α2 provide protection in the reduced state against edge strand initiated aggregation. Formation of the disulfide bond causes the Y53-H103 bond to become distorted and L3 consequently becomes more flexible, leading to slightly increased aggregation propensity in the oxidized state. Complete disruption of the hydrogen bond, as is the case with C49S, may lead to unraveling of α2, and the increased susceptibility to aggregation may be caused by a larger degree of β-sheet exposure. Many other groups have found that the proportion of β-sheet structure increases in protein aggregates, while α-helical content is diminished.52,53 Interestingly, full spectral CD analysis reveals that the majority of β-sheet structure remains intact at the Tm (data not shown). Based on this observation, it is likely that unfurling of the structure at L3 and α2 exposes the β-sheet and may permit edge-strand association to generate aggregates. Because large protein aggregates cannot be observed using solution NMR, the aggregation mechanism will have to be investigated using other methods.
Table 3.
a. Effects of Specific Bonds on PRL-1 Stability
| Decreasing Stability ⇒⇒⇒ | ||||
|---|---|---|---|---|
| C104S | Reduced WT | Oxidized WT | C49S | |
| Y53-H103 hydrogen bond | Yes | Yes | No | No |
| C49-C104 disulfide bond | No | No | Yes | No |
| Active site PO4 binding | Yes | Partial | No | Partial |
| Tm (pH 7.5) | 76.31 | 67.81 | 65.47 | 62.79 |
DISCUSSION
In the current study, low-resolution methods (CD and SLS) were used to rapidly determine whether differences in stability between the various protein forms exist. The resulting hypotheses about protein stabilization or destabilization were then tested using high-resolution solution NMR to correlate the observed changes in stability with specific perturbations in the structure of the folded protein. In the case of PRL-1, we have shown that the integrity of the native PRL-1 structure does not depend on the absence or presence of the disulfide bond directly but rather on the local packing interactions among nearby side chains and coupling between different regions. Each Cys-modulated PRL-1 variant has a unique physical stability based on retention of specific features in its three-dimensional structure, and this concept is summarized in Table 3. Our data indicate the reduced form of the protein compensates for disulfide bond cleavage by a compilation of adjustments focused around the hydrogen bond. Structural instability and aggregation is correlated with flexibility in L3 (containing Y53), and C49S, which lacks both the Y53-H103 hydrogen bond and disulfide bond, is less stable than the oxidized wild type. In this context,15 the disulfide bond increases the stability of the protein by restricting the conformational flexibility of L3 in a different manner.
It is particularly interesting that the hydrogen bond between Y53 and H103 is better able to accomplish stabilization of the protein than the disulfide bond. Other incidents of hydrogen bonds contributing substantially to the stability of proteins have been documented.27,54,55 Polar moieties are accommodated in the hydrophobic core of proteins when their hydrogen bonding potential is satisfied, and the degree of stabilization imparted correlates with the number and strength of the bonds. The methodologies used in these studies, however, could not provide information about cooperative or dynamic influences resulting from disruption of the hydrogen bond and compensation effects could not be observed. The high-resolution approach used here shows that modification of the hydrogen bond causes local perturbations as well as increased dynamics in coupled regions. For example, because disruption of the Y53-H103 bond in PRL-1 indirectly leads to drastic changes in the central β-sheet, packing of these structural features must be interrelated.
Such a drastic change in the protein's hydrophobic core is quite unanticipated. In general, the intrinsic dynamics of a protein predict that changes involving flexible loops or active-site residues occur, and changes of this nature are indeed observed in this study. Rearrangement of a protein's hydrophobic core, however, is rare.56 Even proteins with large conformational changes share similar hydrophobic packing interactions.32,57,58 Recently, a structural change that involves repacking of the protein core has been documented. In this example, virtually all of the tertiary contacts of the lyphotactin fold are replaced with new contacts upon the structural transition.56 Similarly, reduction of the disulfide bond in PRL-1 causes more than 90% of the NMR chemical shifts to change significantly, without drastically altering the secondary structure elements.35 This structural transition likely reflects changes not only to the conformation of the active site but also the interaction of Y53 and H103 and cooperative hydrophobic packing interactions with the β-sheet in the protein core. Additional NMR studies are being pursued to further assess alterations in direct contacts between specific residues that affect stability.
