Abstract
Chiral poly-amido-saccharides (PASs) with a defined molecular weight and narrow polydispersity are synthesized using an anionic ring-opening polymerization of a β-lactam sugar monomer. The PASs have a previously unreported main chain structure that is composed of pyranose rings linked through the 1- and 2-positions by an amide bond with α-stereochemistry. The monomer is synthesized in one-step from benzyl-protected d-glucal and polymerized using mild reaction conditions to give degrees of polymerization ranging from 25 to >150 in high yield. Computational modeling reveals how the monomer’s structure and steric bulk affect the thermodynamics and kinetics of polymerization. Protected and deprotected polymers and model compounds are characterized using a variety of methods (NMR, GPC, IR, DLS, etc.). Reductive debenzylation provides the deprotected, hydrophilic polymers in high yield. Based on circular dichroism, the deprotected polymers possess a regular secondary structure in aqueous solution, which agrees favorably with the prediction of a helical structure using molecular modeling. Furthermore, we provide evidence suggesting that the polymers bind the lectin concanavalin A at the same site as natural carbohydrates, showing the potential of these polymers to mimic natural polysaccharides. PASs offer the advantages associated with synthetic polymers, such as greater control over structure and derivitization, and less batch-to-batch variation. At the same time, they preserve many of the structural features of natural polysaccharides, such as a stereochemically regular, rigid pyranose backbone, that make natural carbohydrate polymers important materials both for their unique properties and useful applications.
INTRODUCTION
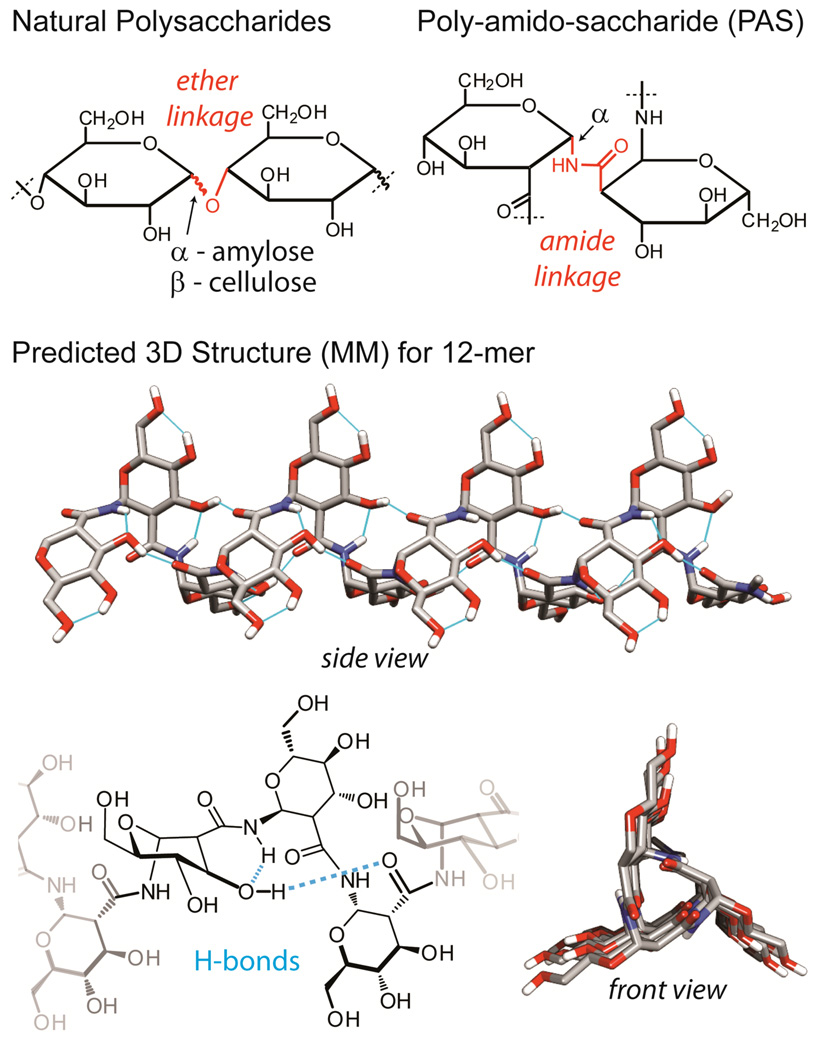
Carbohydrate-based polymers that retain the chiral, cyclic main chain structure of natural polysaccharides (Figure 1, top left) and that can be prepared by controlled synthetic methods are of interest for both basic studies and applications. Specifically, novel polymeric structures having a hydrophilic pyranose backbone not joined with ether linkages1 are interesting because these materials are not found in nature and provide new molecular architectures to be explored. Amide-linked polysaccharides, which we term poly-amido-saccharides (PASs), are an example of one such polymeric structure. However, access to high molecular weight PASs requires the development of new polymerization methods. Here, we present the first synthesis of a 1,2-linked glucose-based poly-amido-saccharide (Figure 1, top right), which is prepared via a robust and controlled anionic ring-opening polymerization of a β-lactam sugar monomer.
Figure 1.
Comparison of natural polysaccharides to poly-amido-saccharides (PASs) and predicted structure of an α-N-1,2-d-glucose poly-amido-saccharide (α-N- 1,2-d-glc PAS) 12-mer based on gas phase minimization with MMFF94s.
Interest in polysaccharides stems from their many varied and essential roles in biological systems, including storing energy (starch), forming rigid structural materials (cellulose), and modulating protein interactions and activity.2 Examples, such as chitosan3 and hyaluronic acid4, are used clinically in their isolated form and with further functionalization as engineered biomaterials.5 However, polysaccharides isolated from natural sources can be polydisperse and show batch-to-batch variations. Additionally, they may require extensive purification and removal of endotoxins prior to use in biomedical applications. The introduction of synthetic methodologies to prepare polymers that mimic natural polysaccharides may give researchers the molecular-level control they are accustomed to with synthetic polymers while taking advantage of the unique chemical and physical properties of natural polysaccharides.
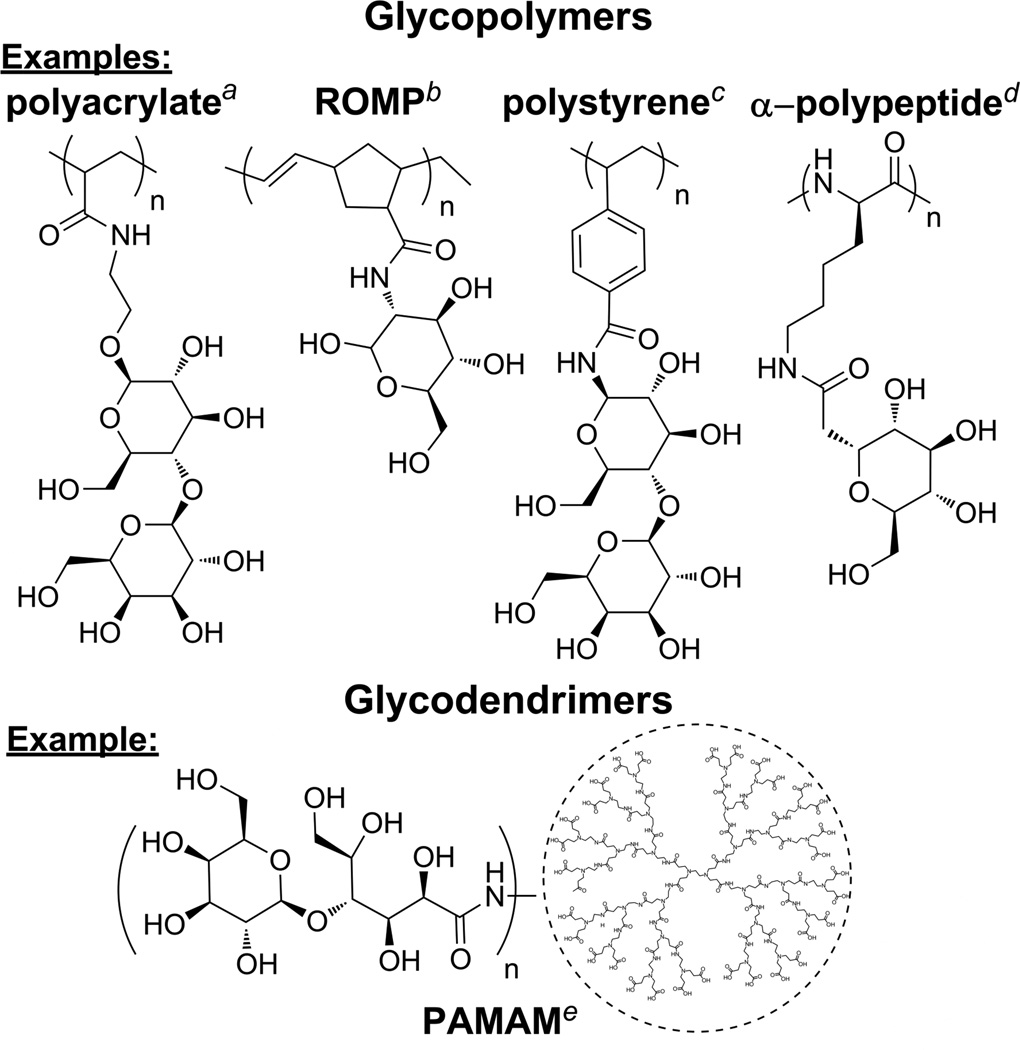
A potential alternative to polysaccharide synthesis is the conjugation of pendent sugar moieties to synthetic polymers and dendrimers to form glycoconjugates (Chart 1). For example, glycopolymers6 and glycodendrimers7 replicate the carbohydrate multivalency commonly found in nature. However, for other biomaterials applications, the lack of a rigid, stereochemically defined pyranose backbone may be a limitation for these materials. Polymer chemists have prepared synthetic polymers with rigid backbones from both carbohydrate and non-carbohydrate starting materials that show varying levels of structural similarity to polysaccharides,8 but many of these materials are not stereochemically defined. Others have used the opening of the carbohydrate ring as a strategy to make polymers that can have defined stereochemistry, but at the expense of losing the rigidity imparted by the pyranose ring.9
Chart 1.
Examples of glycoconjugates
It is a challenge for polymer chemists to synthesize carbohydrate polymers that retain both the cyclic pyranose backbone and the stereochemistry of common natural polysaccharides. Ideally, homopolysaccharides could be accessed using a single polymerization reaction, and such approaches to prepare polymers with a stereochemically defined pyranose backbone are highly desired. However, carbohydrates are challenging synthetic targets because they have a high density of similar functional groups and are sterochemically complex.11 Advances in both solution and solid-phase synthesis provide reliable access to complex oligosaccharides with molecular weights (MW) that are generally less than 2 kDa, but step-wise approaches are not amenable to preparing polysaccharides with high degrees of polymerization (DP).12
Cationic ring-opening polymerization (ROP) works well to synthesize 1,6-linked polysaccharides with high degrees of polymerization (DPs > 100),13 but is less effective in preparing polysaccharides with other linkages, such as 1,4-linked cellulose14 (DP < 20) and chitin15 (DP < 14)).6f,16 Furthermore, cationic ROP is not used to make commercially available polysaccharides, but instead these materials are isolated from natural sources or produced using fermentation.17 The use of isolated enzymes or microorganisms for the controlled synthesis of polysaccharides is a promising alternative to chemical methods.18 Enzymatic approaches can avoid the use of protecting groups, but may require expensive activated monomers. Additionally, the synthesis of nonnatural polysaccharides with unique geometries and linkages, such as those described in this report, may be a challenge for natural enzymes.
