Abstract
Francisella tularensis is a zoonotic intracellular pathogen and the causative agent of the debilitating febrile illness tularemia. Although natural infections by F. tularensis are sporadic and generally localized, the low infectious dose, with the ability to be transmitted to humans via multiple routes and the potential to cause life-threatening infections, has led to concerns that this bacterium could be used as an agent of bioterror and released intentionally into the environment. Recent studies of F. tularensis and other closely related Francisella species have greatly increased our understanding of mechanisms used by this organism to infect and cause disease within the host. Here, we review the intracellular life cycle of Francisella and highlight key genetic determinants and/or pathways that contribute to the survival and proliferation of this bacterium within host cells.
The bacterium Francisella tularensis causes a potentially life-threatening illness. It can be transmitted to humans by multiple routes, and its ability to proliferate within host cells is essential for virulence.
Francisella are nonmotile, encapsulated, Gram-negative coccobacilli and are facultative intracellular pathogens of humans and many animals. The genus consists of three recognized species: Francisella tularensis, Francisella novicida, and Francisella philomiragia. F. tularensis is highly infectious and causes a potentially debilitating febrile illness known as tularemia. F. novicida and F. philomiragia are rarely pathogenic in man and usually only in individuals who are severely immunocompromised. Although F. novicida has a limited ability to cause disease in humans, this organism continues to serve as an important surrogate model to study aspects of F. tularensis pathogenesis and host response to infection owing to its reduced biosafety requirements, the conserved nature of its genome relative to pathogenic F. tularensis derivatives, its apparently similar intracellular life cycle, and its ability to cause a tularemia-like disease in in vivo model systems of infection.
F. tularensis is transmitted from infected animals to humans by multiple routes and can cause disease of varying severities depending on the portal of entry, infectious dose, and subspecies (biovar) of the infecting strain. Person-to-person transmission of F. tularensis has not yet been reported. F. tularensis subspecies tularensis is the most infectious biovar (ID50 < 10 cfu) and is responsible for most cases of tularemia in North America (Saslaw et al. 1961a). This subspecies causes the most severe disease symptoms and has mortality rates approaching 60% if untreated (Saslaw et al. 1961a,b; Dienst 1963). Type A strain Schu S4 is the most commonly studied isolate from this subspecies. F. tularensis subspecies holarctica has an infectious dose <103 cfu and is the primary cause of tularemia in Europe and other regions in the Northern Hemisphere. Infections by this subspecies are generally associated with milder disease symptoms and are rarely fatal. The live vaccine strain (LVS) is an attenuated isolate derived from this subspecies and was developed in the former Soviet Union. However, it is not licensed for use in the United States. The remaining biovar, F. tularensis subspecies mediastica, only occasionally causes disease in humans.
Infection by F. tularensis occurs primarily after inadvertent exposure to infected wildlife species, most frequently rodents, hares, and rabbits. Transmission to humans occurs via direct contact, through arthropod or insect vectors, by ingestion of contaminated material(s), or by inhalation of aerosolized organisms. Regardless of the entry route, F. tularensis can disseminate from the initial infection site to the lungs where it can cause respiratory tularemia, the most severe form of the disease. The low infectious dose, with the ability to be transmitted to humans via multiple routes, and potential to cause life-threatening illness has resulted in the designation of F. tularensis by the United States Centers for Infectious Disease Control and Prevention as a Category A Select Agent with potential to be weaponized and/or intentionally released into the environment. These characteristics have resulted in a renewed interest in the study of Francisella, including characterization of the F. tularensis life cycle, and identification of bacterial and/or host determinants important for aspects of its pathogenesis.
OVERVIEW OF THE Francisella LIFE CYCLE
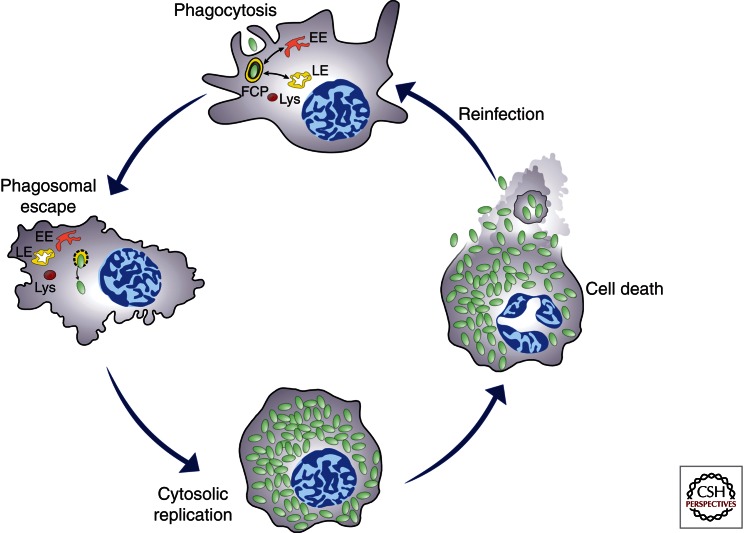
Although Francisella shows an extracellular phase during bacteriemia in mice (Forestal et al. 2007), survival and replication within host cells is thought to be a key aspect of its life cycle. This is exemplified by the ability of various strains of F. tularensis subsp. tularensis and holarctica and of F. novicida to enter, survive, and proliferate within a variety of host-cell types, including macrophages, dendritic cells, polymorphonuclear neutrophils, hepatocytes, endothelial, and type II alveolar lung epithelial cells (Oyston et al. 2004; McCaffrey and Allen 2006; Hall et al. 2007, 2008). Because intracellular proliferation is essential to Francisella virulence, much research has focused on understanding and characterizing specific steps in the intracellular cycle of this bacterium. It has become clear that Francisella survival and proliferation strategies rely on physical escape from its original phagosome and replication in the host-cell cytosol (Fig. 1), making this bacterium a typical cytosol-dwelling pathogen.
Figure 1.
Model of the Francisella intracellular cycle depicting stages that are common to murine and human phagocytes. Upon phagocytosis, bacteria reside in an early phagosome (FCP) that interacts with early (EE) and late (LE) endocytic compartments but not lysosomes (Lys). Bacteria rapidly disrupt the FCP membrane and reach the cytosol where they undergo extensive replication, a process followed by cell death, bacterial release, and subsequent infection.
Francisella ENTRY INTO MAMMALIAN CELLS
Although entry into nonphagocytic cells remains to be further defined, phagocytosis of Francisella by macrophages has been extensively studied and involves the engagement of different phagocytic receptors depending on the bacterium’s opsonization conditions. The mannose receptor (MR) plays a significant role in nonopsonic uptake of F. novicida and F. tularensis strains by either human monocyte-derived macrophages (MDMs), murine bone marrow-derived macrophages (BMMs), or J774A.1 macrophage-like cells (Balagopal et al. 2006; Schulert and Allen 2006; Geier and Celli 2011). Additional, yet-to-be-identified receptors are also likely engaged by nonopsonized Francisella. Serum opsonization, which markedly enhances Francisella uptake (Clemens et al. 2004, 2005; Balagopal et al. 2006; Schulert and Allen 2006; Geier and Celli 2011), mostly redirects the bacterium to the complement receptor CR3 in human and murine macrophages, human neutrophils, and dendritic cells (Balagopal et al. 2006; Ben Nasr et al. 2006; Schulert and Allen 2006; Barker et al. 2009a). The scavenger receptor A (SR-A) (Pierini 2006; Geier and Celli 2011), Fcγ receptors (Balagopal et al. 2006), nucleolin (Barel et al. 2008), and the lung surfactant protein A (SP-A) (Balagopal et al. 2006) have also been implicated to various degrees in uptake of serum-opsonized Francisella by murine or human macrophages. Conversely, Fcγ receptors are the main phagocytic receptors engaged during uptake of antibody-opsonized Francisella, which is also enhanced compared with nonopsonic conditions (Balagopal et al. 2006; Geier and Celli 2011). Opsonophagocytosis of microbes is generally associated with enhanced uptake, as is the case for Francisella. This suggests that this pathogen exposes a limited number of ligands to nonopsonic phagocytic receptors, possibly providing the bacterium with means to limit uptake by bactericidal phagocytes. Additionally, such bacterial ligands may engage receptors that trigger less bactericidal pathways, thereby ensuring better intracellular survival. Consistent with this hypothesis, intracellular proliferation of nonopsonic SchuS4 in murine BMMs is superior to that of opsonized bacteria (Geier and Celli 2011).