Although the solution NMR data reveal that substantial conformational changes occur in solution between the reduced and oxidized states,35 such large differences are not apparent in the crystal structures. The majority of differences are located near the active site when comparing the two crystallized proteins, and these differences are obvious in the NMR data as well. What is not apparent in a comparison of the crystal structures is the importance of the small rearrangements between the beta sheet, L3 and α2 in the two states. This is because the conformation of these regions is ruled by dynamics. Based on the crystal data, one would predict that the hydrophobic packing interactions in the reduced and oxidized states are largely the same,32,36 but the NMR data clearly show otherwise. One explanation for this discrepancy may be the absence of the C-terminus. Crystallized PRL-1 was truncated at residues 156 and 160 for the reduced and oxidized forms, respectively. Studies from our lab previously showed that the missing residues, which compose the polybasic sequence, modulate the structure and activity of PRL-1, such that their removal abrogates disulfide bond formation.35 This result indicates an additional unique coupling exists in the oxidized protein that involves the tail residues, and this interaction is affected by phosphate but in a different manner than the reduced state.
Concluding Remarks
The study shows that stabilization or destabilization of a protein can be achieved in the folded state through the collaboration of specific, local interactions. The strategies presented here illustrate the utility of NMR in pharmaceutical research, as they can be applied to therapeutically relevant proteins to identify stabilizing forces within the structure. Identification of these forces at atomic resolution will facilitate the rational design of improved protein therapeutics and biotechnology products. Furthermore, the work demonstrates that the effects of the oxidation state of cysteine residues on a protein's stability derive from the overall context in which they reside. Understanding this relationship is valuable because it may guide disulfide bond engineering in protein pharmaceuticals.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Drew Vartia for generating the C49S, Y53E and Y53F constructs and helpful discussions, Drs. Lisa Crow and Klaas Hallenga for collecting the C49S NMR assignment data at the Nuclear Magnetic Resonance Facility At Madison (NMRFAM), and Dr. Dave Vander Velde for technical assistance with the NMR spectrometer at the University of Kansas. This publication was made possible by NIH grant number P20 RR-17708 from the National Center for Research Resources and the Kansas University Center for Research. This work was additionally supported by fellowships for Andria Skinner from Amgen, the Edith and Eleta Ernst Cancer Research Fellowship and the Gretta Jean and Gerry D. Goetsch graduate scholarship.
Footnotes
SUPPPORTING INFORMATION AVAILABLE
Supporting information for this work is available via the Internet at http://www.mrw.interscience.wiley.com/suppmat/0022-3549/suppmat/. 1H-15N HSQCs of PRL-1-C49S and Y53E are compared in Figure S1. The molecules in Figures 5D and F are rotated by 180° and shown in Figure S2A and B, respectively.
REFERENCES
- 1.Volkin DB, Mach H, Middaugh CR. Degradative covalent reactions important to protein stability. Molecular biotechnology. 1997;8(2):105–122. doi: 10.1007/BF02752255. [DOI] [PubMed] [Google Scholar]
- 2.Manning MC, Patel K, Borchardt RT. Stability of protein pharmaceuticals. Pharm Res. 1989;6:903–918. doi: 10.1023/a:1015929109894. [DOI] [PubMed] [Google Scholar]
- 3.Hagihara Y, Mine S, Uegaki K. Stabilization of an immunoglobulin fold domain by an engineered disulfide bond at the buried hydrophobic region. The Journal of biological chemistry. 2007;282(50):36489–36495. doi: 10.1074/jbc.M707078200. [DOI] [PubMed] [Google Scholar]
- 4.Saerens D, Conrath K, Govaert J, Muyldermans S. Disulfide bond introduction for general stabilization of immunoglobulin heavy-chain variable domains. Journal of molecular biology. 2008;377(2):478–488. doi: 10.1016/j.jmb.2008.01.022. [DOI] [PubMed] [Google Scholar]