Our approach replaces the ether linkage found in natural polysaccharides with an amide linkage to provide poly-amido-saccharides (PAS) (Figure 1.). Specifically, we report a mild and high-yielding method to synthesize α-N-1,2-d-glucose (α-N-1,2-d-glc) PASs of defined molecular weight with a low polydispersity index (PDI) via the anionic ring-opening polymerization of β-lactam sugar monomer 1 (Scheme 1). Molecules containing pyranose and furanose rings joined via amide linkages have been previously reported, but they differ from the current findings in that they have been prepared via a step-wise approach from sugar amino acids and are oligomers(DP < 10).11b,19 However, oligomers containing the specific α-N-1,2-linkage reported here have not been previously reported. Because α-N-1,2-d-glucose PASs contain a 1,2-peptide linkage they can be considered highly functionalized β-polypeptides. β-polypeptides are a class of synthetic polymers known to form defined secondary structures and are of significant interest for a variety of applications.20 Based on molecular modeling, α-N-1,2-d-glc PASs are predicted to have a helical structure that is promoted by extensive internal hydrogen-bonding and by the rigidity of the pyranose-polyamide backbone (Figure 1).
Scheme 1.
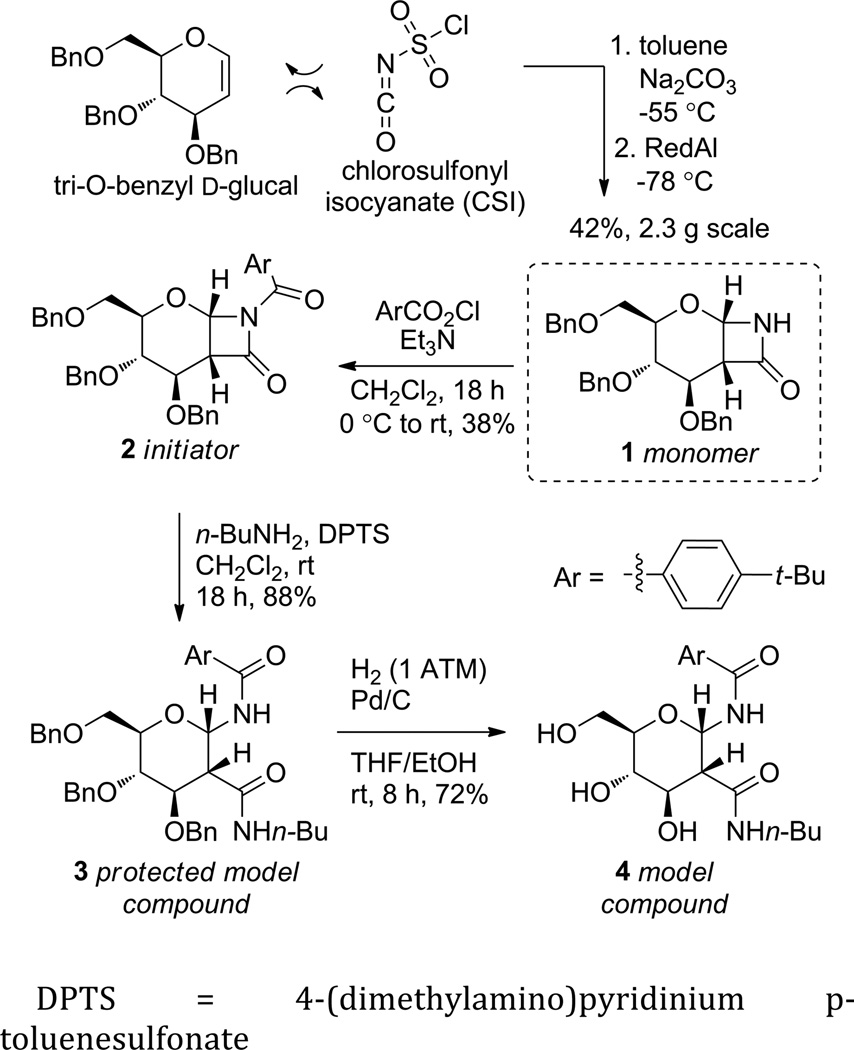
Monomer synthesis.
DPTS = 4-(dimethylamino)pyridinium p-toluenesulfonate
In addition to the synthesis and characterization of α-N-1,2-d-glucose PASs, we discuss how the β-lactam sugar monomer’s structure and steric bulk affect the thermodynamics of polymerization by calculating the monomer’s ring strain and comparing it to other β-lactam monomers. In addition, we comment on how the steric bulk affects the kinetics of the reaction and propose an explanation for why the benzyl-protected monomer polymerizes easily, while the tert-butyldimethylsilyl-protected monomer does not polymerize. We examine the effect of MW on the CD spectrum and solid-state morphology using scanning electron microscopy (SEM). Finally, we report the glucose-dependent binding of higher MW α-N-1,2-glc PASs to the plant lectin concanavalin A using an established aggregation assay,21 showing the potential of PASs to interact with natural carbohydrate receptors.
MATERIALS AND METHODS
Computational Methods
Molecular models were constructed and minimized using the freely available software Avogadro (MMFF94s) and GAMESS 1122 (AM1, DFT[B3LYP/6-31G(d)]). X-ray crystal structures (XRCSs) were used as initial geometries when available (2-azetidinone23, CS424, CS525). For AM1 and DFT methods, minimized structures were verified by confirming that no imaginary frequencies were present. The 12-mer of α-N-1,2-glc PAS was minimized using MMFF94s.
Polymerization. Polymer P1’
In an oven-dried flask under nitrogen, lactam 1 (0.500 g, 1.09 mmol) was dissolved in 9 mL of distilled tetrahydrofuran (THF) dried over 4A molecular sieves. The reaction flask was cooled to 0 °C in an ice bath and initiator 2 (0.027 g, 0.044 mmol, 4.0 mol%) was added as a solution in THF (1 mL). Next, 0.090 mL of a 1.0 M solution of LiHMDS in THF (0.088 mmol, 8.0 mol%) was added and the solution was stirred for 1 h, at which time a drop of sat’d NH4Cl solution was added. Reaction progress was monitored by observing the disappearance of the monomer using TLC. After evaporation of THF, the resulting solid was redissolved in diethyl ether (50 mL) and washed with 1 M HCl, sat’d NaHCO3, and brine. After drying over sodium sulfate, the crude reaction was isolated and then redissolved in the minimum amount of dichloromethane. The polymer was precipitated by adding drop wise into a flask of stirred, cold pentane (50 mL), and then collected by filtration. The solid was redissolved in the minimum amount of dichloromethane and precipitated by adding drop wise into a flask of stirred, cold methanol (50 mL), and then collected by filtration. After drying under high vacuum, 0.444 g (84%) of a white powder was isolated. [α]D = 79.1 (7.1 mg/mL in CH2Cl2, 26 °C); 1H NMR (500 MHz, CDCl3): δ 7.8 (br, 1H), 7.4-6.9 (br, 15H), 5.7 (br, 1H), 4.75-4.25 (br, 5H), 4.2-3.8 (br, 2H), 3.75-3.4 (br, 4H), 2.8 (br, 1H), 1.2 (s, end group, 9H); 13C NMR (126 MHz, CDCl3): δ 170.6, 138.5(2), 138.3, 128.5-127.3, 78.3, 75.0-73.0, 73.1, 68.5, 51.2, 35.0, 31.2; IR (KBr): 1686 cm−1 (C=O); GPC(THF): Mn = 9,500; Mw = 10,500; PDI (Mw/Mn) = 1.1.
Polymer Debenzylation. Polymer P1
Polymer P1’ (0.155 g) and 0.045 g (1.2 equiv) of KOt-Bu were dissolved in 5.0 mL of THF. The polymer solution was added drop wise to a rapidly stirred solution of sodium in anhydrous liquid ammonia (50 mL) at −78 °C under nitrogen. Sodium was washed in toluene and hexane and cut into small pieces before addition. The solution’s deep blue color was maintained by adding additional sodium. After 1 h at −78 °C, sat’d ammonium chloride was added until the blue color disappeared. After evaporation of the ammonia at room temperature, the resulting aqueous layer was washed with diethyl ether twice and then filtered through a 0.22 micron PVDF syringe filter to remove particulates. The solution was dialyzed with 1,000 MWCO tubing for 12 hours with 3 water changes. After lyophilization, the resulting white solid was washed with methanol (10 mL) and collected by decantation after centrifugation a total of three times. Residual methanol was removed under high vacuum. P1 (0.063 g, 98%) was obtained as a white solid. [α]D = 129 (2.0 mg/mL in H20, 24 °C); 1H NMR (500 MHz, D2O): δ 5.75 (d, J = 4.8 Hz, 1H), 4.12 (pseudo t, J = 10.1, 1H), 3.75 (br s, 2H), 3.47 (pseudo t, J = 9.6, 1H), 3.42 (m, J = 10.2, 1H), 3.04 (dd, J = 11.2, 4.8, 1H); 13C NMR (126 MHz, D2O): δ 171.5, 76.0, 74.4, 71.2, 69.7, 61.6, 52.4; IR (ATR): 1682 cm−1(C=O); DLS(H2O, 50 °C) Mw = 5,000; %PD = 26.
Additional experiment details are presented in Supporting Information.
RESULTS AND DISCUSSION
Monomer Design and Synthesis
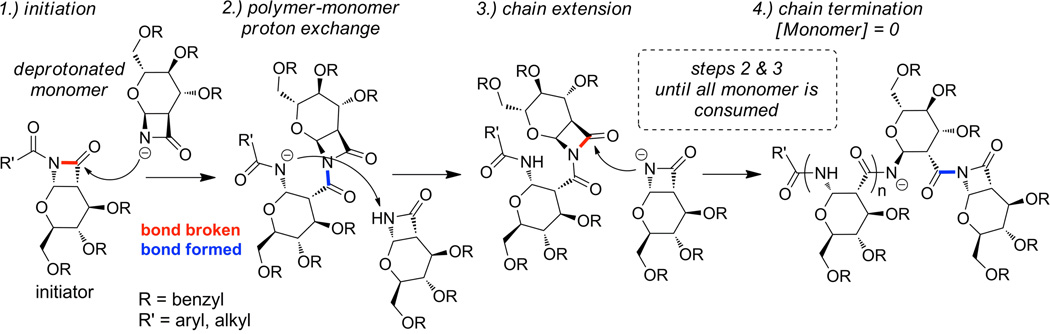
It has been reported that anionic ROP of certain β-lactam monomers can yield polymers of low PDI and controlled length when an appropriate initiator and base are used.26 The proposed polymerization mechanism26 involves cleavage and reformation of an achiral amide bond, and therefore is expected to produce a chiral polymer if a chiral monomer is used.26f,27 The amount of initiator added determines how many polymer chains are grown and hence the polymer length. However, the polymerization of cyclic sugar-derived β-lactam monomers, such as benzyl-protected monomer 1 (Scheme 1), has not been studied. Benzyl ethers are attractive protecting groups for polysaccharides because they can be removed efficiently from large molecules via either Pd-catalyzed hydrogenation15 or metal-ammonia reduction13. Additionally, the use of 1 as a monomer is novel because the polymerization of β-lactams in which the lactam is part of a hemiamidal (a hemiaminal where the amine is replace with an amide) has not been explored. The previously reported β-lactam 128 was accessed on multi-gram scales in moderate yield via the stereoselective cycloaddition of tri-O-benzyl-d-glucal and chlorosulfonyl isocyanate (CSI) followed by in situ reduction29 to remove the sulfonyl group (Scheme 1). Monomer 1 was reacted with 4-tert-butylbenzoyl chloride to provide initiator 2. In anticipation of characterizing the polymers by NMR and IR, a protected model compound (3) was synthesized by opening the β-lactam of 2 with excess n-butylamine and deprotected to form 4. The proposed mechanism for the anionic ROP of monomer 1 is shown in Scheme 2.
