Abstract
Objective
To investigate the association between inherited variation in the estrogen receptor beta (ERβ) gene (ESR2) and ERβ lung tumor expression, a phenotype that possibly affects survival differently in men and women.
Methods
We genotyped 135 lung cancer patients for 22 ESR2 single nucleotide polymorphisms (SNPs) and measured nuclear and cytoplasmic ERβ expression by immunohistochemistry (IHC) in their primary lung tumor. Distributing Allred ERβ IHC scores according to ESR2 genotype classified under a dominant genetic model, we used rank sum tests to identify ESR2 SNPs significantly associated (p<0.05) with ERβ expression.
Results
35%, 35%, and 29% of lung tumors showed no/low (Allred <6), intermediate (Allred 6 to 7), and maximal (Allred 8) cytoplasmic ERβ expression, whereas 13%, 27%, and 60% showed no/low, intermediate, and maximal nuclear ERβ expression. For SNPs rs8021944, rs1256061 and rs10146204, ERβ expression was higher according to the rank sum test in lung tumors from patients with at least one minor allele. For each of these three SNPs, the odds of maximal (Allred 8) relative to no/low (Allred <6) ERβ expression was 3-fold higher in tumors from patients with at least one minor allele than in tumors from patients homozygous for the common allele.
Conclusion
Inherited variability in ESR2 may determine ERβ lung tumor expression.
Keywords: lung cancer, genetic polymorphism, estrogen receptor
Introduction
In 1996, Kuiper et al. [1] described ERβ, an estrogen receptor (ER) isoform coded by the estrogen receptor 2 (ESR2) gene on chromosome 14q23.2. Using immunohistochemistry (IHC), Schwartz et al. detected nuclear ERβ expression in 170 (61%) of 278 lung cancer samples and in 2 (20%) of 10 normal lung samples [2]. Though generally associated with better survival [3–6], nuclear ERβ lung tumor expression, in one study [2], portended poorer survival in women and better survival in men. Having observed both cytoplasmic and nuclear ERβ expression in both normal and subject-matched lung tumor cells, we recently reported poorer outcomes in association with cytoplasmic ERβ lung tumor expression [7]. In a study from Taiwan [6], moderate to strong nuclear ERβ expression occurred less frequently in lung cancer tissue from patients with than patients without a history of cigarette smoking. Genetic variation in ESR2 has been associated with prostate [8, 9], colorectal [10], and breast cancer risk [11–16].
In the same study population we used to study the prognostic importance of lung tumor estrogen receptor expression [7], we examined cytoplasmic and nuclear ERβ lung tumor expression in relation to 22 ESR2 single nucleotide polymorphisms (SNPs). We speculate that inherited variation in ESR2 affects ERβ expression in transformed cells, either directly, or indirectly, by selectively favoring the development of lung tumors with specific expression patterns. To our knowledge, no other study has reported associations between inherited ESR2 gene variation and ERβ protein expression in lung tumors.
Materials and Methods
Study population
The study population, designed as a convenient sample to enable systematic study of a lung tumor marker panel [7], included 204 ≥ 21 year-old patients who received surgery between 1990 and 2006 at a University of Pittsburgh Medical Center hospital for staging or treatment of biopsy-confirmed primary lung cancer. We assembled risk factor and tumor information from several sources, including outpatient paper charts, inpatient and outpatient electronic medical records, and hospital-based cancer registries.
The absence of blood or tissue for DNA extraction reduced the study sample to 185. Low DNA quantity or poor quality further reduced the sample to 172. Excluding 26 subjects with poor genotype call rates (<15 of 18 and <3 of 4 SNP genotypes called on two separate Sequenom multiplex assays) and 11 subjects lacking information about ERβ expression, 135 subjects remained for analysis. This group with available genotype and ERβ tumor expression data included: 54% women, median age 68 years (inter-quartile range 60–75 years), 89% white and 5% black, and 86% current or former cigarette smoker, 10% never smoker, and 4% smoking history unknown. The case series included 10 small cell and 125 non-small cell lung tumors (93% of total; 52% adenocarcinoma, 39% squamous cell, and 9% other non-small cell histology; 60% early (stage I–II), 38% advanced (stage III–IV or recurrent), and 2% unknown stage). The frequency of exclusion did not vary significantly (p>0.1) according to sex, age, smoking status, histology, stage, or lung tumor ERβ expression level. However, relatively high and low proportions of black and unknown race patients, respectively, were excluded (Supplemental Table 1). The University of Pittsburgh Institutional Review Board approved subject recruitment and tissue use protocols.