- 5.Gong R, Vu BK, Feng Y, Prieto DA, Dyba MA, Walsh JD, Prabakaran P, Veenstra TD, Tarasov SG, Ishima R, Dimitrov DS. Engineered human antibody constant domains with increased stability. The Journal of biological chemistry. 2009;284(21):14203–14210. doi: 10.1074/jbc.M900769200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hogg PJ. Disulfide bonds as switches for protein function. Trends in biochemical sciences. 2003;28(4):210–214. doi: 10.1016/S0968-0004(03)00057-4. [DOI] [PubMed] [Google Scholar]
- 7.Betz SF. Disulfide bonds and the stability of globular proteins. Protein Sci. 1993;2(10):1551–1558. doi: 10.1002/pro.5560021002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pace CN, Grimsley GR, Thomson JA, Barnett BJ. Conformational stability and activity of ribonuclease T1 with zero, one, and two intact disulfide bonds. The Journal of biological chemistry. 1988;263(24):11820–11825. [PubMed] [Google Scholar]
- 9.Robinson CR, Sauer RT. Striking stabilization of Arc repressor by an engineered disulfide bond. Biochemistry. 2000;39(40):12494–12502. doi: 10.1021/bi001484e. [DOI] [PubMed] [Google Scholar]
- 10.Wedemeyer WJ, Welker E, Narayan M, Scheraga HA. Disulfide bonds and protein folding. Biochemistry. 2000;39(15):4207–4216. doi: 10.1021/bi992922o. [DOI] [PubMed] [Google Scholar]
- 11.Bhattacharyya R, Pal D, Chakrabarti P. Disulfide bonds, their stereospecific environment and conservation in protein structures. Protein Eng Des Sel. 2004;17(11):795–808. doi: 10.1093/protein/gzh093. [DOI] [PubMed] [Google Scholar]
- 12.Ciaccio NA, Laurence JS. Helicity and disulfide bond formation control solubility and aggregation of ATF5. submitted:Effects of disulfide bond formation and protein helicity on the aggregation of activating transcription factor. 2009. p. 5. [DOI] [PMC free article] [PubMed]
- 13.Zavodszky M, Chen C-W, Huang J-K, Zolkiewski M, Wen L, Krishnamoothi R. Disulfide bond effects on protein stability: designed variants of Cucurbita maxima trypsin inhibitor-V. Protein Sci. 2001;10:149–160. doi: 10.1110/ps.26801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ogawa K, Sonoyama T, Takeda T, Ichiki S, Nakamura S, Kobayashi Y, Uchiyama S, Nakasone K, Takayama SJ, Mita H, Yamamoto Y, Y. S Roles of a short connecting disulfide bond in the stability and function of pyschrophilic Shewanella violacea cytochrome c5. Extremophiles. 2007;11:797–807. doi: 10.1007/s00792-007-0099-5. [DOI] [PubMed] [Google Scholar]
- 15.Santiveri CM, Leon E, Rico M, Jimenez MA. Context-dependence of the contribution of disulfide bonds to beta-hairpin stability. Chemistry (Weinheim an der Bergstrasse, Germany) 2008;14(2):488–499. doi: 10.1002/chem.200700845. [DOI] [PubMed] [Google Scholar]
- 16.Chang SG, Choi KD, Jang SH, Shin HC. Role of disulfide bonds in the structure and activity of human insulin. Molecules and cells. 2003;16(3):323–330. [PubMed] [Google Scholar]
- 17.Fu X, Li W, Mao Q, Chang Z. Disulfide bonds convert small heat shock protein Hsp16.3 from chaperone to a non-chaperone: implications for the evolution of cystein in molecular chaperones. Biochem Biophys Res Commun. 2003;308(3):627–635. doi: 10.1016/s0006-291x(03)01450-5. [DOI] [PubMed] [Google Scholar]
- 18.Matsumura M, Becktel WJ, Levitt M, Matthews BW. Stabilization of phage T4 lysozyme by engineered disulfide bonds. Proc Natl Acad Sci. 1989;86:6562–6566. doi: 10.1073/pnas.86.17.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishikawa H, Kim S, Kwak K, Wakasugi K, Fayer MD. Disulfide bond influence on the protein structural dynamics probed with 2D-IR vibrational echo spectroscopy. Proc Natl Acad Sci. 2007;104(49):19309–19314. doi: 10.1073/pnas.0709760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamerzell TJ, Ramsey JD, Middaugh CR. Immunoglobulin dynamics, conformational fluctuations, and nonlinear elasticity and their effects on stability. J Phys Chem B. 2008;112(10):3240–3250. doi: 10.1021/jp710061a. [DOI] [PubMed] [Google Scholar]