Scheme 2.
Proposed mechanism of anionic ring-opening polymerization.
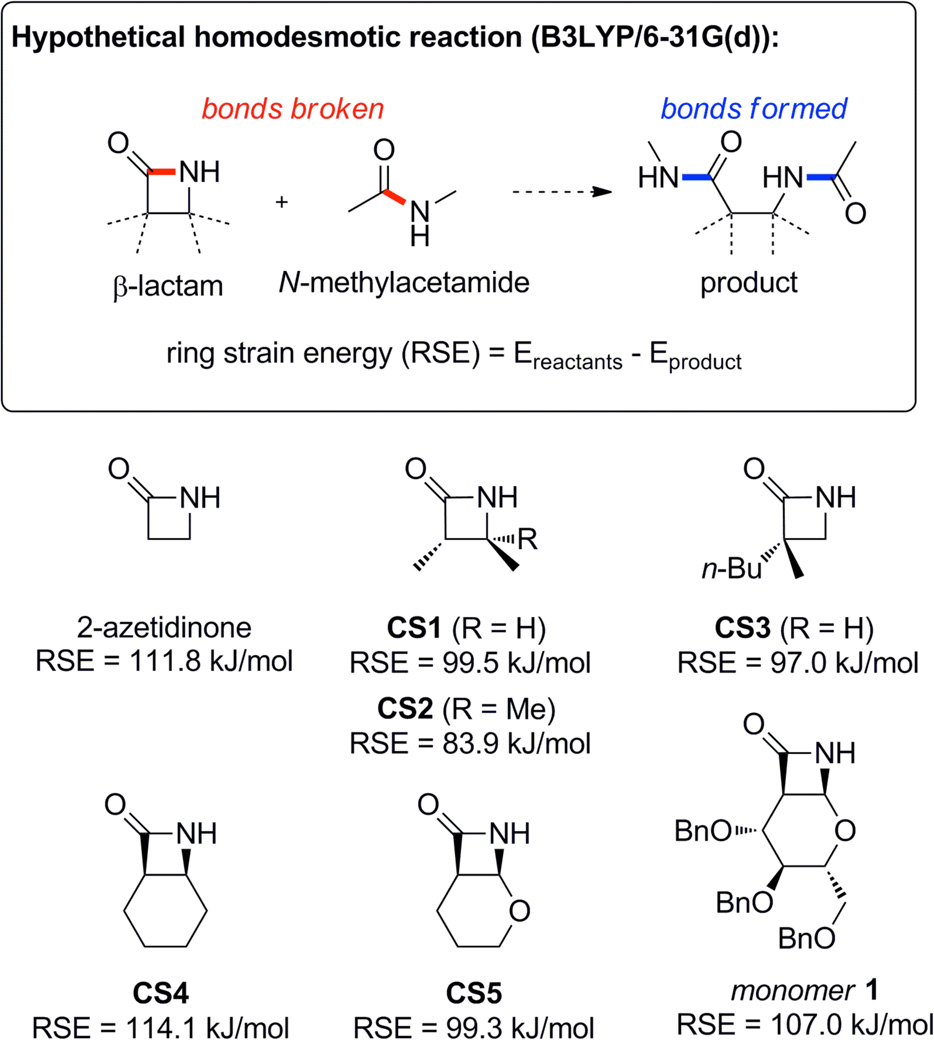
The ring strain of a series of β-lactam monomers was estimated using a homodesmotic reaction as shown in Scheme 3. DFT geometry minimization and energy calculations were performed to determine the energy difference between the reactants and the product for the hypothetical ring-opening reaction.30 All of the ring-opened structures contained an intramolecular hydrogen bond formed by the opened lactam ring (see Figure S1 for structures). The presence of this H-bond in the product but not in the reactants inflates the ring strain energy calculated by this method, but because it is present in all of the products it should not significantly affect the trends.
Scheme 3.
β-lactam ring strain computation.
For the simplest β-lactam, 2-azetidinone, the ring strain energy (RSE) was calculated to be 111.8 kJ/mol. This value is in general agreement with the experimentally determined value of 119.4 kJ/mol, which was described by the authors as an upper limit for the value of the ring strain.31 Additionally, it is similar to the reported experimental value (116 kJ/mol) for the hydrolysis of penicillin G.32 Next, we analyzed two computational structures (CS1 and CS2), which are simplified structures based on monomers reported to readily polymerize.26g–i As would be expected, the addition of substituents to the ring decreases the ring strain by stabilizing the ring-closed form in relation to the ring-open form. The lowering of ring strain energy by the replacement of hydrogen atoms with alkyl groups has been noted for a range of lactam monomers.33 This effect is observed for CS1 (99.5 kJ/mol) relative to 2-azetidinone, and to a larger extent for CS2 (83.9 kJ/mol), which has an additional methyl group. For CS3, the experimentally determined heat of polymerization (ΔHp) for anionic ROP in toluene at 25 °C is reported as 80 kJ/mol.26a The computational method estimates a RSE of 97.0 kJ/mol, which is significantly higher. The higher computational value is likely caused in part by the presence of the intramolecular H-bond in the product. In addition, the computations are for single molecules in the gas phase and do not take into account important factors such as solvent effects.
For the β-lactam cis-fused with a cyclohexane ring (CS4), an increased ring strain (114.1 kJ/mol) was calculated as compared to 2-azetidinone. In this case, the fused bicyclic system induces additional ring strain. This conclusion is supported by the fact that the cyclohexane ring of CS4 is forced to adopt a boat rather than a chair conformation in both the XRCS and the DFT-minimized structure. CS4 readily polymerizes but forms very insoluble homopolymers.26g,26h In contrast to CS4, the calculated ring strain in CS5 (99.3 kJ/mol) is lower than that of 2-azetidinone. The decrease in ring strain in CS5 may be a consequence of stabilization due to a larger anomeric interaction between the ethereal oxygen and the nitrogen in the lactam as compared to the ring-opened form. The polymerization of CS5 has not been reported. The calculated RSE of monomer 1 (107.0 kJ/mol) is less than those of 2-azetidinone and CS4, but more than those of CS1–CS3 and CS5. We attribute the increased RSE of 1 in comparison to CS5 to steric interactions that are relieved upon ring-opening as the pyranose ring relaxes from a twisted-chair to a chair conformation. In general, these results suggest that the polymerization of monomer 1 is highly thermodynamically favored due to the strained β-lactam ring, and that the greater steric bulk of the benzyl ethers increases this strain. These results do not comment on the effect of the monomer’s increased steric bulk on the kinetics of the polymerization. However, the anionic ROP of less strained β-lactams (derivatives of CS1 and CS2, and CS3) is rapid at mild temperatures.26h
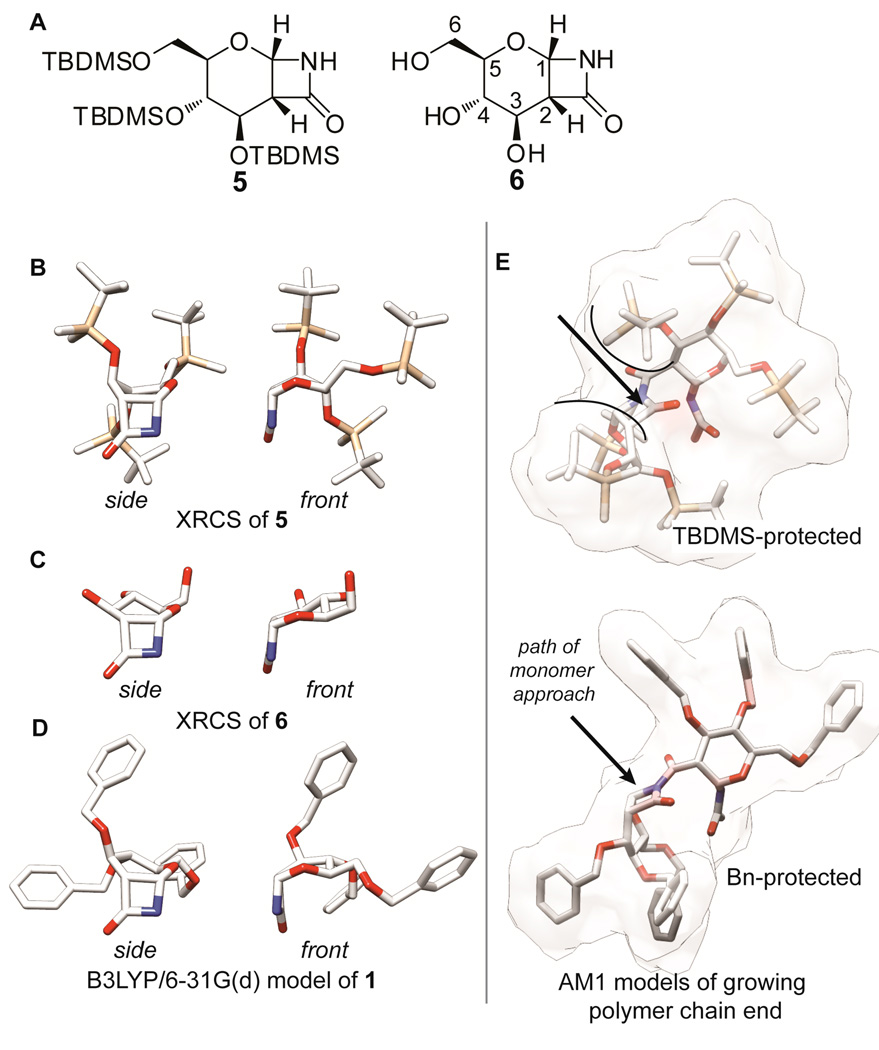
Next, we investigated the conformation adopted by the pyranose ring in 1 by X-ray crystallography and NMR spectroscopy in order to confirm predictions made by modeling. We attempted to grow single crystals of 1 large enough for X-ray analysis, but were unsuccessful. In contrast, the tert-butyldimethylsilyl protected derivative 528 easily formed crystals and an X-ray crystal structure (XRCS) revealed that in the solid state the six-member ring adopts a boat confirmation (Figure 2, ORTEP in Supporting Information). This is in contrast to a previously reported XRCS of 6,24,34 the deprotected form of 1, in which the ring adopts a half-chair conformation. Geometry minimization (B3LYP/6–31G(d)) suggests that 1 adopts a conformation closer to that of a half-chair rather than a boat (Figure 2d). For cyclic sugar derivatives, the J-couplings between adjacent protons provide information about the ring’s conformation because the strength of the coupling varies with the dihedral angle. Therefore, we compared the solution 1H-NMR J-couplings among H3, H4, and H5 for compounds 1, 5, and 6 (Figure S2). The couplings for monomer 1 are approximately the same as those of 634 and differ significantly from those of 5, supporting the hypothesis that 1 has a conformation closer to a half chair rather than a boat, if we assume that the solid-state structures of 5 and 6 are representative of their solution conformations.
Figure 2.
