Abstract
Heme, which is composed of iron and the small organic molecule protoporphyrin, is an essential component of hemoglobin as well as a variety of physiologically important hemoproteins. During erythropoiesis, heme synthesis is induced before, and is essential for, globin synthesis. Although all cells possess the ability to synthesize heme, there are distinct differences between regulation of the pathway in developing erythroid cells and all other types of cells. Disorders that compromise the ability of the developing red cell to synthesize heme can have profound medical implications. The biosynthetic pathway for heme and key regulatory features are reviewed herein, along with specific human genetic disorders that arise from defective heme synthesis such as X-linked sideroblastic anemia and erythropoietic protoporphyria.
All cells can synthesize heme, but regulation of the heme biosynthetic pathway is tissue- and cell-specific. In many disorders of erythroid heme biosynthesis (porphyrias), pathway intermediates accumulate and cause cytotoxicity.
Hemoglobin is recognized for its brilliant red color and ability to reversibly bind oxygen. The molecule owes both of these characteristics to the presence of the small organic iron tetrapyrrole protoheme IX (heme). Indeed, the very name “hemoglobin” derives from the components of the molecule, heme and globin. During erythropoiesis, cells coordinate the synthesis of both heme and globins, not only for efficiency but also because imbalance in the synthesis of either has profoundly negative physiological consequences. Heme-free globin chains are nonfunctional, and free heme is highly toxic to the cell. As is clearly shown in this collection, erythroid cell differentiation is a widely examined issue, with considerable attention given to regulation of globin synthesis, cytoskeleton construction, and mechanisms for the elimination of intracellular organelles. During differentiation of erythroid cells, the induction of heme precedes that of globin synthesis. This is in part attributable to the requirement for heme to inactivate the heme-regulated eukaryotic initiation factor 2α kinase (HRI) and, thereby, allow for protein translation initiation to proceed. In the absence of sufficient cellular heme, HRI is activated and protein synthesis, including that of globin proteins, is prevented. Given the obligatory need for heme, disorders of heme synthesis can have profound implications for the organism. Below we have briefly reviewed heme synthesis, its regulation, and diseases associated with genetic disorders that have particular relevance for erythroid cells.
HEME BIOSYNTHETIC PATHWAY
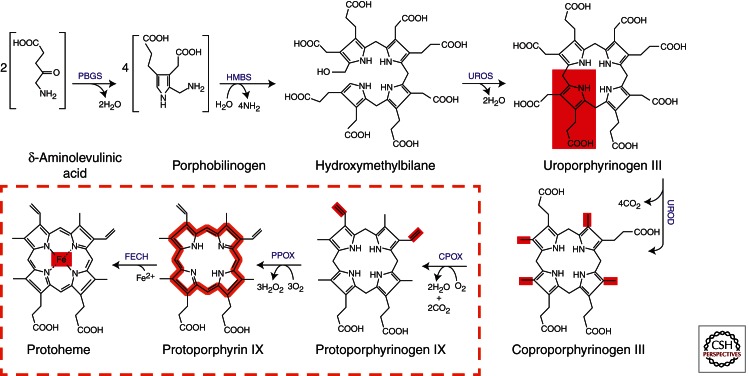
Enzymes of heme biosynthesis are nuclear encoded and cytoplasmically synthesized (see Ajioka et al. 2006; Medlock and Dailey 2009; Layer et al. 2010). The pathway is composed of eight enzymes that are divided between the mitochondria and cytoplasm (Fig. 1). The overall reaction to synthesize heme in metazoan cells is: 8 glycine + 8 succinyl-CoA + 5 O2 + Fe2+ → Heme + 8 CoA + 4 NH4 + 14 CO2 + 3 H2O2 + 2 H+ + 11 H2O. Of the products, 8 CoA (coenzyme A), 8 CO2, 2 H+, and heme are generated in the mitochondrial matrix and 2 CO2, 3 H2O2, and 2 H2O are generated in the mitochondrial inner membrane space. All remaining products are cytosolic. The pathway and the individual enzymes have been well researched and frequently reviewed. Regulation of the pathway is tissue- and cell-specific, subject to modulation by a myriad of developmental and environmental pressures through diverse transcriptional factors, and is beyond the scope of the current review (Medlock and Dailey 2009). In general, however, synthesis of the first committed intermediate, 5-aminolevulinate (ALA), is considered rate-limiting.
Figure 1.
The mammalian heme biosynthetic pathway. The diagram presents the steps and structures of intermediates in the pathway from ALA to heme. Steps that occur in the mitochondrion are enclosed in the dashed box, and those present in the cytosol are outside the box. Synthesis of ALA from glycine and succinyl-CoA, which is the first committed step and occurs in the mitochondrion, is not shown.
Aminolevulinate Synthase
The first step in mammalian heme biosynthesis is catalyzed by the enzyme ALA synthase (ALAS) (EC 2.3.1.37) (Hunter and Ferreira 2011). ALAS, which catalyzes the condensation of glycine with succinyl-CoA to form ALA and CO2, is a homodimeric, pyridoxal phosphate-containing enzyme that is a member of the large and well-characterized α-oxoamine synthase family. The mature protein is located in the mitochondrial matrix. Much is known about the mammalian enzyme from kinetic and site-directed mutagenesis studies, but at present only the crystal structure of ALAS from the bacterium Rhodobacter capsulatus has been determined (Astner et al. 2005). However, it is clear that the mature mammalian ALAS is structurally homologous to the bacterial enzyme except that it possesses an additional carboxy-terminal sequence of approximately 25 residues. Interestingly, recent studies have shown that this region of mammalian ALAS somehow modulates the activity of the enzyme (Whatley et al. 2008). Truncation or selected amino acid mutations in this region result in a form of ALAS that has significantly higher activity than the normal ALAS. Such alterations result in dysfunctional cellular heme biosynthesis (see below).
In mammals, one finds two isozymes of ALAS, one specific for differentiating erythroid cells (ALAS-2 or ALAS-E) and one expressed in all other cell types (ALAS-1 or ALAS-N) (May et al. 1995; Medlock and Dailey 2009; Hunter and Ferreira 2011). The genes for ALAS, as for all heme biosynthetic enzymes, are nuclear, although the final destination for ALAS is the mitochondrion. The genes for ALAS-1 and ALAS-2 are located on human chromosomes 3 and X, respectively, and are differentially regulated (see below). ALAS-1 and -2 clearly represent gene duplication events because the proteins are highly similar in structure and catalytic abilities. For human ALAS-1 and ALAS-2, one finds alternative mRNA splice variants. For ALAS-1, two known splice variants occur in the untranslated region of exon 1 (Roberts and Elder 2001). Via an uncharacterized mechanism, these variants possess altered sensitivity to heme-mediated decay of the message. One splice variant of human ALAS-2 has been found in which exon 4 is absent (Cox et al. 2004). This variant represents 35%–45% of total ALAS2 mRNA in the cell and was shown to encode a protein with slightly reduced activity. This variant ALAS-2 is translocated into the mitochondrial matrix, where it was shown to interact with succinyl CoA synthetase, just as the full-length enzyme does (Furuyama and Sassa 2000), thereby suggesting that it contributes to erythroid heme synthesis.
The precursor proteins of both ALAS-1 and -2 possess mitochondrial targeting sequences that are proteolytically removed after being translocated into the mitochondrial matrix. Both proteins possess three heme regulatory motifs (HRMs) composed of a canonical Cys-Pro sequence (Zhang and Guarente 1995). Two of these HRMs are found in the targeting leader sequence and the third is near the amino terminus of the mature processed ALAS. Evidence from multiple groups has shown that heme binds to the ALAS HRM in vitro and in vivo and inhibits the translocation of the protein into the mitochondrion (Yamauchi et al. 1980; Lathrop and Timko 1993; Dailey et al. 2005). A series of mutagenesis experiments clearly showed the significance of all three ALAS HRMs for heme inhibition of ALAS translocation into mitochondria in vivo (Dailey et al. 2005). Whereas such heme regulation of apoprotein translocation in nonerythroid cells can easily be rationalized, a similar occurrence in differentiating erythroid cells in which massive quantities of heme are being synthesized in a relatively short period of time is less easily justified. Recently, a pair of HRM motifs have been shown to serve as a redox-sensitive switch for heme oxygenase-2 (Yi and Ragsdale 2007), but there is no evidence to suggest such a role with ALAS.
Porphobilinogen Synthase
Once ALA is produced by ALAS, it is exported out of the mitochondrial matrix to reach the second pathway enzyme. The exact mechanism for this transport is not completely defined, but evidence suggests that the mitochondrial inner membrane solute transport protein, SLC25A38, may be responsible for this function (Guernsey et al. 2009). The next enzymatic step is catalyzed by porphobilinogen synthase (PBGS) (EC 4.2.1.24) (previously named ALA dehydratase) (Schubert et al. 2009). PBGS catalyzes the condensation of two molecules of ALA to form one molecule of the monopyrrole PBG. The crystal structure of the soluble, cytoplasmic PBGS reveals a homo-octomer that can best be described as a tetramer of homodimers (Erskine et al. 1997). Each monomer binds one zinc atom for a total of eight zinc atoms. Four metal atoms are essential for catalysis and four are involved in stabilization of tertiary structure. In individuals suffering from chronic lead exposure, one finds that these zinc ions may be replaced by lead, resulting in an inactive enzyme. It has been shown that PBGS exists in alternate quaternary structures named morpheeins (Jaffe and Lawrence 2012). The morpheeins represent a dynamic change in oligomerization of PBGS between the high-activity octomer and a low-activity hexamer. This change in quaternary structure is the basis of allosteric regulation of PBGS. However, given that ALAS is considered rate-limiting to heme synthesis, the role of allosteric regulation at PBGS is something of an enigma.
