Abstract
Ponatinib is a novel tyrosine kinase inhibitor with potent activity against BCR-ABL with mutations including T315I, and also against fms-like tyrosine kinase 3 (FLT3). We tested interactions between ponatinib at pharmacologically relevant concentrations of 50 to 200 nM and the multidrug resistance-associated ATP-binding cassette (ABC) proteins ABCB1, ABCC1 and ABCG2. Ponatinib enhanced uptake of substrates of ABCG2 and ABCB1, but not ABCC1, in cells overexpressing these proteins, with a greater effect on ABCG2 than on ABCB1. Ponatinib potently inhibited [125I]-IAAP binding to ABCG2 and ABCB1, indicating binding to their drug substrate sites, with IC50s of 0.04 μM and 0.63 μM, respectively. Ponatinib stimulated ABCG2 ATPase activity in a concentration-dependent manner and stimulated ABCB1 ATPase activity at low concentrations, consistent with it being a substrate of both proteins at pharmacologically relevant concentrations. The ponatinib IC50s of BCR-ABL-expressing K562 cells transfected with ABCB1 and ABCG2 were approximately the same as and 2-fold higher than that of K562, respectively, consistent with ponatinib being a substrate of both proteins, but inhibiting its own transport, and resistance was also attenuated to a small degree by ponatinib-induced downregulation of ABCB1 and ABCG2 cell surface expression on resistant K562 cells. Ponatinib at pharmacologically relevant concentrations produced synergistic cytotoxicity with ABCB1 and ABCG2 substrate chemotherapy drugs and enhanced apoptosis induced by these drugs, including daunorubicin, mitoxantrone, topotecan and flavopiridol, in cells overexpressing these transport proteins. Combinations of ponatinib and chemotherapy drugs warrant further testing.
Keywords: Ponatinib, ABCG2, ABCB1, multidrug resistance, leukemia
INTRODUCTION
Cancer cell resistance to structurally unrelated drugs, termed multidrug resistance (MDR), is implicated in chemotherapy failure. The ATP-binding cassette (ABC) transport proteins ABCB1 [P-glycoprotein (Pgp), MDR1], ABCC1 [multidrug resistance protein-1 (MRP1)] and ABCG2 [breast cancer resistance protein (BCRP), mitoxantrone resistance protein (MXR)] are strongly implicated in MDR (1). These proteins are expressed on leukemia cells of all subtypes and transport structurally and functionally diverse drugs used to treat leukemias (1).
Diverse novel tyrosine kinase inhibitors (TKIs) used to treat leukemias are ABCB1, ABCC1 and/or ABCG2 substrates and/or inhibitors. These include the BCR-ABL inhibitors imatinib mesylate, nilotinib and dasatinib (2–8), used to treat chronic myelogenous leukemia (CML) and Philadelphia chromosome-positive (Ph+) acute lymphoblastic leukemia (ALL), and the fms-like tyrosine kinase 3 (FLT3) inhibitors midostaurin (9), tandutinib (10), sorafenib (11) and sunitinib (12), in clinical trials in acute myeloid leukemia (AML) with FLT3 internal tandem duplication (ITD), present in 30% of cases and associated with adverse treatment outcome (13). TKI interactions with ABC proteins should be considered in design of treatment regimens, as they may cause resistance to TKIs, sensitization to chemotherapy drugs and/or significant drug interactions.
Ponatinib (AP24534) is a novel TKI, currently in clinical trials, with potent activity in cells with BCR-ABL mutations including T315I, which confers resistance to the approved and available BCR-ABL inhibitors imatinib mesylate, nilotinib and dasatinib (14). Ponatinib inhibits BCR-ABL at concentrations above 40 nM (15), which are achieved with doses of 30 mg and greater (16), and shows promising clinical activity (16,17). Ponatinib also potently inhibits FLT3, and thus may also have a role in AML therapy (18). It also inhibits fibroblast growth factor receptors (FGFRs), vascular endothelial growth factor receptors (VEGFRs) and angiopoietin (Tie2) (14), promising targets in solid tumor therapy (19).
Given that other BCR-ABL inhibitors interact with MDR proteins and given the potential role of ponatinib in treating AML and solid tumors, in addition to CML and Ph+ ALL, we studied interaction of ponatinib with MDR-associated ABC proteins.
MATERIALS AND METHODS
Cell lines
HL60, K562 and MV4–11 leukemia cells were obtained from the American Type Culture Collection (Manassas, VA), vincristine-selected HL60/VCR cells, overexpressing ABCB1 (20), from Dr. Ahmad R. Safa, Indiana University, Indianapolis, IN, and doxorubicin-selected HL60/ADR cells, overexpressing ABCC1 (21), from Dr. Kapil Bhalla, University of Kansas Cancer Center, Kansas City, KS. Parental 8226 myeloma cells and doxorubicin-selected 8226/Dox6 and mitoxantrone-selected 8226/MR20 cells, overexpressing ABCB1 or wild-type (R482) ABCG2, respectively (22), were obtained from Dr. William Dalton, Moffitt Cancer Center, Tampa, FL. Transfected K562/ABCB1 and K562/ABCG2 cells, stably overexpressing ABCB1 (23) or wild-type ABCG2 (24) were gifts from Dr. Michael Gottesman, National Cancer Institute, Bethesda, MD and Dr. Yoshikazu Sugimoto, Kyoritsu University of Pharmacy, Tokyo, Japan, respectively. Doxorubicin and verapamil-selected MCF7/AdrVp breast carcinoma cells, overexpressing ABCG2 with the R482T mutation (25), were obtained from Dr. Douglas Ross, University of Maryland Greenebaum Cancer Center (UMGCC), Baltimore, MD, and flavopiridol-selected MCF-7/Flv1000 cells (26), overexpressing wild-type ABCG2, from Dr. Susan Bates, National Cancer Institute. All cells were cultured in RPMI 1640, pH 7.4, with 10% fetal bovine serum at 37°C in a humidified atmosphere containing 5% CO2. No authentication of cell lines was performed by the authors.
Reagents
Ponatinib (AP24534) was purchased from ChemieTek (Indianapolis, IN) and was stocked at 10 mM in DMSO at −20°C. The fluorescent ABCB1 and ABCC1 substrates 3,3′-diethyloxacarbocyanine iodide [DiOC2(3)] and rhodamine 123 (RH 123) (27) were purchased from Sigma-Aldrich (St Louis, MO), and the fluorescent ABCG2 substrate pheophorbide A (PhA) (28) from Frontier Scientific (Logan, VT). The ABCB1 inhibitor PSC-833 was obtained from Novartis Pharmaceutical Corporation (East Hanover, NJ). The ABCC1 and ABCG2 inhibitors p-[dipropylsulfamoyl] benzoic acid (probenecid) and fumitremorgin C (FTC), respectively, were purchased from Sigma-Aldrich (29). Daunorubicin, mitoxantrone and topotecan were purchased from Sigma-Aldrich, and flavopiridol from Enzo Life Sciences (Farmingdale, NY). MRK16 antibody to an ABCB1 cell surface epitope was purchased from Alexis Biochemicals (San Diego, CA), allophycocyanin conjugate (APC)-tagged 5D3 antibody to an ABCG2 cell surface epitope from BD Biosciences (San Jose, CA), and BXP-21 ABCG2 antibody from Signet Laboratories (Dedham, MA). Fluorescein isothiocyanate (FITC)-conjugated annexin V and propidium iodide (PI) were purchased from Trevigen (Gaithersburg, MD), APC annexin V from BD Biosciences and LIVE/DEAD fixable near-IR dead cell stain from Invitrogen (Carlsbad, CA). Cell proliferation reagent WST-1 was purchased from Roche Diagnostics (Indianapolis, IN) and [125I]Iodoarylazidoprazosin (IAAP) (2200 Ci/mmol) from PerkinElmer Life and Analytical Sciences (Waltham, MA).