SNP selection
We queried Medline®, NCBI Entrez SNP1, Cancer Genome Anatomy Project (CGAP) SNP500Cancer Database2 [17], and FastSNP3 [18] to identify both commonly studied ESR2 SNPs and putative functional ESR2 SNPs located in coding or promoter regions. This procedure identified six SNPs, the AluI SNP (rs4986938) in the 3′-untranslated region of exon 8, the RsaI SNP (rs1256049) in the exon 6 ligand binding domain, and four other SNP500Cancer Database SNPs (rs8006145, rs1256031, rs1256030, and rs3020450). In addition, using data from the International HapMap project (www.hapmap.org; release #24 phase 1 & 2 full dataset; CEU population) and Haploview 4.1 [19] software, we selected 19 tagSNPs to capture common inherited variation in ESR2. TagSNPs capturing common variants [minor allele frequency (MAF)≥0.05] in a region spanning 20 kb upstream and 20 kb downstream of the estrogen receptor beta isoform 2 (NM_001040276) with pairwise correlation r2≥0.80 were chosen by Haploview’s Tagger [20]. Genotyping efforts failed for three SNPs, rs1256031, rs1273196 and rs8018687, leaving 22 SNPs (genotype call rate > 95%) available for analysis. Genotype frequencies in white subjects for one SNP (rs1256120) deviated from Hardy-Weinberg equilibrium (p=0.031). The final 22 SNP set captured (r2 ≥ 0.80) 84 (91%) of the 92 CEU HapMap SNPs with MAF ≥ 0.05.
Genotyping
DNA was extracted using isolation kits from Gentra Systems Inc. (Minneapolis, MN), EASY-DNA Kit from Invitrogen Corporation (Carlsbad, CA), or DNeasy Kit from Qiagen Inc. (Valencia, CA). MassARRAY® iPLEX Gold (Sequenom, Inc., San Diego, CA) was used to determine SNP genotypes. To evaluate genotype data quality, assay runs included sample duplicates, two Centre d’Etude du Polymorphisme Humain (CEPH#7038) positive controls, and two DNA sample-free negative controls. Genotyping results were 100% concordant within duplicates.
Immunohistochemical assay
As previously described [7], the ERβ IHC assay used formalin-fixed and paraffin-embedded tissue specimens, processed on tissue microarrays (n=58), whole tissue sections (n=63), or both (n=14). Slide preparations included deparaffinization and hydration with xylene and ethanol, heat-induced antigen retrieval with 10mM citrate buffer at pH 6, quenching endogenous peroxidase with 3% hydrogen peroxide for 5 min at room temperature, and blocking with non-immune normal serum for 5–20 min at room temperature. ERβ staining used anti-ERβ (MCA1974ST, Serotec) at 1:20 dilution in PBS overnight at 4 C and EnVision™ reagents (DAKO Corp., Carpinteria, CA). Final steps incubated with diaminobenzidine (DAB) chromogenic substrate at room temperature for 5–10 min and counterstained with hematoxylin for 2–2.5 min. Breast cancer tissues, with and without the application of primary antibodies, were used as positive and negative IHC controls. Representative photomicrographs from ERβ IHC can be viewed in our earlier publication [7].
Assessing cytoplasmic and nuclear staining separately, the study pathologist (S.D.) determined the percentage of tumor cells staining and the intensity of staining. Scoring for the percentage of tumor cells staining used a six-level ordinal scale (0 to 5, respectively, for no cells stained, 0–1% cells stained, 2–10% cells stained, 11–33% cells stained, 34–66% cells stained, and 67–100% cells stained). Scoring for intensity of staining used a four-level ordinal scale (0 to 3, respectively, for no, weak, moderate, and strong staining). Data analyses represented IHC expression in terms of the Allred score (range 0 to 8), the sum of the percentage and intensity scores [21], and total IHC expression by averaging the cytoplasmic and nuclear Allred scores.
Statistical analysis
Distributing Allred ERβ IHC scores according to ESR2 genotype classified under ordered and dominant genetic models, we used Jonckheere-Terpstra and Wilcoxon rank sum tests to screen for ESR2 SNPs statistically associated (p<0.05) with ERβ expression. Because the rare variant homozygous genotype was absent or too infrequent for many SNPs, only results from dominant genetic models are shown. For those SNPs statistically associated with ERβ expression according to a rank sum test, we used generalized logistic regression to estimate the strengths of association [odds ratios (OR) and 95% confidence interval (CI)] between ESR2 genotype (binary explanatory variables classified under dominant genetic models) and ERβ expression (three category response variable). To form three ERβ expression categories large enough for statistical analysis, arbitrary Allred cutpoints to define no/low (Allred <6), intermediate (Allred 6 to 7), and maximal (Allred 8) ERβ expression were used. The Wald chi-square test from ordered (cumulative) logistic regression was used to evaluate the statistical significance of the association between ESR2 genotype and three-level ERβ expression category. All analyses used SAS 9.2 (SAS Institute, Inc., Cary, North Carolina) and two-sided p-values.
