Abstract
Personalized medicine is a new framework for medical care that involves modelling and simulation of a disease on the basis of underlying mechanisms. This strategy must replace the 20th century paradigm of defining disease by pathology or associated signs and symptoms, and conducting outcomes research that is based on the presence or absence of the disease syndrome. New technologies, including next-generation sequencing, the ‘omics’, and powerful computers provide massive amounts of accurate data. However, attempts to understand complex disorders such as chronic inflammatory disorders, functional disorders and cancers by applying new technologies within the 20th century framework has failed to produce the expected medical advances. To help physicians embrace a paradigm shift, the limitations of the old framework and major advantages of the new framework must be demonstrated. Chronic pancreatitis is an ideal complex disorder to study, because the organ is so simple, and the advantages of personalized medicine are so profound.
Introduction
Medicine improves in gradual steps and quantum leaps. Medical practice is established within a conceptual framework that is based on the current understanding of a disease and gradually improves as new knowledge fills in the details. After 100 years of incremental progress, new technologies and discoveries in the past few years have revealed major inconsistencies and inadequacies within the existing framework; a new conceptual framework is now required to advance the field. However, no one will transition from the old framework to the new until they know the limitations of the old and the advantages of the new.1
The need for a new framework for Western medicine is clear, as technical breakthroughs, major discoveries and massive spending applied within the 20th century framework has not resulted in the expected improvements in patient outcomes for many complex inflammatory disorders, functional disorders and cancers.2 Indeed, the 20th century approaches cannot conquer these disorders, because the existing framework cannot circumvent two undisputed facts: firstly that most uncured disorders are complex processes, and secondly that each person is fundamentally different from the average of the population.
Personalized medicine is a new conceptual framework that represents the quantum leap from the 20th century paradigm of Western medicine to medicine for the 21st century. The personalized medicine paradigm retains the advances of the 20th century but also effectively addresses complex disorders. What is personalized medicine? How is it different from the current framework? How is the new framework applied to medical practices?
Chronic pancreatitis is a complex chronic inflammatory disorder that illustrates the limitations of the 20th century framework and the advantages of the new personalized medicine framework. A paradigm shift has occurred in the diagnosis and treatment of chronic pancreatitis in Pittsburgh, Pennsylvania, USA, and this program will be used to illustrate how the new paradigm works.
20th century medicine
In the 20th century, the conceptual framework for medicine was based on the germ theory of disease, which was developed during the 19th century. The fundamental assumption was that acquired diseases were caused by a single pathologic factor that results in a complex syndrome. The scientific method that was established for medical education and medical research early in the 20th century was observation, generation of hypotheses and experimental testing to determine whether an association between a single factor and a disease syndrome existed. Koch’s postulates (Box 1) codified the medical scientific method by structured research experiments to test hypotheses within the context of the germ theory of disease. At the same time, disease taxonomy (that is, disease description, nomenclature and classification) was built on tissue pathology, and the diagnosis of a disease was made on the basis of the identification of pathology within a tissue sample or by using surrogate signs and symptoms. If a ‘germ’ or other single aetiological factor was not identified, the disease was defined by the type of pathology (for example, inflammation, metaplasia, or histological abnormality for functional disorders) rather than the aetiology. A treatment was then prescribed on the basis of the diagnosis and evidence that previous patients within a similar disease classification often improved under that therapeutic regimen. This framework was established by the 1910 ‘Flexner Report’.3 The curriculum proposed by Flexner was designed to identify single aetiologies for complex syndromes, and this approach continues to be taught in medical schools today. This framework is also the basis of medical research conducted to generate evidence used to develop practice guidelines.
Box 1 Koch's Postulates.
The microorganism must be found in abundance in all organisms suffering from the disease, but not in healthy organisms.
The microorganism must be isolated from a diseased organism and grown in pure culture.
The cultured microorganism must cause disease when introduced into a healthy organism.
The microorganism must be reisolated from the inoculated, diseased experimental host and identified as being identical to the original specific causative agent.
Chronic pancreatitis is currently defined as a continuing inflammatory disease of the pancreas characterized by irreversible morphologic changes that typically cause pain and/or permanent loss of function.4 Despite considerable research efforts using approaches from the 20th century, by 1995, chronic pancreatitis remained “an enigmatic process of uncertain pathogenesis, unpredictable clinical course, and unclear treatment“.5 A new framework from which to understand and treat chronic pancreatitis is needed.
New technologies
The framework for personalized medicine differs from the 20th century paradigm in many ways (Table 1). The critical issue is that many different aetiological mechanisms can cause the same signs and symptoms in an organ or system. Under the existing framework, multiple disorders are classified as a homogenous disease, and they are evaluated and treated as if there was only one underlying cause. These disorders cannot be resolved using the binary 20th century scientific method of hypothesis testing as no ‘single’ factor is responsible for a complex disease. Furthermore, complex disorders cannot be easily classified using the old hierarchical disease taxonomy,2 and attempts to use disease subclassifications to evaluate multifactor diseases becomes expensive and unwieldy for common disorders and impossible for rare disorders (Figure 1a). The problem is confounded when multiple genetic risk variables are added to the subclassification approach.