Although two distinct genes on separate chromosomes exist for ALAS-1 and -2, only single genes exist for the remaining pathway enzymes. However, for all genes one finds distinct erythroid and housekeeping-specific promoter elements. For PBGS one also finds erythroid versus housekeeping differential splice variants (Kaya et al. 1994). Human PBGS possesses two noncoding exons, 1A and 1B. The translational start site for housekeeping PBGS mRNA is exon 1A, while erythroid PBGS mRNA starts in exon 1B. Thus, an additional 5′ untranslated region is present in the mRNA for the housekeeping variant that is lacking in the erythroid splice variant. Given that these variants are in the noncoding region, the housekeeping and erythroid forms of the PBGS enzymes are identical.
Hydroxymethylbilane Synthase
Following formation of PBG, the enzyme hydroxymethylbilane synthase (HMBS, previously called PBG deaminase or PBGD) (EC 2.5.1.61) catalyzes the head-to-tail synthesis of four PBG molecules to form the linear tetrapyrrole HMB and releases four molecules of ammonium (Schubert et al. 2009). HMBS has been purified from a variety of sources, and the crystal structures of the Escherichia coli (Louie et al. 1992) and human (Gill et al. 2009) enzymes have been determined. The cytoplasmically located monomer is synthesized as an apoprotein that in its first complete catalytic cycle synthesizes a hexameric linear polypyrrole that is covalently bound to the HMBS protein. The distal linear tetrapyrrole, HMB, is cleaved and released, leaving behind a covalently bound dipyrromethane. The dipyrromethane serves as a cofactor for future turnovers. The HMB produced by HMBS is chemically reactive and will spontaneously cyclize to form uroporphyrinogen I in the absence of the next pathway enzyme. Uroporphyrinogen I cannot be converted into protoporphyrin IX. As with ALAS and PBGS, HMBS has erythroid and housekeeping-specific splice variants (Grandchamp et al. 1987). The mRNA of the erythroid form of HMBS skips exon 1 and starts transcription with the noncoding exon 2. The erythroid-specific translational initiation site is in exon 3. The housekeeping HMBS starts with exon 1, which contains the housekeeping-specific initiation site, and skips the noncoding exon 2. The result is that the housekeeping HMBS has an additional 17 amino acid residues at the amino terminus that are lacking in the erythroid form of the protein.
Uroporphyrinogen Synthase
Conversion of HMB to the physiological uroporphyrinogen III isomer requires the action of uroporphyrinogen synthase (UROS) (EC 4.2.1.75) (Schubert et al. 2009). The reaction, catalyzed without a cofactor, is a spiro inversion of the final, or D, ring of HMB followed by cyclization to yield the III isomer of uroporphyrinogen (Fig. 2). UROS is a monomeric protein; the crystal structure has been determined for both the human protein (Mathews et al. 2001) and the bacterial protein from Thermus thermophilus with bound substrate (Schubert et al. 2008). Its structure is a bit unusual because it is composed of two distinct structural domains connected by a flexible linker region. Crystal structures have been obtained for a variety of domain orientations, suggesting that the molecule is highly flexible in solution. Splice variants also exist for UROS (Aizencang et al. 2000). The gene for UROS possesses distinct erythroid and housekeeping promoters. The erythroid promoter elements are located in the intron between exons 1 and 2 and drive transcription from an initiation site in exon 2. The housekeeping promoter region is present upstream of exon 1 and drives transcription from an initiation site in exon 1. However, because exon 1 is noncoding, both housekeeping and erythroid UROS proteins are identical.
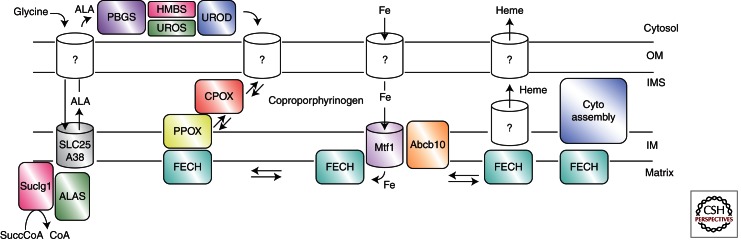
Figure 2.
Proposed model for components involved in heme synthesis. Details are discussed in the text, and in cases in which specific data exist for a particular component, its name is shown. Components for which no solid data exist, but for which there is good reason to expect they exist, are denoted with “?”. IM, Inner membrane; IMS, intramembrane space; OM, outer membrane.
Uroporphyrinogen Decarboxylase
The final cytoplasmic enzyme in the pathway is uroporphyrinogen decarboxylase (UROD) (EC 4.1.1.37) (Shoolingin-Jordan 2003). UROD contains no cofactors or metal ions and catalyzes the decarboxylation of the four pyrrole acetic acid side chains to yield coproporphyrinogen and four molecules of CO2. This homodimeric enzyme has been crystallized (Phillips et al. 2003) and its catalytic mechanism well studied (Lewis and Wolfenden 2008). UROD will use both uroporphyrinogen I and III, converting them into coproporphyrinogen I and III, respectively. In the presence of high concentrations of uroporphyrinogen, UROD will decarboxylate in a random fashion, but it is believed that in situ the reaction starts with the decarboxylation of the D ring acetate and proceeds sequentially in a clockwise fashion (i.e., D, A, B, C). No evidence exists to suggest that mRNA splice variants exist for UROD.
Mitochondrial Import of Coproporphyrinogen
In mammals, coproporphyrinogen III produced by UROD is next transported into the mitochondrial intermembrane space. A series of studies have proposed that ABCB6, a mitochondrial outer membrane ATP-dependent transporter, is responsible for coproporphyrinogen transport (Krishnamurthy et al. 2006; Krishnamurthy and Schuetz 2011). However, this claim must be viewed with skepticism because only the transport of the fully conjugated, planar macrocycle coproporphyrin, not the physiologically relevant nonplanar coproporphyrinogen, has ever been experimentally shown. ABCB6 has also been shown to exist as two different-size variants that localize in not only the mitochondrial outer membrane but also the Golgi and plasma membranes (Paterson et al. 2007; Tsuchida et al. 2008). Two recent reports regarding the role of ABCB6 as a coproporphyrinogen transporter raise additional questions. One identified ABCB6 as the blood group Langereis (Helias et al. 2012), and a second reported that a defect in ABCB6 is the cause of an inherited developmental defect of the eye known as ocular coloboma (Wang et al. 2012). These defects have no known relationship to heme imbalance, and affected individuals do not have free porphyrin accumulations, as are found in porphyrias. However, given that ABCB6 has been observed to be one of a relatively few genes induced at high levels during erythropoiesis (Nilsson et al. 2009), it seems likely that it plays some role in erythropoiesis. Given the size of coproporphyrinogen, the possibility that it transits the outer membrane via a porin rather than specific transporters cannot be ruled out by current data.
Coproporphyrinogen Oxidase
The next enzyme, coproporphyrinogen oxidase (CPOX) (EC 1.3.3.3), is synthesized in the cytoplasm with an unusually long (∼120-amino-acid) mitochondrial leader sequence that targets it to the mitochondrial intermembrane space (Dailey 1990; Akhtar 2003). The enzyme is weakly associated with the outside of the inner membrane, possibly in association with the next pathway enzyme. The mature protein is a homodimer without bound cofactor. The structures of both human (Lee et al. 2005) and yeast (Phillips et al. 2004) (the latter of which is a cytoplasmic protein) CPOX have been solved and were found to possess unique folds. The reaction catalyzed is an unusual oxidative decarboxylation of the A and B ring propionates to yield the vinyl groups of protoporphyrinogen IX. CPOX will use only the coproporphyrinogen III isomer and proceeds in a stepwise fashion that requires two molecules of molecular oxygen and generates two molecules of CO2. The reaction catalyzed by CPOX has been extensively studied both experimentally and in silico (Lash 2005; Silva and Ramos 2011). Studies on the regulation of the CPOX gene indicate that erythroid and housekeeping promoter elements exist (Takahashi et al. 1998), although our knowledge of how these operate is far from complete. Studies in differentiating murine erythroleukemia cell culture have provided data that are difficult to fit within a simple model. Whereas mRNA levels increase rapidly from the onset of erythropoiesis, measured CPOX enzyme activity first decreases (as coproporphyrin concentrations in the culture medium increase) and then increases (Conder et al. 1991). Additionally, constitutive expression of an externally introduced CPOX results in a more rapid and overall greater increase in heme synthesis by cells undergoing erythropoiesis (Taketani et al. 2001). These observations suggest a potential role for CPOX in pathway regulation during erythroid differentiation.
Protoporphyrinogen Oxidase
The penultimate step in heme synthesis is the oxidation of protoporphyrinogen IX to protoporphyrin IX (Dailey 1990; Akhtar 2003). This is catalyzed by protoporphyrinogen oxidase (PPOX) (EC 1.3.3.4) and requires three molecules of molecular oxygen and generates three molecules of hydrogen peroxide. Crystal structures of the plant (Koch et al. 2004), bacterial (Corradi et al. 2006; Qin et al. 2010), and human (Qin et al. 2011) enzyme have been determined and show that the protein is a homodimer with one noncovalently bound FAD per subunit. PPOX is synthesized in the cytoplasm in its mature size and translocated to the outer surface of the inner mitochondrial membrane via a mechanism that requires both an amino-terminal and internal mitochondrial targeting sequence (von und zu Fraunberg et al. 2003; Morgan et al. 2004; Dailey et al. 2005; Davids et al. 2006). Interestingly, the active site is proposed to be situated in the middle of a tunnel that passes through the protein. The gene for PPOX, like that for CPOX, has been modestly characterized and found to possess both erythroid and housekeeping promoter elements (Taketani et al. 1995; Dailey et al. 2002). There is no evidence to suggest that mRNA splice variants exist for PPOX. Although details are lacking, available data suggest that PPOX, along with the terminal pathway enzyme, are regulated in a fashion distinct from the earlier pathway enzyme genes during erythroid differentiation (Conder et al. 1991). Full erythroid induction of these proteins appears to require the presence of elevated cellular heme (Yin and Dailey 1998).