Results
With expression values skewed toward higher Allred scores (Figure) and moderately correlated expression levels consistently equal or higher in the nucleus than the cytoplasm (Table 1; Spearman correlation coefficient 0.68), a roughly equal number of lung tumors (35%, 35%, and 29%) showed no/low (Allred <6), intermediate (Allred 6 to 7), and maximal (Allred 8) cytoplasmic ERβ expression, whereas 13%, 27%, and 60% showed no/low, intermediate, and maximal nuclear ERβ expression. Apart from possibly higher cytoplasmic ERβ expression in tumors from black subjects and higher nuclear ERβ expression in early stage tumors and tumors from older subjects, ERβ expression appeared independent of sex, race, age, smoking status, histology, and stage (Supplemental Table 2). As shown in Table 1 Parts B–D, tumors with no/low total ERβ expression showed no/low expression in the cytoplasm, but a range of expression in the nucleus, tumors with intermediate total ERβ expression uniformly showed at least intermediate expression in the nucleus, and tumors with maximal total ERβ expression showed, by definition, maximal expression in both the cytoplasm and nucleus.
Figure 1.
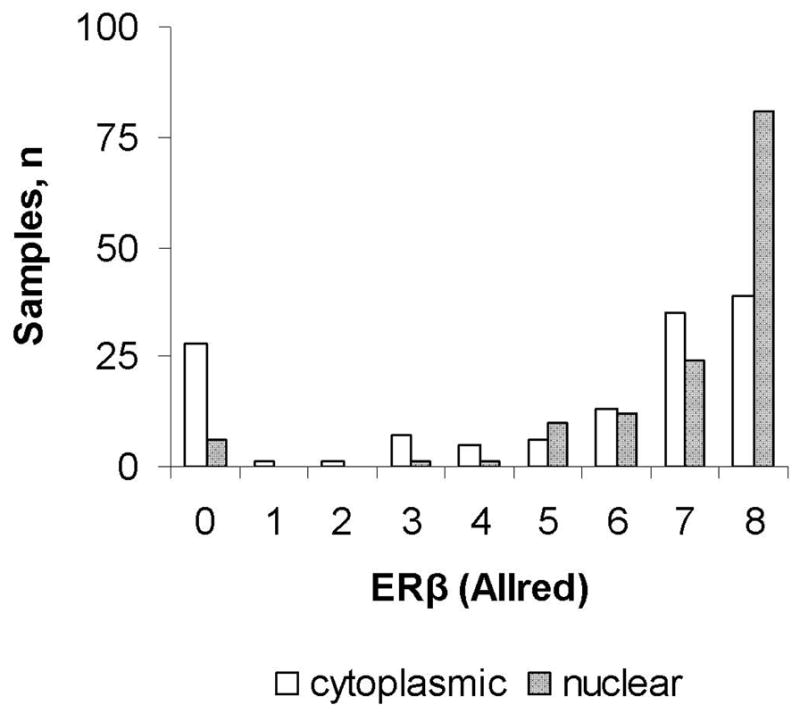
Figure Distribution of cytoplasmic and nuclear ERβ lung tumor expression scores (n=135).
Table 1.
Lung tumors cross-tabulated according to cytoplasmic and nuclear ERβ expression (n=135).
| Part A: All tumors | |||
|---|---|---|---|
| Cytoplasmic ERβ expression | Nuclear ERβ expression | ||
| no/low | intermediate | maximal | |
| no/low | 18 | 19 | 11 |
| intermediate | 0 | 16 | 32 |
| maximal | 0 | 1 | 38 |
| Part B: Tumors with no/low total ERβ expression (n=42) | |||
|---|---|---|---|
| Cytoplasmic ERβ expression | Nuclear ERβ expression | ||
| no/low | intermediate | maximal | |
| no/low | 18 | 17 | 7 |
| intermediate | 0 | 0 | 0 |
| maximal | 0 | 0 | 0 |
| Part C: Tumors with intermediate total ERβ expression (n=55) | |||
|---|---|---|---|
| Cytoplasmic ERβ expression | Nuclear ERβ expression | ||
| no/low | intermediate | maximal | |
| no/low | 0 | 2 | 4 |
| intermediate | 0 | 16 | 32 |
| maximal | 0 | 1 | 0 |
| Part D: Tumors with maximal total ERβ expression (n=38) | |||
|---|---|---|---|
| Cytoplasmic ERβ expression | Nuclear ERβ expression | ||
| no/low | intermediate | maximal | |
| no/low | 0 | 0 | 0 |
| intermediate | 0 | 0 | 0 |
| maximal | 0 | 0 | 38 |
Table 2 uses percentile cutpoints to summarize cytoplasmic, nuclear, and total ERβ Allred score distributions according to ESR2 genotype. Genotype-specific differences in ERβ expression were most evident in the nucleus, where statistically significant (p<0.05) differences were observed for three SNPs (rs8021944, rs1256061, and rs10146204). For the three SNPs associated with nuclear ERβ expression, differences in cytoplasmic and nuclear ERβ expression uniformly achieved at least borderline significance (p<0.10), with cytoplasmic, nuclear, and total ERβ expression higher in tumors from subjects with minor allele-containing genotypes.
Table 2.