Table 1.
Medicine in the twentieth and twenty-first centuries
| Domain | 20th century | 21st century |
|---|---|---|
| Overarching goal | Treatment of disease | Prevention of disease |
| Enabling technology | Microscope, culture techniques, biopsies | NGS, biomarkers, computers |
| Disease model | Germ theory | Complex risk, variant response to stress |
| Paradigm-shifting force | Flexner Report of 1910 | Economics |
| Education focus | Disease diagnosis and classification | Normal responses, assessment of variants |
| Scientific focus | Determine associations | Determine mechanisms |
| Scientific approach | Koch’s Postulates, global statistics | Modelling and simulation, performance characteristics |
| Disease classification | Tissue pathology, syndromes | Genetic and environmental risks, surrogate endpoints |
| Disease time frame | Static, cross-sectional | Dynamic, longitudinal |
| Physician focus | Overall organ dysfunction | Activity and trajectory of dysfunctional systems |
| Assessment | Disease classification | Outcome prediction |
| Treatment | Trial and error | Targeted, optimized |
| Success measures | Population based | Individual based |
| Utility of the paradigm | Infectious diseases, Mendelian genetics, single agent disorders, cancer detection | Inflammatory disease, complex genetics, functional disorders, cancer control |
Abbreviation: NGS, next generation sequencing.
Figure 1.
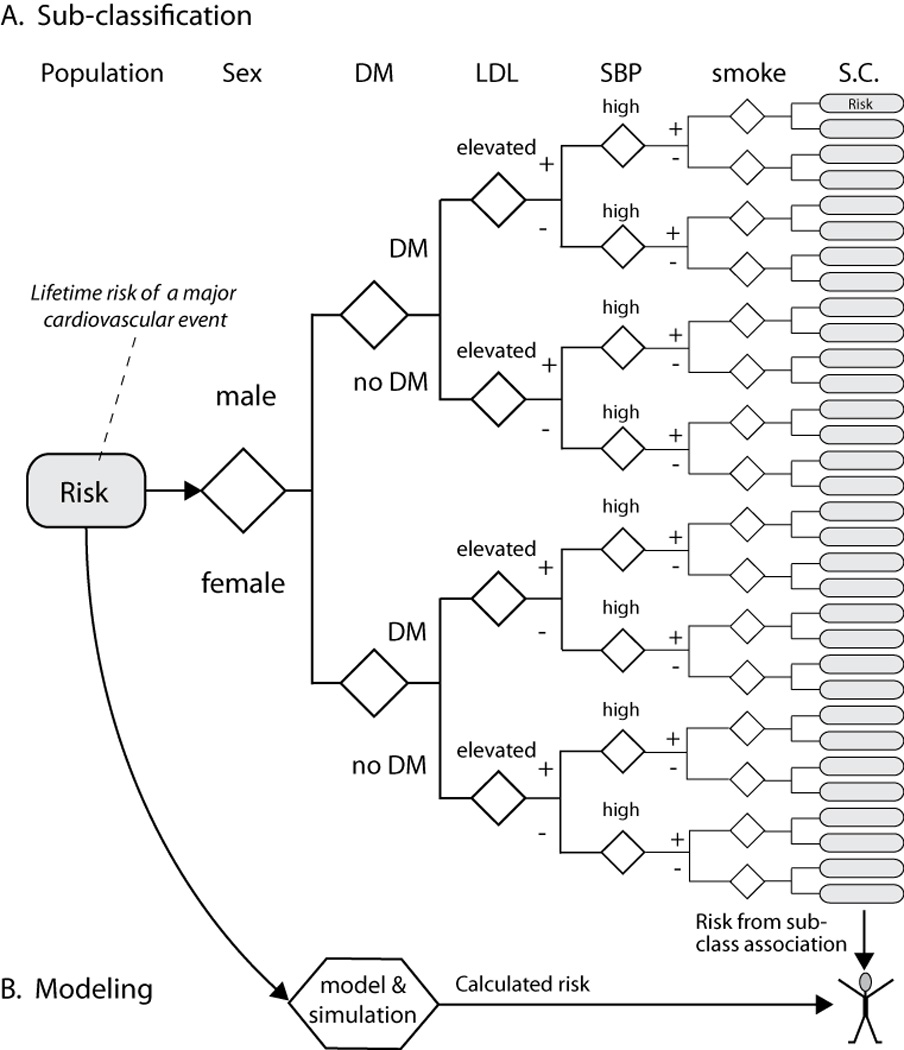
Two approaches to assess individual risk on the basis of the presence or absence of six variables. a | Traditional subclassification approaches divide the population into 32 categories, record the outcomes and then estimate the individual’s risk based on the category with characteristics most like the individual. b | Modelling and simulation approaches can start with results from subclassification approaches and improve on them using statistical and mechanistic (deterministic) approaches based on knowledge of biology. Risk estimates for independent or combined variables can be calculated to replicate the results of the population study and provide new insights (for example, Archimedes14). Modelling is useful for simulation population studies, and also for an individual patient. For an individual patient, modelling provides prognostic information that can be more accurate when it uses continuous variables rather than arbitrary cut-off values, when the patient is in a poorly-populated subclassification with limited outcomes data, when deterministic relationships based on biological mechanisms are included, when new variables of known mechanistic effect are added (for example, genetic risk) or when their disease falls outside of current recommendations and population studies to determine complex risk subcategories cannot be done owing to small patient numbers or high costs. Rounded rectangles, patient populations; diamonds, decision points; hexagon, risk calculator; DM, diabetes mellitus; LDL, low density lipoproteins; SBP, systolic blood pressure; smoke, history of smoking; SC, subclassification.