- 21.Ramsey JD, Gill ML, Kamerzell TJ, Price ES, Joshi SB, Biship SM, Oliver CN, Middaugh CR. Using empirical phase diagrams to understand the role of intramolecular dynamics in immunoglobulin G stability. J Pharm Sci. 2009;98(7):2432–2447. doi: 10.1002/jps.21619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamerzell TJ, Middaugh CR. The complex inter-relationships between protein flexibility and stability. J Pharm Sci. 2008;97(9):3494–3517. doi: 10.1002/jps.21269. [DOI] [PubMed] [Google Scholar]
- 23.Thirumangalathu R, Krishnan S, Ricci MS, Brems DN, Randolph TW, Carpenter JF. Silicone oil- and agitation-induced aggregation of a monoclonal antibody in aqueous solution. J Pharm Sci. 2009;98(9):3167–3181. doi: 10.1002/jps.21719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Esfandiary R, Kickhoefer VA, Rome LH, Joshi SB, Middaugh CR. Structural stability of vault proteins. J Pharm Sci. 2009;98(4):1376–1386. doi: 10.1002/jps.21508. [DOI] [PubMed] [Google Scholar]
- 25.Fan HF, Ralston J, Dibiase M, Faulkner E, Middaugh CR. Solution behavior of IFN-Beta- 1a: An empirical phase diagram based approach. J Pharm Sci. 2005;94(9):1893–1911. doi: 10.1002/jps.20410. [DOI] [PubMed] [Google Scholar]
- 26.Fontana A. Analysis and modulation of protein stability. Current opinion in biotechnology. 1991;2(4):551–560. doi: 10.1016/0958-1669(91)90080-o. [DOI] [PubMed] [Google Scholar]
- 27.Thurlkill RL, Grimsley GR, Scholtz JM, Pace CN. Hydrogen bonding markedly reduces the pK of buried carboxyl groups in proteins. Journal of molecular biology. 2006;362:594–604. doi: 10.1016/j.jmb.2006.07.056. [DOI] [PubMed] [Google Scholar]
- 28.Kuwata K, Hoshino M, Era S, Batt CA, Goto Y. Alpha to beta transition of betalactogobulin as evidenced by heteronuclear NMR. Journal of molecular biology. 1998;283:731–739. doi: 10.1006/jmbi.1998.2117. [DOI] [PubMed] [Google Scholar]
- 29.Kuwata K, Shastry R, Cheng H, Hoshino M, Batt C, Goto Y, Roder H. Structural and kinetic characterization of early folding events in beta-lactoglobulin. Nature Struct Biol. 2001;8(2):151–155. doi: 10.1038/84145. [DOI] [PubMed] [Google Scholar]
- 30.Sakurai K, Konuma T, Yagi M, Goto Y. Structural dynamics and folding of beta-lactoglobulin probed by heteronuclear NMR. Biochim Biophys Acta: General Stubjects. 2009;1790:527–537. doi: 10.1016/j.bbagen.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 31.Chiarugi P, Buricchi F. Protein tyrosin phosphorylation and reversible oxidation: two cross-talking posttranslation modifications. Antiox and Redox Signal. 2007;9(1):1–24. doi: 10.1089/ars.2007.9.1. [DOI] [PubMed] [Google Scholar]
- 32.Sun J-P, Wang W-Q, Yang H, Liu S, Liang F, Fedorov AA, Almo SC, Zhang Z-Y. Structure and biochemical properties of PRL-1, a phosphatase implicated in cell growth, differentiation, and tumor invasion. Biochem. 2005;44:12009–12021. doi: 10.1021/bi0509191. [DOI] [PubMed] [Google Scholar]
- 33.Laurence JS, Hallenga K, Stauffacher CV. 1H, 15N, 13C resonance assignments of the human protein tyrosine phosphatase PRL-1. J Biomol NMR. 2004;29:417–418. doi: 10.1023/B:JNMR.0000032506.16792.c6. [DOI] [PubMed] [Google Scholar]
- 34.Skinner A, Laurence JS. 1H, 15N, 13C resonance assignments of the reduced and active form of human protein tyrosine phosphatase, PRL-1. Biomol NMR Assign online first. 2009 doi: 10.1007/s12104-008-9142-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skinner AL, Vartia AA, Williams TD, Laurence JS. Enzyme activity of phosphatase of regenerating liver is controlled by the redox environment and its C-terminal residues. Biochem. 2009 doi: 10.1021/bi900241k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jeong DG, Kim SJ, Kim JH, Son JH, Park MR, Lim SM, Yoon T-S, Ryu SE. Trimeric structure of PRL-1 phosphatase reveals an active enzyme conformation and regulation mechanisms. Journal of molecular biology. 2005;345:401–413. doi: 10.1016/j.jmb.2004.10.061. [DOI] [PubMed] [Google Scholar]
- 37.Lepe-Zuniga JL, Zigler JS, Jr., Gery I. Toxicity of light-exposed Hepes media. J Immunol Methods. 1987;103(1):145. doi: 10.1016/0022-1759(87)90253-5. [DOI] [PubMed] [Google Scholar]