Genotype distributions under a dominant genetic model for 22 ESR2 single nucleotide polymorphisms (SNPs) and genotype-specific ERβ Allred score percentile cutpointsa (35th and 70th percentile cutpoints for cytoplasmic ERβ, 13th and 40th percentile cutpoints for nuclear ERβ, 30th and 70th percentile cutpoints for total ERβ).
| Cytoplasmic ERβ | Nuclear ERβ | Total ERβ | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| SNP | Genotype | nb | P35 | P70 | p-valuec | P13 | P40 | p-valuec | P30 | P70 | p-valuec |
| rs8021944 | TT | 118 | 5.5 | 7.7 | 0.081 | 5.5 | 7.8 | 0.029 | 5.5 | 7.8 | 0.053 |
| TG+GG | 15 | 7.0 | 8.0 | 7.9 | 8.0 | 7.5 | 8.0 | ||||
|
| |||||||||||
| rs968257 | AA | 44 | 5.5 | 7.8 | 0.576 | 5.0 | 7.9 | 0.439 | 5.5 | 7.9 | 0.500 |
| AG+GG | 77 | 5.8 | 7.8 | 6.5 | 8.0 | 6.2 | 7.9 | ||||
|
| |||||||||||
| rs1152589 | AA | 31 | 6.0 | 8.0 | 0.864 | 7.0 | 8.0 | 0.064 | 6.5 | 8.0 | 0.628 |
| AT+TT | 85 | 5.8 | 7.8 | 5.5 | 7.8 | 5.5 | 7.9 | ||||
|
| |||||||||||
| rs1255998 | CC | 100 | 6.0 | 8.0 | 0.259 | 5.5 | 8.0 | 0.185 | 6.3 | 8.0 | 0.214 |
| CG+GG | 33 | 5.0 | 7.3 | 6.5 | 7.6 | 5.5 | 7.5 | ||||
|
| |||||||||||
| rs8006145 | CC | 61 | 5.6 | 7.5 | 0.570 | 5.5 | 7.7 | 0.068 | 5.5 | 7.8 | 0.390 |
| CA+AA | 60 | 5.9 | 8.0 | 6.5 | 8.0 | 6.3 | 8.0 | ||||
|
| |||||||||||
| rs4986938 AluI | GG | 44 | 5.0 | 7.3 | 0.462 | 5.0 | 7.6 | 0.137 | 5.0 | 7.5 | 0.335 |
| GA+AA | 77 | 5.8 | 8.0 | 6.5 | 8.0 | 6.4 | 8.0 | ||||
|
| |||||||||||
| rs1256063 | CC | 108 | 5.6 | 8.0 | 0.756 | 6.0 | 8.0 | 0.271 | 5.9 | 8.0 | 0.500 |
| CT+TT | 13 | 6.5 | 7.3 | 0.0 | 7.5 | 4.8 | 7.5 | ||||
|
| |||||||||||
| rs1256061 | CC | 33 | 3.0 | 7.3 | 0.054 | 5.0 | 7.4 | 0.022 | 3.3 | 7.5 | 0.039 |
| CA+AA | 88 | 6.0 | 8.0 | 6.5 | 8.0 | 6.5 | 8.0 | ||||
|
| |||||||||||
| rs1952585 | TT | 96 | 6.0 | 8.0 | 0.130 | 5.5 | 8.0 | 0.173 | 6.4 | 8.0 | 0.127 |
| TC+CC | 25 | 5.0 | 7.0 | 6.5 | 7.5 | 5.5 | 7.4 | ||||
|
| |||||||||||
| rs17766755 | GG | 46 | 5.5 | 7.3 | 0.375 | 5.0 | 7.6 | 0.140 | 5.0 | 7.5 | 0.286 |
| GA+AA | 74 | 5.8 | 8.0 | 6.1 | 8.0 | 6.4 | 8.0 | ||||
|
| |||||||||||
| rs1256049 RsaI | GG | 112 | 6.0 | 8.0 | 0.421 | 5.8 | 8.0 | 0.584 | 5.5 | 8.0 | 0.340 |
| GA+AA | 8 | 4.0 | 7.0 | 6.5 | 7.6 | 6.0 | 7.5 | ||||
|
| |||||||||||
| rs8003490 | GG | 110 | 6.0 | 8.0 | 0.054 | 5.5 | 8.0 | 0.119 | 6.6 | 8.0 | 0.065 |
| GA+AA | 23 | 4.0 | 7.0 | 6.1 | 7.5 | 5.0 | 7.5 | ||||
|
| |||||||||||
| rs12435284 | CC | 109 | 5.5 | 7.7 | 0.073 | 5.8 | 7.8 | 0.087 | 5.5 | 7.9 | 0.055 |
| CT+TT | 12 | 7.0 | 8.0 | 7.9 | 8.0 | 7.5 | 8.0 | ||||
|
| |||||||||||
| rs1256036 | AA | 33 | 5.0 | 8.0 | 0.774 | 6.5 | 8.0 | 0.434 | 5.5 | 8.0 | 0.969 |
| AG+GG | 88 | 6.0 | 7.8 | 5.5 | 8.0 | 6.2 | 7.9 | ||||