Monomer Structure. (a.) Structures of 5 and 6. (b.) XRCS of 5. (c.) XRCS of 6. (d.) Structure of 1 geometry minimized using B3LYP/6–31G(d). (e.) Models of benzyl and TBDMS protected chain ends minimized using AM1 showing the effect of protecting group size on steric barriers to polymerization.
In addition to studying the monomer’s ring strain, we used modeling to predict how the steric bulk of the benzyl protecting groups would affect the kinetics of polymerization. Zhang and co-workers attributed a slower reaction rate for a CS2 derivative as compared to a CS1 derivative to the additional steric bulk of the added methyl group in CS2.26g,26h In this case, the proximity of the methyl group to the lactam clearly suggests a steric influence. However, the model of 1 reveals that the steric effects of the benzyl groups may be relatively modest because they are positioned so that one face of the monomer is clearly open for reaction, even in a model of the growing polymer chain (Figure 2e.). In contrast, the tert-butyldimethylsilyl groups pose a steric impediment to polymerization of 5. As predicted, our attempts to polymerize 5 under the conditions used for 1 (and with heating up to 40 °C) were unsuccessful, suggesting that additional steric bulk beyond that of the benzyl group can reduce the reactivity of the β-lactam in anionic ROP by increasing kinetic barriers to reaction.
Polymer Synthesis and Characterization
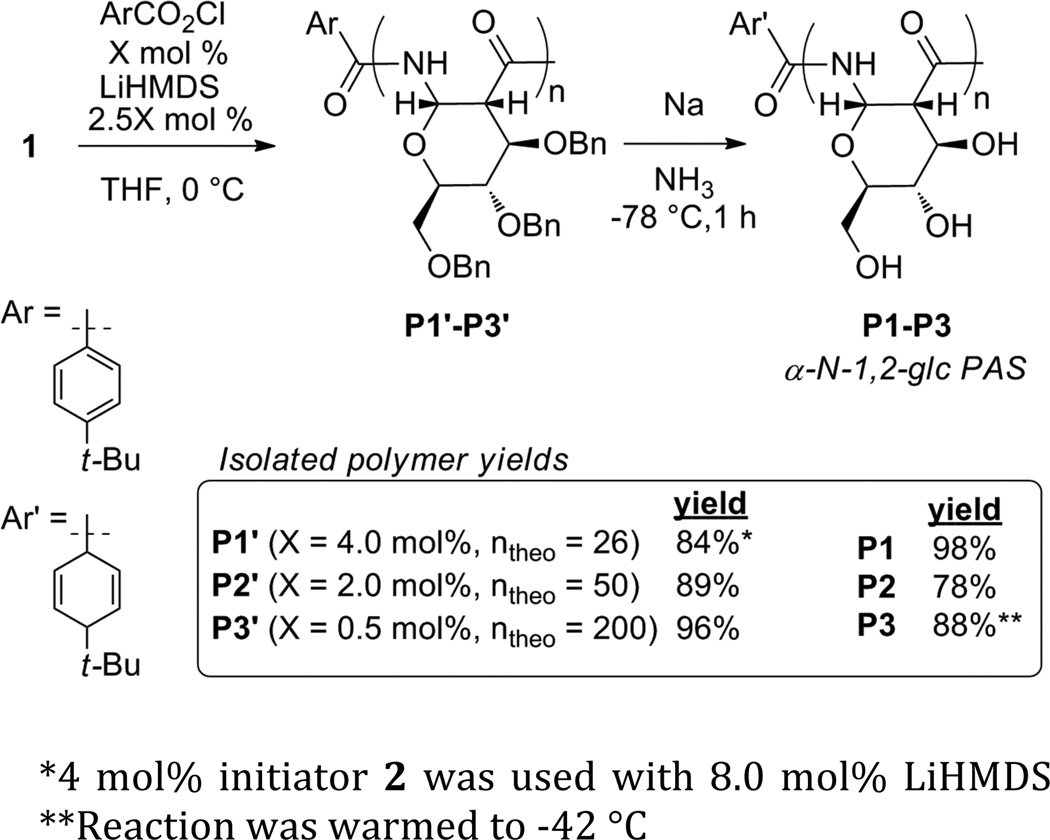
Monomer 1 was polymerized with 4 mol% initiator to obtain polymer P1’ (DPtheo = 26) (Scheme 4). For polymers P2’ (DPtheo = 50) and P3’ (DPtheo = 200), the initiator was formed in situ by adding the appropriate amount of 4-tert-butylbenzoyl chloride. Polymerization conditions were based on those reported by Zhang et al with modifications.26g–i Polymers P1’–P3’ were characterized with 1H-NMR (Figure S3) and 13C-NMR, GPC, polarimetry, and IR. The DP was estimated by comparing the integration of the initiator’s tert-butyl signal at 1.2 ppm to the polymer integration. In the 13C-NMR spectra, signals at 170 ppm (amide), 73 ppm (C1), and 51 ppm (C2) are clearly present. Characterization of P1’–P3’ with GPC(THF) and polystyrene standards indicated that P1’–P3’ had low levels of polydispersity (PDI = 1.1). For P2’ and P3’, the DP as measured by GPC is lower than the theoretical value and the value measured by NMR. This discrepancy may be a consequence of the polymer possessing a different solution structure than polystyrene of a similar molecular weight. The specific rotations of P1’–P3’ measured in CH2Cl2 increased slightly with polymer length ([α]D = 79.1 (P1’), 80.5 (P2’), 83.3 (P3’)). The IR spectra of P1’–P3’ show a single strong amide stretch (≈ 1690 cm−1), are nearly identical to each other, and are in good agreement with the spectrum of the protected model compound 3 (Figure S4).
Scheme 4.
Polymer synthesis
*4 mol% initiator 2 was used with 8.0 mol% LiHMDS
**Reaction was warmed to −42 °C
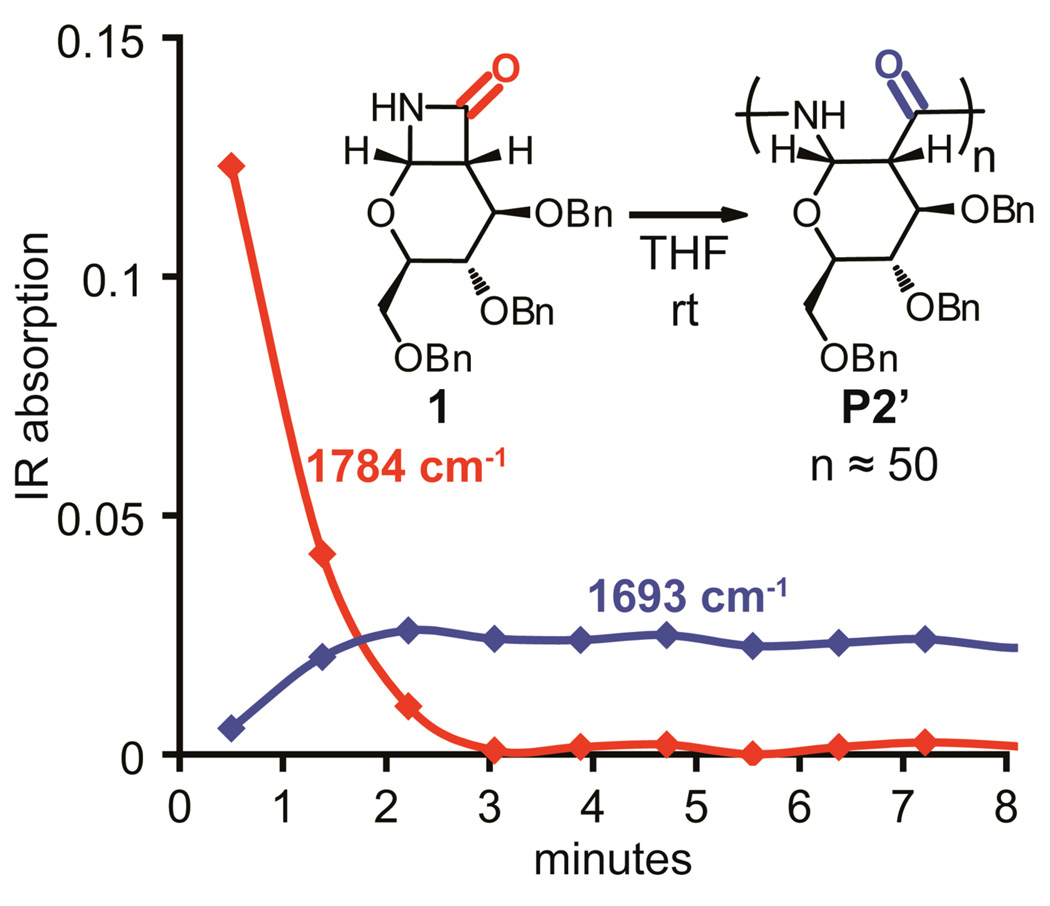
Based on TLC, the monomer was completely consumed in less than 30 minutes at either 0 °C or 25 °C for all of the polymerizations. To better observe the reaction progress, we performed the polymerization of P2’ while monitoring the IR signal with an in situ probe (Figure 3). The monomer was consumed in less than 5 minutes at 25 °C, based on the decrease in the signal from the carbonyl stretch of the β-lactam (1784 cm−1, red). We also observed an increase in the amide carbonyl stretch of the polymer (1693 cm−1, blue) within the first 5 minutes. Future kinetic studies of the polymerization will be performed in order to make quantitative comparisons between the rate of polymerization of monomer 1 and the rates reported for other monomers.26h Our observations confirm that the polymerization of 1 is rapid and that the steric bulk of the benzyl protecting groups does not significantly hinder polymerization.
Figure 3.
The progress of the polymerization was monitored by observing the decrease in the IR absorbance of the monomer β-lactam carbonyl stretch (1784 cm−1, red) and the increase in the polymer amide carbonyl stretch (1693 cm−1, blue). Reaction conditions: THF, rt, [1] = 0.15 M, 2 mol% of 4-tert-butylbenzoyl chloride, and 5 mol% LiHMDS. Data recording began 30 sec after addition of the base.
Attempts at polymer debenzylation using Pd-catalyzed hydrogenation at room temperature and elevated pressure (3 atm) failed. At elevated temperature (70 °C) in dimethylacetamide, the hydrogenation was successful for shorter polymers, but resulted in low yields. Hence, sodium metal in ammonia (Birch reduction) was used for deprotection. Prior to reduction, the polymers were treated with potassium tert-butoxide to deprotonate and therefore protect the amide groups from reduction. At −78 °C, the debenzylation of P3 was incomplete. Repeating the procedure with warming to −42 °C provided complete removal of the protecting groups based on 1H-NMR and IR. Deprotected polymers (P1–P3) were purified by dialysis and then lyophilized. Further washing with methanol, in which the polymers are insoluble, removed trace salts.