THE Francisella-CONTAINING PHAGOSOME
Following uptake, Francisella resides within a phagosome, the Francisella-containing phagosome (FCP), an initial vacuolar compartment along the endocytic degradative pathway that is normally subjected to progressive maturation into a bactericidal phagolysosome. Newly formed FCPs sequentially acquire markers of early endosomes and late endosomes, such as EEA-1, CD63, LAMP-1, LAMP-2, and Rab7 (Clemens et al. 2004, 2009; Santic et al. 2005a, 2008; Checroun et al. 2006; Chong et al. 2008; Wehrly et al. 2009), indicative of a normal maturation process. Yet, such interactions do not proceed toward fusion with lysosomes, as FCPs do not seem to accumulate lysosomal luminal hydrolases, such as cathepsin D, or lysosomal tracers (Anthony et al. 1991; Clemens et al. 2004; Santic et al. 2005b; Bonquist et al. 2008) and bacteria physically disrupt the phagosomal membrane and escape into the host-cell cytosol (see below). Another important feature of phagosomal maturation is the progressive acidification of the phagosomal lumen, via the recruitment of the vacuolar ATPase (v-ATPase), which is both a requirement and a consequence of phagosomal maturation (Huynh and Grinstein 2007). Whether the FCP becomes acidified or not before disruption remains contentious. Some studies have shown that FCPs containing either LVS, Schu S4, or a F. tularensis clinical isolate resist acidification and acquire limited amounts of v-ATPase (Clemens et al. 2004; Bonquist et al. 2008; Cremer et al. 2009), whereas others report that FCPs containing either F. novicida strain U112 or SchuS4 become acidified and acquire the v-ATPase proton pump before phagosomal disruption (Chong et al. 2008; Santic et al. 2008). Additional controversy resides in the fact that studies that found that FCPs become acidified show a role for acidification in optimal phagosomal escape (Chong et al. 2008; Santic et al. 2008), whereas a study in which FCPs were found to resist acidification did not observe any effect of acidification inhibitors (Clemens et al. 2009). It is noteworthy that acidified FCPs are observed using nonopsonic infection conditions, whereas nonacidified FCPs are generated using serum-opsonized Francisella, suggesting that the mode of uptake may affect acidification of the FCP and account for discrepancy in the literature. Although FCP intraluminal pH remains to be measured under different opsonization conditions, these studies nonetheless suggest that physicochemical cues within the FCP contribute to Francisella intracellular fate. This reinforces the importance of the FCP in the Francisella intracellular cycle, in agreement with the fact that it constitutes the site of intracellular induction of virulence genes encoded within the Francisella pathogenicity island (FPI), a genetic loci that has been predicted to encode a type VI secretion system (Nano et al. 2004; Chong et al. 2008; Wehrly et al. 2009).
REACTIVE OXYGEN SPECIES AND INHIBITION OF NADPH OXIDASE ACTIVATION
A major bactericidal function of phagosomes is production of reactive oxygen species (ROS) during the oxidative burst, a process mediated by recruitment and assembly of a multiprotein complex called the NADPH oxidase onto the phagosomal membrane. This enzyme mediates the conversion of molecular oxygen to superoxide (O2−) anions, an ROS that can be subsequently converted to other reactive species including hydrogen peroxide (H2O2), hypochlorous acid (bleach), and peroxynitrite (ONOO−) within certain cell types or under certain conditions (Zahrt and Deretic 2002). The NADPH oxidase complex consists of two integral membrane proteins (gp91phox and p22phox) that together form flavocytochrome b558, and various soluble components that are translocated to the phagosomal membrane on activation. Flavocytochrome b558 contains the redox center of NADPH oxidase and is the docking site for cytosolic subunits of the enzyme complex including p47phox, p40phox, and p67phox. Translocation of these determinants to flavocytochrome b558 requires phosphorylation of p47phox and p40phox; activation of NADPH oxidase also requires association with a small G protein such as Rac1 or Rac2 (Babior 2004).
To ensure their survival, many bacterial pathogens have evolved strategies to resist and/or interfere with ROS generation. For Francisella, these survival strategies have been best elucidated in human polymorphonuclear neutrophils (PMNs) (McCaffrey and Allen 2006; Allen and McCaffrey 2007; Buchan et al. 2009; Schulert et al. 2009; McCaffrey et al. 2010; Mohapatra et al. 2010), a cell type normally encountered by the bacterium in vivo and a potent producer of NADPH oxidase-generated O2− (Allen and McCaffrey 2007). In these cells, F. tularensis and F. novicida have been shown to actively disrupt ROS generation by altering at least two different steps in NADPH oxidase activation: (1) by inhibiting assembly of flavocytochrome b558 in the phagosomal membrane (McCaffrey et al. 2010), and/or (2) by inhibiting the phosphorylation of p47phox and p40phox (McCaffrey et al. 2010; Mohapatra et al. 2010). Inhibition of NADPH oxidase assembly by F. tularensis is dependent on opsonization, and is observed with strains from both F. tularensis subspecies tularensis and holarctica, including the LVS (McCaffrey et al. 2010; Geier and Celli 2011). Interestingly, F. tularensis also possesses the unique ability to inhibit NADPH oxidase activity postcomplex assembly, an event that limits subsequent PMN activation by other stimuli and diminishes ROS production following phagocytosis of antibody-opsonized F. tularensis (McCaffrey et al. 2010). Regardless of mechanism(s), inhibition of NADPH oxidase activity enhances survival of both F. tularensis and F. novicida in this cell type (McCaffrey and Allen 2006; McCaffrey et al. 2010; Mohapatra et al. 2010; Geier and Celli 2011). Similarly, monocytes or MDMs that have been infected with F. tularensis or F. novicida also fail to trigger a robust and/or productive oxidative burst (Schulert et al. 2009; Mohapatra et al. 2010; Geier and Celli 2011). However, in contrast to PMNs, the inability of monocytes and MDMs to produce ROS following infection is likely owing to Francisella engagement of and/or internalization by receptors not coupled to NADPH oxidase activation (Schulert et al. 2009). Consistent with this observation, intracellular growth of F. tularensis or F. novicida is similar following ex vivo infection of monocytes and/or MDMs from wild-type mice compared with p47phox−/− mice (Lindgren et al. 2005; Mohapatra et al. 2010). Furthermore, IgG-opsonization of F. tularensis results in NADPH oxidase-dependent ROS generation in wild-type murine BMMs, an affect that is not observed in macrophages lacking gp91phox (Geier and Celli 2011).
Although the ability of Francisella to inhibit NADPH oxidase activity in neutrophils in vitro has been well established, the importance of neutrophils and/or NADPH oxidase activity in this and other cell types in vivo remains less clear. Antibody-mediated depletion of neutrophils from mice, or targeted recruitment of neutrophils to the lungs using intranasally delivered recombinant MIP-2, a neutrophil-chemotactic chemokine, does not affect bacterial burden or time-to-death kinetics in mice that have been infected with F. tularensis compared with untreated control animals (KuoLee et al. 2011). Similarly, gp91phox−/− mice show only slight differences in time-to-death and overall bacterial burden in target organs of infection compared with wild-type animals following respiratory infection with F. tularensis (KuoLee et al. 2011). Thus, although Francisella may possess specific strategies to inhibit, counteract, or bypass the production of ROS by neutrophils and other phagocytes, the importance of the NADPH oxidase in controlling infection by pathogenic biovars of Francisella appears limited in murine tularemia.