Instead, the framework for medicine in the 21st century must begin with mechanistic and predictive modelling of normal biological systems before determining which genetic, environmental and structural factors, or a combination of these factors, alter ‘normal’ beyond the capacity of the system to adapt and therefore results in disease. A model is a simplified representation of the system that is designed to help researchers understand the real system. The model should define all of the critical elements of the real system such that manipulation of the model predicts the outcome of manipulating the system in patients. Simulation involves modelling the processes of the system over time and enables the investigator to understand the interaction of the parts and predict outcomes on the basis of results obtained by manipulating the model. Modelling and simulation of complex disorders is critical because complex interactions and evolving processes cannot be understood by studying a single variable or even multiple but independent variables. Simulating the disease risk or trajectory enables the effect of interventions to be evaluated in silico, on the basis of the combination of risk factors specific to each individual patient (Figure 1b).
A new paradigm is required
Four advances in technology illustrate the limitations of the outdated 20th century framework. The advances are critical to ensure that patients with complex disorders receive appropriate care.
Imaging and diagnostic biopsies
A diagnosis based on tissue pathology was the foundation of medicine in the 20th century. However, advances in obtaining high quality biopsy samples from an anatomical location using exploratory surgery, laparoscopy, endoscopy and advanced imaging have not led to increased effectiveness of treatments for complex inflammatory disorders or cancers. In the case of cancer, where the character of the disease and patient outcomes are determined by aberrant genetics within the tumour, a personalized medicine approach is needed to identify the genetic signatures that define the mechanisms underlying the pathology in a particular patient. This approach means that the behaviour of a tumour can be predicted, and appropriate targets for therapy identified.6,7 However, for complex inflammatory disorders such as chronic pancreatitis, tissue evaluation tells us about disease activity, but not about aetiology, mechanisms,8,9 or why some people develop pancreatitis whereas others do not.10
Evidence-based medicine
The explosion of clinical and basic research publications in the last half of the 20th century resulted in data overload for physicians. Evidence-based medicine is an analysis framework developed by clinical epidemiologists in the early 1990s to evaluate, synthesize and present clinical research reports in a standardized fashion that could be understood and acted upon by clinicians and policy makers.11 Although successful in achieving these goals, evidence-based medicine is not a new paradigm, as it exists within the 20th century framework as an information literacy model.12 In addition, evidence-based medicine that utilizes retrospective analysis of old clinical studies that approached complex medical disorders using a binary (germ theory) method and disease classifications cannot generate fundamentally new insights or mechanistic breakthroughs, and actually diminishes the usefulness of existing information. For example, classification of cardiovascular risk for systolic blood pressure (Figure 1a) requires a cut-off value to generate a binary operator; such as <140 mmHg equating to no risk and >140 mmHg equating to full risk, implying that lowering systolic blood pressure from 142 mmHg to 138 mmHg is more important than lowering systolic blood pressure from 160 mmHg to 142 mmHg. Mathematical modelling can use continuous variables to provide more accurate risk and outcome predictions (Figure 1b). New computational tools, such as Archimedes,13 are needed for guiding clinical decisions in complex disorders14 and especially for uncommon diseases for which large populations of patients are not widely available. Thus, the ‘next generation’ of evidence-based methods is needed to evaluate the mechanistic evidence underlying complex disorders and the performance of new computational tools.
Genome-wide association studies
A genome-wide association study (GWAS) uses thousands to millions of common single nucleotide polymorphisms (SNPs) in the human genome as chromosome location markers to pinpoint the genetic location of a gene that contributes to a particular disease. Genetic data from large cohorts of patients with and without a defined disease feature are compared through an unbiased statistical approach to discovering the genetic basis of an inherited trait.15 The underlying assumptions are that humans are related at some level, that pieces of chromosomes that carry disease risk are inherited in subsets of people, and that the culprit gene can be discovered through a nearby genetic marker that can be tracked through various populations of patients with the disease. This approach has been useful for identifying genes responsible for disorders with a fairly simple genetic basis, but the inherent statistical challenges are further weakened upon application to conditions with high disease complexity. Huge numbers of patients with the global, syndrome-defined phenotype of interest are required. As independent effects are small, multiple testing of SNPs (for example, n = 1,000,000) requires very high P values to reduce false positive findings (for example, P <0.00000001), and large variance occurs as a result of vague phenotypic measures and mixed populations.