Uptake of fluorescent ABC protein substrates
To measure ponatinib effect on uptake of fluorescent ABC protein substrates, HL60/VCR, 8226/Dox6 and K562/ABCB1 cells (1 × 106) were incubated for 30 minutes at 37° C with DiOC2(3) (0.6 ng/mL) and ponatinib (0–200 nM) or PSC-883 (2.5 μM) as a control, HL60/ADR cells with RH 123 (0.5 μg/mL) and ponatinib (0–200 nM) or probenecid (1 mM) as a control and 8226/MR20, K562/ABCG2 and MCF7/AdrVp cells with PhA (1 μM) and ponatinib (0–200 nM) or FTC (10 μM) as a control. The cells were then washed twice, resuspended in phosphate-buffered saline (PBS), then acquired on a FACSCanto II flow cytometer (BD Biosciences) and analyzed using FlowJo software (Tree Star, Inc., Ashland, OR). Substrate content after uptake with and without modulator was compared by the Kolmogorov-Smirnov statistic, expressed as a D-value ranging from 0 (no difference) to 1 (no overlap), with D-values ≥0.2 indicating significant modulation (27).
Photoaffinity labeling of ABCB1 and ABCG2 with [125I]IAAP
High-Five insect cell membrane vesicles expressing ABCB1 and crude membranes from MCF-7/Flv1000 cells (30 μg) expressing ABCG2 were incubated with 0–10 μM ponatinib for 5 minutes at 21–23°C in 50 mM Tris-HCl, pH 7.5. [125I]-IAAP (2200 Ci/mmole), 3–6 nM, was added and photoaffinity labeling of ABCB1 and ABCG2 by [125I]-IAAP was measured as previously described (30,31).
ABCB1 and ABCG2 ATPase assay
Crude membrane protein (100 μg protein/ml) from ABCB1- and ABCG2-expressing High-Five insect cells was incubated at 37°C with ponatinib, in varying concentrations, with and without 0.3 mM sodium orthovanadate, for ABCB1, or BeFx (0.2 mM beryllium sulfate and 2.5 mM sodium fluoride), for ABCG2, and the amount of inorganic phosphate released and the Vi- or BeFx-sensitive ATPase activity were measured as previously described (32).
MDR protein cell surface expression
To detect ABCG2 cell surface expression, cells were incubated with APC-conjugated 5D3 antibody at room temperature for 30 minutes, then washed twice with PBS. To detect ABCB1 cell surface expression, cells were incubated with MRK16 antibody for 1 hour, washed twice with PBS, then incubated with phycoerythrin (PE)-conjugated anti-human antibody for 30 minutes. Cells were acquired on a FACSCanto II (BD Biosciences) and analyzed with FlowJo. Replicate measurements of mean fluorescence intensity under different conditions were compared using the Student t-test.
Cell viability assay
1×104 log-phase cells were seeded per well in 96-well tissue culture plates and incubated with ponatinib (0–10 μM) or chemotherapy drugs at a range of concentrations at 37°C in 5% CO2 for 96 hours. Viable drug-treated cells were quantified using the WST-1 assay (29). Experiments were performed in triplicate at least three times.
Drug interactions and statistical analysis
For cell lines for which ponatinib was cytotoxic at pharmacologically relevant concentrations in cell viability assays, ponatinib effects on chemotherapy drug cytotoxicity were evaluated in drug combination studies. Combination experiments were designed by the maximal power design (33,34), using SynStat version 1.beta software (35). The method maximizes the power of the F-test to detect departures from the additive action of drugs. It does not assume a constant relative potency of the two drugs. With information from single-agent experiments, SynStat derives mixtures of two drugs and replicates of each mixture based on the pooled variations in single-agent experiments, which have 80% statistical power to detect at least a 15% difference in viability between the predicted additive values and the observed values at a significance level of 5%. Cells are then exposed to these multiple mixtures, and the cytotoxicity of these combinations is determined. Upon completion of the experiments, the F-statistic (33) is used to test the hypothesis of the additive action of two drugs and calculate the p-value of the F-test. If the p-value is greater than 0.05, the hypothesis of additive action is accepted. Otherwise, we calculate the interaction index (τ) (36) as where, for a given cytotoxic effect, xA and xB are the concentrations of drugs A and B in the combination, and XA and XB are the concentrations of drugs A and B that achieve the same cytotoxic effect when given alone. A τ value of 1 indicates additivity, τ <1 indicates synergy, and τ >1 indicates antagonism. The combination index surface is then fitted using the two-dimensional B-spline method (34), and the contour plot shows the dose-mixture areas of additive action, synergy and antagonism for the joint action of the two drugs.
Curve shift assay
MCF7/AdrVP cells, for which ponatinib was not cytotoxic at pharmacologically relevant concentrations in cell viability assays, were plated with mitoxantrone at a range of concentrations in a cell viability assay in the presence and absence of ponatinib at several concentrations, with analysis by the WST-1 colorimetric assay, as described above.
Measurement of apoptosis
8226/MR20 cells, overexpressing ABCG2, were incubated with mitoxantrone, topotecan or flavopiridol for 48 hours in the presence and absence of ponatinib, and apoptosis and necrosis were measured by staining with annexin V-FITC and PI. HL60/VCR and 8226/Dox6 cells, overexpressing ABCB1, were incubated with daunorubicin for 48 hours in the presence and absence of ponatinib, and apoptosis and necrosis were measured using APC annexin V and LIVE/DEAD fixable near-IR dead cell stain, to avoid spectral overlap with daunorubicin. Post treatment, cells (2–3 × 105) were washed with PBS, resuspended in annexin V binding buffer (1x), stained with annexin V-FITC (1 μL) and PI (2 μL) or APC annexin V (2.5 μL) and LIVE/DEAD fixable near-IR dead cell stain (0.5 μL), incubated at room temperature in the dark, then washed and acquired on a FACSCanto II and analyzed with FlowJo.
Flow cytometric cell cycle analysis
1 × 105 HL60/VCR, 8226/MR20, K562 and MV4–11 cells were treated with 0, 1, 5, 50 and 100 nM ponatinib for 24 and 48 hours, fixed in chilled ethanol (70%), washed with PBS, then treated with DNase-free RNase (200 μg/ml) for 1 hour at 37°C, stained with PI (40 μg/ml) and kept in the dark for 15 minutes at 20–25°C. Staining was measured on a FACScan, and percentages of cells in different cell cycle phases were determined using FlowJo.