|
| |||||||||||
| rs1887994 | GG | 102 | 5.6 | 7.8 | 0.584 | 5.8 | 8.0 | 0.981 | 5.5 | 7.9 | 0.667 |
| GT+TT | 19 | 6.0 | 8.0 | 6.5 | 7.8 | 6.5 | 8.0 | ||||
|
| |||||||||||
| rs3020450 | GG | 52 | 6.0 | 7.8 | 0.886 | 5.5 | 7.9 | 0.727 | 6.0 | 7.9 | 0.842 |
| GA+AA | 69 | 5.8 | 8.0 | 6.1 | 8.0 | 5.5 | 8.0 | ||||
|
| |||||||||||
| rs3020449 | TT | 38 | 4.0 | 7.4 | 0.156 | 5.0 | 7.6 | 0.114 | 4.0 | 7.5 | 0.117 |
| TC+CC | 82 | 6.0 | 8.0 | 6.5 | 8.0 | 6.2 | 8.0 | ||||
|
| |||||||||||
| rs10137185 | CC | 106 | 5.5 | 7.8 | 0.080 | 5.5 | 7.8 | 0.149 | 5.0 | 7.9 | 0.065 |
| CT+TT | 15 | 7.0 | 8.0 | 7.6 | 8.0 | 7.4 | 8.0 | ||||
|
| |||||||||||
| rs3020443 | AA | 66 | 5.6 | 7.5 | 0.433 | 5.5 | 7.8 | 0.109 | 5.9 | 7.8 | 0.322 |
| AC+CC | 54 | 6.0 | 8.0 | 6.5 | 8.0 | 6.5 | 8.0 | ||||
|
| |||||||||||
| rs1256120 | TT | 100 | 5.3 | 7.8 | 0.567 | 5.6 | 8.0 | 0.843 | 5.3 | 7.9 | 0.561 |
| TC+CC | 19 | 6.8 | 7.9 | 6.5 | 7.9 | 7.0 | 7.9 | ||||
|
| |||||||||||
| rs10146204 | GG | 42 | 4.0 | 7.0 | 0.051 | 5.0 | 7.5 | 0.025 | 4.0 | 7.5 | 0.029 |
| GA+AA | 79 | 6.0 | 8.0 | 6.5 | 8.0 | 6.2 | 8.0 | ||||
|
| |||||||||||
| rs1256108 | TT | 30 | 3.0 | 7.4 | 0.119 | 5.0 | 7.6 | 0.211 | 3.8 | 7.6 | 0.103 |
| TC+CC | 101 | 6.0 | 8.0 | 6.5 | 8.0 | 6.6 | 8.0 | ||||
Percentile cutpoints distinguish no/low (Allred <6) from intermediate (Allred 6 to 7) and intermediate from maximal (Allred 8) ERβ expression in the entire (n=135) subject set.
ESR2 genotypes evaluable for n=133 subjects at four SNPs (rs8021944, rs1255998, rs8003490, and rs1256108) measured on one Sequenom multiplex and evaluable for n=121 subjects at 18 other SNPs measured on a separated multiplex.
Statistical significance (Wilcoxon test) of genotype-specific differences in the Allred scores.
For the three ESR2 SNPs significantly associated with ERβ expression, Table 3 uses the odds ratio (OR) to express associations between genotype and cytoplasmic, nuclear, and total ERβ Allred score categories. For each SNP, the odds of maximal (Allred 8) relative to no/low (Allred <6) ERβ expression was approximately 3-fold higher in tumors from subjects with a minor allele-containing genotype than in tumors from subjects homozygous for the common allele. For two SNPs (rs1256061 and rs10146204), statistically significant association (p<0.05) persisted, with or without adjustments for age, in analyses restricted to white subjects and/or non-small cell histology tumors (data not shown). For rs1256061, statistically significant association (p<0.05) persisted, with or without adjustment for age, for tumors with adenocarcinoma histology (data not shown).
Table 3.
Association between three ESR2 SNPs and ERβ Allred scores (n=135).