Early results from GWAS confirm that no single gene causes complex syndromes but instead reveals that dozens and dozens of factors are associated with complex disorders, as illustrated with at least 47 confirmed loci for ulcerative colitis16 and 71 for Crohn’s disease.17 Thus, GWAS for more complex and traditionally defined disorders requires multi-million dollar projects with tens of thousands of patients and even more controls, which must often involve international consortia.16–19 Furthermore, the conceptual foundations of the 20th century medical paradigm are inadequate to interpret these results and provide direction for clinical decision-making. Instead, incorporating genes identified in GWAS into mechanistic models within a new personalized medicine framework will guide improved diagnosis and treatment.
Next-generation sequencing
Next-generation sequencing is a rapidly evolving technology that uses massive parallel sequencing with computer reconstruction of DNA sequence fragments to generate a partial or full genomic sequence. It is a major advance over GWAS, because it reveals disease-causing genetic variants rather than nearby markers and is faster and costs less than traditional Sanger sequencing of a single large gene. Clinical application of next-generation sequencing reveals a tremendous number of genetic variations among individuals, with many new mutations and multiple susceptibly risk factors seen in each patient.20,21 These realities highlight the near impossibility of resolving the complexity of a patient’s genome using epidemiology, statistics and patient subclassification strategies within the framework of 20th century medicine. However, application of next-generation sequencing into new paradigms will be the foundation of medicine in the 21st century.
Using expensive technology to diagnose patients on the basis of the old framework is inefficient and ineffective, because the complex data sets cannot be adequately interpreted. What is needed is a new framework so that new types of data relevant to individual patients can be integrated into a treatment plan that targets the true aetiology precisely.
Personalized medicine
In contrast to current practice, health care should now not focus on disease, but on health; not on disease status but on trajectory; not on treatment based on pathology but on avoiding pathology; not on treatment trial and error but on selection of the best treatment with continuous optimization (Table 1).
The process of developing disease models is challenging but can be accomplished using reverse engineering, an approach used to understand other complex systems. The system is broken into its component parts, the function of each component is modelled, the modelled components are reintegrated, and the effects of the integrated components on overall processes are simulated under multiple conditions. In medicine, we know a lot about the components of complex disorders, including the specialized cells, organs, signalling and regulation under normal and stress conditions, which we can model and simulate. The advantage of such modelling and simulation is that it can be done on a patient-by-patient basis, incorporating the unique variables that each patient possesses.
Fortunately, we do not need to model each biological step in every pathway to develop useful disease models. Through detailed evaluation of a series of patients with a complex disease involving a known system, and by isolating and evaluating individual parts, the components that are commonly dysfunctional can be identified. Then, the dysfunctional mechanism can be simulated and brought back to the larger model. At this point, the effect of any combination of variables in a single patient can be calculated, and the effect of a therapeutic intervention can be simulated. Our application of this approach to the problems of early diagnosis and management of chronic pancreatitis provides a useful illustrative model.
Pancreatitis—a good model?
Pancreatitis can be modelled because the exocrine pancreas is so simple, with only two cell types (acinar and duct), each of which is responsible for only one function (synthesize digestive enzymes and flush enzymes out of the pancreas, respectively). Few external risk factors affect the pancreas, as the organ is not directly exposed to the environment and does not metabolize or concentrate toxins. Furthermore, accurate mathematical models of the duct have already been developed22 and have been able to predict both emerging biology23 and a new subclass of disease-causing mutations.24
Modelling pancreatitis involves only one major mechanism of injury: trypsinogen activation. Furthermore, five major susceptibility genes linked to trypsin are known and have been replicated throughout the world, with additional key genes under evaluation.25,26 The first phase of the North American Pancreatitis Study 2 (NAPS2) evaluated 1,000 patients with recurrent acute and chronic pancreatitis, revealing the heterogeneity of a disorder previously classified as one entity (ICD9 code 577.1). Only 15% of patients developed pancreatitis with heavy alcohol consumption alone (long presumed to be the primary cause of pancreatitis). We found that 24% of cases had a genetic aetiology (14% CFTR, 3% CFTR and SPINK1, 4% SPINK1, 3% PRSS1); 4% were attributable to hyperlipidaemia or autoimmune causes; 9% were caused by obstruction; 3% resulted from severe acute pancreatitis (usually with pancreatic necrosis); and 42% were idiopathic.27 Alcohol had a surprisingly weak independent effect (relative risk [RR] 1.37), with smoking surprisingly strong as an independent risk factor (RR 2.35). The two together were found to act synergistically (RR 8.07).28 These data provide support for a new framework for a personalized approach to chronic pancreatitis in the clinic.