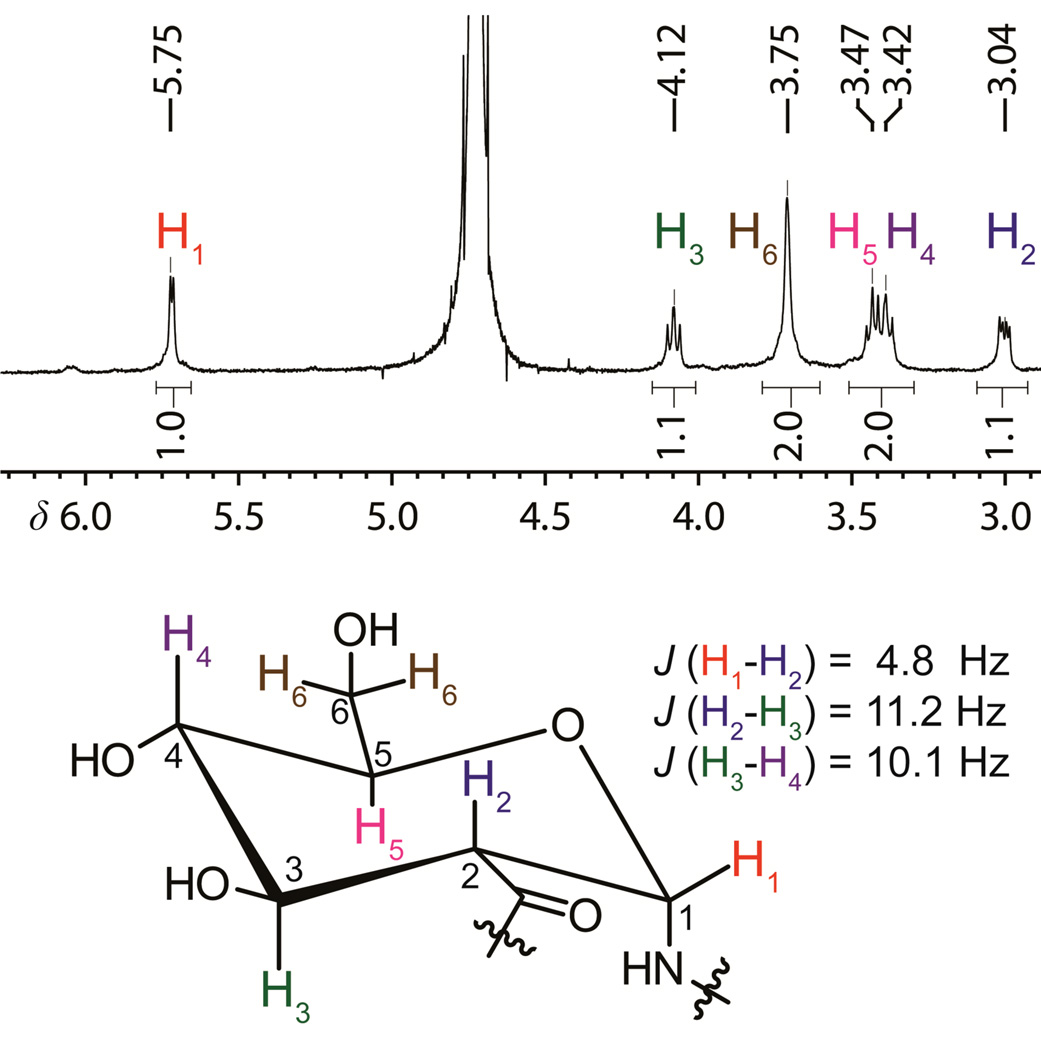
Polymers P1–P3 were characterized with 1H-NMR (Figure 4 and S5), dynamic light scattering (DLS) (Table 1), IR (Figure S4), and circular dichroism (CD) (Figure 5). The proton NMR spectra of P1–P3 were well-resolved (Figure 4) and a gCOSY spectrum of P1 was collected to confirm coupling assignments. In regards to the structure of the end group after debenzylation, aromatic amides with a hydrogen on the nitrogen do not undergo amide cleavage under Birch reduction conditions, but rather the aromatic ring is partially reduced to several products with a preference for the 1,4-dihydro product (Ar’ in Scheme 4).35 Integration of the tert-butyl signals of the reduced aromatic end-group suggest a DP of 30 which is in good agreement with the DP of 26 measured by DLS. A 13C-NMR spectrum of freshly dissolved P1 showed the expected 7 signals. Polymers P2 and P3 gave 1H-NMR spectra similar to P1 with attenuated signals due to the lower solubility of the higher molecular weight polymers. For P2, a DP of 47 was estimated by integration of the tert-butyl signals, but for P3 the signals could not be distinguished from the baseline. Beyond estimating the DP, the 1H-NMR spectra (Figure 4, S5) of P1–P3 suggest that in aqueous solution at room temperature the polymer backbone has a well-defined chair conformation with the bond between C1 and the nitrogen of the amide linkage in an axial position (α). This is confirmed by the smaller J-coupling between protons H1 and H2 (J(H1–H2) = 4.8 Hz) versus the larger couplings between protons H2 and H3, and between protons H3 and H4 (Figure 4). The spectra of all three polymers show that the stereochemistry of the polymer is not affected by the reductive debenzylation. Based on DLS, P2 and P3 do not exist as individual polymer chains in solution, but rather as aggregates with radii greater than 50 nm. The specific rotation of P1 measured in H2O was positive ([α]D = 129), but the specific rotations of P2 and P3 were not measured due to their aggregated structures. IR spectra of P1–P3 show a single peak in the amide region and the complete removal of the benzyl groups as demonstrated by the disappearance of the aromatic C-H signals between 3000 to 3100 cm−1 (see Figure S4).
Figure 4.
Proton NMR spectrum of P1. The coupling constant (J) of 4.8 Hz between protons H1 and H2 indicates that they are in an equatorial-axial relationship. The larger coupling constants between H2–H3 (11.2 Hz) and H3–H4 (10.1 Hz) indicate that they are in an axial-axial relationship. These relationships suggest that the polymer adopts a chair configuration with the C1-N bond axial and the other ring substituents equatorial.
Table 1.
Polymer Characterization
| Mn(theo) (kDa) |
Mn(NMR)a (kDa) |
Mn(GPC)b (kDa) |
Mw(GPC)b (kDa) |
PDIc | DPtheo | DPNMRa | DPGPCb | [α]D (CH2Cl2) |
|
|---|---|---|---|---|---|---|---|---|---|
| P1’ | 11.9 | 11.0 | 9.5 | 10.5 | 1.1 | 26 | 24 | 21 | 79.1 |
| P2’ | 23.0 | 27.5 | 16.7 | 19.0 | 1.1 | 50 | 60 | 36 | 80.5 |
| P3’ | 91.8 | 82.6 | 56.2 | 64.5 | 1.1 | 200 | 180 | 122 | 83.3 |
| Mn(theo) (kDa) | Mn(NMR)d (kDa) | Radiuse (nm) | Mw(DLS)f (kDa) | DPtheo | DPNMR | DPDLS | [α]D (H2O) |
||
| P1 | 4.7 | 5.7 | 1.6± 0.4 | 5.0 | 26 | 26 | 30f | 26 | 129 |
| P2 | 9.5 | 8.9 | >50g | n.d. | n.d. | 50 | 47f | n.d. | n.d. |
| P3 | 37.8 | n.d. | >50g | n.d. | n.d. | 200 | n.d. | n.d. | n.d. |
Determined by integration of the 1H-NMR signal from the t-butyl group on the initiator.
THF GPC with polystyrene standards.
Mw/Mn.
Determined by integrating t-butyl groups of reduced t-butyl benzamide end group.
Determined by dynamic light scattering, T = 50 °C (average of 30 measurements).
Based on model for linear polysaccharides, %PD = percent polydispersity.
The large radius is indicative of aggregates in solution. n.d. = not determined.
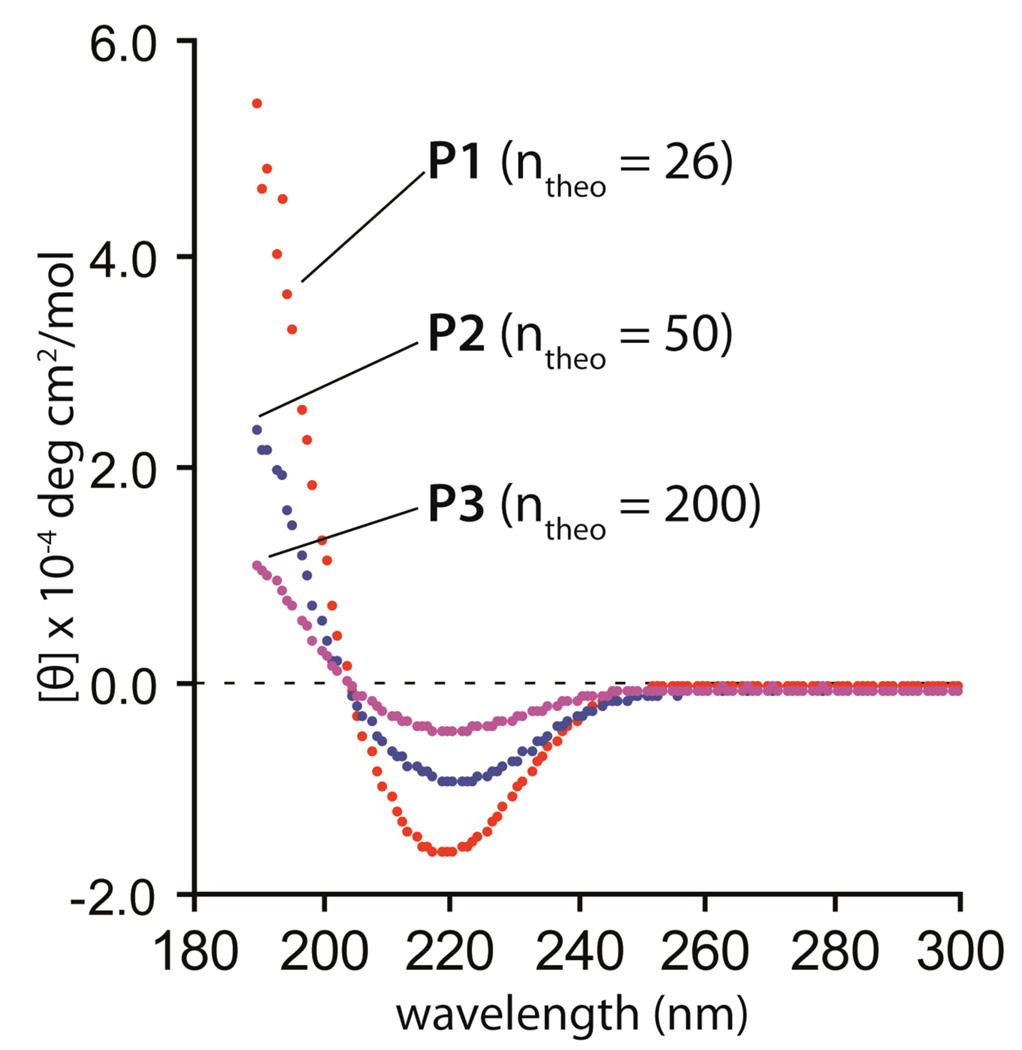
Figure 5.
CD spectra. [P] = 0.030 mg/mL in H20, rt.
The CD spectra of freshly dissolved samples of P1–P3 in water at room temperature suggest that the polymers adopt a regular secondary structure (Figure 5).36 The polymers have a minimum at 221 nm (P1, −16,000 deg cm2/mol; P2, −9,800 deg cm2/mol; P3, −5,000 deg cm2/mol) and a maximum at 190 nm (where recording was stopped) (P1, 54,000 deg cm2/mol; P2, 23,000 deg cm2/mol; P3, 10,000 deg cm2/mol). At the same concentration of 0.030 mg/mL, the shortest polymer, P1, has the strongest signal and the longest polymer, P3, has the weakest. Molecular modeling suggests that the polymer structure can form a right-handed helix (Figure 1) and the CD spectra are in general agreement. The formation of helical secondary structures by oligo- and poly-β-peptides has been previously reported and studied with CD.37 The trend in CD signal also suggests that the shorter polymer length promotes secondary structure formation. This trend may be a consequence of P2 and P3 forming aggregates in solution, as suggested by DLS (Table 1).