| SNP | Genotype | <6
|
6 to 7
|
8
|
p-valuea | ||||
|---|---|---|---|---|---|---|---|---|---|
| n | n | OR | 95% CI | n | OR | 95% CI | |||
| Part A. Tumors according to cytoplasmic ERβ Allred score
|
|||||||||
| rs8021944 | TT | 44 | 41 | Ref | 33 | Ref | |||
| TG+GG | 2 | 7 | 3.76 | 0.74–19.1 | 6 | 4.00 | 0.76–21.1 | 0.123 | |
| rs1256061 | CC | 15 | 13 | Ref | 5 | Ref | |||
| CA+AA | 28 | 30 | 1.24 | 0.50–3.05 | 30 | 3.21 | 1.03–10.0 | 0.056 | |
| rs10146204 | GG | 17 | 18 | Ref | 7 | Ref | |||
| GA+AA | 26 | 25 | 0.91 | 0.38–2.15 | 28 | 2.62 | 0.93–7.32 | 0.102 | |
| Part B. Tumors according to nuclear ERβ Allred score
|
|||||||||
| rs8021944 | TT | 17 | 33 | Ref | 68 | Ref | |||
| TG+GG | 1 | 1 | 0.52 | 0.03–8.76 | 13 | 3.25 | 0.40–26.6 | 0.048 | |
| rs1256061 | CC | 7 | 11 | Ref | 15 | Ref | |||
| CA+AA | 9 | 20 | 1.41 | 0.41–4.85 | 59 | 3.06 | 0.98–9.55 | 0.026 | |
| rs10146204 | GG | 9 | 13 | Ref | 20 | Ref | |||
| GA+AA | 7 | 18 | 1.78 | 0.53–6.02 | 54 | 3.47 | 1.14–10.6 | 0.017 | |
| Part C. Tumors according to total ERβ Allred score.
|
|||||||||
| rs8021944 | TT | 39 | 47 | Ref | 32 | Ref | |||
| TG+GG | 1 | 8 | 6.64 | 0.80–55.4 | 6 | 7.31 | 0.84–63.9 | 0.087 | |
| rs1256061 | CC | 14 | 14 | Ref | 5 | Ref | |||
| CA+AA | 23 | 35 | 1.52 | 0.61–3.78 | 30 | 3.65 | 1.15–11.6 | 0.029 | |
| rs10146204 | GG | 15 | 20 | Ref | 7 | Ref | |||
| GA+AA | 22 | 29 | 0.99 | 0.42–2.36 | 28 | 2.73 | 0.95–7.84 | 0.080 | |
Legend: SNP – single nucleotide polymorphism; OR – odds ratio; CI – confidence interval; Ref –reference category
Statistical significance of association between genotype and ERβ class (Wald chi-square test from ordered logistic regression).
Discussion
We observed higher ERβ expression in lung tumors from patients with a minor-allele-containing ESR2 genotype for three SNPs, rs8021944, rs1256061, and rs10146204 (Tables 2 and 3). ERβ expression differences observed in relation to two SNPs (rs1256061 and rs10146204) were independent of race, age, and tumor histology. With respect to rs1256061, differences remained statistically significant for the subset of lung tumors with adenocarcinoma histology.
The three SNPs associated with ERβ expression were selected as tagSNPs. SNP rs8021944 resides in an intron of an ESR2 gene neighbor (spectrin repeat containing nuclear envelope 2, SYNE2). SYNE2 codes for a nuclear outer membrane protein (nesprin-2) that binds cytoplasmic F-actin. In a follow-up study, our laboratory used the Illumina whole genome DASL HT Assay to profile mRNA expression in a subset of lung tumors included in the current report. We retrieved SYNE2 and ESR2 mRNA expression data available for 43 lung tumors, including 13 tumors with a high ERβ / low progesterone receptor (PR) IHC expression pattern and 30 tumors with a low ERβ / high PR IHC expression pattern. As reported in 2011 [7], these expression patterns distinguish lung tumors with less and more favorable outcomes, respectively. We observed positive correlation between the mRNA expression values of the SYNE2 and ESR2 genes (Spearman correlation coefficient = 0.39, p-value = 0.010). SNP rs1256061 resides in an intron located toward the 3′ end of ESR2. Finally, SNP rs10146204 resides 5′ of ESR2 in a genomic region between ESR2 and MTHFD1. In our white sample, these SNPs mutually showed low linkage disequilibrium (r2<0.3).
We used the National Institute of Environmental Health Sciences (NIEHS) SNP Function Prediction (FuncPred) tool4 to evaluate possible functional significance [22]. FuncPred placed rs10146204 in a transcription factor binding site. Given its location in 5′ of ESR2, genetic variation in rs10146204 may affect transcription factor binding directly and ESR2 expression secondarily. In this context, we noted a not quite statistically significant (p=0.07) association between rs10146204 and tumor stage at diagnosis among lung tumors with non-small cell histology (data not shown). FuncPred did not predict functional effects for rs8021944 or rs1256061. These SNPs may be linked to other unknown, but functional genetic variants.
Our panel included two often studied SNPs (rs1256049 [RsaI] and rs4986938 [AluI]), previously examined in relation to cancer at various sites, including colon or rectum [10], endometrium [23], ovary [24], prostate [9, 25, 26], and breast [11–14, 16, 27, 28], though implicated only in rectal (rs1256049 (RsaI); [10]) and breast cancer (rs4986938 (AluI); [12]). Though rs1256049 [RsaI] showed moderate linkage with rs1256061 (r2=0.55), differences in lung tumor ERβ expression in relation to rs1256049 [RsaI] were not statistically significant (Table 2).