Several bacterial determinants have been implicated in the resistance of Francisella to ROS. For example, Francisella encodes a number of putative acid phosphatases, enzymes that hydrolyze monoesters at acidic pH, that may inhibit the respiratory burst. Purified AcpA, the major contributor of acid phosphatase activity in Francisella, is able to inhibit the oxidative burst in porcine neutrophils that have been activated with exogenous stimulants (Reilly et al. 1996). Recent evidence suggests that AcpA is secreted by F. tularensis and F. novicida strains in vitro (Dai et al. 2011), and is detectable in the cytosol of macrophages infected with F. novicida shortly after infection (Dai et al. 2011). These findings indicate that AcpA would be in a physical location (near the phagosomal membrane or in the cytosol) to interact with membrane-bound or soluble components of the NADPH oxidase (Dai et al. 2011). Consistent with these observations, an F. novicida mutant lacking AcpA is partially compromised in its ability to suppress the oxidative burst following infection of human neutrophils or MDMs (Mohapatra et al. 2010), a phenotype that becomes more pronounced as additional predicted acid phosphatases (acpB, acpC, and hap) are deleted from the bacterium (Mohapatra et al. 2010). Furthermore, loss of AcpA and/or other acid phosphatases reduces the ability of F. novicida to survive following infection of these and other cell types in vitro (Mohapatra et al. 2007, 2008, 2010), and in vivo following intraperitoneal or intranasal infection of mice (Mohapatra et al. 2007, 2008). The importance of acid phosphatases in resistance of F. novicida to ROS can also be seen following treatment of neutrophils and MDMs with reagents that block assembly and/or function of NADPH oxidase, or in macrophages from p47phox−/− mice, where the ability of F. novicida ΔacpA and/or ΔacpABCH mutants to survive intracellularly is improved (Mohapatra et al. 2010). Additionally, ascorbate treatment of murine or human macrophages infected with the LVS blocks acid phosphatase production and inhibits intracellular growth of the bacterium, a phenotype that is dependent on production of AcpA (McRae et al. 2009). Thus, AcpA and other predicted acid phosphatases appear to play an important role in NADPH oxidase inhibition and resistance to ROS following infection by F. novicida.
It is, however, important to note that AcpA and/or the other acid phosphatases appear to play little if any role in the inhibition of NADPH oxidase activity or resistance to ROS in phagocytes that have been infected with human-virulent strains of F. tularensis. An acpA mutant generated in Schu S4 does not trigger a respiratory burst and shows similar growth characteristics to the wild-type parent following infection of human PMNs, even though this bacterium lacks detectable acid phosphatase activity (McCaffrey et al. 2010). Similarly, deletion of acpA, acpB, or acpC alone or sequentially does not alter growth and/or trafficking characteristics of Schu S4 in vitro in murine BMMs or human MDMs, or in vivo following intranasal infection of mice (Child et al. 2010). Thus, acid phosphatases may play a different role in F. novicida compared with F. tularensis, or strains of Francisella that are pathogens of humans may possess/use additional determinants/strategies for ROS resistance that are not conserved or functional in F. novicida.
Apart from acid phosphatases, other determinants have also been implicated in Francisella resistance to ROS. A number of determinants involved in pyrimidine biosynthesis (carA, carB, and pyrB) mediate resistance to ROS in F. tularensis LVS, although it is unclear whether these genes act indirectly by simply enhancing the fitness of the bacterium (Schulert et al. 2009). FevR, a transcription factor that regulates expression of ∼100 genes including those from the FPI (Brotcke and Monack 2008; Meibom et al. 2009), is also required for the inhibition of NADPH oxidase activation and subsequent ROS production in PMNs by both F. tularensis LVS (Buchan et al. 2009) and F. tularensis subspecies tularensis (McCaffrey et al. 2010). FTN1133 (FTL0803) encodes a hypothetical gene product that is required for resistance of F. novicida and F. tularensis LVS to organic hydroperoxides (Llewellyn et al. 2011). Mutants lacking this enzyme are attenuated for survival in macrophages in vitro and show reduced burdens in mice following intradermal infection, a phenotype that is partially or completely ablated in cells or animals lacking gp91phox (Llewellyn et al. 2011). F. tularensis also encodes at least two superoxide dismutases that mediate the detoxification of superoxide to H2O2 and O2. sodB, encoding an FeSOD, and sodC, encoding CuZn SOD, mediate resistance to paraquat, H2O2, and/or pyrogallol in F. tularensis LVS (Bakshi et al. 2006; Melillo et al. 2009), and are required for virulence of F. tularensis LVS in macrophages and mice (Bakshi et al. 2006). The attenuation in virulence observed with these strains in vitro and in vivo is attributable to NADPH oxidase, as addition of NADPH oxidase inhibitors or utilization of cells or mice unable to make this enzyme complex fail to control infection (Melillo et al. 2009). Finally, F. tularensis LVS mutants lacking KatG, a catalase, show increased sensitivity to H2O2 in vitro and are attenuated for virulence in mice in vivo (Lindgren et al. 2007). Interestingly, a similar phenotype is not observed in strains from subspecies tularensis (Lindgren et al. 2007). Thus, Francisella likely uses multiple strategies and a variety of determinants to mediate resistance to ROS.
Francisella PHAGOSOMAL ESCAPE
Several laboratories have described phagosomal escape by various strains of F. tularensis or F. novicida in either human or murine macrophages, and other cell types (Golovliov et al. 2003; Clemens et al. 2004; Santic et al. 2005a, 2008; Checroun et al. 2006; Chong et al. 2008), clearly indicating that this process is conserved and is a hallmark of the Francisella intracellular life cycle. The study of phagosomal escape kinetics have nonetheless brought controversy to the field, as early studies have reported phagosomal disruption and bacterial access to the cytosol that range anywhere from 1 h to 8 h postinfection (Golovliov et al. 2003; Clemens et al. 2004; Santic et al. 2005a; Checroun et al. 2006; McCaffrey and Allen 2006). Such large variations in kinetics are likely attributed to technical differences in the assays and criteria used to assess phagosomal disruption, and also to variations in the models studied between laboratories where different species and strains of Francisella, and different macrophage models were examined. In support of the latter, more recent side-by-side comparisons of phagosomal escape kinetics of F. tularensis and F. novicida strains have highlighted differences in this process (Chong et al. 2008). For example, more rapid phagosomal escape is observed under nonopsonic conditions (Golovliov et al. 2003; Lindgren et al. 2004; Checroun et al. 2006; Chong et al. 2008; Santic et al. 2008; Barker et al. 2009a; Wehrly et al. 2009; Child et al. 2010; Edwards et al. 2010), whereas slower escape kinetics occur when opsonic conditions are used (Clemens et al. 2004; McCaffrey and Allen 2006; Schulert et al. 2009). Recent systematic comparisons of phagosomal escape kinetics by nonopsonic and opsonized SchuS4 in murine macrophages have clarified this issue further by demonstrating that opsonization of Francisella with either complement or antibodies targets bacteria to phagocytic pathways that restrict the extent and timing of phagosomal escape (Geier and Celli 2011). Although the basis for restricted phagosomal escape on serum opsonization is unclear, that occurring on antibody opsonization and bacterial targeting to Fcγ receptors depends on the rapid and transient NADPH oxidase-dependent oxidative burst (Geier and Celli 2011) that the bacteria do not seem to be able to prevent, further confirming that conditions encountered by phagocytosed Francisella within the FCP can influence its intracellular fate.