RESULTS
Ponatinib increases substrate uptake in cells overexpressing ABCB1 and ABCG2
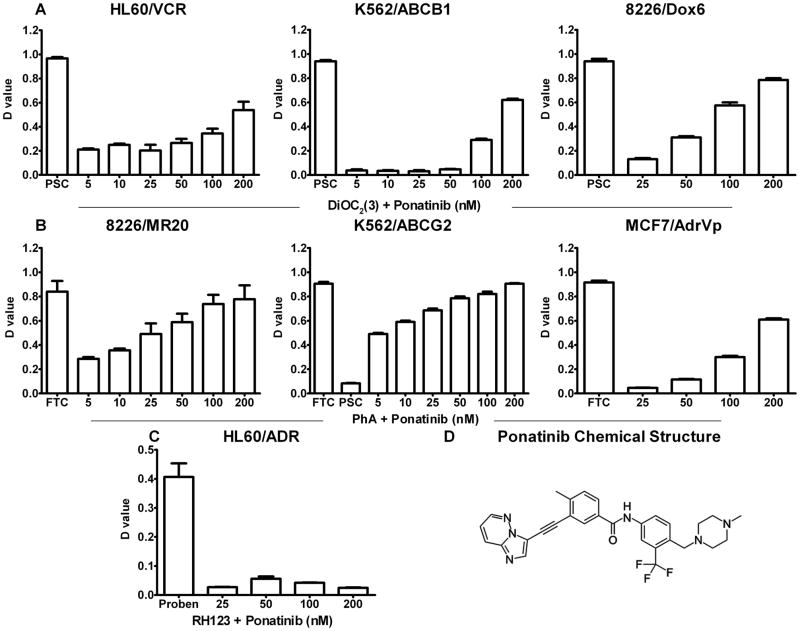
Ponatinib produced a significant concentration-dependent increase in uptake of the ABCB1 substrate DiOC2(3) in ABCB1-overexpressing HL60/VCR, K562/ABCB1 and 8226/Dox6 cells, and of the ABCG2 substrate PhA in ABCG2-overexpressing 8226/MR20, K562/ABCG2 and MCF7/AdrVP cells, with greater inhibition of ABCG2 than of ABCB1 (Figure 1). The effect in MCF7/AdrVp was less than in 8226/MR20 and K562/ABCG2, likely due to greater degree of resistance in solid tumor, in relation to hematopoietic, cell lines, rather than to presence of the R482T mutation in MCF7/AdrVp, though the latter is also possible. Since the R482T ABCG2 mutation is not clinically relevant, we did not pursue this distinction. Ponatinib had no effect on RH 123 uptake in ABCC1-overexpressing HL60/ADR cells.
Figure 1. Ponatinib enhances uptake of substrates of ABCG2 and ABCB1, but not ABCC1, in cells overexpressing these proteins.
Ponatinib effect on transport mediated by ABCB1 (A), ABCG2 (B) and ABCC1 (C) was measured by comparing cellular fluorescence after uptake of their fluorescent substrates DiOC2(3), pheophorbide A (PhA) and rhodamine 123 (RH 123), respectively, in the presence and absence of ponatinib in relevant cell lines, with specific modulators 2.5 μM PSC-833, 10 μM fumitremorgin C (FTC) and 1 mM probenecid (proben) as positive controls. Each bar represents the mean ± SEM of three individual experiments. D-value is the Kolmogorov-Smirnov statistic. The chemical structure of ponatinib is shown in D.
Ponatinib inhibits [125I]-IAAP photolabeling of ABCB1 and ABCG2
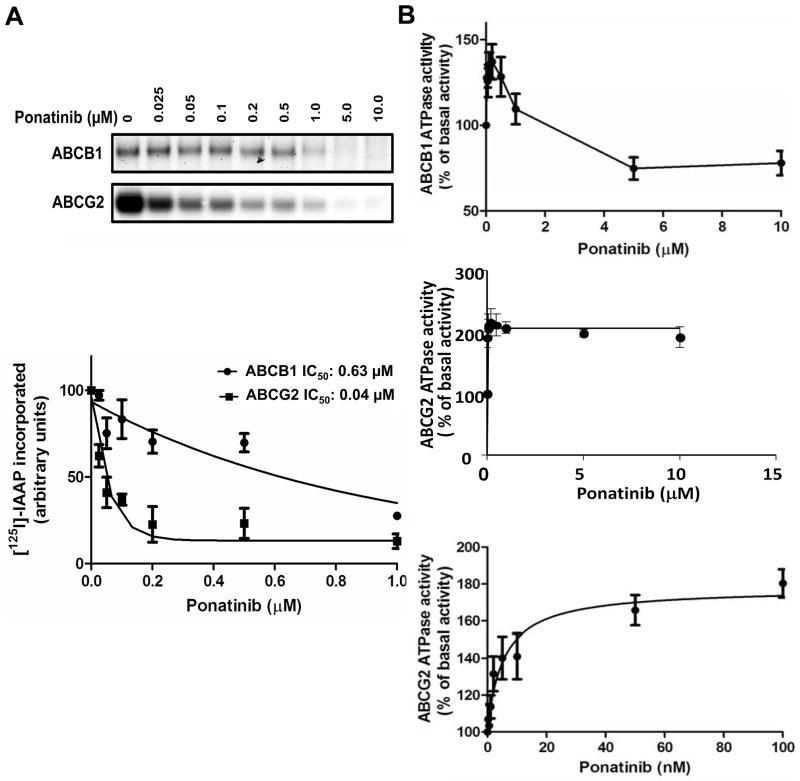
Given that ponatinib inhibited transport by ABCB1 and ABCG2, we studied its binding to their drug substrate sites by measuring its effect on their photolabeling with [125I]-IAAP. Crude membranes from High-Five cells expressing ABCB1 and MCF-7 FLV1000 cells expressing ABCG2 were photo-crosslinked with 3–6 nM [125I]-IAAP (2200 Ci/mmole) in the presence of 0–10 μM ponatinib. Ponatinib inhibited [125I]-IAAP binding to ABCG2 and ABCB1 with IC50’s of 0.04 μM and 0.63 μM, respectively (Figure 2A), indicating strong and weaker binding, respectively, to the ABCG2 and ABCB1 drug substrate sites.
Figure 2.
A. Ponatinib decreases [125I]-IAAP photolabeling of ABCB1 and ABCG2 Crude membranes from High-Five cells expressing ABCB1 and MCF-7 FLV1000 cells expressing ABCG2 were incubated with 0–10 μM ponatinib and [125I]-IAAP. Representative autoradiograms are shown in the upper panel. In the lower panel, incorporation of [125I]-IAAP into the ABCB1 and ABCG2 bands was plotted as a function of ponatinib concentration. Ponatinib inhibits [125I]-IAAP binding to ABCG2 and ABCB1 with IC50’s of 0.04 μM and 0.63 μM, respectively. Values are mean ± SD from three independent experiments. B. Ponatinib increases ABCB1 and ABCG2 ATPase activity. Crude membrane protein from High-Five cells expressing ABCB1 or ABCG2 was incubated with ponatinib at a range of concentrations in the presence or absence of sodium orthovanadate or beryllium sulfate and sodium fluoride, respectively. Average ABCB1 ATPase activity (upper panel), ABCG2 ATPase activity at the same ponatinib concentrations as for ABCB1 (middle panel) and ABCG2 ATPase activity at low ponatinib concentrations (lower panel) from independent duplicate experiments are shown, with standard errors.
Ponatinib stimulates ABCB1 and ABCG2 ATPase activity
Given that ponatinib exhibited binding to the ABCG2 and ABCB1 drug substrate sites, we determined its effect on their ATPase activity, as drug-stimulated ATPase activity is a useful measure of substrate interaction at the drug-binding sites of these transporters (37). Ponatinib stimulated ABCG2 ATPase activity in a concentration-dependent manner (Figure 2B), and stimulated ABCB1 ATPase activity at low, pharmacologically relevant, concentrations, but not at higher concentrations (Figure 2B). Thus ponatinib significantly increased ABCG2 and ABCB1 ATPase activity at lower concentrations, indicating direct interaction at these transporters’ drug substrate binding sites, similarly to other TKIs (38).