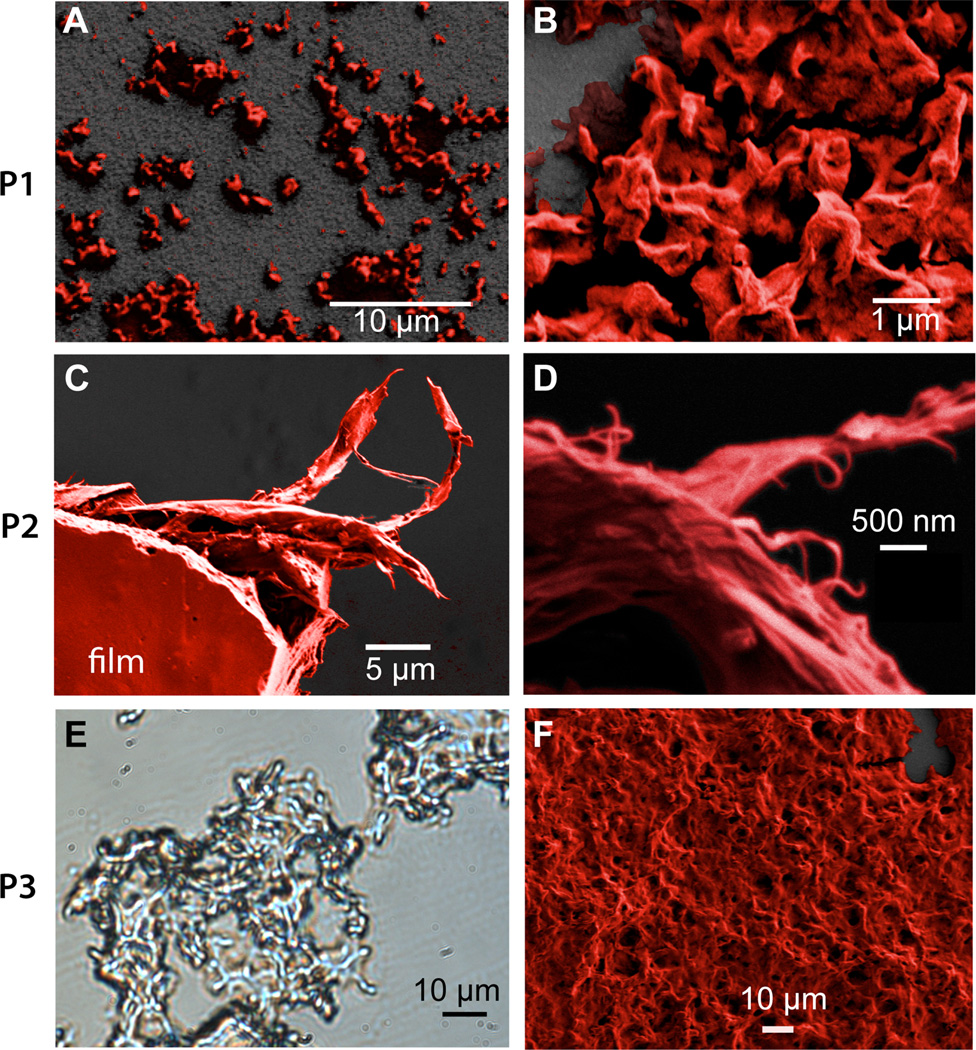
To better understand how the molecular weights of P1–P3 affect their aggregated structures, we used both light and electron microscopy to visualize the solid-state structure of the polymers. The samples for observation were prepared by allowing a saturated aqueous solution of the polymers to precipitate over 24 hours. The lowest MW polymer, P1, gave an amorphous structure with high surface roughness that showed little tendency to form films when viewed by SEM (Figure 6, a & b). In contrast to P1, polymer P2 formed smooth films that, when cracked during SEM sample preparation, revealed fibrillar alignment of the polymers (Figure 6, c & d). For P3, what appeared to be fibrils of approximately 1 µm in diameter were observed using phase contrast light microscopy (Figure 6, e). In SEM, P3’s surface appeared amorphous and rough, but well-defined fibrils could not be observed (Figure 6, f).
Figure 6.
Electron and optical microscopy. (a. & b.) SEM micrographs of P1 showing absence of film formation. (c. & d.) SEM micrograph of P2 showing a piece of polymer film that tore apart during sample preparation leaving behind fibrous shreds at two magnifications. (e.) phase contrast light micrograph of P3 showing complex morphology. (f.) SEM micrograph of P3 showing an amorphous morphology. SEM Images have been colorized and the dark gray back-ground is the substrate.
Polymers P1–P3 preserve some important properties of natural polysaccharides. Like natural polysaccharides, P1–P3 are chiral and are joined via a linkage of α-stereochemistry. The solubility behavior of P1–P3 is in broad agreement with Whistler’s observations on the solubility of natural polysaccharides.38 He generalized that polysaccharides composed of a single type of sugar repeat unit are relatively insoluble in water even at low degrees of polymerization (DP = 20–30), and that longer polysaccharides generally exist as aggregates rather than individual polymers in solution. When first isolated, P1 (DPtheo = 26) had a solubility of > 5.0 mg/mL, which decreased over time as a white precipitate formed. Polymers P2 and P3 had lower initial solubilities, 2.0 mg/mL and 1.0 mg/mL, respectively, but precipitation over time was less clear as compared to P1. However, the solubility of P1 always remained higher than P2 and P3, as expected.
Biological activity
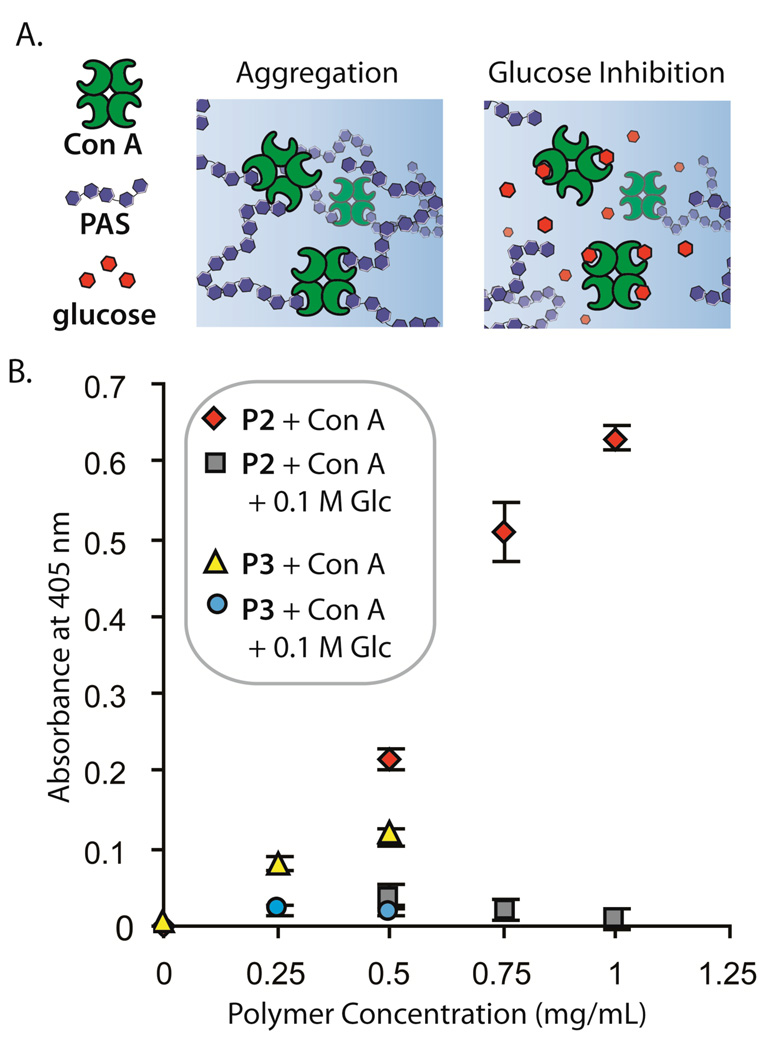
Because PASs P1–P3 are based on a unique structure not found in nature, we next determined whether these PASs are recognized by a carbohydrate binding protein. Concanavalin A (Con A), a readily available lectin, is known to bind glucose or mannose groups with an α-orientation at C1 by interacting predominantly with the C3, C4, and C6 alcohols.21a We reasoned that the polymers may bind in the same pocket as natural glucose derivatives because P1–P3 are joined via a linkage of α-stereochemistry and the 1,2-linkage does not disturb the alcohols at C3, C4, and C6. Using an established assay, the binding of Con A to polysaccharides can be measured by the increased turbidity of a solution due to the aggregated structures (Figure 7a) formed by multivalent polysaccharides and the tetravalent lectin.21b,21c On a mass concentration basis, P2 showed the largest increase in turbidity in the presence of Con A (Figure 7b, red diamonds). No significant response was observed for P1. It has previously been noted that polymers that cannot span the distance of approximately 6.5 nm between adjacent carbohydrate binding sites in a Con A tetramer show lower binding affinities because they cannot readily engage in multivalent interactions.39 Based on measurements made from modeling, P1 would be between 6–7 nm long if fully extended and therefore may not be able to benefit from significant multivalent interactions when it binds. P2 showed more intense scattering when bound as compared to P3 (yellow triangles), when evaluated on a mass concentration basis. To confirm that P2 and P3 bind Con A at the same site as natural carbohydrates, 0.1 M glucose was shown to inhibit aggregation (grey squares, blue circles, respectively).21b Glucose inhibition of binding was nearly complete for both P2 and P3. A minimal amount of scattering remains in the presence of glucose which is also observed when the binding of glycogen to Con A is inhibited with 0.1 M glucose using these assay conditions.
Figure 7.
Concanavalin A binding. (a.) Schematic representation of the binding of the lectin concanavalin A with PASs. (b.) The turbidity was measured based on the scattering at 405 nm for samples of P2 and P3 in the presence of Con A and with Con A and 0.1 M glucose. [Con A] = 1 mg/mL; Tris buffer, pH = 7.2; each data point is the average of three samples and error bars show 1 standard deviation.
We suggest two explanations for the higher response from P2 as compared to P3. First, Con A may have a preference for binding at the end of polymer chains rather than at inner residues due to steric issues.7c At a given mass concentration (mg/mL), a solution of P2 will contain more polymer ends for binding because P2 is significantly shorter and weighs less than P3. Secondly, the solution aggregation state of P3 may differ significantly from that of P2. P3 may be aggregated in a manner than makes its sugar residues less available for binding. We infer this difference in aggregation behavior from the lower solubility of P3 as compared to P2 and the less intense CD spectrum of P3 as compared to P2.
When taken together, the changes in solubility, CD spectrum, solid-state structure as viewed by microscopy, and lectin binding confirm that we are able to prepare, from a single monomer and a single polymerization method, a series of polymers that have chain-length dependent properties. Our ability to readily observe these changes while making relatively minor changes in DP (P2 is only twice as long as P1) highlights the controlled nature of the polymerization. Controlled polymerizations to make glycopolymers are established and have been used in the past to create a wide-range of glycoconjugates.6 We contend that PASs are not meant to replace glycoconjugates in applications where glycopolymers and glycodendrimers have proven highly effective, but rather as carbohydrate polymers with complimentary properties that are derived from the rigid pyranose backbone and stereochemically defined amide linkage.