Francisella phagosomal escape is requisite to intracellular proliferation, as exemplified by the inability of numerous phagosomal escape-deficient mutants to grow within macrophages (Lindgren et al. 2004; Santic et al. 2005b; Bonquist et al. 2008; Barker et al. 2009b; Wehrly et al. 2009; Broms et al. 2012). Given the important role of phagosomal escape in the Francisella intracellular cycle, much effort has been made to identify bacterial factors that contribute to this process. Unlike Listeria monocytogenes or Shigella flexneri, Francisella does not encode lipid hydrolases or phospholipases classically involved in phagosomal membrane disruption (Larsson et al. 2005). However, F. tularensis does appear to express a type VI-like secretion system (encoded by the FPI) (Nano et al. 2004), and numerous genes that are expressed from this genetic locus or that regulate expression from this locus are important for this phagosomal egress (reviewed in Chong et al. 2008). These include iglC and iglD (Lindgren et al. 2004; Santic et al. 2005b; Bonquist et al. 2008; Chong et al. 2008), pdpA (Schmerk et al. 2009), iglI and iglJ (Barker et al. 2009b; McCaffrey et al. 2010), vgrG (Barker et al. 2009b; Broms et al. 2012), dotU (Broms et al. 2012), mglA (Baron and Nano 1998; Santic et al. 2005b; Bonquist et al. 2008), fevR (Brotcke and Monack 2008; Buchan et al. 2009; Wehrly et al. 2009), and migR (Buchan et al. 2009). However, other determinants not linked to the FPI have also been implicated in phagosomal escape by Francisella including the acid phosphatases of F. novicida (AcpA, AcpB, AcpC, and Hap) (Mohapatra et al. 2008), pyrimidine biosynthetic genes (carA, carB, and pyrB) (Schulert et al. 2009), as well as several genes of unknown function including FTT1103 (Qin and Mann 2006; Qin et al. 2009) and FTT1676 (Wehrly et al. 2009). Thus, it remains unclear whether Francisella use a single mechanism for escape from the phagosome; further delineation of this process, including the genes required, remains a top priority in the field given its importance for subsequent stages of Francisella survival.
Francisella REPLICATION IN THE CYTOSOL
Besides phagosomal escape, the ability of Francisella to proliferate within the host-cell cytosol is another key aspect of its intracellular life cycle, as exemplified by the avirulence in mice of the large number of identified replication-deficient mutants (Brotcke et al. 2006; Tempel et al. 2006; Su et al. 2007; Weiss et al. 2007; Alkhuder et al. 2009; Wehrly et al. 2009). Although the cytosol may be viewed as permissive for bacterial proliferation, compared with a vacuole along the endocytic degradative pathway, only cytosol-adapted pathogens can readily proliferate within this host-cell compartment (O’Riordan and Portnoy 2002), through the expression of dedicated factors and interactions with host components that either favor or antagonize bacterial growth. Because phagosomal escape is a prerequisite to cytosolic replication, identifying bacterial factors that are specifically required for growth in the cytosol (and not for phagosomal escape) via mutagenesis and in vitro and/or in vivo screens has been difficult. In general, identification of specific Francisella cytosolic replication-defective mutants has required use of more specialized secondary readouts including those based on electron and/or immunofluorescence microscopy. To date, only purine biosynthetic genes (purMCD) (Pechous et al. 2006, 2008), a γ-glutamyl transpeptidase (ggt) (Alkhuder et al. 2009), and several genes of unknown function including FTT0369c/dipA (Wehrly et al. 2009; Chong et al. 2012), FTT0989 (Brotcke et al. 2006), and ripA (Fuller et al. 2008), have been identified as being specifically required for cytosolic replication by Francisella. However, it is likely that there remain a large number of additional gene products that also contribute to this process (reviewed in Chong et al. 2008).
Although Francisella have evolved mechanisms to adapt to a cytosolic lifestyle, some host factors have been identified that either contribute or interfere with cytosolic proliferation. Recently, Akimana et al. have identified through a genome-wide RNAi screen in Drosophila melanogaster S2R+ cells the type III PI4-kinase α subunit PI4KC and the ubiquitin-specific peptidase USP22 as required for cytosolic replication of F. novicida (Akimana et al. 2010). Whether the same host proteins are required for replication of virulent F. tularensis and what their function in bacterial replication is remains to be established. Nonetheless, these findings argue that this bacterium modulates specific host pathways associated with lipid signaling and ubiquitin systems to foster replication.
Not surprisingly, innate immune mechanisms have been shown to negatively control intracellular proliferation of Francisella. The cytokine interferon (IFN)-γ is essential to control primary infections (Leiby et al. 1992; Elkins et al. 1996), and has long been known to restrict intracellular growth of various Francisella strains in different host-cell models (Anthony et al. 1992; Fortier et al. 1992; Polsinelli et al. 1994), although the effector mechanisms are unclear. IFN-γ control of either LVS or Schu S4 intracellular growth in murine peritoneal exudate cells (PECs) seems to depend on generation of nitric oxide by the inducible nitric oxide synthase (iNOS) (Lindgren et al. 2005, 2007), but not in murine alveolar macrophages (Polsinelli et al. 1994), nor in either human blood-derived or murine bone marrow-derived primary macrophages (Edwards et al. 2010). Nonetheless, independent studies have established that cytosolic replication rather than phagosomal escape of F. tularensis is the target of IFN-γ, as this cytokine does not affect phagosomal escape of either LVS or SchuS4 in either J774A.1 cells and human or murine primary macrophages (Bonquist et al. 2008; Edwards et al. 2010), but restricts cytosolic proliferation (Edwards et al. 2010). These findings are, however, inconsistent with those of Santic et al. (2005a), who reported that IFN-γ activation of human blood-derived macrophages prevented phagosomal escape of F. novicida. Although variations in the experimental designs of these studies may account for such contrasting conclusions, and further work is required to reconcile these results and clarify the effector mechanisms induced by IFN-γ, these findings clearly illustrate that phagocytes express specific mechanisms that control cytosolic proliferation of Francisella. In addition to the effect of IFN-γ on Francisella intracellular growth, alternative activation of macrophages by mast cells via interleukin-4 (IL-4) controls intramacrophage growth of LVS (Ketavarapu et al. 2008). Although increased ATP production, prolonged FCP acidification, and up-regulation of the MR have been invoked in this control (Rodriguez et al. 2010), whether IL-4 affects Francisella phagosomal escape and/or cytosolic replication remains to be examined in more depth.
Francisella AND INNATE IMMUNE RECOGNITION
Francisella phagocytosis by macrophages and intracellular trafficking through vacuolar and cytosolic compartments likely subjects the bacterium to innate immune recognition by various pattern recognition receptors (PRRs) either located on the cell surface or endosomes, such as the Toll-like receptor (TLRs), or in the cytosolic compartment such as Nod-like receptors (NLRs). TLRs or NLRs typically recognize pathogen-associated molecular patterns (PAMPs) or danger-associated molecular patterns (DAMPs) to trigger proinflammatory responses, among which the activation of the inflammasome, a cytosolic molecular complex, orchestrates caspase-1 activation, processing and release of the proinflammatory cytokines IL-1β and IL-18, and cell death via pyroptosis.
Because of the unique structure of its lipid A moiety (Hajjar et al. 2006), Francisella LPS shows very low endotoxicity and the bacterium does not induce TLR4-mediated signaling. Instead, LVS significantly stimulates TLR2-mediated signaling resulting in proinflammatory cytokine production (Katz et al. 2006; Cole et al. 2007), although it is also capable of subsequently suppressing TLR-mediated signaling and secretion of proinflammatory cytokines induced by other agonists (Telepnev et al. 2003). This ability of Francisella to suppress or dampen proinflammatory responses has been extended to the virulent subspecies tularensis in human dendritic cells, where infection impairs their activation and function (Bosio and Dow 2005; Chase et al. 2009), further supporting the notion that Francisella interferes with immune detection and responses.