Ponatinib cytotoxicty in K562, K562/ABCB1 and K562/ABCG2 cells
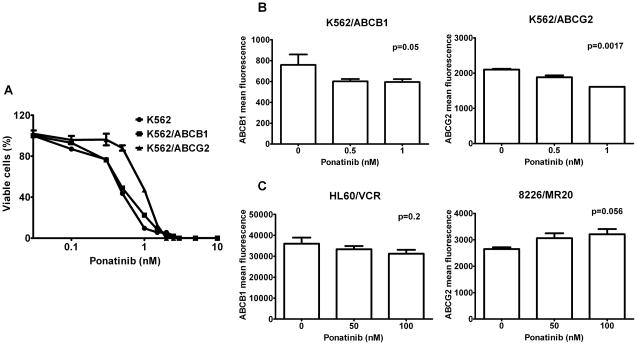
K562, K562/ABCB1 and K562/ABCG2 cells, with the BCR-ABL translocation, were incubated with ponatinib in cell viability assays, yielding IC50s of 0.46, 0.5 and 0.92 nM, respectively (Figure 3A). Thus ABCG2 confers resistance, albeit low-level, to ponatinib, attributable to efflux of this drug by this transporter, but the resistance was modest and was likely attenuated by inhibition of ABCG2-mediated ponatinib transport by ponatinib itself. ABCB1-mediated ponatinib transport was also likely attenuated by ponatinib. An additional possible mechanism of attenuation of resistance is ponatinib-induced decreased cell surface expression of ABCG2 and ABCB1.
Figure 3.
A. Ponatinib cytotoxicity in K562/ABCB1 and K562/ABCG2 cells.
K562, K562/ABCB1 and K562/ABCG2 cells were incubated with ponatinib (0–10 nM) for 96 hours and cell viability was measured using the WST-1 assay. Each data point represents the mean ± SEM of at least three experiments in triplicate. IC50s were 0.46 nM, 0.51 nM and 0.92 nM for K562, K562/ABCB1 and K562/ABCG2 cells, respectively. B. Ponatinib decreases ABCG2 and ABCB1 surface expression on K562/ABCG2 and K562/ABCB1 cells. K562/ABCB1 and K562/ABCG2 cells were treated with ponatinib (0, 0.5 and 1 nM) for 48 hours, then stained with MRK16 and 5D3 antibodies, respectively. Each bar represents the mean ± SEM of fluorescence intensity in at least two experiments in duplicate. C. Ponatinib does not decrease ABCB1 and ABCG2 surface expression on HL60/VCR and 8226/MR20 cells. HL60/VCR and 8226/MR20 cells were treated with ponatinib (0, 50 and 100 nM) for 48 hours, then stained as above.
Ponatinib decreases ABCG2 and ABCB1 cell surface expression
To determine whether ponatinib might also decrease ABCG2 and ABCB1 cell surface expression on K562/ABCG2 and K562/ABCB1 cells, respectively, cells were incubated for 48 hours with ponatinib at 0, 0.5, and 1.0 nM. Surface ABCG2 [mean ± standard error of the mean (SEM), 1612 ± 2 vs. 2103 ± 21; p=0.0017] and ABCB1 (mean ± SEM, 596 ± 16 vs. 759 ± 58; p=0.05) expression decreased on K562/ABCG2 and K562/ABCB1 cells incubated with 1 nM ponatinib, in relation to control (Figure 3B). This effect might contribute to sensitization of K562/ABCG2 and K562/ABCB1 cells to ponatinib, albeit minimally, since decreases in expression were small. In contrast, ABCG2 (mean ± SEM, 2650 ± 70 vs. 3210 ± 197; p=0.056) and ABCB1 (mean ± SEM, 31224 ± 1865 vs. 36030 ± 2898; p=0.2) cell surface expression did not decrease on 8226/MR20 and HL60/VCR cells, which do not express BCR-ABL, incubated for 48 hours with vs. without ponatinib (Figure 3C). ABCG2 and ABCB1 cell surface expression on cells expressing BCR-ABL may decrease due to inhibition of Akt downstream of BCR-ABL (39). Alternatively, 5D3 binding is altered by ABCG2 conformational changes induced by effects on function (40), though this does not explain different effects in K562/ABCG2 and 8226/MR20.
Ponatinib and chemotherapy drug interactions
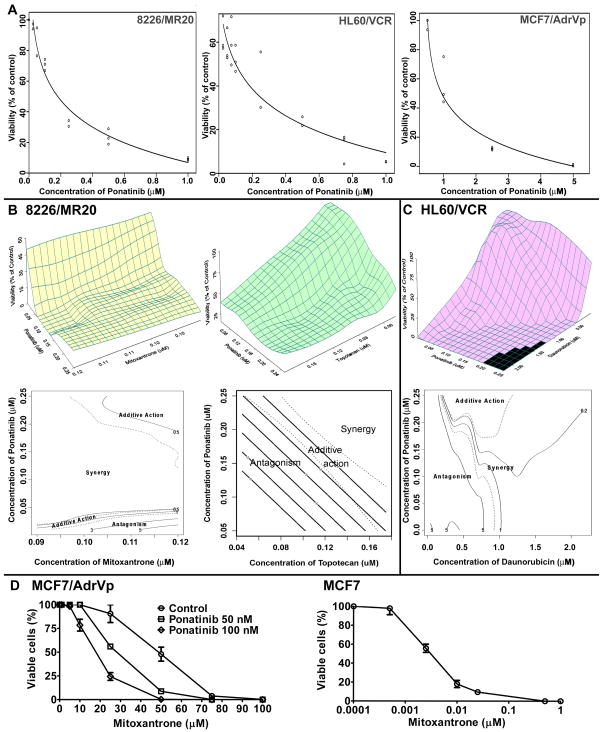
Since ponatinib was cytotoxic to 8226/MR20 and HL60/VCR cells at pharmacologically relevant concentrations, with IC50s of 160 nM and 180 nM, respectively (Figure 4A, left and middle panels), ABCG2 and ABCB1 substrate chemotherapy drug cytotoxicity in the presence of ponatinib was analyzed in drug interaction studies in these cells.
Figure 4.
A. Ponatinib cytotoxicity in HL60/VCR, 8226/MR20 and MCF7/AdrVp cells 8226/MR20, HL60/VCR and MCF7/AdrVp cells were incubated with ponatinib at a range of concentrations for 96 hours and cell viability was measured using the WST-1 assay. Ponatinib was cytotoxic to HL60/VCR and 8226/MR20, but not MCF7/AdrVP, cells at pharmacologically relevant concentrations of 50 to 200 nM. B. Ponatinib and mitoxantrone or topotecan interactions in 8226/MR20 cells. Upper panels, response surfaces of ponatinib with mitoxantrone and with topotecan in 8226/MR20 cells, and lower panels, corresponding contour plots of the interaction index surfaces. Dashed lines indicate 95% confidence surface for additive action (interaction index =1) as described in Results. C. Ponatinib and daunorubicin interactions in HL60/VCR cells. Upper panel, response surface of ponatinib with daunorubicin in HL60/VCR cells, and lower panel, corresponding contour plot of the interaction index surface. D. Mitoxantrone was studied with ponatinib at 0, 50, and 100 nM in MCF7/AdrVP cells, yielding IC50s of 44, 28 and 18 μM, respectively (left panel), while the IC50 of mitoxantrone in parental MCF7 cells was 3.2 nM (right panel).