Ferrochelatase
The terminal step of heme synthesis is the insertion of ferrous iron into the protoporphyrin IX macrocycle to produce protoheme IX (heme). This is catalyzed by the enzyme ferrochelatase (FECH) (EC 4.99.1.1) (Dailey and Dailey 2003). This enzyme is synthesized in the cytoplasm as a preprotein and is translocated to the mitochondrial matrix, where it is associated with the inner mitochondrial membrane. The mature, processed protein is a homodimer with each subunit possessing a [2Fe–2S] cluster (Wu et al. 2001). The presence of the [2Fe–2S] cluster is necessary for enzyme activity, but there is no evidence to suggest that it participates directly in catalysis. Nevertheless, the fact that the cluster must be present for activity makes the cluster a de facto iron regulatory feature for the enzyme (Crooks et al. 2010). Interestingly, recent studies suggest that the cluster may serve a regulatory or sensory role (DI Shah, N Takahashi-Makise, JD Cooney, et al., unpubl.). The crystal structures of human ferrochelatase with and without substrate and product have been determined, and it has been shown that the molecule undergoes considerable active-site remodeling during catalysis (Medlock et al. 2007a,b, 2009). Next to the ALAS and HMBS genes, regulatory features of the FECH gene are probably the best characterized in the pathway. Distinct erythroid and housekeeping elements exist in the promoter region (Tugores et al. 1994; Magness et al. 1998, 2000), and there is evidence for alternate splicing in the noncoding 3′ region of the mouse mRNA (Chan et al. 1993).
HEME BIOSYNTHESIS PATHWAY REGULATION
As noted above, pathway regulation is generally ascribed to modulation of ALAS activity. The ALAS-1 gene is subject to tissue-specific regulation by diverse factors frequently associated with xenobiotic, hormone, and drug metabolism (May et al. 1995; Medlock and Dailey 2009). It has an estimated half-life of only a few hours and may be induced as much as 100-fold in some cells, thereby allowing for rapid and vigorous response to cellular heme needs. ALAS-1 is present in erythroid precursor cells before they begin erythroid differentiation, but the gene is turned off as ALAS-2 is turned on when these cells begin to synthesize heme for hemoglobin (Conder et al. 1991; Yin and Dailey 1998). Thus, ALAS-1 cannot substitute for a deficiency of ALAS-2 and vice versa.
ALAS-2 is transcriptionally regulated by erythroid-specific factors such as GATA-1 and possesses an iron regulatory element (IRE) located in the mRNA 5′ untranslated region that allows for translational regulation by cellular iron concentration (Dierks 1990; Bhasker et al. 1993; Melefors et al. 1993). As with similarly regulated systems, such as for ferritin synthesis, the IRE binds the iron-free form of the iron regulatory protein (IRP), thereby preventing translation of the ALAS-2 message when cellular iron concentrations are insufficient for optimal heme synthesis. Thus, maximal ALAS-2 activity requires gene induction by erythroid-specific transcription factors as well as sufficient cellular iron levels to support heme synthesis. Of note are studies (Schranzhofer et al. 2006) that suggest the IRE–IRP system may not be entirely applicable during the later, accelerated hemoglobinization phase of erythropoiesis because elevated iron would be expected to diminish transferrin receptor expression and increase ferritin synthesis, both of which would diminish iron availability for heme synthesis. The data gathered with differentiating erythroblasts showed that ALAS-2 translational regulation is “uncoupled” from that of transferrin receptors (which are increased, not decreased) and ferritin (whose protein levels do not increase). Additionally, it was postulated that during maximal hemoglobinization, ALAS2 mRNA increases disproportionately to IRP synthesis, thus circumventing the IRE–IRP regulatory mechanism at this time. It is suggested that the “kiss and run” model (Zhang and Guarente 1995) may offer a possible explanation. In this hypothesis, during terminal erythropoiesis, vesicular iron acquired by receptor-mediated endocytosis is targeted directly to the mitochondrion, thereby bypassing the cytosolic iron pool and the IRE–IRP system. Unlike ALAS-2, ALAS-1 is not regulated via the IRE–IRP system.
Some reviews have presented tables and figures based on compilations of published data suggesting that the activity of pathway enzymes from PBGS to FECH are in excess over ALAS activity (see Ponka 1999; Puy et al. 2010). However, these calculations are based on in vitro assays of individual enzymes under what are considered optimal conditions and then extrapolated back to in vivo cell conditions. Interestingly, if these calculations are to be believed, the relative enzyme activities for pathway enzymes appear to vary considerably. Whereas there is no clear evidence for the induction of biosynthetic pathway enzymes other than ALAS-1 in nonerythroid cells, there is up-regulation of all pathway enzymes during erythroid differentiation. Even so, current dogma relegates pathway regulation in all cells to ALAS-1 or -2 with the proposition that all later pathway enzymes are present in excess. This model conveniently ignores the observation that coproporphyrin is routinely excreted by mammals (Kappas et al. 1995), and that in cancer cells one finds an elevation of CPOX and diminishment of FECH that results in the accumulation of protoporphyrin during ALA-based phototherapy (Sopena et al. 2008; Takahashi et al. 2011). Additional recent data may cause a reevaluation of this model for differentiating erythroid cells since it has been discovered that a mutation in the carboxyl terminus of ALAS-2, which results in a hyperactive enzyme, results in vivo in the accumulation of free protoporphyrin and zinc-bound protoporphyrin (Whatley et al. 2008). This is thus similar to what is found in the disease erythropoietic protoporphyria (EPP), although generally protoporphyrin is in the free form. The name currently given this disorder is X-linked protoporphyria (XLPP) to reflect the X chromosome location of ALAS-2. The observation of protoporphyrin accumulation in XLPP is particularly interesting because levels of ferrochelatase have been generally assumed to be present in considerable excess over those of ALAS in the cell. Indeed, EPP occurs only when ferrochelatase levels decrease to approximately one-fourth of normal (Elder et al. 2009). This suggests that either the erythroid pathway is designed so that the amount of ferrochelatase available (or iron transport mechanisms for heme synthesis) is normally just slightly above the maximum possible levels of ALAS-2, or that additional regulatory steps or limiting mechanisms exist at the end of the pathway that we currently do not recognize or understand.
Interestingly, it has been proposed that during the course of normal erythropoiesis, heme synthesis in developing erythroid cells overproduces heme to an extent that it is toxic to the cell unless it is exported by the plasma membrane heme transporter feline leukemia virus subtype C receptor (FLVCR) (Khan and Quigley 2011). Although it is clear that disabling mutations in FLVCR result in cell death, presumably as a result of the toxic effects of excessive heme, it seems counterintuitive that nature would have evolved such intricate regulatory mechanisms for erythroid heme synthesis (i.e., IRE–IRP, erythroid-specific transcription factors, and complex iron supply regulatory schemes) that all allow for excessive synthesis of heme so that it necessitates export and degradation by macrophages and liver. Indeed, given the “cost” of heme synthesis, it seems more likely that the “excessive” heme produced is a planned synthesis of heme by erythroid cells (which must be considered the ultimate heme-synthesizing factories in the body) for orderly export and transit to other organs and cells whose heme-synthesizing capabilities may be physiologically limited. The presence of specific heme transporters, such as hemopexin (Shipulina et al. 2000), and the observation that exogenous administration of heme to acute porphyric patients both down-regulates heme synthesis and is used for hemoproteins by the patient receiving the infusion (Anderson et al. 2005) argue for this hypothesis.
MULTIENZYME COMPLEXES IN HEME BIOSYNTHESIS
Given the reactivity of the pathway intermediates, it is highly unlikely that the intracellular concentrations of any of them attain the micromolar concentrations of the measured enzyme Kms. Thus it is reasonable to assume that “free intermediates” do not exist in the cell. This topic was first approached experimentally in the 1980s when data were presented in support of the hypothesis that the terminal three pathway enzymes, which are all mitochondrial membrane associated, form a transient complex to facilitate the transfer of intermediates (Ferreira et al. 1988; Dailey 1990; Proulx et al. 1993). Biochemical data clearly showed that while obligate, tight substrate channeling, such as occurs in tryptophan synthesis, does not exist, under normal circumstances equilibration of products/substrates with the bulk medium does not occur. For the potential interaction between the terminal two pathway enzymes, data gleaned from PPOX and FECH crystal structures provide good support for the possible interaction (Koch et al. 2004; Medlock et al. 2007b). The presence and nature of multienzyme complexes involving the terminal three membrane-associated enzymes is considerably advanced over what is known, or not known, about the earlier pathway enzymes, but is still extremely rudimentary. Radiolabeling experiments using isolated mitochondrial fragments clearly support transient interactions in situ, although reconstitution of this process with purified components was not accomplished (Proulx et al. 1993). In silico structural studies show that an interaction across the inner mitochondrial membrane to transport protoporphyrin from PPOX to FECH is feasible and highly likely (Koch et al. 2004). Interestingly, in the docked PPOX–FECH complex, the openings of the active-site tunnels of the PPOX dimer not only coincide with the position of the openings to the active-site pockets of the dimeric FECH, but are spatially juxtaposed to surface-bound porphyrin molecules observed in some FECH crystal structures. The observation that binding of substrate and product induce changes in the surface contour and charge distribution around the active-site pocket opening of FECH provide an explanation for how PPOX and postcatalytic heme-accepting proteins “recognize” the appropriate form of FECH with which to interact (Medlock et al. 2007b). Given the need to acquire coproporphyrinogen from the cytosol and movement of at least some heme out of the mitochondrion, the existence of a complex of an outer membrane coproporphyringen transporter, CPOX, PPOX, the inner membrane iron transporter mitoferrin (Shaw et al. 2006), FECH, and a heme chaperone/outer membrane heme transporter at a mitofilin-mediated junction between outer and inner mitochondrial membranes is an intriguing possibility (Fig. 2).