INFLAMMASOME ACTIVATION
Given its cytosolic location, Francisella has become a model pathogen to study the function of inflammasomes, which has also expanded our understanding of how this bacterium may interfere with immune recognition. Release of F. novicida in the macrophage cytosol is sensed in a type I IFN-dependent manner (Henry et al. 2007), and activates the absent in melanoma 2 (AIM2)-containing inflammasome (Fernandes-Alnemri et al. 2010; Rathinam et al. 2010; Jones et al. 2011), a type I IFN-inducible complex whose activation induces secretion of the proinflammatory cytokines IL1β and pro-IL18 and pyroptosis (Mariathasan et al. 2005). AIM2 inflammasome activation is potentiated by TLR2 signaling (Jones et al. 2010) and is required in vivo in mice to control bacterial burden by eliminating infected macrophages that constitute the Francisella proliferation niche (Mariathasan et al. 2005; Fernandes-Alnemri et al. 2010; Jones et al. 2011). Consistent with AIM2 recognizing cytosolic double-stranded DNA (Rathinam et al. 2010), F. novicida activation of the AIM2 inflammasome is associated with bacterial release of DNA through lysis in the cytosol (Jones et al. 2011). These studies have focused on murine systems and the nonhuman pathogenic F. novicida species, so an important question that remains unanswered is whether the conclusions drawn from these studies also apply to humans and virulent strains. For example, recent evidence indicates that infection of human cells with F. novicida, LVS, and the highly virulent F. tularensis Schu S4 strains activate the NLRP3 inflammasome in addition to the AIM2 inflammasome (Atianand et al. 2011), suggesting some functional differences in the innate immune complexes involved in Francisella recognition between mice and humans. Moreover, several studies using F. tularensis subspecies suggest that infections of BMMs with the LVS strain (Huang et al. 2010; Ulland et al. 2010), or of human primary macrophages with virulent Schu S4 (Lindemann et al. 2010) do induce little cytotoxicity compared with F. novicida, raising the question of relevance of cytotoxicity in infections with virulent tularensis strains.
Given the high adaptation of Francisella to the cytosol and the potential of proinflammatory pathways to restrict its proliferation, this bacterium has likely evolved mechanisms to counteract inflammasome activation to preserve its replication niche. In support of this concept, many laboratories have identified hypercytotoxic mutants of F. novicida (Brotcke et al. 2006; Hager et al. 2006; Weiss et al. 2007; Lai et al. 2010), LVS (Huang et al. 2010; Ulland et al. 2010), and Schu S4 (Lindemann et al. 2010), which typically induced increased inflammasome-dependent cell death and secretion of proinflammatory cytokines in either mouse or human macrophages. Intriguingly, the hypercytotoxic phenotype results from the interruption of genes encoding a variety of proteins, such as the oligopeptide permease OppB (Brotcke et al. 2006), the metallopeptidase PepO (Brotcke et al. 2006; Hager et al. 2006), the inner membrane protein RipA (Huang et al. 2010), the IclR family transcriptional factor FTT0784 and an unknown protein FTT0584 (Weiss et al. 2007), and several proteins involved in biosynthesis of type IV pili, the LPS and the O-antigen polysaccharidic capsule (Huang et al. 2010; Ulland et al. 2010), bringing some confusion as to whether Francisella specifically suppress inflammasome activation. The hypothesis of Francisella evasion of inflammasome activation was recently challenged by Peng et al., who showed that many previously identified hypercytotoxic mutants of F. novicida and LVS are deficient in membrane-associated proteins and lyse more extensively in the cytosol than wild-type bacteria, consequently triggering increased inflammasome activation, secretion of proinflammatory cytokines, and pyroptosis (Peng et al. 2011). Although this study does not completely rule out the existence of Francisella inflammasome suppression factors, it elegantly argues that most genes identified as potential suppressors of inflammasome activation do not act specifically and that the hypercytotoxic phenotype is in many cases an indirect consequence of bacterial cell wall fragilization and enhanced release of PAMPs. Because this study has been performed in F. novicida and LVS, it will also be interesting to verify that these conclusions also apply to virulent strains.
XENOPHAGIC CAPTURE
Another intracellular innate immune defense mechanism is xenophagy, a selective process of capture of intracellular microorganisms within a double-membrane-bound vacuole, the autophagosome, for delivery to and degradation into the lysosomal compartment (Levine et al. 2011). Francisella cytosolic location makes this pathogen an ideal substrate for selective autophagic recognition and capture, a process that shares many molecular machineries with nonselective, canonical autophagy. Yet, surprisingly, this bacterium is able to replicate within murine and human macrophages over long periods of time (>18 h) without evidence of a xenophagic response, suggesting it is either capable of avoiding recognition or inhibiting autophagic processes. A belated autophagic response has nonetheless been observed in murine BMMs that encloses a fraction of cytosolic Francisella into large vacuoles with autophagic features (Checroun et al. 2006), but the role of these vacuoles and their relevance is unclear, as they do not form in human macrophages (Akimana et al. 2010; Edwards et al. 2010) and may not constitute a bona fide stage of the bacterium’s intracellular cycle. Current evidence that Francisella may interfere with the autophagy pathway stems from the down-regulation of several autophagy-related genes, such as beclin1, ATG5, ATG12, ATG16L, ATG7, and ATG4a in either Schu S4- or F. novicida-infected human monocytes (Butchar et al. 2008; Cremer et al. 2009), which suggests that Francisella may suppress an autophagic response at the gene expression level. Yet, it remains to be shown whether down-regulation of some autophagy gene during infection is sufficient to block this constitutive pathway rapidly enough to prevent bacteria that have reached the cytosol from being targeted. Interestingly, a ΔdipA (FTT0369c) deletion mutant of SchuS4, which is deficient in cytosolic replication, is eventually captured and cleared by autophagy following a loss of viability in the cytosol (Chong et al. 2012), suggesting that viable Francisella may possess mechanisms that prevent their recognition by the autophagic machinery. Additionally, whether Francisella infection negatively modulates nonselective autophagy also needs to be examined to further understand Francisella potential interference with this degradative pathway.
CONCLUDING REMARKS
The ability of Francisella to survive and replicate within phagocytes and other host cells following infection is a key aspect of its life cycle and represents an essential step required by this bacterium to cause disease within the host. Alterations in mechanisms that mediate this ability, including the engagement of appropriate receptors, resistance to ROS, escape from the phagosome, replication within the cytosol, and/or avoidance of innate immune recognition or other defense mechanisms, can lead to altered survival characteristics and potential attenuation of F. tularensis in virulence in vitro and/or in vivo. Continued study of the genetic determinants contributing to these mechanisms will be essential for the development of therapeutics or vaccines that are able to specifically inhibit or prevent disease caused by this bacterium.
ACKNOWLEDGMENTS
The authors apologize to those investigators in the field whose excellent work we were not able to reference. We are grateful to Anita Mora at the Rocky Mountain Laboratory for her assistance with graphic art. T.C.Z. acknowledges support from the National Institutes of Health, National Institute of Allergy and Infectious Diseases (NIAID) (1R21 AI097597-01A1), and the NIAID’s Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research Program. Membership within and support from the Region V Great Lakes Regional Center of Excellence (National Institutes of Health Award 2-U54-AI-057153) is also acknowledged. J.C. acknowledges support from the Intramural Research Program of the National Institutes of Health, National Institute of Allergy and Infectious Diseases.