Study limitations included 1) a subject sample, with limited racial heterogeneity, too small for adequate subset analysis, 2) reliance on lung tissue as a DNA source resulting in subject losses due to poor DNA quality and a potential for somatic mutation contributing to measured genetic variability, and 3) the inherent subjective and semi-quantitative nature of immunohistochemistry as a measure of protein expression. In particular, skewing of immunohistochemistry results toward higher ERβ expression limited the number of samples with no or very low expression. A study strength included mutually blind assessments of ESR2 genotype and ERβ expression.
Some studies [2–6], but not all [7], identify ERβ expression as a favorable lung cancer prognostic factor. Our study results suggest that host genetic variation in ESR2 may determine lung tumor ERβ expression. To our knowledge, no other study has evaluated inherited ESR2 genetic variation in relation to lung tumor ERβ expression. Considering the possibly specific association involving ESR2 rs1256061 and adenocarcinoma, we speculate that an ESR2 genotype and ERβ expression association may depend on tumor histology. These findings require replication.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grants P50 CA090440, R25 CA057703, and P30 CA047904. Study sponsors did not play any role in the study design, in the collection, analysis or interpretation of data, in the writing of the manuscript, or in the decision to submit the manuscript for publication.
Footnotes
Conflict of interest statement
The authors have no conflicts of interest to report.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kuiper GG, Enmark E, Pelto-Huikko M, Nilsson S, Gustafsson JA. Cloning of a novel receptor expressed in rat prostate and ovary. Proc Natl Acad Sci U S A. 1996;93(12):5925–30. doi: 10.1073/pnas.93.12.5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schwartz AG, Prysak GM, Murphy V, Lonardo F, Pass H, Schwartz J, et al. Nuclear estrogen receptor beta in lung cancer: expression and survival differences by sex. Clin Cancer Res. 2005;11(20):7280–7. doi: 10.1158/1078-0432.CCR-05-0498. [DOI] [PubMed] [Google Scholar]
- 3.Kawai H, Ishii A, Washiya K, Konno T, Kon H, Yamaya C, et al. Estrogen receptor alpha and beta are prognostic factors in non-small cell lung cancer. Clin Cancer Res. 2005;11(14):5084–9. doi: 10.1158/1078-0432.CCR-05-0200. [DOI] [PubMed] [Google Scholar]
- 4.Nose N, Sugio K, Oyama T, Nozoe T, Uramoto H, Iwata T, et al. Association between estrogen receptor-beta expression and epidermal growth factor receptor mutation in the postoperative prognosis of adenocarcinoma of the lung. J Clin Oncol. 2009;27(3):411–7. doi: 10.1200/JCO.2008.18.3251. [DOI] [PubMed] [Google Scholar]
- 5.Skov BG, Fischer BM, Pappot H. Oestrogen receptor beta over expression in males with non-small cell lung cancer is associated with better survival. Lung Cancer. 2008;59(1):88–94. doi: 10.1016/j.lungcan.2007.07.025. [DOI] [PubMed] [Google Scholar]
- 6.Wu CT, Chang YL, Shih JY, Lee YC. The significance of estrogen receptor beta in 301 surgically treated non-small cell lung cancers. J Thorac Cardiovasc Surg. 2005;130(4):979–86. doi: 10.1016/j.jtcvs.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 7.Stabile LP, Dacic S, Land SR, Lenzner DE, Dhir R, Acquafondata M, et al. Combined analysis of estrogen receptor β-1 and progesterone receptor expression identifies lung cancer patients with poor outcome. Clin Cancer Res. 2011;17(1):154–64. doi: 10.1158/1078-0432.CCR-10-0992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thellenberg-Karlsson C, Lindstrom S, Malmer B, Wiklund F, Augustsson-Balter K, Adami H-O, et al. Estrogen receptor beta polymorphism is associated with prostate cancer risk. Clin Cancer Res. 2006;12(6):1936–41. doi: 10.1158/1078-0432.CCR-05-0269. [DOI] [PubMed] [Google Scholar]
- 9.Chen Y-C, Kraft P, Bretsky P, Ketkar S, Hunter DJ, Albanes D, et al. Sequence variants of estrogen receptor beta and risk of prostate cancer in the National Cancer Institute Breast and Prostate Cancer Cohort Consortium. Cancer Epidemiol Biomarkers Prev. 2007;16(10):1973–81. doi: 10.1158/1055-9965.EPI-07-0431. [DOI] [PubMed] [Google Scholar]
- 10.Slattery ML, Sweeney C, Murtaugh M, Ma KN, Wolff RK, Potter JD, et al. Associations between ERalpha, ERbeta, and AR genotypes and colon and rectal cancer. Cancer Epidemiol Biomarkers Prev. 2005;14(12):2936–42. doi: 10.1158/1055-9965.EPI-05-0514. [DOI] [PubMed] [Google Scholar]