Other than data for the terminal, membrane-associated enzymes, there exists little experimental evidence to support multienzyme complexes of earlier pathway enzymes. For the first step, an interaction of ALAS-2 with succinyl CoA synthetase on the inner mitochondrial membrane has been demonstrated by two groups (Furuyama and Sassa 2000; Cox et al. 2004). With the recent identification of SLC25A38 as the putative glycine/ALA transporter (Guernsey et al. 2009), it is reasonable to anticipate the existence of transient complexes between ALAS, succinyl CoA synthetase, and SLC25A38 on the mitochondrial inner membrane. The possibility for a stable, rigid complex for these and most other heme synthesis enzymes seems unlikely given that they possess active-site pockets with a single entrance/exit. The necessity for substrate(s) and product(s) to enter and exit via a single route would require movement of one enzyme between donor and acceptor molecules in the complex, much as cytochrome c physically cycles between electron donors and acceptors. The only enzyme for which this may not be the case is PPOX, which appears to have a channel through the protein, with the active site being located within this feature.
For the synthesis of uroporphyrinogen III from PBG, individuals have long believed that close proximity of HMBS and UROS would be likely given the chemical reactivity of the linear tetrapyrrolic intermediate, HMB. Unfortunately, at present, no data exist to support the presence of a multienzyme complex involving PBGS and HMBS. Given that ALA must be exported out and coproporphyrinogen imported into the mitochondria, logic would suggest that not only PBGS and HMBS but also UROS and UROD exist in a supramolecular complex that is spatially close to the mitochondrion. To date, high-resolution microscopic or biochemical/biophysical studies that may identify the intracellular distribution of these enzymes are lacking. Approaches that rely on cell disruption and physical isolation of complexes or in vitro reconstitution of complexes from isolated components assume a strength of protein–protein interaction in solution that may not exist and may not be necessary in the highly concentrated cytosolic milieu. One intriguing possibility is that PBGS, which forms a large complex best categorized as a tetramer of homodimers, could, because of its size, serve as a scaffold for a multienzyme complex. The observation that this octomer can also assume a hexameric shape may hint at its plastic role as a supramolecular assembly nucleation site. Resolution of these issues will require masterful research, but the results should be interesting.
Although tetrapyrrole synthesis and iron metabolism must be intimately linked to prevent the inappropriate accumulation of either molecule, there have been limited studies that address this issue at either the genetic or molecular level. The role of the iron-responsive element has been examined by a number of groups (Dierks 1990; Bhasker et al. 1993; Melefors et al. 1993; Wang and Pantopoulos 2011), and the cellular machinery for iron sulfur cluster assembly (Lill and Kispal 2000) has likewise garnered significant interest. Whole-body and intercellular iron trafficking is fairly well understood, but only recently has there been significant advancement with the identification of mitoferrin as the mitochondrial inner membrane iron transporter that supplies iron to ferrochelatase for heme synthesis (Shaw et al. 2006; Troadec et al. 2011). Specifically, mitoferrin 1 is responsible for iron transport for heme synthesis during erythropoiesis, but the presence of another inner membrane transport protein, ABCB10, is required to stabilize mitoferrin 1 (Chen et al. 2009). The exact role that ABCB10 plays has yet to be elucidated. Evidence has also been presented showing that mitoferrin forms a complex with ferrochelatase that might make possible a direct transfer of transported ferrous iron for heme synthesis to ferrochelatase (Chen et al. 2010). If this does occur, structural data from human ferrochelatase and its orientation relative to the inner membrane would indicate that iron would enter the active site of ferrochelatase via a solvent-filled channel whose outside entrance is on the back side of the enzyme (Sellers et al. 2001; Medlock et al. 2007b). It will be of interest to learn the molecular details of this process and how it is regulated.
Transit of heme away from its site of synthesis is unexplored territory. It has been shown that in in vitro assays the release of heme from the enzyme postmetalation is the rate-limiting step (Hoggins et al. 2007). This probably reflects the absence of the native heme acceptor in the in vitro assay. Given that heme is used throughout the cell in a variety of compartments, it is clear that a variety of heme chaperones may exist. For transit from the mitochondrion to other membranous compartments, it appears likely that transfer may occur at direct contact points rather than by diffusion through the cytoplasm (Schultz et al. 2010), but even that will require moving heme from FECH to the site of transfer between membranes. As yet unknown is what molecule first accepts heme from ferrochelatase. Given its location on the inner membrane, it would seem that for respiratory cytochrome maturation direct transfer from FECH to cytochrome assembly machinery could occur. Some experimental systems hint strongly at this possibility (Richard-Fogal and Kranz 2010), but definitive data are still lacking. For cytoplasmic hemoproteins, including hemoglobin, there are no identified chaperones. Putative heme/porphyrin binding/carrier proteins have been suggested and even biochemically characterized (Blackmon et al. 2002; Dias et al. 2006), but none of these is supported by data as in vivo players in heme trafficking.
DISORDERS OF ERYTHROID HEME BIOSYNTHESIS
In discussing disorders of heme metabolism, one may consider both heme synthesis and degradation. However, with particular regard to red cell metabolism, the only concern is heme synthesis because heme degradation is not a significant property of red cells. Thus, below we briefly consider only disorders of synthesis and focus only on those disorders that have significant red cell involvement. The chemical intermediates in tetrapyrrole synthesis (typically porphyrinogens in their oxidized porphyrin state) are relatively reactive and can be cytotoxic, through their inherent photoreactive and chemical properties. The physiological proof of this statement is amply shown by the fact that a deficiency in any one of the pathway enzymes results in a clinically distinct disease in animals (Elder et al. 2009) and inhibition of some later steps can result in photo-induced death of plants or microorganisms. Other than a defect in ALAS-2, which results in X-linked sideroblastic anemia, the problem in these disorders, named the porphyrias, is generally not anemia, but is attributable to the accumulation of heme pathway intermediates proximal to the deficiency. Inheritance of these diseases may be recessive or dominant depending on the natural level of enzyme activity. However, most dominantly inherited porphyrias have variable penetrence so that only about 30%–40% of individuals with a gene defect have symptoms. The two X-linked diseases (see below) are dominant in males, but variable in females owing to lyonization of the X chromosome. In the case of early pathway enzyme deficiencies, the porphyrias are neurological in nature and are believed to result from the accumulation of ALA and/or PBG. For the enzymatic steps after the synthesis of the first tetrapyrrole, the most profound symptoms are cutaneous in nature because of the chemical reactivity and photoreactivity of the tetrapyrrole intermediates (Table 1). These diseases and their molecular basis have been well studied and frequently reviewed by others. Interestingly, most heme biosynthetic disorders can be considered nonerythroid, and indeed, early publications categorized the porphyrias as hepatic versus erythroid. Below we have focused only on those diseases associated with erythroid cells.
Table 1.
Summary of the heme biosynthetic enzymes and associated porphyrias, noting their predominant mode of inheritance and clinical features
| Enzyme | Disorder | Inheritance | Clinical features |
|---|---|---|---|
| Aminolevulinate synthase | X-linked protoporphyria | X-linked | Immediate photosensitivity, liver disease secondary to protoporphyrin accumulation |
| X-linked sideroblastic anemia | X-linked | Anemia with ringed sideroblasts, some pyridoxine responsive | |
| Porphobilinogen synthase | Aminolevulinate dehydratase porphyria | Recessive | Acute neurological attacks |
| Hydroxymethylbilane synthase | Acute intermittent porphyria | Dominant | Acute neurological attacks |
| Uroporphyrinogen-III synthase | Congenital erythropoietic porphyria | Recessive | Severe vesiculoerosive skin disease |
| Uroporphyrinogen decarboxylase | Porphyria cutanea tarda | Acquired, but may be associated with a dominantly inherited mutation in up to approximately 30% | Vesiculoerosive skin disease; usually coexists with iron overload and a number of precipitating factors, particularly alcohol exposure, liver disease, and renal failure |
| Coproporphyrinogen oxidase | Hereditary coproporphyria | Dominant | Acute neurological attacks and vesiculoerosive skin disease |
| Protoporphyrinogen oxidase | Variegate porphyria | Dominant | Acute neurological attacks and vesiculoerosive skin disease |
| Ferrochelatase | Erythropoietic protoporphyria | Recessive, but more common than most recessive disorders because the disease-associated mutation is frequently inherited with a low-expression polymorphism that is common in populations of European extraction | Immediate photosensitivity |
Erythropoietic Protoporphyria (EPP)
EPP results from a partial deficiency of FECH (Elder et al. 2009). This manifests biochemically as an increased concentration of the heme precursor protoporphyrin (in its free, unbound-to-iron form) in erythrocytes and a secondary protoporphyrin accumulation in the skin and liver. This buildup of protoporphyrin may have clinical sequelae in the form of burning, stinging photocutaneous sensitivity, most often from infancy, and in approximately 3% of patients, liver disease.
Inheritance of EPP is a bit unusual in that about 94% of cases have a primary FECH mutation in trans to a hypomorphic low-expression FECH polymorphism (termed IVS3-48C) (Gouya et al. 2002). The IVS3-48C polymorphism increases the use of an aberrant splice site and is present in normal populations with a frequency varying between 1% and 45% (Gouya et al. 2006). This form of inheritance is most appropriately termed recessive because there is a required coinheritance of two molecular defects that result in a decrease in FECH activity to 25%–35% of normal activity and in overt clinical disease. Patients who have inherited only one such FECH defect generally remain asymptomatic throughout their lives. Approximately 4% of EPP cases are due to true homozygous or compound heterozygous FECH mutations (Gouya et al. 2006). Acquired EPP has been reported in the setting of hematological malignancy and cytogenetic FECH deletions associated with clonal expansion of FECH-deficient bone marrow cells (Aplin et al. 2001; Bharati et al. 2006; Goodwin et al. 2006).