Clinical application
At the University of Pittsburgh, to create clinically useful models, we had to develop an aetiology-based risk classification system in which the aetiological factors were organized and evaluated.4 Development of a disease progression model29 enabled the factors that were driving disease progression to be identified in human studies30 and the mechanism validated in animal models,31 resulting in the recognition that early intervention can change outcome for some patients (for example, cessation of alcohol consumption32 or smoking33) but not others (for example, susceptibility gene mutations plus SPINK1 mutations – Whitcomb, unpublished 2012). A growing understanding of genetics and gene–environment interactions also means that patients can be classified by mechanism.
An academic medical centre is useful for organizing dedicated physicians–scientists into teams to discover the components of the complex systems that lead to the chronic inflammatory disorders left uncured from the 20th century.27 At the University of Pittsburgh, we have developed an evaluation and treatment work-flow that is based on a personalized patient pancreatitis paradigm (Figure 2). Most important is that genetic testing and complex risk analysis comes at the beginning, not the end. The initial analysis (Figure 2b) immediately stratifies the patients into low and high probability categories on the basis of integrated risk and biomarker evidence of injury (for example, biomarker A). Diagnostic work-ups are rapid, specific and efficient. If the genetic or other combinations of risk and biomarker tests are positive, diagnostic work-up stops. This strategy is in contrast to current approaches to work-up of patients with chronic pancreatitis using repeated CT scans, endoscopic ultrasound, pancreatic function testing, endoscopic retrograde cholangiopancreatography with sphincter of Oddi manometry, and so on to identify irreversible structural and functional damage for a surrogate histological diagnosis. Failure to pinpoint risk factors or biomarkers of pancreatitis in our system does not exclude the possibility of a complex pancreatitis disorder, but, as new genetic factors are discovered and incorporated into the models, the number of patients with true idiopathic pancreatitis shrinks and the likelihood that the symptoms arise from the pancreas when the initial evaluation reveals no aetiology, diminishes. Furthermore, this approach immediately classifies the patients as having simple or complex genetic syndromes, autoimmune disorders, or mechanistic problems that require different treatments (Figure 2c).
Figure 2.
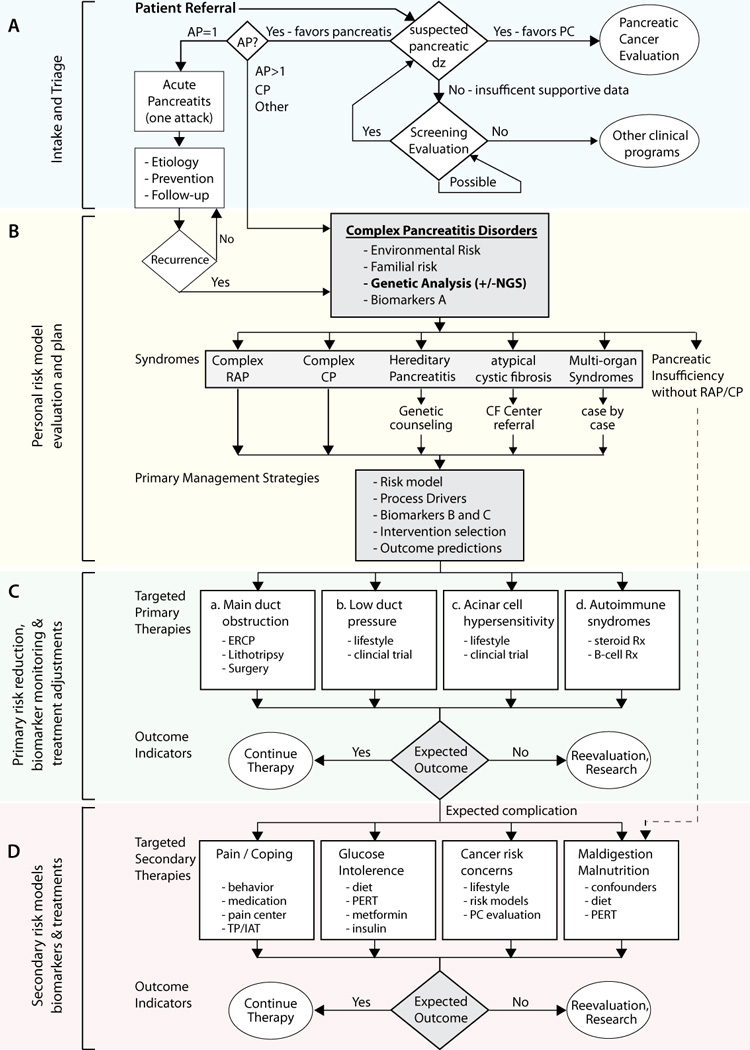
Clinical pathway of patients being evaluated for suspected pancreatic disease. a | Referred patients undergo initial evaluation and triage. b | Patients with idiopathic or complicated pancreatic diseases undergo genetic testing and risk modelling. c | Patients are classified into general disease mechanism groups and treatment effectiveness is monitored. d | Common complications are also anticipated and addressed, with appropriate reference to sequencing data results. Patients with unexpected outcomes are re-evaluated and additional assessment is done as needed. Abbreviations: AP, acute pancreatitis; CF, cystic fibrosis; CP, chronic pancreatitis; dz, disease; ERCP, endoscopic retrograde cholangiopancreatography; NGS, next-generation sequencing; PERT, pancreatic enzyme replacement therapy; PC, pancreatic cancer; RAP, recurrent acute pancreatitis; Rx, treatment; TP/IAT, Total pancreatectomy with islet autotransplantation.