Footnotes
Editors: Pascale Cossart and Stanley Maloy
Additional Perspectives on Bacterial Pathogenesis available at www.perspectivesinmedicine.org
REFERENCES
- Akimana C, Al-Khodor S, Abu Kwaik Y 2010. Host factors required for modulation of phagosome biogenesis and proliferation of Francisella tularensis within the cytosol. PLoS ONE 5: e11025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkhuder K, Meibom KL, Dubail I, Dupuis M, Charbit A 2009. Glutathione provides a source of cysteine essential for intracellular multiplication of Francisella tularensis. PLoS Pathog 5: e1000284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen LA, McCaffrey RL 2007. To activate or not to activate: Distinct strategies used by Helicobacter pylori and Francisella tularensis to modulate the NADPH oxidase and survive in human neutrophils. Immunol Rev 219: 103–117 [DOI] [PubMed] [Google Scholar]
- Anthony LD, Burke RD, Nano FE 1991. Growth of Francisella spp. in rodent macrophages. Infect Immun 59: 3291–3296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony LS, Morrissey PJ, Nano FE 1992. Growth inhibition of Francisella tularensis live vaccine strain by IFN-γ-activated macrophages is mediated by reactive nitrogen intermediates derived from L-arginine metabolism. J Immunol 148: 1829–1834 [PubMed] [Google Scholar]
- Atianand MK, Duffy EB, Shah A, Kar S, Malik M, Harton JA 2011. Francisella tularensis reveals a disparity between human and mouse NLRP3 inflammasome activation. J Biol Chem 286: 39033–39042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babior BM 2004. NADPH oxidase. Curr Opin Immunol 16: 42–47 [DOI] [PubMed] [Google Scholar]
- Bakshi CS, Malik M, Regan K, Melendez JA, Metzger DW, Pavlov VM, Sellati TJ 2006. Superoxide dismutase B gene (sodB)-deficient mutants of Francisella tularensis demonstrate hypersensitivity to oxidative stress and attenuated virulence. J Bacteriol 188: 6443–6448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balagopal A, MacFarlane AS, Mohapatra N, Soni S, Gunn JS, Schlesinger LS 2006. Characterization of the receptor-ligand pathways important for entry and survival of Francisella tularensis in human macrophages. Infect Immun 74: 5114–5125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barel M, Hovanessian AG, Meibom K, Briand JP, Dupuis M, Charbit A 2008. A novel receptor-ligand pathway for entry of Francisella tularensis in monocyte-like THP-1 cells: Interaction between surface nucleolin and bacterial elongation factor Tu. BMC Microbiol 8: 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker JH, McCaffrey RL, Baman NK, Allen LA, Weiss JP, Nauseef WM 2009a. The role of complement opsonization in interactions between F. tularensis subsp. novicida and human neutrophils. Microbes Infect 11: 762–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker JR, Chong A, Wehrly TD, Yu JJ, Rodriguez SA, Liu J, Celli J, Arulanandam BP, Klose KE 2009b. The Francisella tularensis pathogenicity island encodes a secretion system that is required for phagosome escape and virulence. Mol Microbiol 74: 1459–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron GS, Nano FE 1998. MglA and MglB are required for the intramacrophage growth of Francisella novicida. Mol Microbiol 29: 247–259 [DOI] [PubMed] [Google Scholar]
- Ben Nasr A, Haithcoat J, Masterson JE, Gunn JS, Eaves-Pyles T, Klimpel GR 2006. Critical role for serum opsonins and complement receptors CR3 (CD11b/CD18) and CR4 (CD11c/CD18) in phagocytosis of Francisella tularensis by human dendritic cells (DC): Uptake of Francisella leads to activation of immature DC and intracellular survival of the bacteria. J Leukoc Biol 80: 774–786 [DOI] [PubMed] [Google Scholar]
- Bonquist L, Lindgren H, Golovliov I, Guina T, Sjostedt A 2008. MglA and Igl proteins contribute to the modulation of Francisella tularensis live vaccine strain-containing phagosomes in murine macrophages. Infect Immun 76: 3502–3510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosio CM, Dow SW 2005. Francisella tularensis induces aberrant activation of pulmonary dendritic cells. J Immunol 175: 6792–6801 [DOI] [PubMed] [Google Scholar]
- Broms JE, Meyer L, Lavander M, Larsson P, Sjostedt A 2012. DotU and VgrG, core components of type VI secretion systems, are essential for Francisella LVS pathogenicity. PLoS ONE 7: e34639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brotcke A, Monack DM 2008. Identification of fevR, a novel regulator of virulence gene expression in Francisella novicida. Infect Immun 76: 3473–3480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brotcke A, Weiss DS, Kim CC, Chain P, Malfatti S, Garcia E, Monack DM 2006. Identification of MglA-regulated genes reveals novel virulence factors in Francisella tularensis. Infect Immun 74: 6642–6655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan BW, McCaffrey RL, Lindemann SR, Allen LA, Jones BD 2009. Identification of migR, a regulatory element of the Francisella tularensis live vaccine strain iglABCD virulence operon required for normal replication and trafficking in macrophages. Infect Immun 77: 2517–2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butchar JP, Cremer TJ, Clay CD, Gavrilin MA, Wewers MD, Marsh CB, Schlesinger LS, Tridandapani S 2008. Microarray analysis of human monocytes infected with Francisella tularensis identifies new targets of host response subversion. PLoS ONE 3: e2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase JC, Celli J, Bosio CM 2009. Direct and indirect impairment of human dendritic cell function by virulent Francisella tularensis Schu S4. Infect Immun 77: 180–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Checroun C, Wehrly TD, Fischer ER, Hayes SF, Celli J 2006. Autophagy-mediated reentry of Francisella tularensis into the endocytic compartment after cytoplasmic replication. Proc Natl Acad Sci 103: 14578–14583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Child R, Wehrly TD, Rockx-Brouwer D, Dorward DW, Celli J 2010. Acid phosphatases do not contribute to the pathogenesis of type A Francisella tularensis. Infect Immun 78: 59–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong A, Wehrly TD, Nair V, Fischer ER, Barker JR, Klose KE, Celli J 2008. The early phagosomal stage of Francisella tularensis determines optimal phagosomal escape and Francisella pathogenicity island protein expression. Infect Immun 76: 5488–5499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong A, Wehrly TD, Child R, Hansen B, Hwang S, Virgin HW, Celli J 2012. Cytosolic clearance of replication-deficient mutants reveals Francisella tularensis interactions with the autophagic pathway. Autophagy 8: 1342–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens DL, Lee BY, Horwitz MA 2004. Virulent and avirulent strains of Francisella tularensis prevent acidification and maturation of their phagosomes and escape into the cytoplasm in human macrophages. Infect Immun 72: 3204–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens DL, Lee BY, Horwitz MA 2005. Francisella tularensis enters macrophages via a novel process involving pseudopod loops. Infect Immun 73: 5892–5902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens DL, Lee BY, Horwitz MA 2009. Francisella tularensis phagosomal escape does not require acidification of the phagosome. Infect Immun 77: 1757–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole LE, Shirey KA, Barry E, Santiago A, Rallabhandi P, Elkins KL, Puche AC, Michalek SM, Vogel SN 2007. Toll-like receptor 2-mediated signaling requirements for Francisella tularensis live vaccine strain infection of murine macrophages. Infect Immun 75: 4127–4137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer TJ, Amer A, Tridandapani S, Butchar JP 2009. Francisella tularensis regulates autophagy-related host cell signaling pathways. Autophagy 5: 125–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai S, Mohapatra NP, Schlesinger LS, Gunn JS 2011. The acid phosphatase AcpA is secreted in vitro and in macrophages by Francisella spp. Infect Immun 80: 1088–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dienst FT Jr 1963. Tularemia: A perusal of three hundred thirty-nine cases. J La State Med Soc 115: 114–127 [PubMed] [Google Scholar]