Typically, porphyrin-induced phototoxicity is reported seasonally and acutely, that is, within minutes of sun exposure. Mild edema and erythema may arise immediately after sun exposure, and chronic lesions such as thickening of the skin on the hands and waxlike scarring on the face may occur. Manifest liver disease is a rare complication, occurring in approximately 3% of EPP patients (Anstey and Hift 2007). Free protoporphyrin is capable of diffusing out of red cells, with reticulocytes containing proportionally more protoporphyrin than mature red cells (Piomelli et al. 1975). Protoporphyrin is excreted into bile by the liver and undergoes extensive enterohepatic circulation because of its lipophilic nature. Increasing concentrations may lead to protoporphyrin insolubility, cholestasis, and cholelithiasis. Hepatocyte injury is manifest with inflammation, which may eventually lead to cirrhosis (Poh-Fitzpatrick 1986). In the terminal phase of EPP-related liver disease, increasing cholestasis impairs hepatic protoporphyrin excretion. Oxidative stress-induced hemolysis occurs with increasing protoporphyrin concentrations, driving erythropoiesis and increasing protoporphyrin load presented to the liver—propagating a cycle of clinical deterioration (Key et al. 1992; Anstey and Hift 2007). The precise pathogenesis of hepatopathy in EPP, however, remains to be fully elucidated. Although photosensitivity may be painful, distressing, or disruptive to an affected individual’s lifestyle (Todd 1994), liver disease may be fatal and only liver transplant may effect control of the disease progression. Avoidance of sunlight is the backbone of the management of skin symptoms, with some forms of medical treatment, such as oral β-carotene, reportedly effective in about a third of patients.
Apart from the clinical characteristics, which are diagnostic of EPP, the hallmark of the biochemical diagnosis is elevated free protoporphyrin in the red cells and stool (and/or bile). Further, many patients may have a slight microcytic, hypochromic anemia, although this is not invariant. Interestingly, trace amounts of zinc-chelated protoporphyrin are normally formed during heme biosynthesis, but during systemic iron deficiency, zinc protoporphyrin accumulates in erythrocytes (Langer et al. 1972). Similarly, during diverse states of de facto intramitochondrial iron deficiency, as occurs in lead poisoning (Lamola and Yamane 1974), in the anemia of chronic disorders (Hastka et al. 1993), and during accelerated (especially in hemolytic anemia) or ineffective erythropoiesis (Finch 1994), zinc protoporphyrin may increase.
X-Linked Protoporphyria (XLPP)
Recently, a small number of families in Europe and South Africa with an atypical protoporphyria have been investigated. Inexplicably these families presented with a highly penetrant and severe form of protoporphyria, with a relatively high incidence of protoporphyria-associated liver disease and extremely elevated protoporphyrin, with 30%–40% zinc protoporphyrin. Yet no abnormalities in FECH, its gene, or its regulatory components were apparent. There was no anemia or evidence of iron overload. The inheritance appeared X-linked. A collaborative study showed that these families are suffering from a novel form of porphyria, now termed X-linked protoporphyria (XLPP; also known as X-linked dominant protoporphyria, or XLDPP) (Whatley et al. 2008). This arises pathogenetically from a gain-of-function mutation in the erythroid form of ALAS-2. This disorder is, thus, similar, but not identical, to that observed when ALAS-2 is overproduced in Ireb2–/– mice (Cooperman et al. 2005). All of the eight families studied displayed one of two ALAS2 deletions, either c.1706–1709 delAGTG or c.1699–1700 delAT, resulting in frameshifts that lead to replacement or deletion of the 19–20 carboxy-terminal residues of the enzyme. The c.1706–1709 delAGTG mutation is predicted to cause a frameshift in 24 codons distal to the 4-bp mutation (p.E569GfsX24), resulting in the extension of ALAS-2 by four amino acids. In contrast, the c.1699–1700 delAT mutation leads to a frameshift mutation in two codons (p.M567EfsX2) and a truncation of ALAS-2. It is interesting that a single-base-pair substitution in this same region (S568G) reportedly causes a loss of function in ALAS-2, manifesting as X-linked sideroblastic anemia (Harigae et al. 1999) (see below). This mutation is located between two XLPP-causing mutations. It is surprising that mutations in such close proximity can have diametrically opposite effects.
The elevated ALAS-2 activity in XLPP results in a greatly increased flux of porphyrin(ogen)s through the heme pathway, FECH becomes rate-limiting (probably at the level of iron availability), and free and zinc protoporphyrin accumulate. This accounts for the resemblance to classic EPP and presence of photosensitive skin disease and potentially severe liver disease. Management is similar to that of EPP, and investigations are under way to establish a potential role for iron therapy in these patients.
X-Linked Sideroblastic Anemia (XLSA)
Sideroblastic anemia (SA) is characterized by anemia with the presence of “ringed” sideroblasts in the bone marrow. This morphology results from iron-loaded mitochondria in the erythroblasts (Fleming 2002). The resulting mitochondrial iron toxicity results in ineffective erythropoiesis and is a feature of severe forms of SA. In SA, intestinal iron absorption is increased and the affected patients may develop liver and systemic iron overload (Bottomley 2006). For this reason SA is considered an iron-loading anemia.
Inherited SA is a rare and heterogeneous disease caused by mutations of genes involved in heme biosynthesis (ALAS2) (Cotter et al. 1992; Bottomley 2006), Fe–S cluster biogenesis (ABCB7) (Pondarre et al. 2006), Fe–S cluster transport (Wingert et al. 2005), or mitochondrial transporters (SLC25A38) (Kannengiesser et al. 2011). The most common form of SA, X-linked SA (XLSA), results from one of a number of mutations in the ALAS2 gene (Cotter et al. 1992; Bottomley 2006). A much rarer form, XLSA with ataxia, has also been described in which the X-linked ATP-binding cassette transporter B7 gene, ABCB7 (Allikmets et al. 1999), is defective. ABCB7 is involved in Fe–S cluster transport, and its functionality is essential for erythroid cells and the central nervous system—hence the presence of anemia and spinocerebellar ataxia and cerebellar hypoplasia in these patients (Pondarre et al. 2007). Interestingly a form of non-X-linked SA resulting from mutations of the GLRX5 gene, which encodes a mitochondrial protein involved in iron–sulfur cluster biogenesis, has relatively recently been described (Rouault and Tong 2008).
Most XLSA-associated mutations in ALAS2 are missense substitutions resulting in loss of ALAS-2 functionality, but there are rarer ALAS2 promoter mutations (Bergmann et al. 2010; Ducamp et al. 2011). XLSA prevalently affects males, but anemia may also be present in females because of either X chromosome inactivation (Cazzola et al. 2000) or excessive skewing that occurs in hematopoietic tissue with aging (Aivado et al. 2006). Thus, XLSA may present early in infancy or not until later in adult life (Camaschella 2009). A number of ALAS2 mutations decrease the binding of the pyridoxal phosphate cofactor, which neatly accounts for the pyridoxine responsiveness in patients carrying those mutations (Cox et al. 1994; May and Bishop 1998). Patients unresponsive to pyridoxine possess a mutation in an area not connected with pyridoxal phosphate cofactor binding (Furuyama et al. 1997) or impaired interaction with succinyl CoA synthetase (Furuyama and Sassa 2000). Asymptomatic or mildly anemic patients seldom require intervention, although oral pyridoxine supplementation should be considered because in some cases partial to full restoration of hemoglobin levels may be achieved. Severely pyridoxine-unresponsive individuals may require transfusions of red blood cell concentrates in conjunction with iron chelation. Iron status should be monitored even in nontransfused patients and appropriate treatment should be started if iron overload is observed. Iron overload can be treated by phlebotomy (Bottomley 2006; Camaschella 2009). Interestingly, the study of these rare diseases has enlightened our understanding of the relationship between the two main pathways of mitochondrial iron metabolism—those of heme and iron–sulfur biosynthesis (Camaschella 2009).
ACKNOWLEDGMENTS
H.A.D. is supported by grant DK32303 from the National Institutes of Health. P.N.M. is supported by funding from the SA Medical Research Council, the National Research Foundation, the Harry Crossley Foundation, and the University of Cape Town Research Committee. The authors acknowledge T. A. Dailey (University of Georgia) and C. E. Haumann (University of Cape Town) for reviewing and editing the manuscript.