With knowledge of a disease mechanism that is linked to biomarkers of activity (biomarker B) or progression (biomarker C) patient-specific strategies for risk reduction and treatment can be prescribed or tested. The mechanism of disease can then be used to identify possible treatments or treatment trials for particular disorders (Figure 2c). It follows that: main duct obstruction caused by pancreas divisum, stones or tumours should be treated with mechanical solutions; low hydrostatic duct pressure caused by CFTR or other mutations could be treated with CFTR correctors,34 distal duct resistance reduction at the down-stream sphinctors, or treatments to facilitate duct cell function; acinar cell hyperstimulation risk, hypersensitivity linked to PRSS1 mutations or calcium dysregulation could be treated with targeted therapies such as calcium channel blockers35 or more targeted calcium regulatory mechanisms; autoimmune pancreatitis such as IgG4-related pancreatitis should be treated with glucocorticoids and immunosuppression.36
Targeted therapy does not diminish the importance of general public health policies to reduce obesity and stop smoking, but build on these approaches. Factors that drive inflammation and fibrosis, such as alcohol and smoking, are also addressed with counselling or referral to specialized programs. Knowledge of the mechanism, proper selection of biomarkers to monitor treatment effectiveness and early signals of progression are making it possible to manage strategies to prevent or delay mechanisms of susceptibility.
Highly variable complications of pancreatic injury and inflammation include pain, diabetes mellitus, metaplasia and pancreatic exocrine failure (Figure 2d). Many patients are seen after these problems are well established, but personalized approaches might still be valuable. Pain, for example, is stratified as episodic or continuous, as the type of pain strongly affects quality of life.37 Early and aggressive treatment plans are also needed as delays in treatment diminish the likelihood of success for endoscopic and surgical approaches.38 Anxiety, depression and poor coping skills are also addressed. Patients with complex genotypes and severe symptoms who develop an unrelenting course are evaluated as candidates for total pancreatectomy and islet autotransplantation in an attempt to avoid chronic pain syndromes and diabetes mellitus from destruction of islet cells and to improve quality of life.39,40
Using new technologies
At the University of Pittsburgh, we are exploring the use of next-generation sequencing in the initial evaluation of patients with complex pancreatic diseases. The cost of whole-exome sequencing and some other sequencing technologies is already less than the cost of traditional sequencing of the known susceptibility genes for pancreatitis and continues to drop. Although whole-exome sequencing provides sequence data on thousands of genes, we ‘mask’ all of the data except for the five pancreatitis susceptibility genes (PRSS1, CFTR, SPINK1, CTRC, CASR), which we evaluate.41 Genetic variants in these genes are verified in a certified clinical laboratory. The remaining sequence data are saved for research purposes and to develop new and improved genetic risk models. As the sequencing only needs to be done once, the cost of additional genetic variation data is minimal, and the time frame to answer additional clinical questions in the future is rapid.
Better care at a lower cost
The personalized medicine approach for chronic pancreatitis not only provides better care than other approaches, it probably saves money. Savings are made by minimizing diagnostic costs, avoiding high-risk diagnostic procedures, limiting the use of therapies to patients who are most likely to benefit, and addressing the aetiology to minimize the rate of disease progression, and thereby avoiding the high cost, suffering and disability associated with advanced disease.
Diagnosis of chronic pancreatitis has traditionally been challenging, as it focuses on detecting irreversible damage to the pancreas. Early genetic testing and susceptibility evaluation can largely replace the current use of insensitive and expensive diagnostic testing in patients with pancreatitis-like symptoms. Our new approach not only saves the cost of multiple diagnostic tests, but it also avoids the risk of actually causing pancreatitis from endoscopic retrograde cholangiopancreatography (ERCP – as above), sphincter of Oddi manometry, and other invasive diagnostic procedures. Furthermore, genetic testing costs less than one endoscopic procedure and many sophisticated imaging methods. In addition, the results are definitive and enduring (they never need to be repeated); they reveal underlining mechanisms, and they direct future treatment strategies.