CONCLUSION
In summary, α-N-1,2-d-glucose poly-amido-saccharides (PASs) are novel carbohydrate-derived polymers in which the ether linkage found in natural polysaccharides is replaced with an amide linkage. They are synthesized using the anionic ring-opening polymerization of a chiral β-lactam monomer derived from benzyl-protected d-glucal. The mild and high-yielding polymerization method provides materials of controlled molecular weight and narrow polydispersity. After debenzylation, the resulting hydrophilic β-polypeptides contain a rigid pyranose ring in the main chain with a 1,2-linkage of defined stereochemistry that molecular modeling suggests may promote a right-handed helical structure. Computational modeling results suggest that the monomer is highly reactive in part because of additional ring strain induced by the half-chair conformation of the pyranose ring. In addition, modeling reveals that negative steric effects due to the benzyl groups appear to be minimized because of the monomer’s geometry and the flexibility of the ether bond. Characterization of model compounds and polymers using NMR, GPC, DLS, and IR confirms that the polymers have the desired molecular structure. In addition, the effect of chain length on solubility, CD spectrum, and solid-state morphology further evinces the controlled nature of the polymerization. Finally, glucose-dependent binding of higher MW α-N-1,2-d-glc PASs to the plant lectin concanavalin A demonstrates the potential of PAS’s to interact with natural carbohydrate receptors. Future studies will focus on expanding the range of structures that can be synthesized, understanding the solution and solid-state aggregation structures, and identifying biomedical applications where α-N-1,2-d-glc PASs may offer unique advantages.
Supplementary Material
ACKNOWLEDGMENT
E.L.D acknowledges receipt of an NIH/NIGMS Postdoctoral Fellowship (1F32GM097781). The authors thank Aaron Colby for help obtaining SEM images, Andrew Lynch and Prof. Karen Allen for help obtaining DLS data, and Dr. Jeffrey Bacon for collecting X-ray crystal diffraction data and solving the structure.
Footnotes
ASSOCIATED CONTENT
Supporting Information. Experimental details, supplemental figures, IR spectra, 1HNMR, gCOSY, 13C spectra, XRCS details, and xyz coordinates and energies for computational models. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Galbis JA, García-Martín MG. In: Monomers, Polymers and Composites from Renewable Resources. Mohamed Naceur B, Alessandro G, editors. Amsterdam: Elsevier; 2008. p. 89. [Google Scholar]
- 2.(a) Dwek RA. Chem. Rev. 1996;96:683. doi: 10.1021/cr940283b. [DOI] [PubMed] [Google Scholar]; (b) Dumitriu S. Polysaccharides: Structural Diversity and Functional Versatility. 2nd ed. New York: Marcel Dekker, Inc; 2005. [Google Scholar]; (c) Stern R, Jedrzejas M. J. Chem. Rev. 2008;108:5061. doi: 10.1021/cr078240l. [DOI] [PubMed] [Google Scholar]; (d) Ernst B, Magnani JL. Nat. Rev. Drug Discov. 2009;8:661. doi: 10.1038/nrd2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kumar MNVR, Muzzarelli RAA, Muzzarelli C, Sashiwa H, Domb A. J. Chem. Rev. 2004;104:6017. [Google Scholar]
- 4.(a) Lapčík L, De Smedt S, Demeester J, Chabreček P. Chem. Rev. 1998;98:2663. doi: 10.1021/cr941199z. [DOI] [PubMed] [Google Scholar]; (b) Burdick JA, Prestwich GD. Adv. Mater. 2011;23:H41. doi: 10.1002/adma.201003963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.(a) Marchessault RH, Ravenelle F, Zhu XX. Polysaccharides for drug delivery and pharmaceutical applications. New York, NY: Oxford University Press; 2006. [Google Scholar]; (b) Rinaudo M. Polym. Int. 2008;57:397. [Google Scholar]; (c) Burdick JA, Mauck RL. Biomaterials for Tissue Engineering Applications. New York, NY: Springer- Verlag/Wien; 2011. [Google Scholar]
- 6.(a) Nishimura S, Matsuoka K, Furuike T, Ishii S, Kurita K, Nishimura KM. Macromolecules. 1991;24:4236. [Google Scholar]; (b) Mortell KH, Gingras M, Kiessling LL. J. Am. Chem. Soc. 1994;116:12053. [Google Scholar]; (c) Fraser C, Grubbs RH. Macromolecules. 1995;28:7248. [Google Scholar]; (d) Havard JM, Vladimirov N, Fréchet JMJ, Yamada S, Willson CG, Byers JD. Macromolecules. 1998;32:86. [Google Scholar]; (e) Grande D, Baskaran S, Chaikof EL. Macromolecules. 2001;34:1640. [Google Scholar]; (f) Wang Q, Dordick JS, Linhardt R. J. Chem. Mater. 2002;14:3232. [Google Scholar]; (g) Baskaran S, Grande D, Sun X-L, Yayon A, Chaikof EL. Bioconjugate Chem. 2002;13:1309. doi: 10.1021/bc0255485. [DOI] [PubMed] [Google Scholar]; (h) Sun X-L, Grande D, Baskaran S, Hanson SR, Chaikof EL. Biomacromolecules. 2002;3:1065. doi: 10.1021/bm025561s. [DOI] [PubMed] [Google Scholar]; (i) Ladmiral V, Melia E, Haddleton DM. Eur. Polym. J. 2004;40:431. [Google Scholar]; (j) Papp I, Dernedde J, Enders S, Haag R. Chem. Comm. 2008:5851. doi: 10.1039/b813414f. [DOI] [PubMed] [Google Scholar]; (k) Becer CR, Gibson MI, Geng J, Ilyas R, Wallis R, Mitchell DA, Haddleton DM. J. Am. Chem. Soc. 2010;132:15130. doi: 10.1021/ja1056714. [DOI] [PMC free article] [PubMed] [Google Scholar]; (l) Lee S-G, Brown JM, Rogers CJ, Matson JB, Krishnamurthy C, Rawat M, Hsieh-Wilson LC. Chem. Sci. 2010;1:322. doi: 10.1039/C0SC00271B. [DOI] [PMC free article] [PubMed] [Google Scholar]; (m) Kramer JR, Deming TJ. J. Am. Chem. Soc. 2010;132:15068. doi: 10.1021/ja107425f. [DOI] [PubMed] [Google Scholar]; (n) Kramer JR, Deming TJ. J. Am. Chem. Soc. 2012;134:4112. doi: 10.1021/ja3007484. [DOI] [PubMed] [Google Scholar]
- 7.(a) Roy R, Zanini D, Meunier SJ, Romanowska A. J. Chem. Soc., Chem. Commun. 1993:1869. [Google Scholar]; (b) Aoi K, Itoh K, Okada M. Macromolecules. 1995;28:5391. [Google Scholar]; (c) Woller EK, Walter ED, Morgan JR, Singel DJ, Cloninger MJ. J. Am. Chem. Soc. 2003;125:8820. doi: 10.1021/ja0352496. [DOI] [PubMed] [Google Scholar]; (d) Wolfenden ML, Cloninger MJ. J. Am. Chem. Soc. 2005;127:12168. doi: 10.1021/ja053008n. [DOI] [PubMed] [Google Scholar]; (e) Chabre YM, Roy R. Adv. Carbohydrate. Chem. Biochem. 2010;63:165. doi: 10.1016/S0065-2318(10)63006-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.(a) Kurita K, Masuda N, Aibe S, Murakami K, Ishii S, Nishimura S-I. Macromolecules. 1994;27:7544. [Google Scholar]; (b) Chen X, Gross RA. Macromolecules. 1998;32:308. [Google Scholar]; (c) Shen Y, Chen X, Gross RA. Macromolecules. 1999;32:2799. [Google Scholar]; (d) Shen Y, Chen X, Gross RA. Macromolecules. 1999;32:3891. [Google Scholar]; (e) Meier S, Reisinger H, Haag R, Mecking S, Mulhaupt R, Stelzer F. Chem. Comm. 2001:855. [Google Scholar]; (f) Kumar R, Gao W, Gross RA. Macromolecules. 2002;35:6835. [Google Scholar]; (g) Haba O, Tomizuka H, Endo T. Macromolecules. 2005;38:3562. [Google Scholar]; (h) Gao W, Hagver R, Shah V, Xie W, Gross RA, Ilker MF, Bell C, Burke KA, Coughlin EB. Macromolecules. 2006;40:145. [Google Scholar]; (i) Lienkamp K, Kins CF, Alfred SF, Madkour AE, Tew GN. J. Poly. Sci. A: Polymer Chemistry. 2009;47:1266. [Google Scholar]; (j) Wathier M, Stoddart SS, Sheehy MJ, Grinstaff MW. J. Am. Chem. Soc. 2010;132:15887. doi: 10.1021/ja106488h. [DOI] [PubMed] [Google Scholar]
- 9.(a) García- Martín MdG, de Paz Báñez MV, Galbis JA. J. Carbohyd. Chem. 2000;19:805. [Google Scholar]; (b) García- Martín MdG, Báñez MVdP, Galbis JA. J. Carbohyd. Chem. 2001;20:145. [Google Scholar]; (c) Metzke M, Bai JZ, Guan Z. J. Am. Chem. Soc. 2003;125:7760. doi: 10.1021/ja0349507. [DOI] [PubMed] [Google Scholar]; (d) Metzke M, Guan Z. Biomacromolecules. 2007;9:208. doi: 10.1021/bm701013y. [DOI] [PubMed] [Google Scholar]; (e) Urakami H, Guan Z. Biomacromolecules. 2008;9:592. doi: 10.1021/bm701180r. [DOI] [PubMed] [Google Scholar]; (f) Liao SW, Yu T-B, Guan Z. J. Am. Chem. Soc. 2009;131:17638. doi: 10.1021/ja907097t. [DOI] [PubMed] [Google Scholar]
- 10.Kobayashi K, Tsuchida A, Usui T, Akaike T. Macromolecules. 1997;30:2016. [Google Scholar]
- 11.(a) Seeberger PH, Haase W-C. Chem. Rev. 2000;100:4349. doi: 10.1021/cr9903104. [DOI] [PubMed] [Google Scholar]; (b) Gruner SAW, Locardi E, Lohof E, Kessler H. Chem. Rev. 2002;102:491. doi: 10.1021/cr0004409. [DOI] [PubMed] [Google Scholar]
- 12.Stick RV, Williams SJ. Carbohydrates The Essential Molecules of Life. 2nd ed. Oxford: Elsevier; 2009. p. 203. [Google Scholar]
- 13.(a) Frechet J, Schuerch C. J. Am. Chem. Soc. 1969;91:1161. [Google Scholar]; (b) Zachoval J, Schuerch C. J. Am. Chem. Soc. 1969;91:1165. [Google Scholar]; (c) Schuerch C. Acc. Chem. Res. 1973;6:184. [Google Scholar]