- Edwards JA, Rockx-Brouwer D, Nair V, Celli J 2010. Restricted cytosolic growth of Francisella tularensis subsp. tularensis by IFN-γ activation of macrophages. Microbiology 156: 327–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkins KL, Rhinehart-Jones TR, Culkin SJ, Yee D, Winegar RK 1996. Minimal requirements for murine resistance to infection with Francisella tularensis LVS. Infect Immun 64: 3288–3293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes-Alnemri T, Yu JW, Juliana C, Solorzano L, Kang S, Wu J, Datta P, McCormick M, Huang L, McDermott E, et al. 2010. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat Immunol 11: 385–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forestal CA, Malik M, Catlett SV, Savitt AG, Benach JL, Sellati TJ, Furie MB 2007. Francisella tularensis has a significant extracellular phase in infected mice. J Infect Dis 196: 134–137 [DOI] [PubMed] [Google Scholar]
- Fortier AH, Polsinelli T, Green SJ, Nacy CA 1992. Activation of macrophages for destruction of Francisella tularensis: Identification of cytokines, effector cells, and effector molecules. Infect Immun 60: 817–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller JR, Craven RR, Hall JD, Kijek TM, Taft-Benz S, Kawula TH 2008. RipA, a cytoplasmic membrane protein conserved among Francisella species, is required for intracellular survival. Infect Immun 76: 4934–4943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geier H, Celli J 2011. Phagocytic receptors dictate phagosomal escape and intracellular proliferation of Francisella tularensis. Infect Immun 79: 2204–2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovliov I, Baranov V, Krocova Z, Kovarova H, Sjostedt A 2003. An attenuated strain of the facultative intracellular bacterium Francisella tularensis can escape the phagosome of monocytic cells. Infect Immun 71: 5940–5950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager AJ, Bolton DL, Pelletier MR, Brittnacher MJ, Gallagher LA, Kaul R, Skerrett SJ, Miller SI, Guina T 2006. Type IV pili-mediated secretion modulates Francisella virulence. Mol Microbiol 62: 227–237 [DOI] [PubMed] [Google Scholar]
- Hajjar AM, Harvey MD, Shaffer SA, Goodlett DR, Sjostedt A, Edebro H, Forsman M, Bystrom M, Pelletier M, Wilson CB, et al. 2006. Lack of in vitro and in vivo recognition of Francisella tularensis subspecies lipopolysaccharide by Toll-like receptors. Infect Immun 74: 6730–6738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JD, Craven RR, Fuller JR, Pickles RJ, Kawula TH 2007. Francisella tularensis replicates within alveolar type II epithelial cells in vitro and in vivo following inhalation. Infect Immun 75: 1034–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JD, Woolard MD, Gunn BM, Craven RR, Taft-Benz S, Frelinger JA, Kawula TH 2008. Infected-host-cell repertoire and cellular response in the lung following inhalation of Francisella tularensis Schu S4, LVS, or U112. Infect Immun 76: 5843–5852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry T, Brotcke A, Weiss DS, Thompson LJ, Monack DM 2007. Type I interferon signaling is required for activation of the inflammasome during Francisella infection. J Exp Med 204: 987–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang MT, Mortensen BL, Taxman DJ, Craven RR, Taft-Benz S, Kijek TM, Fuller JR, Davis BK, Allen IC, Brickey WJ, et al. 2010. Deletion of ripA alleviates suppression of the inflammasome and MAPK by Francisella tularensis. J Immunol 185: 5476–5485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh KK, Grinstein S 2007. Regulation of vacuolar pH and its modulation by some microbial species. Microbiol Mol Biol Rev 71: 452–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JW, Kayagaki N, Broz P, Henry T, Newton K, O’Rourke K, Chan S, Dong J, Qu Y, Roose-Girma M, et al. 2010. Absent in melanoma 2 is required for innate immune recognition of Francisella tularensis. Proc Natl Acad Sci 107: 9771–9776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JW, Broz P, Monack DM 2011. Innate immune recognition of Francisella tularensis: Activation of type-I interferons and the inflammasome. Front Microbiol 2: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz J, Zhang P, Martin M, Vogel SN, Michalek SM 2006. Toll-like receptor 2 is required for inflammatory responses to Francisella tularensis LVS. Infect Immun 74: 2809–2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketavarapu JM, Rodriguez AR, Yu JJ, Cong Y, Murthy AK, Forsthuber TG, Guentzel MN, Klose KE, Berton MT, Arulanandam BP 2008. Mast cells inhibit intramacrophage Francisella tularensis replication via contact and secreted products including IL-4. Proc Natl Acad Sci 105: 9313–9318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- KuoLee R, Harris G, Conlan JW, Chen W 2011. Role of neutrophils and NADPH phagocyte oxidase in host defense against respiratory infection with virulent Francisella tularensis in mice. Microbes Infect 13: 447–456 [DOI] [PubMed] [Google Scholar]
- Lai XH, Shirley RL, Crosa L, Kanistanon D, Tempel R, Ernst RK, Gallagher LA, Manoil C, Heffron F 2010. Mutations of Francisella novicida that alter the mechanism of its phagocytosis by murine macrophages. PLoS ONE 5: e11857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson P, Oyston PC, Chain P, Chu MC, Duffield M, Fuxelius HH, Garcia E, Halltorp G, Johansson D, Isherwood KE, et al. 2005. The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nat Genet 37: 153–159 [DOI] [PubMed] [Google Scholar]
- Leiby DA, Fortier AH, Crawford RM, Schreiber RD, Nacy CA 1992. In vivo modulation of the murine immune response to Francisella tularensis LVS by administration of anticytokine antibodies. Infect Immun 60: 84–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Mizushima N, Virgin HW 2011. Autophagy in immunity and inflammation. Nature 469: 323–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindemann SR, Peng K, Long ME, Hunt JR, Apicella MA, Monack DM, Allen LA, Jones BD 2010. Francisella tularensis Schu S4 O-antigen and capsule biosynthesis gene mutants induce early cell death in human macrophages. Infect Immun 79: 581–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren H, Golovliov I, Baranov V, Ernst RK, Telepnev M, Sjostedt A 2004. Factors affecting the escape of Francisella tularensis from the phagolysosome. J Med Microbiol 53: 953–958 [DOI] [PubMed] [Google Scholar]
- Lindgren H, Stenman L, Tarnvik A, Sjostedt A 2005. The contribution of reactive nitrogen and oxygen species to the killing of Francisella tularensis LVS by murine macrophages. Microbes Infect 7: 467–475 [DOI] [PubMed] [Google Scholar]
- Lindgren H, Shen H, Zingmark C, Golovliov I, Conlan W, Sjostedt A 2007. Resistance of Francisella tularensis strains against reactive nitrogen and oxygen species with special reference to the role of KatG. Infect Immun 75: 1303–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llewellyn AC, Jones CL, Napier BA, Bina JE, Weiss DS 2011. Macrophage replication screen identifies a novel Francisella hydroperoxide resistance protein involved in virulence. PLoS ONE 6: e24201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariathasan S, Weiss DS, Dixit VM, Monack DM 2005. Innate immunity against Francisella tularensis is dependent on the ASC/caspase-1 axis. J Exp Med 202: 1043–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey RL, Allen LA 2006. Francisella tularensis LVS evades killing by human neutrophils via inhibition of the respiratory burst and phagosome escape. J Leukoc Biol 80: 1224–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey RL, Schwartz JT, Lindemann SR, Moreland JG, Buchan BW, Jones BD, Allen LA 2010. Multiple mechanisms of NADPH oxidase inhibition by type A and type B Francisella tularensis. J Leukoc Biol 88: 791–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRae S, Pagliai FA, Mohapatra NP, Gener A, Mahmou AS, Gunn JS, Lorca GL, Gonzalez CF 2009. Inhibition of AcpA phosphatase activity with ascorbate attenuates Francisella tularensis intramacrophage survival. J Biol Chem 285: 5171–5177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meibom KL, Barel M, Charbit A 2009. Loops and networks in control of Francisella tularensis virulence. Future Microbiol 4: 713–729 [DOI] [PubMed] [Google Scholar]