Aetiology-based care is also a money saver. Early diagnosis enables immediate implementation of a plan to minimize the factors driving the pancreatitis symptoms and to slow progression through targeted means, including lifestyle changes (such as no smoking or alcohol). This strategy also immediately identifies patients with genetic syndromes that have implications for family members (for example, hereditary pancreatitis) or who might benefit from attention to the risk of problems in other organs (such as lung disease in patients with mild cystic fibrosis). Aetiology-based care might also eliminate unnecessary procedures, such as performing a cholecystectomy on everyone with pancreatitis just in case the aetiology is gallbladder dysfunction. Furthermore, the ‘reverse engineering‘ approach recognizes that complications do not occur in parallel (for example, a physician cannot tell from a CT scan showing moderate fibrosis whether or not the patient has pain, maldigestion or diabetes mellitus), and provides direction for atypical presentations (such as minimal change pancreatitis with no fibrosis but considerable pain). Finally, aetiology-based interventions will probably slow disease progression and will minimize the high cost of treating complications of pancreatitis, including endoscopic treatment of duct obstructions, pancreatic enzyme replacement therapy for maldigestion, management of diabetes mellitus and pain management costs associated with medicines, endoscopies, surgeries and disability. An ounce of prevention is worth a pound of cure.
Aetiology-based care does not preclude the use of interventions that are currently available. In some cases, the combination of genetic risks and uncontrollable disease activity result in a horrible quality of life, including the inability to work or attend school, and a real threat of a chronic pain syndrome or diabetes mellitus. Well-designed modelling and simulation approaches, such as those described for selecting patients for intervention in cardiovascular disease,14 might also be useful for clinical decision making where no evidence-based guidelines exist. Such models provide the combined benefit of improving outcomes through optimal timing of irreversible interventions (for example, total pancreatectomy with islet autotransplantation) and applying these to the right patients.
Conclusions
The framework from which medicine operated in the 20th century was useful for infectious disease and disorders with a single aetiology. Medical care for complex inflammatory disorders, functional disorders and cancers has not improved to the same degree, and additions of powerful new technologies have not appreciably improved patient outcomes. A new framework that is effective in individual patients is needed. The pancreatitis program at the University of Pittsburgh Medical Center capitalizes on both a new framework and new technologies to provide a new approach to this complex disorder and serves as a roadmap to personalized medicine that delivers much better care at a much lower cost.
Acknowledgements
Dr Whitcomb would like to thank the following people for critical review of the manuscript and helpful discussion: Michelle Kienholz, Christopher Langmead PhD, Mark S. Roberts MD MMP and Adam Slivka MD PhD. The work from Dr Whitcomb’s group reported in this publication was supported by the following: National Institutes of Health (NIH) DK054709, DK061451, DK077906, the University of Pittsburgh Genomics and Proteomics Core Laboratory and the National Center for Research Resources (NCRR), a component of the and NIH Roadmap for Medical Research (UL1 RR024153), the UPCI Clinical Genomics Immunoproteomics and Sequencing Facility (NIH P30CA047904), The Frieda G. and Saul F. Shapira BRCA Cancer Research Program and the Wayne Fusaro Pancreatic Cancer Research Fund (DCW).
Footnotes
Competing interests
D. C. Whitcomb declares associations with the following company: Ambry Genetics. See the article online for full details of the relationships.
Online statement
D. C. Whitcomb owns stocks in Ambry Genetics and also the US patent 6406846 entitled “Method for determining whether a human patient is susceptible to hereditary pancreatitis, and primers therefore”, which has been licensed and provides royalty income.
Dr Whitcomb is the Giant Eagle Professor of Cancer Genetics, Professor of Medicine (with tenure), Cell Biology and Physiology, and Human Genetics at the University of Pittsburgh, USA. Dr Whitcomb received his PhD in physiology in 1983 and MD in 1985 from The Ohio State University, Columbus, Ohio, USA. He completed an Internal Medicine residency in 1988 and Gastroenterology fellowship in 1992 at Duke University, Durham, North Carolina, USA. He joined the Gastroenterology and Hepatology Division, Department of Medicine, University of Pittsburgh, in 1991, becoming Chief of Gastroenterology, Hepatology and Nutrition. Dr Whitcomb specializes in pancreatic disease research related to acute pancreatitis, chronic pancreatitis, pancreatic effects of cystic fibrosis, and pancreatic cancer with an emphasis on translational research. Dr Whitcomb has received dozens of honours and awards, written over 200 scientific articles and edited eight books. He is among the most-cited authors in the field: over a dozen of his articles have been referenced by his peers more than 100 times.