Footnotes
Editors: David Weatherall, Alan N. Schechter, and David G. Nathan
Additional Perspectives on Hemoglobin and Its Diseases available at www.perspectivesinmedicine.org
REFERENCES
- Aivado M, Gattermann N, Rong A, Giagounidis AA, Prall WC, Czibere A, Hildebrandt B, Haas R, Bottomley SS 2006. X-linked sideroblastic anemia associated with a novel ALAS2 mutation and unfortunate skewed X-chromosome inactivation patterns. Blood Cells Mol Dis 37: 40–45 [DOI] [PubMed] [Google Scholar]
- Aizencang G, Solis C, Bishop DF, Warner C, Desnick RJ 2000. Human uroporphyrinogen-III synthase: Genomic organization, alternative promoters, and erythroid-specific expression. Genomics 70: 223–231 [DOI] [PubMed] [Google Scholar]
- Ajioka RS, Phillips JD, Kushner JP 2006. Biosynthesis of heme in mammals. Biochim Biophys Acta 1763: 723–736 [DOI] [PubMed] [Google Scholar]
- Akhtar M 2003. Coproporphyrinogen III and protoporphyrinogen IX oxidase. In The porphyrin handbook (ed. Kadish KM, Smith KM, Guilard R), pp. 75–92 Academic, San Diego [Google Scholar]
- Allikmets R, Raskind WH, Hutchinson A, Schueck ND, Dean M, Koeller DM 1999. Mutation of a putative mitochondrial iron transporter gene (ABC7) in X-linked sideroblastic anemia and ataxia (XLSA/A). Hum Mol Genet 8: 743–749 [DOI] [PubMed] [Google Scholar]
- Anderson KE, Bloomer JR, Bonkovsky HL, Kushner JP, Pierach CA, Pimstone NR, Desnick RJ 2005. Recommendations for the diagnosis and treatment of the acute porphyrias. Ann Intern Med 142: 439–450 [DOI] [PubMed] [Google Scholar]
- Anstey AV, Hift RJ 2007. Liver disease in erythropoietic protoporphyria: Insights and implications for management. Gut 56: 1009–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aplin C, Whatley SD, Thompson P, Hoy T, Fisher P, Singer C, Lovell CR, Elder GH 2001. Late-onset erythropoietic porphyria caused by a chromosome 18q deletion in erythroid cells. J Invest Dermatol 117: 1647–1649 [DOI] [PubMed] [Google Scholar]
- Astner I, Schulze JO, van den Heuvel J, Jahn D, Schubert WD, Heinz DW 2005. Crystal structure of 5-aminolevulinate synthase, the first enzyme of heme biosynthesis, and its link to XLSA in humans. EMBO J 24: 3166–3177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann AK, Campagna DR, McLoughlin EM, Agarwal S, Fleming MD, Bottomley SS, Neufeld EJ 2010. Systematic molecular genetic analysis of congenital sideroblastic anemia: Evidence for genetic heterogeneity and identification of novel mutations. Pediatr Blood Cancer 54: 273–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharati A, Badminton MN, Whatley SD, O’Brien DV, Bell HK 2006. Late-onset erythropoietic protoporphyria in association with haematological malignancy. Clin Exp Dermatol 31: 668–670 [DOI] [PubMed] [Google Scholar]
- Bhasker CR, Burgiel G, Neupert B, Emery-Goodman A, Kuhn LC, May BK 1993. The putative iron-responsive element in the human erythroid 5-aminolevulinate synthase mRNA mediates translational control. J Biol Chem 268: 12699–12705 [PubMed] [Google Scholar]
- Blackmon JB, Dailey TA, Lianchun X, Dailey HA 2002. Characterization of a human and mouse tetrapyrrole-binding protein. Arch Biochem Biophys 407: 196–201 [DOI] [PubMed] [Google Scholar]
- Bottomley SS 2006. Congenital sideroblastic anemias. Curr Hematol Rep 5: 41–49 [PubMed] [Google Scholar]
- Camaschella C 2009. Hereditary sideroblastic anemias: Pathophysiology, diagnosis, and treatment. Semin Hematol 46: 371–377 [DOI] [PubMed] [Google Scholar]
- Cazzola M, May A, Bergamaschi G, Cerani P, Rosti V, Bishop DF 2000. Familial-skewed X-chromosome inactivation as a predisposing factor for late-onset X-linked sideroblastic anemia in carrier females. Blood 96: 4363–4365 [PubMed] [Google Scholar]
- Chan RY, Schulman HM, Ponka P 1993. Expression of ferrochelatase mRNA in erythroid and non-erythroid cells. Biochem J 292: 343–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Paradkar PN, Li L, Pierce EL, Langer NB, Takahashi-Makise N, Hyde BB, Shirihai OS, Ward DM, Kaplan J, et al. 2009. Abcb10 physically interacts with mitoferrin-1 (Slc25a37) to enhance its stability and function in the erythroid mitochondria. Proc Natl Acad Sci 106: 16263–16268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Dailey HA, Paw BH 2010. Ferrochelatase forms an oligomeric complex with mitoferrin-1 and Abcb10 for erythroid heme biosynthesis. Blood 116: 628–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conder LH, Woodard SI, Dailey HA 1991. Multiple mechanisms for the regulation of haem synthesis during erythroid cell differentiation. Possible role for coproporphyrinogen oxidase. Biochem J 275: 321–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooperman SS, Meyron-Holtz EG, Olivierre-Wilson H, Ghosh MC, McConnell JP, Rouault TA 2005. Microcytic anemia, erythropoietic protoporphyria, and neurodegeneration in mice with targeted deletion of iron-regulatory protein 2. Blood 106: 1084–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradi HR, Corrigall AV, Boix E, Mohan CG, Sturrock ED, Meissner PN, Acharya KR 2006. Crystal structure of protoporphyrinogen oxidase from Myxococcus xanthus and its complex with the inhibitor acifluorfen. J Biol Chem 281: 38625–38633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter PD, Baumann M, Bishop DF 1992. Enzymatic defect in “X-linked” sideroblastic anemia: Molecular evidence for erythroid δ-aminolevulinate synthase deficiency. Proc Natl Acad Sci 89: 4028–4032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox TC, Bottomley SS, Wiley JS, Bawden MJ, Matthews CS, May BK 1994. X-linked pyridoxine-responsive sideroblastic anemia due to a Thr388-to-Ser substitution in erythroid 5-aminolevulinate synthase. N Engl J Med 330: 675–679 [DOI] [PubMed] [Google Scholar]
- Cox TC, Sadlon TJ, Schwarz QP, Matthews CS, Wise PD, Cox LL, Bottomley SS, May BK 2004. The major splice variant of human 5-aminolevulinate synthase-2 contributes significantly to erythroid heme biosynthesis. Int J Biochem Cell Biol 36: 281–295 [DOI] [PubMed] [Google Scholar]
- Crooks DR, Ghosh MC, Haller RG, Tong WH, Rouault TA 2010. Posttranslational stability of the heme biosynthetic enzyme ferrochelatase is dependent on iron availability and intact iron-sulfur cluster assembly machinery. Blood 115: 860–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dailey HA 1990. Conversion of coproporphyrinogen to protoheme in higher eukaryotes and bacteria: Terminal three enzymes. In Biosynthesis of heme and chlorophylls (ed. Dailey HA), pp. 123–161 McGraw-Hill, New York [Google Scholar]
- Dailey HA, Dailey TA 2003. Ferrochelatase. In The porphyrin handbook (ed. Kadish KM, Smith KM, Guilard R), pp. 93–122 Academic, San Diego [Google Scholar]
- Dailey TA, McManus JF, Dailey HA 2002. Characterization of the mouse protoporphyrinogen oxidase gene. Cell Mol Biol (Noisy-le-grand) 48: 61–69 [PubMed] [Google Scholar]
- Dailey TA, Woodruff JH, Dailey HA 2005. Examination of mitochondrial protein targeting of haem synthetic enzymes: In vivo identification of three functional haem-responsive motifs in 5-aminolaevulinate synthase. Biochem J 386: 381–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davids LM, Corrigall AV, Meissner PN 2006. Mitochondrial targeting of human protoporphyrinogen oxidase. Cell Biol Int 30: 416–426 [DOI] [PubMed] [Google Scholar]
- Dias JS, Macedo AL, Ferreira GC, Peterson FC, Volkman BF, Goodfellow BJ 2006. The first structure from the SOUL/HBP family of heme-binding proteins, murine P22HBP. J Biol Chem 281: 31553–31561 [DOI] [PubMed] [Google Scholar]
- Dierks P 1990. Molecular biology of eukaryotic 5-aminolevulinate synthase. In Biosynthesis of heme and chlorophylls (ed. Dailey HA), pp. 201–233 McGraw-Hill, New York [Google Scholar]
- Ducamp S, Kannengiesser C, Touati M, Garcon L, Guerci-Bresler A, Guichard JF, Vermylen C, Dochir J, Poirel HA, Fouyssac F, et al. 2011. Sideroblastic anemia: Molecular analysis of the ALAS2 gene in a series of 29 probands and functional studies of 10 missense mutations. Hum Mutat 32: 590–597 [DOI] [PubMed] [Google Scholar]
- Elder GH, Gouya L, Whatley SD, Puy H, Badminton MN, Deybach JC 2009. The molecular genetics of erythropoietic protoporphyria. Cell Mol Biol (Noisy-le-grand) 55: 118–126 [PubMed] [Google Scholar]
- Erskine PT, Senior N, Awan S, Lambert R, Lewis G, Tickle IJ, Sarwar M, Spencer P, Thomas P, Warren MJ, et al. 1997. X-ray structure of 5-aminolaevulinate dehydratase, a hybrid aldolase. Nat Struct Biol 4: 1025–1031 [DOI] [PubMed] [Google Scholar]
- Ferreira GC, Andrew TL, Karr SW, Dailey HA 1988. Organization of the terminal two enzymes of the heme biosynthetic pathway. Orientation of protoporphyrinogen oxidase and evidence for a membrane complex. J Biol Chem 263: 3835–3839 [PubMed] [Google Scholar]
- Finch C 1994. Regulators of iron balance in humans. Blood 84: 1697–1702 [PubMed] [Google Scholar]
- Fleming MD 2002. The genetics of inherited sideroblastic anemias. Semin Hematol 39: 270–281 [DOI] [PubMed] [Google Scholar]
- Furuyama K, Sassa S 2000. Interaction between succinyl CoA synthetase and the heme-biosynthetic enzyme ALAS-E is disrupted in sideroblastic anemia. J Clin Invest 105: 757–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama K, Fujita H, Nagai T, Yomogida K, Munakata H, Kondo M, Kimura A, Kuramoto A, Hayashi N, Yamamoto M 1997. Pyridoxine refractory X-linked sideroblastic anemia caused by a point mutation in the erythroid 5-aminolevulinate synthase gene. Blood 90: 822–830 [PubMed] [Google Scholar]