In the ponatinib and mitoxantrone combination experiments in 8226/MR20 cells, mitoxantrone concentrations were 0.09 to 0.12 μM and ponatinib concentrations 0.001 to 0.25 μM. Based on the maximal power design, 18 mixtures were chosen, with 5 replicates of each mixture, yielding 108 total observations. The maximum and minimum viabilities (% of control) were 55% and 1.28%, mean 12.77% and standard deviation (SD) 13.111. Figure 4B, upper left, shows the response surface of the combination of ponatinib and mitoxantrone in 8226/MR20 cells. With the observations from the combination experiments, the F-test (34) shows that we cannot accept that ponatinib and mitoxantrone have additive activity [F(16, 90)=40.20, p<0.0001]. The contour plot of the interaction index surface (Figure 4B, lower left) shows that the combination of ponatinib and mitoxantrone in 8226/MR20 cells is antagonistic or additive at lower ponatinib concentrations (less than 40 nM, approximately), synergistic at ponatinib concentrations above approximately 40 nM and additive at the highest concentrations of both drugs studied.
In the ponatinib and topotecan combination experiments in 8226/MR20 cells, topotecan concentrations were 0.045 μM to 0.175 μM, and ponatinib concentrations 0.05 μM to 0.25 μM. Based on the maximal power design, 16 mixtures were chosen, with 5 replicates of each mixture, yielding 96 total observations. Maximum and minimum viabilities were 100% and 15%, mean 50.85% and SD 23.00%. Figure 4B, upper right, shows the response surface of the combination of ponatinib and topotecan against 8226/MR20 cells. With the observations from the combination experiments, the F-test (34) shows that we cannot accept that topotecan and ponatinib have additive action [F(14, 78)=14.55, p<0.0001]. The contour plot of the interaction index surface (Figure 4B, lower right) shows that the combination of topotecan with ponatinib is antagonistic in 8226/MR20 cells at lower concentrations of both drugs and synergistic at the higher concentrations tested.
Ponatinib and daunorubicin combination experiments were performed in HL60/VCR cells, overexpressing ABCB1. Daunorubicin concentrations were 0.04 to 2.20 μM and ponatinib concentrations 0.001 to 0.25 μM. Based on the maximal power design, 19 mixtures were chosen, with 4 replicates of each mixture, for 95 total observations. Maximum and minimum viabilities were 100% and 0.1%, mean 53.60% and SD 40.027. Figure 4C, upper panel, shows the response surface of the combination of ponatinib and daunorubicin in HL60/VCR cells. With the observations from the combination experiments, the F-test (34) shows that we cannot accept that ponatinib and daunorubicin have additive effects in HL60/VCR cells [F(17, 76)=85.70, p<0.0001]. The contour plot of the interaction index surface (Figure 4C, lower panel) shows that the combination of ponatinib and daunorubicin is antagonistic in HL60/VCR cells for daunorubicin concentrations less than approximately 0.25 μM for all concentrations of ponatinib studied, but that daunorubicin and ponatinib are synergistic at daunorubicin concentrations of 0.25 to 1.1 μM and ponatinib concentrations greater than approximately 0.18 μM.
Sensitization to chemotherapy drug
As the IC50 of ponatinib in ABCG2-overexpressing MCF7/AdrVP cells was 1.2 μM, and ponatinib was not cytotoxic at the pharmacologically relevant concentrations of 50 nM and 100 nM (Figure 4A, right panel), its effect on sensitivity of these cells to the ABCG2 substrate chemotherapy drug mitoxantrone was studied in a curve shift assay. Mitoxantrone IC50s in the presence of 0, 50 and 100 nM ponatinib were 44, 28 and 18 μM (Figure 4D, left panel), demonstrating concentration-dependent ponatinib sensitization of MCF7/AdrVP cells to mitoxantrone. In contrast, mitoxantrone was cytotoxic to parental MCF7 cells in nanomolar range, with an IC50 of 3.2 nM (Figure 4D, right panel).
Ponatinib enhances apoptosis induction by ABCG2 or ABCB1 substrate chemotherapy drugs in cell lines overexpressing these proteins
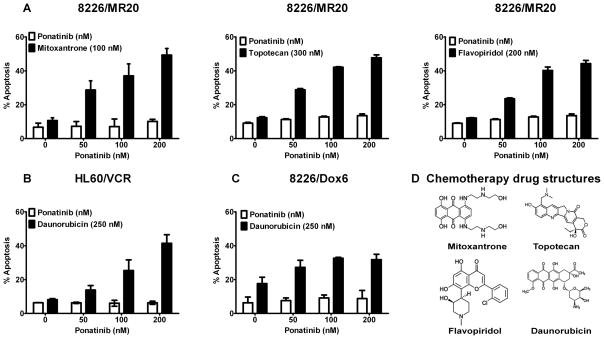
Ponatinib at pharmacologically relevant concentrations of 50, 100 and 200 nM sensitized 8226/MR20 cells, overexpressing ABCG2, to apoptosis induction by 100 nM mitoxantrone, 300 nM topotecan and 200 nM flavopiridol (Figure 5A), and also increased apoptosis induced by daunorubucin (250 nM) in HL60/VCR (Figure 5B) and 8226/Dox6 (Figure 5C) cells, overexpressing ABCB1, in a concentration-dependent manner.
Figure 5. Ponatinib enhances apoptosis in combination with ABCG2 and ABCB1 substrate chemotherapy drugs.
(A) ABCG2-overexpressing 8226/MR20 cells were treated with the ABCG2 substrates mitoxantrone, topotecan and flavopiridol, and ABCB1-overexpressing HL60/VCR (B) and 8226/Dox6 (C) cells were treated with the ABCB1 substrate daunorubicin at fixed concentrations, alone and in combination with 50, 100 and 200 nM ponatinib for 48 hours. Percentages of apoptotic cells are shown. Each bar represents the mean ± SEM of at least three experiments. Mitoxantrone, topotecan, flavopiridol and daunorubicin chemical structures are shown in D.
Cell cycle effects of ponatinib
Finally, TKIs can cause cell cycle arrest, which can result in kinetic resistance when they are combined with chemotherapy drugs. We therefore studied the effect of ponatinib on cell cycle in HL60/VCR and 8226/MR20 cells. Ponatinib had no effect on cell cycle parameters in these drug-resistant cells without BCR-ABL rearrangement or FLT3-ITD (Figure 6). In contrast, as expected, it caused cell cycle arrest and apoptosis in K562 cells, with BCR-ABL rearrangement, and in MV4–11 cells, with FLT3-ITD (Figure 6).
Figure 6. Effect of ponatinib on cell cycle distribution.
HL60/VCR, 8226/MR20, K562 and MV4–11 cells treated with ponatinib at the indicated concentrations for the indicated times were harvested and fixed in 70% ethanol, then stained with propidium iodide and analyzed by flow cytometry.
DISCUSSION
We have demonstrated that the novel BCR-ABL and FLT3 inhibitor ponatinib is a potent inhibitor of drug transport by ABCG2 at pharmacologically relevant concentrations, and synergizes with ABCG2 substrate chemotherapy drugs in inducing cytotoxicity and apoptosis in cells overexpressing ABCG2. Ponatinib also inhibits drug transport by ABCB1, albeit less potently, and synergizes with ABCB1 substrate drugs.