- Melillo AA, Mahawar M, Sellati TJ, Malik M, Metzger DW, Melendez JA, Bakshi CS 2009. Identification of Francisella tularensis live vaccine strain CuZn superoxide dismutase as critical for resistance to extracellularly generated reactive oxygen species. J Bacteriol 191: 6447–6456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra NP, Balagopal A, Soni S, Schlesinger LS, Gunn JS 2007. AcpA is a Francisella acid phosphatase that affects intramacrophage survival and virulence. Infect Immun 75: 390–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra NP, Soni S, Reilly TJ, Liu J, Klose KE, Gunn JS 2008. Combined deletion of four Francisella novicida acid phosphatases attenuates virulence and macrophage vacuolar escape. Infect Immun 76: 3690–3699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra NP, Soni S, Rajaram MV, Dang PM, Reilly TJ, El-Benna J, Clay CD, Schlesinger LS, Gunn JS 2010. Francisella acid phosphatases inactivate the NADPH oxidase in human phagocytes. J Immunol 184: 5141–5150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nano FE, Zhang N, Cowley SC, Klose KE, Cheung KK, Roberts MJ, Ludu JS, Letendre GW, Meierovics AI, Stephens G, et al. 2004. A Francisella tularensis pathogenicity island required for intramacrophage growth. J Bacteriol 186: 6430–6436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Riordan M, Portnoy DA 2002. The host cytosol: Front-line or home front? Trends Microbiol 10: 361–364 [DOI] [PubMed] [Google Scholar]
- Oyston PC, Sjostedt A, Titball RW 2004. Tularaemia: Bioterrorism defence renews interest in Francisella tularensis. Nat Rev Microbiol 2: 967–978 [DOI] [PubMed] [Google Scholar]
- Pechous R, Celli J, Penoske R, Hayes SF, Frank DW, Zahrt TC 2006. Construction and characterization of an attenuated purine auxotroph in a Francisella tularensis live vaccine strain. Infect Immun 74: 4452–4461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechous RD, McCarthy TR, Mohapatra NP, Soni S, Penoske RM, Salzman NH, Frank DW, Gunn JS, Zahrt TC 2008. A Francisella tularensis Schu S4 purine auxotroph is highly attenuated in mice but offers limited protection against homologous intranasal challenge. PLoS ONE 3: e2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng K, Broz P, Jones J, Joubert LM, Monack D 2011. Elevated AIM2-mediated pyroptosis triggered by hypercytotoxic Francisella mutant strains is attributed to increased intracellular bacteriolysis. Cell Microbiol 13: 1586–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierini LM 2006. Uptake of serum-opsonized Francisella tularensis by macrophages can be mediated by class A scavenger receptors. Cell Microbiol 8: 1361–1370 [DOI] [PubMed] [Google Scholar]
- Polsinelli T, Meltzer MS, Fortier AH 1994. Nitric oxide-independent killing of Francisella tularensis by IFN-γ-stimulated murine alveolar macrophages. J Immunol 153: 1238–1245 [PubMed] [Google Scholar]
- Qin A, Mann BJ 2006. Identification of transposon insertion mutants of Francisella tularensis tularensis strain Schu S4 deficient in intracellular replication in the hepatic cell line HepG2. BMC Microbiol 6: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin A, Scott DW, Thompson JA, Mann BJ 2009. Identification of an essential Francisella tularensis subsp. tularensis virulence factor. Infect Immun 77: 152–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam VA, Jiang Z, Waggoner SN, Sharma S, Cole LE, Waggoner L, Vanaja SK, Monks BG, Ganesan S, Latz E, et al. 2010. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol 11: 395–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly TJ, Baron GS, Nano FE, Kuhlenschmidt MS 1996. Characterization and sequencing of a respiratory burst-inhibiting acid phosphatase from Francisella tularensis. J Biol Chem 271: 10973–10983 [DOI] [PubMed] [Google Scholar]
- Rodriguez AR, Yu JJ, Murthy AK, Guentzel MN, Klose KE, Forsthuber TG, Chambers JP, Berton MT, Arulanandam BP 2010. Mast cell/IL-4 control of Francisella tularensis replication and host cell death is associated with increased ATP production and phagosomal acidification. Mucosal Immunol 4: 217–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santic M, Molmeret M, Abu Kwaik Y 2005a. Modulation of biogenesis of the Francisella tularensis subsp. novicida-containing phagosome in quiescent human macrophages and its maturation into a phagolysosome upon activation by IFN-γ. Cell Microbiol 7: 957–967 [DOI] [PubMed] [Google Scholar]
- Santic M, Molmeret M, Klose KE, Jones S, Kwaik YA 2005b. The Francisella tularensis pathogenicity island protein IglC and its regulator MglA are essential for modulating phagosome biogenesis and subsequent bacterial escape into the cytoplasm. Cell Microbiol 7: 969–979 [DOI] [PubMed] [Google Scholar]
- Santic M, Asare R, Skrobonja I, Jones S, Abu Kwaik Y 2008. Acquisition of the vacuolar ATPase proton pump and phagosome acidification are essential for escape of Francisella tularensis into the macrophage cytosol. Infect Immun 76: 2671–2677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saslaw S, Eigelsbach HT, Prior JA, Wilson HE, Carhart S 1961a. Tularemia vaccine study. II. Respiratory challenge. Arch Intern Med 107: 702–714 [DOI] [PubMed] [Google Scholar]
- Saslaw S, Eigelsbach HT, Wilson HE, Prior JA, Carhart S 1961b. Tularemia vaccine study. I. Intracutaneous challenge. Arch Intern Med 107: 689–701 [DOI] [PubMed] [Google Scholar]
- Schmerk CL, Duplantis BN, Howard PL, Nano FE 2009. A Francisella novicida pdpA mutant exhibits limited intracellular replication and remains associated with the lysosomal marker LAMP-1. Microbiology 155: 1498–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulert GS, Allen LA 2006. Differential infection of mononuclear phagocytes by Francisella tularensis: Role of the macrophage mannose receptor. J Leukoc Biol 80: 563–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulert GS, McCaffrey RL, Buchan BW, Lindemann SR, Hollenback C, Jones BD, Allen LA 2009. Francisella tularensis genes required for inhibition of the neutrophil respiratory burst and intramacrophage growth identified by random transposon mutagenesis of strain LVS. Infect Immun 77: 1324–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J, Yang J, Zhao D, Kawula TH, Banas JA, Zhang JR 2007. Genome-wide identification of Francisella tularensis virulence determinants. Infect Immun 75: 3089–3101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telepnev M, Golovliov I, Grundstrom T, Tarnvik A, Sjostedt A 2003. Francisella tularensis inhibits Toll-like receptor-mediated activation of intracellular signalling and secretion of TNF-α and IL-1 from murine macrophages. Cell Microbiol 5: 41–51 [DOI] [PubMed] [Google Scholar]
- Tempel R, Lai XH, Crosa L, Kozlowicz B, Heffron F 2006. Attenuated Francisella novicida transposon mutants protect mice against wild-type challenge. Infect Immun 74: 5095–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulland TK, Buchan BW, Ketterer MR, Fernandes-Alnemri T, Meyerholz DK, Apicella MA, Alnemri ES, Jones BD, Nauseef WM, Sutterwala FS 2010. Cutting edge: Mutation of Francisella tularensis mviN leads to increased macrophage absent in melanoma 2 inflammasome activation and a loss of virulence. J Immunol 185: 2670–2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrly TD, Chong A, Virtaneva K, Sturdevant DE, Child R, Edwards JA, Brouwer D, Nair V, Fischer ER, Wicke L, et al. 2009. Intracellular biology and virulence determinants of Francisella tularensis revealed by transcriptional profiling inside macrophages. Cell Microbiol 11: 1128–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss DS, Brotcke A, Henry T, Margolis JJ, Chan K, Monack DM 2007. In vivo negative selection screen identifies genes required for Francisella virulence. Proc Natl Acad Sci 104: 6037–6042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahrt TC, Deretic V 2002. Reactive nitrogen and oxygen intermediates and bacterial defenses: Unusual adaptations in Mycobacterium tuberculosis. Antioxid Redox Signal 4: 141–159 [DOI] [PubMed] [Google Scholar]