- Gill R, Kolstoe SE, Mohammed F, Al d-Bass A, Mosely JE, Sarwar M, Cooper JB, Wood SP, Shoolingin-Jordan PM 2009. Structure of human porphobilinogen deaminase at 2.8 Å: The molecular basis of acute intermittent porphyria. Biochem J 420: 17–25 [DOI] [PubMed] [Google Scholar]
- Goodwin RG, Kell WJ, Laidler P, Long CC, Whatley SD, McKinley M, Badminton MN, Burnett AK, Williams GT, Elder GH 2006. Photosensitivity and acute liver injury in myeloproliferative disorder secondary to late-onset protoporphyria caused by deletion of a ferrochelatase gene in hematopoietic cells. Blood 107: 60–62 [DOI] [PubMed] [Google Scholar]
- Gouya L, Puy H, Robreau AM, Bourgeois M, Lamoril J, Da Silva V, Grandchamp B, Deybach JC 2002. The penetrance of dominant erythropoietic protoporphyria is modulated by expression of wildtype FECH. Nat Genet 30: 27–28 [DOI] [PubMed] [Google Scholar]
- Gouya L, Martin-Schmitt C, Robreau AM, Austerlitz F, Da Silva V, Brun P, Simonin S, Lyoumi S, Grandchamp B, Beaumont C, et al. 2006. Contribution of a common single-nucleotide polymorphism to the genetic predisposition for erythropoietic protoporphyria. Am J Hum Genet 78: 2–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandchamp B, De Verneuil H, Beaumont C, Chretien S, Walter O, Nordmann Y 1987. Tissue-specific expression of porphobilinogen deaminase. Two isoenzymes from a single gene. Eur J Biochem 162: 105–110 [DOI] [PubMed] [Google Scholar]
- Guernsey DL, Jiang H, Campagna DR, Evans SC, Ferguson M, Kellogg MD, Lachance M, Matsuoka M, Nightingale M, Rideout A, et al. 2009. Mutations in mitochondrial carrier family gene SLC25A38 cause nonsyndromic autosomal recessive congenital sideroblastic anemia. Nat Genet 41: 651–653 [DOI] [PubMed] [Google Scholar]
- Harigae H, Furuyama K, Kimura A, Neriishi K, Tahara N, Kondo M, Hayashi N, Yamamoto M, Sassa S, Sasaki T 1999. A novel mutation of the erythroid-specific δ-aminolaevulinate synthase gene in a patient with X-linked sideroblastic anaemia. Br J Haematol 106: 175–177 [DOI] [PubMed] [Google Scholar]
- Hastka J, Lasserre JJ, Schwarzbeck A, Strauch M, Hehlmann R 1993. Zinc protoporphyrin in anemia of chronic disorders. Blood 81: 1200–1204 [PubMed] [Google Scholar]
- Helias V, Saison C, Ballif BA, Peyrard T, Takahashi J, Takahashi H, Tanaka M, Deybach JC, Puy H, Le Gall M, et al. 2012. ABCB6 is dispensable for erythropoiesis and specifies the new blood group system Langereis. Nat Genet 44: 170–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoggins M, Dailey HA, Hunter CN, Reid JD 2007. Direct measurement of metal ion chelation in the active site of human ferrochelatase. Biochemistry 46: 8121–8127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter GA, Ferreira GC 2011. Molecular enzymology of 5-aminolevulinate synthase, the gatekeeper of heme biosynthesis. Biochim Biophys Acta 1814: 1467–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe EK, Lawrence SH 2012. Allostery and the dynamic oligomerization of porphobilinogen synthase. Arch Biochem Biophys 519: 144–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannengiesser C, Sanchez M, Sweeney M, Hetet G, Kerr B, Moran E, Fuster Soler JL, Maloum K, Matthes T, Oudot C, et al. 2011. Missense SLC25A38 variations play an important role in autosomal recessive inherited sideroblastic anemia. Haematologica 96: 808–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappas A, Sassa S, Galbraith RA, Nordmann Y 1995. The porphyrias. In The metabolic and molecular basis of inherited disease (ed. Scriver CR, Beaudet AL, Sly WS, Valle D), pp. 2103–2159 McGraw-Hill, New York [Google Scholar]
- Kaya AH, Plewinska M, Wong DM, Desnick RJ, Wetmur JG 1994. Human δ-aminolevulinate dehydratase (ALAD) gene: Structure and alternative splicing of the erythroid and housekeeping mRNAs. Genomics 19: 242–248 [DOI] [PubMed] [Google Scholar]
- Key NS, Rank JM, Freese D, Bloomer JR, Hammerschmidt DE 1992. Hemolytic anemia in protoporphyria: Possible precipitating role of liver failure and photic stress. Am J Hematol 39: 202–207 [DOI] [PubMed] [Google Scholar]
- Khan AA, Quigley JG 2011. Control of intracellular heme levels: Heme transporters and heme oxygenases. Biochim Biophys Acta 1813: 668–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch M, Breithaupt C, Kiefersauer R, Freigang J, Huber R, Messerschmidt A 2004. Crystal structure of protoporphyrinogen IX oxidase: A key enzyme in haem and chlorophyll biosynthesis. EMBO J 23: 1720–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy P, Schuetz JD 2011. The role of ABCG2 and ABCB6 in porphyrin metabolism and cell survival. Curr Pharm Biotechnol 12: 647–655 [DOI] [PubMed] [Google Scholar]
- Krishnamurthy PC, Du G, Fukuda Y, Sun D, Sampath J, Mercer KE, Wang J, Sosa-Pineda B, Murti KG, Schuetz JD 2006. Identification of a mammalian mitochondrial porphyrin transporter. Nature 443: 586–589 [DOI] [PubMed] [Google Scholar]
- Lamola AA, Yamane T 1974. Zinc protoporphyrin in the erythrocytes of patients with lead intoxication and iron deficiency anemia. Science 186: 936–938 [DOI] [PubMed] [Google Scholar]
- Langer EE, Haining RG, Labbe RF, Jacobs P, Crosby EF, Finch CA 1972. Erythrocyte protoporphyrin. Blood 40: 112–128 [PubMed] [Google Scholar]
- Lash TD 2005. The enigma of coproporphyrinogen oxidase: How does this unusual enzyme carry out oxidative decarboxylations to afford vinyl groups? Bioorg Med Chem Lett 15: 4506–4509 [DOI] [PubMed] [Google Scholar]
- Lathrop JT, Timko MP 1993. Regulation by heme of mitochondrial protein transport through a conserved amino acid motif. Science 259: 522–525 [DOI] [PubMed] [Google Scholar]
- Layer G, Reichelt J, Jahn D, Heinz DW 2010. Structure and function of enzymes in heme biosynthesis. Protein Sci 19: 1137–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DS, Flachsova E, Bodnarova M, Demeler B, Martasek P, Raman CS 2005. Structural basis of hereditary coproporphyria. Proc Natl Acad Sci 102: 14232–14237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis CA Jr, Wolfenden R 2008. Uroporphyrinogen decarboxylation as a benchmark for the catalytic proficiency of enzymes. Proc Natl Acad Sci 105: 17328–17333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill R, Kispal G 2000. Maturation of cellular Fe-S proteins: An essential function of mitochondria. Trends Biochem Sci 25: 352–356 [DOI] [PubMed] [Google Scholar]
- Louie GV, Brownlie PD, Lambert R, Cooper JB, Blundell TL, Wood SP, Warren MJ, Woodcock SC, Jordan PM 1992. Structure of porphobilinogen deaminase reveals a flexible multidomain polymerase with a single catalytic site. Nature 359: 33–39 [DOI] [PubMed] [Google Scholar]
- Magness ST, Tugores A, Diala ES, Brenner DA 1998. Analysis of the human ferrochelatase promoter in transgenic mice. Blood 92: 320–328 [PubMed] [Google Scholar]
- Magness ST, Tugores A, Brenner DA 2000. Analysis of ferrochelatase expression during hematopoietic development of embryonic stem cells. Blood 95: 3568–3577 [PubMed] [Google Scholar]
- Mathews MA, Schubert HL, Whitby FG, Alexander KJ, Schadick K, Bergonia HA, Phillips JD, Hill CP 2001. Crystal structure of human uroporphyrinogen III synthase. EMBO J 20: 5832–5839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- May A, Bishop DF 1998. The molecular biology and pyridoxine responsiveness of X-linked sideroblastic anaemia. Haematologica 83: 56–70 [PubMed] [Google Scholar]
- May BK, Dogra SC, Sadlon TJ, Bhasker CR, Cox TC, Bottomley SS 1995. Molecular regulation of heme biosynthesis in higher vertebrates. Prog Nucleic Acid Res Mol Biol 51: 1–51 [DOI] [PubMed] [Google Scholar]
- Medlock AE, Dailey HA 2009. Regulation of mammalian heme biosynthesis. In Tetrapyrroles: Birth, life, and death (ed. Warren MJ, Smith AG), pp. 116–127 Landes Bioscience and Springer Science+Business Media, New York [Google Scholar]
- Medlock A, Swartz L, Dailey TA, Dailey HA, Lanzilotta WN 2007a. Substrate interactions with human ferrochelatase. Proc Natl Acad Sci 104: 1789–1793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medlock AE, Dailey TA, Ross TA, Dailey HA, Lanzilotta WN 2007b. A π-helix switch selective for porphyrin deprotonation and product release in human ferrochelatase. J Mol Biol 373: 1006–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medlock AE, Carter M, Dailey TA, Dailey HA, Lanzilotta WN 2009. Product release rather than chelation determines metal specificity for ferrochelatase. J Mol Biol 393: 308–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melefors O, Goossen B, Johansson HE, Stripecke R, Gray NK, Hentze MW 1993. Translational control of 5-aminolevulinate synthase mRNA by iron-responsive elements in erythroid cells. J Biol Chem 268: 5974–5978 [PubMed] [Google Scholar]
- Morgan RR, Errington R, Elder GH 2004. Identification of sequences required for the import of human protoporphyrinogen oxidase to mitochondria. Biochem J 377: 281–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson R, Schultz IJ, Pierce EL, Soltis KA, Naranuntarat A, Ward DM, Baughman JM, Paradkar PN, Kingsley PD, Culotta VC, et al. 2009. Discovery of genes essential for heme biosynthesis through large-scale gene expression analysis. Cell Metab 10: 119–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson JK, Shukla S, Black CM, Tachiwada T, Garfield S, Wincovitch S, Ernst DN, Agadir A, Li X, Ambudkar SV, et al. 2007. Human ABCB6 localizes to both the outer mitochondrial membrane and the plasma membrane. Biochemistry 46: 9443–9452 [DOI] [PubMed] [Google Scholar]