Ponatinib inhibition of drug transport by ABCG2 and ABCB1 appears to occur by direct interaction with these transporters. The results of the [125I]-IAAP photolabeling assay indicated strong binding of ponatinib to the drug substrate site of ABCG2, and weaker binding to that of ABCB1. Additionally, ponatinib stimulated ABCG2 ATPase activity in a concentration-dependent manner and stimulated ABCB1 ATPase activity at low, pharmacologically relevant, concentrations. Thus ponatinib directly interacts at the substrate-binding sites of ABCG2 and ABCB1 at pharmacologically relevant low concentrations. Ambudkar et al. (41) defined three classes of ABCB1 inhibitors based on their effects on ABCB1 ATPase activity. Class I agents stimulate ATPase activity at low concentrations but inhibit it at high concentrations, whereas Class II compounds stimulate ATPase activity in a concentration-dependent manner without any inhibition, and Class III compounds inhibit ATPase activity. We found ponatinib to be a Class I ABCB1 inhibitor.
All currently available BCR-ABL inhibitors are ABCB1 and ABCG2 substrates and/or inhibitors (2–8), and the relative order of potency was recently demonstrated to be nilotinib, then imatinib, then dasatinib (8). Two BCR-ABL inhibitors in current development have also been studied. Bosutinib was found not to be a substrate of ABCB1 or ABCG2 (42), while danusertib was found to be susceptible to resistance mediated by ABCG2 (43). Of note, it is important that drugs be studied at pharmacologically relevant concentrations.
Mechanism and concentration-dependence of inhibition of transport have been most extensively studied for imatinib. Using [125I]-IAAP photolabeling and ATPase assays, Shukla et al. demonstrated that imatinib mesylate is a substrate of both ABCB1 and ABCG2, and that it interacts with both transporters at low micromolar concentrations, indicating relatively high affinity (5). Imatinib appeared not to be transported at higher concentrations, likely because it inhibits its own transport at those concentrations (5), an observation that resolved seemingly contradictory earlier findings (3,4). Nakanishi et al. also demonstrated that imatinib decreased cell surface expression of ABCG2 on K562/BCRP-MX10 cells, expressing BCR-ABL, but not on ABCG2-overexpressing cells without BCR-ABL rearrangement, likely by inhibition of Akt downstream of BCR-ABL (39). We found that ponatinib also decreases surface ABCB1 and ABCG2 expression in cells with, but not without, BCR-ABL rearrangement.
While BCR-ABL inhibitors are used as single agents to treat CML, in the treatment of Ph+ ALL they are administered in combination with chemotherapy drugs including the ABCB1 substrates doxorubicin and vincristine and the ABCG2 substrates 6-mercaptopurine (44) and methotrexate (45). ABCB1 and ABCG2 inhibition by BCR-ABL inhibitors might therefore be beneficial in combination regimens, but might also cause clinically significant drug interactions.
Ponatinib also potently inhibits FLT3, and may thus be applicable in AML therapy (18). FLT3 inhibitors are being combined with chemotherapy to treat AML with FLT3 mutations (9,46–50), and also have activity in AML with wild-type FLT3 (46). Initial FLT3 inhibitors tested included the staurosporine derivatives lestaurtinib and midostaurin, but their use is complicated by high plasma protein binding, cell cycle inhibition (47) and multikinase inhibition potentially causing off-target effects and toxicities. Lestaurtinib was not efficacious following chemotherapy in patients with AML with FLT3 mutations in first relapse in a randomized trial (48), and results of a randomized trial of midostaurin in newly diagnosed AML patients with FLT3 mutations are awaited. Sorafenib has been combined with idarubicin and infusional high-dose cytarabine (49). The selective FLT3 inhibitor AC220 (50) has not yet been combined with chemotherapy. Ponatinib has favorable features, including low plasma protein binding (15), good tolerability (16,17) and, as shown here, no induction of cell cycle arrest in cells with wild-type FLT3. Inhibition of ABCB1 and ABCG2 makes it attractive for further testing in combination with chemotherapy in AML.
Acknowledgments
Grant support: Leukemia and Lymphoma Society Translational Research Award (M.R. Baer), University of Maryland, Baltimore UMMG Cancer Research Grant #CH 649 CRF, State of Maryland Department of Health and Mental Hygiene (DHMH) under the Cigarette Restitution Fund Program (M.R. Baer), NCI Cancer Center Support Grant P30 CA134274 (UMGCC), and NIH Intramural Research Program, National Cancer Institute, Center for Cancer Research (S. Shukla, S.V. Ambudkar).
Footnotes
The authors have no conflicts of interest to disclose.
References
- 1.Szakács G, Paterson JK, Ludwig JA, Booth-Genthe C, Gottesman MM. Targeting multidrug resistance in cancer. Nat Rev Drug Discov Nat Rev Drug Discov. 2006;5:219–34. doi: 10.1038/nrd1984. [DOI] [PubMed] [Google Scholar]
- 2.Illmer T, Schaich M, Platzbecker U, Freiberg-Richter J, Oelschlägel U, von Bonin M, et al. P-glycoprotein-mediated drug efflux is a resistance mechanism of chronic myelogenous leukemia cells to treatment with imatinib mesylate. Leukemia. 2004;18:401–8. doi: 10.1038/sj.leu.2403257. [DOI] [PubMed] [Google Scholar]
- 3.Burger H, van Tol H, Boersma AW, Brok M, Wiemer EA, Stoter G, et al. Imatinib mesylate (STI571) is a substrate for the breast cancer resistance protein (BCRP)/ABCG2 drug pump. Blood. 2004;104:2940–2. doi: 10.1182/blood-2004-04-1398. [DOI] [PubMed] [Google Scholar]
- 4.Houghton PJ, Germain GS, Harwood FC, Schuetz JD, Stewart CF, Buchdunger E, et al. Imatinib mesylate is a potent inhibitor of the ABCG2 (BCRP) transporter and reverses resistance to topotecan and SN-38 in vitro. Cancer Res. 2004;64:2333–7. doi: 10.1158/0008-5472.can-03-3344. [DOI] [PubMed] [Google Scholar]
- 5.Shukla S, Sauna ZE, Ambudkar SV. Evidence for the interaction of imatinib at the transport-substrate site(s) of the multidrug-resistance-linked ABC drug transporters ABCB1 (P-glycoprotein) and ABCG2. Leukemia. 2008;22:445–7. doi: 10.1038/sj.leu.2404897. [DOI] [PubMed] [Google Scholar]