- 11.Breast and Prostate Cancer Cohort Consortium. Haplotypes of the estrogen receptor beta gene and breast cancer risk. Int J Cancer. 2008;122(2):387–92. doi: 10.1002/ijc.23127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gallicchio L, Berndt SI, McSorley MA, Newschaffer CJ, Thuita LW, Argani P, et al. Polymorphisms in estrogen-metabolizing and estrogen receptor genes and the risk of developing breast cancer among a cohort of women with benign breast disease. BMC Cancer. 2006;6:173. doi: 10.1186/1471-2407-6-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gold B, Kalush F, Bergeron J, Scott K, Mitra N, Wilson K, et al. Estrogen receptor genotypes and haplotypes associated with breast cancer risk. Cancer Res. 2004;64(24):8891–900. doi: 10.1158/0008-5472.CAN-04-1256. [DOI] [PubMed] [Google Scholar]
- 14.Maguire P, Margolin S, Skoglund J, Sun X-F, Gustafsson J-A, Borresen-Dale A-L, et al. Estrogen receptor beta (ESR2) polymorphisms in familial and sporadic breast cancer. Breast Cancer Res Treat. 2005;94(2):145–52. doi: 10.1007/s10549-005-7697-7. [DOI] [PubMed] [Google Scholar]
- 15.Tsezou A, Tzetis M, Gennatas C, Giannatou E, Pampanos A, Malamis G, et al. Association of repeat polymorphisms in the estrogen receptors alpha, beta (ESR1, ESR2) and androgen receptor (AR) genes with the occurrence of breast cancer. Breast. 2008;17(2):159–66. doi: 10.1016/j.breast.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 16.Zheng SL, Zheng W, Chang B-l, Shu X-O, Cai Q, Yu H, et al. Joint effect of estrogen receptor beta sequence variants and endogenous estrogen exposure on breast cancer risk in Chinese women. Cancer Res. 2003;63(22):7624–9. [PubMed] [Google Scholar]
- 17.Packer B, Yeager M, Burdett L, Welch R, Beerman M, Qi L, et al. SNP500Cancer: a public resource for sequence validation, assay development, and frequency analysis for genetic variation in candidate genes. Nucleic Acids Res. 2006;34(Database issue):D617–D21. doi: 10.1093/nar/gkj151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan H-Y, Chiou J-J, Tseng W-H, Liu C-H, Liu C-K, Lin Y-J, et al. FASTSNP: An always up-to-date and extendable service for SNP function analysis and prioritization. Nucleic Acids Res. 2006;34:W635–W41. doi: 10.1093/nar/gkl236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barrett J, Fry B, JM, MJD Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 20.de Bakker P, Yelensky R, Pe’er I, Gabriel S, Daly M, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–23. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 21.Allred DC, Harvey JM, Berardo M, Clark GM. Prognostic and predictive factors in breast cancer by immunohistochemical analysis. Mod Pathol. 1998;11(2):155–68. [PubMed] [Google Scholar]
- 22.Xu Z, Taylor JA. SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res. 2009;37(Web Server issue):W600–5. doi: 10.1093/nar/gkp290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Setiawan VW, Hankinson SE, Colditz GA, Hunter DJ, De Vivo I. Estrogen receptor beta (ESR2) polymorphisms and endometrial cancer (United States) Cancer Cause Control. 2004;15(6):627–33. doi: 10.1023/B:CACO.0000036170.28502.5f. [DOI] [PubMed] [Google Scholar]
- 24.Leigh Pearce C, Near AM, Butler JL, Van Den Berg D, Bretsky P, Conti DV, et al. Comprehensive evaluation of ESR2 variation and ovarian cancer risk. Cancer Epidemiol Biomarkers Prev. 2008;17(2):393–6. doi: 10.1158/1055-9965.EPI-07-2512. [DOI] [PubMed] [Google Scholar]
- 25.Sun Y-h, Yang B, Wang X-h, Xu C-l, Gao X-f, Gao X, et al. Association between single-nucleotide polymorphisms in estrogen receptor beta gene and risk of prostate cancer. Chung-Hua Wai Ko Tsa Chih [Chinese Journal of Surgery] 2005;43(14):948–51. [PubMed] [Google Scholar]
- 26.McIntyre MH, Kantoff PW, Stampfer MJ, Mucci LA, Parslow D, Li H, et al. Prostate cancer risk and ESR1 TA, ESR2 CA repeat polymorphisms. Cancer Epidemiol Biomarkers Prev. 2007;16(11):2233–6. doi: 10.1158/1055-9965.EPI-07-0481. [DOI] [PubMed] [Google Scholar]
- 27.Forsti A, Zhao C, Israelsson E, Dahlman-Wright K, Gustafsson J-A, Hemminki K. Polymorphisms in the estrogen receptor beta gene and risk of breast cancer: no association. Breast Cancer Res Treat. 2003;79(3):409–13. doi: 10.1023/a:1024020609833. [DOI] [PubMed] [Google Scholar]
- 28.Georgopoulos NA, Adonakis GL, Fotopoulos A, Koika V, Spinos N, Saltamavros A, et al. Estrogen receptor polymorphisms in tamoxifen-treated women with breast cancer. Gynecol Endocrinol. 2006;22(4):185–9. doi: 10.1080/09513590600645767. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