- 38.Masson JF, Gauda E, Mizaikoff B, Kranz C. The interference of HEPES buffer during amperometric detection of ATP in clinical applications. Anal Bioanal Chem. 2008;390(8):2067–2071. doi: 10.1007/s00216-008-2015-y. [DOI] [PubMed] [Google Scholar]
- 39.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182(319-326) doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 40.Gastiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Delaglio F, Grezesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensonal spectra processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 43.Goddard TD, Kneller DG. SPARKY. University of California; San Francisco: 2004. [Google Scholar]
- 44.Jaren OR, Kranz JK, Sorensen BR, Wand J, Shea MA. Calcium iduced conformational switching of paramecium calmodulin provides evidence for domain coupling. Biochem. 2002;41(48):14158–14166. doi: 10.1021/bi026340+. [DOI] [PubMed] [Google Scholar]
- 45.Eghbalnia HR, Bahrami A, Wang L, Assadi A, Markley JL. Probabilistic identification of spin systems and their assignments including coil-helix interference as output (PISTACHIO). J Biomol NMR. 2005;32:219–233. doi: 10.1007/s10858-005-7944-6. [DOI] [PubMed] [Google Scholar]
- 46.Wishart DS, Bigam CG, Yoa J, Abildgaard F, Dyson HH, Oldfield E, Markley JL, Sykes BD. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J Biomol NMR. 1995;6:135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Z-Y. Chemical and mechanistic approaches to the study of protein tyrosine phosphatases. Acc Chem Res. 2003;36:385–392. doi: 10.1021/ar020122r. [DOI] [PubMed] [Google Scholar]
- 48.Stickle DF, Presta LG, Dill KA, Rose GD. Hydrogen bonding in globular proteins. Journal of molecular biology. 1992;226:1143–1159. doi: 10.1016/0022-2836(92)91058-w. [DOI] [PubMed] [Google Scholar]
- 49.Cavanagh J, Fairbrother WJ, Palmer AG, III, Rance M, Skelton N. Protein NMR Spectroscopy: Principles and Practice. 2nd ed. Academic Press; Boston: 2007. [Google Scholar]
- 50.Sun J-P, Luo Y, Yu X, Wang W-Q, Zhou B, Liang F, Zhang Z-Y. Phosphatase activity, trimerization, and the C-terminal polybasic region are all required for PRL1-mediated cell growth and migration. The Journal of biological chemistry. 2007;282(39):29043–29051. doi: 10.1074/jbc.M703537200. [DOI] [PubMed] [Google Scholar]
- 51.Richardson JS, Richardson DC. Natural beta-sheet proteins use negative design to avoid edge-to-edge aggregation. Proc Natl Acad Sci. 2002;99(5):2754–2759. doi: 10.1073/pnas.052706099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Petsko GA, Dagmar R. Protein Structure and Function. New Science Press Ltd; 2004. pp. 160–161. [Google Scholar]
- 53.Pan K-M, Baldwin M, Nguyen J, Gasset M, Serban A, Groth D, Mehlhorn I, Huang Z, Fletterick RJ, Cohen FE, Prusiner SB. Conversion of alpha-helices into beta sheets features in the formation of the scrapie prion protein. Proc Natl Acad Sci. 1993;90:10962–10966. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shirley BA, Stanssens P, Hahn U, Pace CN. Contribution of hydrogen bonding to the conformational stability of ribonuclease T1. Biochem. 1992;31:725–732. doi: 10.1021/bi00118a013. [DOI] [PubMed] [Google Scholar]
- 55.Pace CN, Hebert EJ, Shaw KL, Schell D, Both V, Krajcikova D, Sevcik J, Wilson KS, Dauter Z, Hartley RW, Grimsley GR. Conformational stability and thermodynamics of folding of ribonucleases Sa, Sa2 and Sa3. Journal of molecular biology. 1998;279(1):271–286. doi: 10.1006/jmbi.1998.1760. [DOI] [PubMed] [Google Scholar]
- 56.Tuinstra RL, Peterson FC, Kutlesa S, Elgin ES, Kron MA, Volkman BF. Interconversion between two unrelated protein folds in the lyphotactin native state. Proc Natl Acad Sci. 2008;105:5057–5062. doi: 10.1073/pnas.0709518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cordes MHJ, Burton RE, Walsh NP, McKnight J, Sauer RT. An evolutionary bridge to a new protein fold. Nature Struct Biol. 2000;7(12):1129–1132. doi: 10.1038/81985. [DOI] [PubMed] [Google Scholar]
- 58.Luo X, Tang Z, Xia G, Wassmann K, Matsumoto T, Riza J, Yu H. The Mad2 spindle checkpoint protein has two distinct natively folded states. Nature Struct Biol. 2004;11(4):338–345. doi: 10.1038/nsmb748. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.