- 6.Tiwari AK, Sodani K, Wang SR, Kuang YH, Ashby CR, Jr, Chen X, et al. Nilotinib (AMN107, Tasigna) reverses multidrug resistance by inhibiting the activity of the ABCB1/Pgp and ABCG2/BCRP/MXR transporters. Biochem Pharmacol. 2009;78:153–61. doi: 10.1016/j.bcp.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 7.Lagas JS, van Waterschoot RA, van Tilburg VA, Hillebrand MJ, Lankheet N, Rosing H, et al. Brain accumulation of dasatinib is restricted by P-glycoprotein (ABCB1) and breast cancer resistance protein (ABCG2) and can be enhanced by elacridar treatment. Clin Cancer Res. 2009;15:2344–51. doi: 10.1158/1078-0432.CCR-08-2253. [DOI] [PubMed] [Google Scholar]
- 8.Dohse M, Scharenberg C, Shukla S, Robey RW, Volkmann T, Deeken JF, et al. Comparison of ATP-binding cassette transporter interactions with the tyrosine kinase inhibitors imatinib, nilotinib, and dasatinib. Drug Metab Dispos. 2010;38:1371–80. doi: 10.1124/dmd.109.031302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hunter HM, Pallis M, Seedhouse CH, Grundy M, Gray C, Russell NH. The expression of P-glycoprotein in AML cells with FLT3 internal tandem duplications is associated with reduced apoptosis in response to FLT3 inhibitors. Br J Haematol. 2004;127:26–33. doi: 10.1111/j.1365-2141.2004.05145.x. [DOI] [PubMed] [Google Scholar]
- 10.Yang JJ, Milton MN, Yu S, Liao M, Liu N, Wu JT, et al. P-glycoprotein and breast cancer resistance protein affect disposition of tandutinib, a tyrosine kinase inhibitor. Drug Metab Lett. 2010;4:201–12. doi: 10.2174/187231210792928279. [DOI] [PubMed] [Google Scholar]
- 11.Lagas JS, van Waterschoot RA, Sparidans RW, Wagenaar E, Beijnen JH, Schinkel AH. Breast cancer resistance protein and P-glycoprotein limit sorafenib brain accumulation. Mol Cancer Ther. 2010;9:319–26. doi: 10.1158/1535-7163.MCT-09-0663. [DOI] [PubMed] [Google Scholar]
- 12.Shukla S, Robey RW, Bates SE, Ambudkar SV. Sunitinib (Sutent, SU11248), a small-molecule receptor tyrosine kinase inhibitor, blocks function of the ATP-binding cassette (ABC) transporters P-glycoprotein (ABCB1) and ABCG2. Drug Metab Dispos. 2009;37:359–65. doi: 10.1124/dmd.108.024612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schlenk RF, Döhner K, Krauter J, Fröhling S, Corbacioglu A, Bullinger L, et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358:1909–1918. doi: 10.1056/NEJMoa074306. [DOI] [PubMed] [Google Scholar]
- 14.O’Hare T, Shakespeare WC, Zhu X, Eide CA, Rivera VM, Wang F, et al. AP24534, a pan-BCR-ABL inhibitor for chronic myeloid leukemia, potently inhibits the T315I mutant and overcomes mutation-based resistance. Cancer Cell. 2009;16:401–12. doi: 10.1016/j.ccr.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang WS, Metcalf CA, Sundaramoorthi R, Wang Y, Zou D, Thomas RM, et al. Discovery of 3-[2-(imidazo[1,2-b]pyridazin-3-yl)ethynyl]-4-methyl-N-{4-[(4-methylpiperazin-1yl)methyl]-3-(trifluoromethyl)phenyl}benzamide (AP24534), a potent, orally active pan-inhibitor of breakpoint cluster region-abelson (BCR-ABL) kinase including the T315I gatekeeper mutant. J Med Chem. 2010;53:4701–19. doi: 10.1021/jm100395q. [DOI] [PubMed] [Google Scholar]
- 16.Cortes J, Talpaz M, Bixby D, Deininger M, Shah N, Flinn IW, et al. A phase 1 trial of oral ponatinib (AP24534) in patients with refractory chronic myelogenous leukemia (CML) and other hematologic malignancies: Emerging safety and clinical response findings. Blood. 2010;116:210. (abstract) [Google Scholar]
- 17.Cortes JE, Kim D-W, Pinilla-Ibarz J, Le Coutre PD, Chuah C, Nicolini FE, et al. Initial findings from the PACE trial: A pivotal phase 2 study of ponatinib in patients with CML and Ph+ ALL resistant or intolerant to dasatinib or nilotinib, or with the T315I mutation. Blood. 2011;118:109. (abstract) [Google Scholar]
- 18.Gozgit JM, Wong MJ, Wardwell S, Tyner JW, Loriaux MM, Mohemmad QK, et al. Potent activity of Ponatinib (AP24534) in models of FLT3-driven acute myeloid leukemia and other hematologic malignancies. Mol Cancer Ther. 2011;10:1028–35. doi: 10.1158/1535-7163.MCT-10-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gozgit JM, Wong MJ, Moran L, Wardwell S, Mohemmad QK, Narasimhan NI, et al. Ponatinib (AP24534), a multi-targeted pan-FGFR inhibitor with activity in multiple FGFR-amplified or mutated cancer models. Mol Cancer Ther. 2012;11:690–9. doi: 10.1158/1535-7163.MCT-11-0450. [DOI] [PubMed] [Google Scholar]
- 20.Ogretmen B, Safa AR. Identification and characterization of the MDR1 promoter-enhancing factor 1 (MEF1) in the multidrug resistant HL60/VCR human acute myeloid leukemia cell line. Biochemistry. 2000;39:194–204. doi: 10.1021/bi991943f. [DOI] [PubMed] [Google Scholar]
- 21.Marsh W, Sicheri D, Center MS. Isolation and characterization of adriamycin-resistant HL60 cells which are not defective in the initial accumulation of the drug. Cancer Res. 1986;46:4053–7. [PubMed] [Google Scholar]
- 22.Hazlehurst LA, Foley NE, Gleason-Guzman MC, Hacker MP, Cress AE, Greenberger LW, et al. Multiple mechanisms confer drug resistance to mitoxantrone in the human 8226 myeloma cell line. Cancer Res. 1999;59:1021–8. [PubMed] [Google Scholar]
- 23.Hafkemeyer P, Licht T, Pastan I, Gottesman MM. Chemoprotection of hematopoietic cells by a mutant P-glycoprotein resistant to a potent chemosensitizer of multidrug-resistant cancers. Hum Gene Ther. 2000;11:555–65. doi: 10.1089/10430340050015743. [DOI] [PubMed] [Google Scholar]
- 24.Yanase K, Tsukahara S, Asada S, Ishikawa E, Imai Y, Sugimoto Y. Gefitinib reverses breast cancer resistance protein-mediated drug resistance. Mol Cancer Ther. 2004;3:1119–25. [PubMed] [Google Scholar]
- 25.Chen YN, Mickley LA, Schwartz AM, Acton EM, Hwang JL, Fojo AT. Characterization of adriamycin-resistant human breast cancer cells which display overexpression of a novel resistance-related membrane protein. J Biol Chem. 1990;265:10073–80. [PubMed] [Google Scholar]
- 26.Robey RW, Medina-Pérez WY, Nishiyama K, Lahusen T, Miyake K, Litman T, et al. Overexpression of the ATP-binding cassette half-transporter, ABCG2 (Mxr/BCrp/ABCP1), in flavopiridol-resistant human breast cancer cells. Clin Cancer Res. 2001;7:145–52. [PubMed] [Google Scholar]
- 27.Minderman H, Suvannasankha A, O’Loughlin KL, Scheffer GL, Scheper RJ, Robey RW, et al. Flow cytometric analysis of breast cancer resistance protein expression and function. Cytometry. 2002;48:59–65. doi: 10.1002/cyto.10111. [DOI] [PubMed] [Google Scholar]