- 14.Nakatsubo F, Kamitakahara H, Hori M. J. Am. Chem. Soc. 1996;118:1677. [Google Scholar]
- 15.Kadokawa J- i, Kasai S, Watanabe Y, Karasu M, Tagaya H, Chiba K. Macromolecules. 1997;30:8212. [Google Scholar]
- 16.(a) Penczek S. Models of Biopolymers By Ring- Opening Polymerization. Boca Raton, Fl: CRC Press; 1989. [Google Scholar]; (b) Klemm D, Heublein B, Fink H-P, Bohn A. Angew. Chem., Int. Ed. 2005;44:3358. doi: 10.1002/anie.200460587. [DOI] [PubMed] [Google Scholar]
- 17.Lichtenthaler FW. Ullmann's Encyclopedia of Industrial Chemistry. Wiley-VCH Verlag; 2000. [Google Scholar]
- 18.(a) Weigel PH, DeAngelis PL. J. Biol. Chem. 2007;282:36777. doi: 10.1074/jbc.R700036200. [DOI] [PubMed] [Google Scholar]; (b) Kadokawa J-i. Chem. Rev. 2011;111:4308. doi: 10.1021/cr100285v. [DOI] [PubMed] [Google Scholar]
- 19.(a) Fuchs E-F, Lehmann J. Carbohydr. Res. 1976;49:267. [Google Scholar]; (b) Suhara Y, Hildreth JEK, Ichikawa Y. Tet. Lett. 1996;37:1575. [Google Scholar]; (c) Grafvon Roedern E, Lohof E, Hessler G, Hoffmann M, Kessler H. J. Am. Chem. Soc. 1996;118:10156. [Google Scholar]; (d) Suhara Y, Ichikawa M, Hildreth JEK, Ichikawa Y. Tet. Lett. 1996;37:2549. [Google Scholar]; (e) Timmers CM, Turner JJ, Ward CM, van der Marel GA, Kouwijzer MLCE, Grootenhuis PDJ, vanBoom JH. Chem. Eur. J. 1997;3:920. [Google Scholar]; (f) Nishimura S-I. Chem. Comm. 1998:617. [Google Scholar]; (g) Szabo L, Smith BL, McReynolds KD, Parrill AL, Morris ER, Gervay J. J. Org. Chem. 1998;63:1074. [Google Scholar]; (h) Suhara Y, Yamaguchi Y, Collins B, Schnaar RL, Yanagishita M, Hildreth JEK, Shimada I, Ichikawa Y. Bioorg. Med. Chem. 2002;10:1999. doi: 10.1016/s0968-0896(02)00020-2. [DOI] [PubMed] [Google Scholar]; (i) Gervay- Hague J, Weathers JTM. J. Carbohyd. Chem. 2002;21:867. [Google Scholar]; (j) Gruner SAW, Truffault V, Voll G, Locardi E, Stöckle M, Kessler H. Chem. Eur. J. 2002;8:4365. doi: 10.1002/1521-3765(20021004)8:19<4365::AID-CHEM4365>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]; (k) Sharma GVM, Reddy KR, Krishna PR, Sankar AR, Narsimulu K, Kumar SK, Jayaprakash P, Jagannadh B, Kunwar AC. J. Am. Chem. Soc. 2003;125:13670. doi: 10.1021/ja035752i. [DOI] [PubMed] [Google Scholar]; (l) Gregar TQ, Gervay-Hague J. J. Org. Chem. 2003;69:1001. doi: 10.1021/jo035312+. [DOI] [PubMed] [Google Scholar]; (m) Fujimura F, Hirata T, Morita T, Kimura S, Horikawa Y, Sugiyama J. Biomacromolecules. 2006;7:2394. doi: 10.1021/bm060415y. [DOI] [PubMed] [Google Scholar]; (n) Saludes JP, Ames JB, Gervay-Hague J. J. Am. Chem. Soc. 2009;131:5495. doi: 10.1021/ja808286x. [DOI] [PMC free article] [PubMed] [Google Scholar]; (o) Soengas RG, Estévez AM, Estévez JC, Estévez RJ. C. R. Chim. 2011;14:313. [Google Scholar]
- 20.(a) Appella DH, Christianson LA, Klein DA, Powell DR, Huang X, Barchi JJ, Gellman SH. Nature. 1997;387:381. doi: 10.1038/387381a0. [DOI] [PubMed] [Google Scholar]; (b) Gellman SH. Acc. Chem. Res. 1998;31:173. [Google Scholar]; (c) Cheng RP, Gellman SH, DeGrado WF. Chem. Rev. 2001;101:3219. doi: 10.1021/cr000045i. [DOI] [PubMed] [Google Scholar]; (d) García-Martín MdG, de Paz Báñez MV, García-Alvarez M, Muñoz-Guerra S, Galbis JA. Macromolecules. 2001;34:5042. [Google Scholar]; (e) Hill DJ, Mio MJ, Prince RB, Hughes TS, Moore JS. Chem. Rev. 2001;101:3893. doi: 10.1021/cr990120t. [DOI] [PubMed] [Google Scholar]; (f) Horne WS, Gellman SH. Acc. Chem. Res. 2008;41:1399. doi: 10.1021/ar800009n. [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Seebach D, Gardiner J. Acc. Chem. Res. 2008;41:1366. doi: 10.1021/ar700263g. [DOI] [PubMed] [Google Scholar]; (h) Lee M-R, Stahl SS, Gellman SH, Masters KSJ. J Am. Chem. Soc. 2009;131:16779. doi: 10.1021/ja9050636. [DOI] [PMC free article] [PubMed] [Google Scholar]; (i) Mowery BP, Lindner AH, Weisblum B, Stahl SS, Gellman SH. J. Am. Chem. Soc. 2009;131:9735. doi: 10.1021/ja901613g. [DOI] [PubMed] [Google Scholar]; (j) Dohm MT, Mowery BP, Czyzewski AM, Stahl SS, Gellman SH, Barron AE. J. Am. Chem. Soc. 2010;132:7957. doi: 10.1021/ja909734n. [DOI] [PMC free article] [PubMed] [Google Scholar]; (k) Zhang J, Markiewicz MJ, Mowery BP, Weisblum B, Stahl SS, Gellman SH. Biomacromolecules. 2011;13:323. doi: 10.1021/bm2013058. [DOI] [PMC free article] [PubMed] [Google Scholar]; (l) Liu R, Masters KS, Gellman SH. Biomacromolecules. 2012;13:1100. doi: 10.1021/bm201847n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.(a) Goldstein IJ, Hollerman CE, Smith EE. Biochemistry. 1965;4:876. doi: 10.1021/bi00881a013. [DOI] [PubMed] [Google Scholar]; (b) Doyle RJ, Woodside EE, Fishel CW. Biochem. J. 1968;106:35. doi: 10.1042/bj1060035. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Colonna P, Biton V, Mercier C. Carbohydr. Res. 1985;137:151. [Google Scholar]
- 22.Schmidt MW, Baldridge KK, Boatz JA, Elbert ST, Gordon MS, Jensen JH, Koseki S, Matsunaga N, Nguyen KA, Su S, Windus TL, Dupuis M, Montgomery JA. J. Comput. Chem. 1993;14:1347. [Google Scholar]
- 23.Yang Q-C, Seiler P, Dunitz JD. Acta Crystallogr., Sect. C: Cryst. Struct. Commun. 1987;43:565. [Google Scholar]
- 24.Urbancyzk-Lipkowska Z, Suwinska K, Luboradzki R, Mostowicz D, Kaluza Z, Grodner J, Chmielewski M. Carbohydr. Res. 1994;256:1. [Google Scholar]
- 25.Argay G, Fabian L, Kalman A, Bernath G, Gyarmati ZC. Acta Crystallogr., Sect.E:Struct.Rep.Online. 2004;60:o170. doi: 10.1107/S0108768104024553. [DOI] [PubMed] [Google Scholar]
- 26.(a) Šebenda J, Hauer J, Biroš J. J. Poly. Sci.: Polym. Chem. Ed. 1976;14:2357. [Google Scholar]; (b) Šebenda J, Hauer J. Polym. Bull. 1981;5:529. [Google Scholar]; (c) Hashimoto K, Oi T, Yasuda J, Hotta K, Okada M. J. Polym. Sci., Part A: Polym. Chem. 1997;35:1831. [Google Scholar]; (d) Hashimoto K, Yasuda J, Kobayashi M. J. Polym. Sci., Part A: Polym. Chem. 1999;37:909. [Google Scholar]; (e) Hashimoto K. Prog. Polym. Sci. 2000;25:1411. [Google Scholar]; (f) Cheng J, Deming TJ. J. Am. Chem. Soc. 2001;123:9457. doi: 10.1021/ja0110022. [DOI] [PubMed] [Google Scholar]; (g) Zhang J, Kissounko DA, Lee SE, Gellman SH, Stahl SS. J. Am. Chem. Soc. 2009;131:1589. doi: 10.1021/ja8069192. [DOI] [PubMed] [Google Scholar]; (h) Zhang J, Gellman SH, Stahl SS. Macromolecules. 2010;43:5618. [Google Scholar]; (i) Zhang J, Markiewicz MJ, Weisblum B, Stahl SS, Gellman SH. ACS Macro Lett. 2012:714. doi: 10.1021/mz300172y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hashimoto K, Mori K, Okada M. Macromolecules. 1992;25:2592. [Google Scholar]
- 28.Chmielewski M, Kaluza Z. Carbohydr. Res. 1987;167:143. [Google Scholar]
- 29.Kałuza Z, Abramski W, Bełzecki C, Grodner J, Mostowicz D, Urbański R, Chmielewski M. Synlett. 1994;1994:539. [Google Scholar]
- 30.Wheeler SE, Houk KN, Schleyer PvR, Allen WD. J. Am. Chem. Soc. 2009;131:2547. doi: 10.1021/ja805843n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roux MV, Jiménez P, Dávalos JZ, Castaño O, Molina MT, Notario R, Herreros M, Abboud JLM. J. Am. Chem. Soc. 1996;118:12735. [Google Scholar]
- 32.Kishore N, Tewari YB, Yap WT, Goldberg RN. Biophys. Chem. 1994;49:163. doi: 10.1016/0301-4622(93)e0067-f. [DOI] [PubMed] [Google Scholar]
- 33.(a) Cubbon RCP. Makromol. Chem. 1964;80:44. [Google Scholar]; (b) Šebenda J. Pure & Appl. Chem. 1976;48:329. [Google Scholar]
- 34.Chmielewski M, Kaluza Z, Dodziuk H, Suwinska K, Rosenbaum D, Duddeck H, Magnus PD, Huffman JC. Carbohydr. Res. 1990;203:183. [Google Scholar]
- 35.Dickson L, Matuszak CA, Qazi AH. J. Org. Chem. 1978;43:1007. [Google Scholar]
- 36.Venyaminov SY, Yang JT. In: Circular Dichroism and the Conformational Analysis of Biomolecules. Fasman GD, editor. New York: Plenum Press; 1996. p. 69. [Google Scholar]
- 37.(a) Seebach D, Overhand M, Kühnle FNM, Martinoni B, Oberer L, Hommel U, Widmer H. Helv. Chim. Acta. 1996;79:913. [Google Scholar]; (b) Cheng J, Deming T. J. Macromolecules. 2001;34:5169. [Google Scholar]
- 38.Whistler RL. Carbohydrates in Solution. Vol. 117. American Chemical Society; 1973. p. 242. [Google Scholar]
- 39.(a) Mortell KH, Weatherman RV, Kiessling LL. J. Am. Chem. Soc. 1996;118:2297. [Google Scholar]; (b) Kanai M, Mortell KH, Kiessling LL. J. Am. Chem. Soc. 1997;119:9931. [Google Scholar]; (c) Mann DA, Kanai M, Maly DJ, Kiessling LL. J. Am. Chem. Soc. 1998;120:10575. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.