- Phillips JD, Whitby FG, Kushner JP, Hill CP 2003. Structural basis for tetrapyrrole coordination by uroporphyrinogen decarboxylase. EMBO J 22: 6225–6233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JD, Whitby FG, Warby CA, Labbe P, Yang C, Pflugrath JW, Ferrara JD, Robinson H, Kushner JP, Hill CP 2004. Crystal structure of the oxygen-dependant coproporphyrinogen oxidase (Hem13p) of Saccharomyces cerevisiae. J Biol Chem 279: 38960–38968 [DOI] [PubMed] [Google Scholar]
- Piomelli S, Lamola AA, Poh-Fitzpatrick MF, Seaman C, Harber LC 1975. Erythropoietic protoporphyria and lead intoxication: The molecular basis for difference in cutaneous photosensitivity. I. Different rates of disappearance of protoporphyrin from the erythrocytes, both in vivo and in vitro. J Clin Invest 56: 1519–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poh-Fitzpatrick MB 1986. The erythropoietic porphyrias. Dermatol Clin 4: 291–296 [PubMed] [Google Scholar]
- Pondarre C, Antiochos BB, Campagna DR, Clarke SL, Greer EL, Deck KM, McDonald A, Han AP, Medlock A, Kutok JL, et al. 2006. The mitochondrial ATP-binding cassette transporter Abcb7 is essential in mice and participates in cytosolic iron–sulfur cluster biogenesis. Hum Mol Genet 15: 953–964 [DOI] [PubMed] [Google Scholar]
- Pondarre C, Campagna DR, Antiochos B, Sikorski L, Mulhern H, Fleming MD 2007. Abcb7, the gene responsible for X-linked sideroblastic anemia with ataxia, is essential for hematopoiesis. Blood 109: 3567–3569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponka P 1999. Cell biology of heme. Am J Med Sci 318: 241–256 [DOI] [PubMed] [Google Scholar]
- Proulx KL, Woodard SI, Dailey HA 1993. In situ conversion of coproporphyrinogen to heme by murine mitochondria: Terminal steps of the heme biosynthetic pathway. Protein Sci 2: 1092–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puy H, Gouya L, Deybach JC 2010. Porphyrias. Lancet 375: 924–937 [DOI] [PubMed] [Google Scholar]
- Qin X, Sun L, Wen X, Yang X, Tan Y, Jin H, Cao Q, Zhou W, Xi Z, Shen Y 2010. Structural insight into unique properties of protoporphyrinogen oxidase from Bacillus subtilis. J Struct Biol 170: 76–82 [DOI] [PubMed] [Google Scholar]
- Qin X, Tan Y, Wang L, Wang Z, Wang B, Wen X, Yang G, Xi Z, Shen Y 2011. Structural insight into human variegate porphyria disease. FASEB J 25: 653–664 [DOI] [PubMed] [Google Scholar]
- Richard-Fogal C, Kranz RG 2010. The CcmC:heme:CcmE complex in heme trafficking and cytochrome c biosynthesis. J Mol Biol 401: 350–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AG, Elder GH 2001. Alternative splicing and tissue-specific transcription of human and rodent ubiquitous 5-aminolevulinate synthase (ALAS1) genes. Biochim Biophys Acta 1518: 95–105 [DOI] [PubMed] [Google Scholar]
- Rouault TA, Tong WH 2008. Iron–sulfur cluster biogenesis and human disease. Trends Genet 24: 398–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schranzhofer M, Schifrer M, Cabrera JA, Kopp S, Chiba P, Beug H, Mullner EW 2006. Remodeling the regulation of iron metabolism during erythroid differentiation to ensure efficient heme biosynthesis. Blood 107: 4159–4167 [DOI] [PubMed] [Google Scholar]
- Schubert HL, Phillips JD, Heroux A, Hill CP 2008. Structure and mechanistic implications of a uroporphyrinogen III synthase–product complex. Biochemistry 47: 8648–8655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert HL, Erskine PT, Cooper JB 2009. 5-Aminolaevulinic acid dehydratase, porphobilinogen deaminase and uroporphyrinogen III synthase. In Tetrapyrroles: Birth, life and death (ed. Warren MJ, Smith AG), pp. 43–73 Landes Bioscience and Springer Science+Business Media, New York [Google Scholar]
- Schultz IJ, Chen C, Paw BH, Hamza I 2010. Iron and porphyrin trafficking in heme biogenesis. J Biol Chem 285: 26753–26759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellers VM, Wu CK, Dailey TA, Dailey HA 2001. Human ferrochelatase: Characterization of substrate–iron binding and proton-abstracting residues. Biochemistry 40: 9821–9827 [DOI] [PubMed] [Google Scholar]
- Shaw GC, Cope JJ, Li L, Corson K, Hersey C, Ackermann GE, Gwynn B, Lambert AJ, Wingert RA, Traver D, et al. 2006. Mitoferrin is essential for erythroid iron assimilation. Nature 440: 96–100 [DOI] [PubMed] [Google Scholar]
- Shipulina N, Smith A, Morgan WT 2000. Heme binding by hemopexin: Evidence for multiple modes of binding and functional implications. J Protein Chem 19: 239–248 [DOI] [PubMed] [Google Scholar]
- Shoolingin-Jordan PM 2003. The biosynthesis of coproporphyrinogen III. In The porphyrin handbook (ed. Kadish KM, Smith KM, Guilard R), pp. 33–92 Academic, San Diego [Google Scholar]
- Silva PJ, Ramos MJ 2011. Computational characterization of the substrate-binding mode in coproporphyrinogen III oxidase. J Phys Chem B 115: 1903–1910 [DOI] [PubMed] [Google Scholar]
- Sopena YE, Ferramola de Sancovich AM, Sancovich HA 2008. Hexachlorobenzene treatment on hepatic mitochondrial function parameters and intracellular coproporphyrinogen oxidase location. Int J Toxicol 27: 455–465 [DOI] [PubMed] [Google Scholar]
- Takahashi S, Taketani S, Akasaka JE, Kobayashi A, Hayashi N, Yamamoto M, Nagai T 1998. Differential regulation of coproporphyrinogen oxidase gene between erythroid and nonerythroid cells. Blood 92: 3436–3444 [PubMed] [Google Scholar]
- Takahashi K, Ikeda N, Nonoguchi N, Kajimoto Y, Miyatake S, Hagiya Y, Ogura S, Nakagawa H, Ishikawa T, Kuroiwa T 2011. Enhanced expression of coproporphyrinogen oxidase in malignant brain tumors: CPOX expression and 5-ALA-induced fluorescence. Neuro Oncol 13: 1234–1243 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Taketani S, Inazawa J, Abe T, Furukawa T, Kohno H, Tokunaga R, Nishimura K, Inokuchi H 1995. The human protoporphyrinogen oxidase gene (PPOX): Organization and location to chromosome 1. Genomics 29: 698–703 [DOI] [PubMed] [Google Scholar]
- Taketani S, Furukawa T, Furuyama K 2001. Expression of coproporphyrinogen oxidase and synthesis of hemoglobin in human erythroleukemia K562 cells. Eur J Biochem 268: 1705–1711 [PubMed] [Google Scholar]
- Todd DJ 1994. Erythropoietic protoporphyria. Br J Dermatol 131: 751–766 [DOI] [PubMed] [Google Scholar]
- Troadec MB, Warner D, Wallace J, Thomas K, Spangrude GJ, Phillips J, Khalimonchuk O, Paw BH, Ward DM, Kaplan J 2011. Targeted deletion of the mouse Mitoferrin1 gene: From anemia to protoporphyria. Blood 117: 5494–5502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida M, Emi Y, Kida Y, Sakaguchi M 2008. Human ABC transporter isoform B6 (ABCB6) localizes primarily in the Golgi apparatus. Biochem Biophys Res Commun 369: 369–375 [DOI] [PubMed] [Google Scholar]
- Tugores A, Magness ST, Brenner DA 1994. A single promoter directs both housekeeping and erythroid preferential expression of the human ferrochelatase gene. J Biol Chem 269: 30789–30797 [PubMed] [Google Scholar]
- von und zu Fraunberg M, Nyroen T, Kauppinen R 2003. Mitochondrial targeting of normal and mutant protoporphyrinogen oxidase. J Biol Chem 278: 13376–13381 [DOI] [PubMed] [Google Scholar]
- Wang J, Pantopoulos K 2011. Regulation of cellular iron metabolism. Biochem J 434: 365–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, He F, Bu J, Liu X, Du W, Dong J, Cooney JD, Dubey SK, Shi Y, Gong B, et al. 2012. ABCB6 mutations cause ocular coloboma. Am J Hum Genet 90: 40–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whatley SD, Ducamp S, Gouya L, Grandchamp B, Beaumont C, Badminton MN, Elder GH, Holme SA, Anstey AV, Parker M, et al. 2008. C-terminal deletions in the ALAS2 gene lead to gain of function and cause X-linked dominant protoporphyria without anemia or iron overload. Am J Hum Genet 83: 408–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingert RA, Galloway JL, Barut B, Foott H, Fraenkel P, Axe JL, Weber GJ, Dooley K, Davidson AJ, Schmid B, et al. 2005. Deficiency of glutaredoxin 5 reveals Fe–S clusters are required for vertebrate haem synthesis. Nature 436: 1035–1039 [DOI] [PubMed] [Google Scholar]
- Wu CK, Dailey HA, Rose JP, Burden A, Sellers VM, Wang BC 2001. The 2.0 Å structure of human ferrochelatase, the terminal enzyme of heme biosynthesis. Nat Struct Biol 8: 156–160 [DOI] [PubMed] [Google Scholar]
- Yamauchi K, Hayashi N, Kikuchi G 1980. Translocation of δ-aminolevulinate synthase from the cytosol to the mitochondria and its regulation by hemin in the rat liver. J Biol Chem 255: 1746–1751 [PubMed] [Google Scholar]
- Yi L, Ragsdale SW 2007. Evidence that the heme regulatory motifs in heme oxygenase-2 serve as a thiol/disulfide redox switch regulating heme binding. J Biol Chem 282: 21056–21067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Dailey HA 1998. Erythroid 5-aminolevulinate synthase is required for erythroid differentiation in mouse embryonic stem cells. Blood Cells Mol Dis 24: 41–53 [DOI] [PubMed] [Google Scholar]
- Zhang L, Guarente L 1995. Heme binds to a short sequence that serves a regulatory function in diverse proteins. EMBO J 14: 313–320 [DOI] [PMC free article] [PubMed] [Google Scholar]