- 28.Robey RW, Steadman K, Polgar O, Morisaki K, Blayney M, Mistry P, et al. Pheophorbide a is a specific probe for ABCG2 function and inhibition. Cancer Res. 2004;64:1242–6. doi: 10.1158/0008-5472.can-03-3298. [DOI] [PubMed] [Google Scholar]
- 29.Minderman H, O’Loughlin KL, Pendyala L, Baer MR. VX-710 (biricodar) increases drug retention and enhances chemosensitivity in resistant cells overexpressing P-glycoprotein, multidrug resistance protein, and breast cancer resistance protein. Clin Cancer Res. 2004;10:1826–34. doi: 10.1158/1078-0432.ccr-0914-3. [DOI] [PubMed] [Google Scholar]
- 30.Sauna ZE, Ambudkar SV. Evidence for a requirement for ATP hydrolysis at two distinct steps during a single turnover of the catalytic cycle of human P-glycoprotein. Proc Natl Acad Sci U S A. 2000;97:2515–20. doi: 10.1073/pnas.97.6.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shukla S, Robey RW, Bates SE, Ambudkar SV. The calcium channel blockers, 1,4-dihydropyridines, are substrates of the multidrug resistance-linked ABC drug transporter, ABCG2. Biochemistry. 2006;45:8940–51. doi: 10.1021/bi060552f. [DOI] [PubMed] [Google Scholar]
- 32.Ambudkar SV. Drug-stimulatable ATPase activity in crude membranes of human MDR1-transfected mammalian cells. Methods Enzymol. 1998;292:504–14. doi: 10.1016/s0076-6879(98)92039-0. [DOI] [PubMed] [Google Scholar]
- 33.Tan M, Fang HB, Tian GL, Houghton PJ. Experimental design and sample size determination for testing synergism in drug combination studies based on uniform measures. Stat Med. 2003;22:2091–2100. doi: 10.1002/sim.1467. [DOI] [PubMed] [Google Scholar]
- 34.Fang HB, Ross DD, Sausville E, Tan M. Experimental design and interaction analysis of combination studies of drugs with log-linear dose responses. Stat Med. 2008;27:3071–83. doi: 10.1002/sim.3204. [DOI] [PubMed] [Google Scholar]
- 35.http://www.umgcc.org/research/biostat_software.htm
- 36.Berenbaum MC. Synergy, additivism and antagonism in immunosuppression. A critical review Clin Exp Immunol. 1977;28:1–18. [PMC free article] [PubMed] [Google Scholar]
- 37.Evans GL, Ni B, Hrycyna CA, Chen D, Ambudkar SV, Pastan I, et al. Heterologous expression systems for P-glycoprotein: E. coli, yeast and baculovirus. J Bioenerg and Biomembr. 1995;27:43–52. doi: 10.1007/BF02110330. [DOI] [PubMed] [Google Scholar]
- 38.Brózik A, Hegedüs C, Erdei Z, Hegedus T, Özvegy-Laczka C, Szakács G, et al. Tyrosine kinase inhibitors as modulators of ATP binding cassette multidrug transporters: substrates, chemosensitizers or inducers of acquired multidrug resistance? Expert Opin Drug Metab Toxicol. 2011;7:623–42. doi: 10.1517/17425255.2011.562892. [DOI] [PubMed] [Google Scholar]
- 39.Nakanishi T, Shiozawa K, Hassel BA, Ross DD. Complex interaction of BCRP/ABCG2 and imatinib in BCR-ABL-expressing cells: BCRP-mediated resistance to imatinib is attenuated by imatinib-induced reduction of BCRP expression. Blood. 2006;108:678–84. doi: 10.1182/blood-2005-10-4020. [DOI] [PubMed] [Google Scholar]
- 40.Ozvegy-Laczka C, Várady G, Köblös G, Ujhelly O, Cervenak J, Schuetz JD, et al. Function-dependent conformational changes of the ABCG2 multidrug transporter modify its interaction with a monoclonal antibody on the cell surface. J Biol Chem. 2005;280:4219–27. doi: 10.1074/jbc.M411338200. [DOI] [PubMed] [Google Scholar]
- 41.Ambudkar SV, Dey S, Hrycyna CA, Ramachandra M, Pastan I, Gottesman MM. Biochemical, cellular, and pharmacological aspects of the multidrug transporter. Annu Rev Pharmacol Toxicol. 1999;39:361–98. doi: 10.1146/annurev.pharmtox.39.1.361. [DOI] [PubMed] [Google Scholar]
- 42.Hegedus C, Ozvegy-Laczka C, Apati A, Magocsi M, Nemet K, Orfi L, et al. Interaction of nilotinib, dasatinib and bosutinib with ABCB1 and ABCG2: implications for altered anti-cancer effects and pharmacological properties. Br J Pharmacol. 2009;158:1153–64. doi: 10.1111/j.1476-5381.2009.00383.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Balabanov S, Gontarewicz A, Keller G, Raddrizzani L, Braig M, Bosotti R, et al. Abcg2 overexpression represents a novel mechanism for acquired resistance to the multi-kinase inhibitor danusertib in BCR-ABL-positive cells in vitro. PLoS One. 2011;6:e19164. doi: 10.1371/journal.pone.0019164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Wolf C, Jansen R, Yamaguchi H, de Haas M, van de Wetering K, Wijnholds J, et al. Contribution of the drug transporter ABCG2 (breast cancer resistance protein) to resistance against anticancer nucleosides. Mol Cancer Ther. 2008;7:3092–102. doi: 10.1158/1535-7163.MCT-08-0427. [DOI] [PubMed] [Google Scholar]
- 45.Volk EL, Farley KM, Wu Y, Li F, Robey RW, Schneider E. Overexpression of wild-type breast cancer resistance protein mediates methotrexate resistance. Cancer Res. 2002;62:5035–40. [PubMed] [Google Scholar]
- 46.Fischer T, Stone RM, Deangelo DJ, Galinsky I, Estey E, Lanza C, et al. Phase IIB trial of oral Midostaurin (PKC412), the FMS-like tyrosine kinase 3 receptor (FLT3) and multi-targeted kinase inhibitor, in patients with acute myeloid leukemia and high-risk myelodysplastic syndrome with either wild-type or mutated FLT3. J Clin Oncol. 2010;28(28):4339–45. doi: 10.1200/JCO.2010.28.9678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Levis M, Pham R, Smith BD, Small D. In vitro studies of a FLT3 inhibitor combined with chemotherapy: sequence of administration is important to achieve synergistic cytotoxic effects. Blood. 2004;104:1145–50. doi: 10.1182/blood-2004-01-0388. [DOI] [PubMed] [Google Scholar]
- 48.Levis M, Ravandi F, Wang ES, Baer MR, Perl A, Coutre S, et al. Results from a randomized trial of salvage chemotherapy followed by lestaurtinib for patients with FLT3 mutant AML in first relapse. Blood. 2011;117:3294–301. doi: 10.1182/blood-2010-08-301796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ravandi F, Cortes JE, Jones D, Faderl S, Garcia-Manero G, Konopleva MY, et al. Phase I/II study of combination therapy with sorafenib, idarubicin, and cytarabine in younger patients with acute myeloid leukemia. J Clin Oncol. 2010;28:1856–62. doi: 10.1200/JCO.2009.25.4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zarrinkar PP, Gunawardane RN, Cramer MD, Gardner MF, Brigham D, Belli B, et al. AC220 is a uniquely potent and selective inhibitor of FLT3 for the treatment of acute myeloid leukemia (AML) Blood. 2009;114:2984–92. doi: 10.1182/blood-2009-05-222034. [DOI] [PMC free article] [PubMed] [Google Scholar]