References
- 1.Kuhn TS. The Structure of Scientific Revolutions. 1st. ed ed. Chicago: Univ. of Chicago; 1962. [Google Scholar]
- 2.Committee on a Framework for Development a New Taxonomy of Disease; National Research Council. Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease. Washington, DC: National Academies Press; [PubMed] [Google Scholar]
- 3.Flexner A. Medical Education in the United States and Candida: A report to the Carnegie Foundation for the Advancement of teaching. Boston: Mass; 1910. [Google Scholar]
- 4.Etemad B, Whitcomb DC. Chronic pancreatitis: Diagnosis, classification, and new genetic developments. Gastroenterology. 2001;120:682–707. doi: 10.1053/gast.2001.22586. [DOI] [PubMed] [Google Scholar]
- 5.Steer ML, Waxman I, Freedman S. Chronic pancreatitis. New England Journal of Medicine. 1995;332(22):1482–1490. doi: 10.1056/NEJM199506013322206. [DOI] [PubMed] [Google Scholar]
- 6.Pao W, Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol. 2011 Feb;12(2):175–180. doi: 10.1016/S1470-2045(10)70087-5. [DOI] [PubMed] [Google Scholar]
- 7.Russnes HG, Navin N, Hicks J, Borresen-Dale AL. Insight into the heterogeneity of breast cancer through next-generation sequencing. The Journal of Clinical Investigation. 2011 Oct;121(10):3810–3818. doi: 10.1172/JCI57088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friess H, Ding J, Kleeff J, Liao Q, Berberat PO, Hammer J, et al. Identification of disease-specific genes in chronic pancreatitis using DNA array technology. Ann Surg. 2001 Dec;234(6):769–778. doi: 10.1097/00000658-200112000-00008. discussion 78-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shrikhande SV, Martignoni ME, Shrikhande M, Kappeler A, Ramesh H, Zimmermann A, et al. Comparison of histological features and inflammatory cell reaction in alcoholic, idiopathic and tropical chronic pancreatitis. Br J Surg. 2003;90(12):1565–1572. doi: 10.1002/bjs.4353. [DOI] [PubMed] [Google Scholar]
- 10.Whitcomb DC. Mechanisms of disease: Advances in understanding the mechanisms leading to chronic pancreatitis. Nat Clin Pract Gastroenterol Hepatol. 2004 Nov;1(1):46–52. doi: 10.1038/ncpgasthep0025. [DOI] [PubMed] [Google Scholar]
- 11.Evidence-based medicine. A new approach to teaching the practice of medicine. JAMA : the journal of the American Medical Association. 1992 Nov 4;268(17):2420–2425. doi: 10.1001/jama.1992.03490170092032. [DOI] [PubMed] [Google Scholar]
- 12.Wyer PC, Silva SA. Where is the wisdom? A conceptual history of evidence-based medicine. Journal of Evaluation in Clinical Practice. 2009 Dec;15(6):891–898. doi: 10.1111/j.1365-2753.2009.01323.x. [DOI] [PubMed] [Google Scholar]
- 13.Krishna R. Model-based benefit-risk assessment: Can Archimedes help? Clin Pharmacol Ther. 2009 Mar;85(3):239–240. doi: 10.1038/clpt.2008.240. [DOI] [PubMed] [Google Scholar]
- 14.Eddy DM, Adler J, Patterson B, Lucas D, Smith KA, Morris M. Individualized guidelines: the potential for increasing quality and reducing costs. Annals of Internal Medicine. 2011 May 3;154(9):627–634. doi: 10.7326/0003-4819-154-9-201105030-00008. [DOI] [PubMed] [Google Scholar]
- 15.Witte JS. Genome-wide association studies and beyond. Annu Rev Public Health. 2010 Apr 21;31:9–20. doi: 10.1146/annurev.publhealth.012809.103723. 4 p following. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anderson CA, Boucher G, Lees CW, Franke A, D'Amato M, Taylor KD, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nature Genetics. 2011 Mar;43(3):246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nature Genetics. 2010 Dec;42(12):1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wheeler E, Barroso I. Genome-wide association studies and type 2 diabetes. Brief Funct Genomics. 2011 Mar;10(2):52–60. doi: 10.1093/bfgp/elr008. [DOI] [PubMed] [Google Scholar]
- 19.Voight BF, Scott LJ, Steinthorsdottir V, Morris AP, Dina C, Welch RP, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet. 2010 Jul;42(7):579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Depristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011 May;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKernan KJ, Peckham HE, Costa GL, McLaughlin SF, Fu Y, Tsung EF, et al. Sequence and structural variation in a human genome uncovered by short-read, massively parallel ligation sequencing using two-base encoding. Genome Research. 2009 Sep;19(9):1527–1541. doi: 10.1101/gr.091868.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whitcomb DC, Ermentrout GB. A mathematical model of the pancreatic duct cell generating high bicarbonate concentrations in pancreatic juice. Pancreas. 2004;29(2):E30–E40. doi: 10.1097/00006676-200408000-00016. [DOI] [PubMed] [Google Scholar]
- 23.Park HW, Nam JH, Kim JY, Namkung W, Yoon JS, Lee JS, et al. Dynamic regulation of CFTR bicarbonate permeability by [Cl-]i and its role in pancreatic bicarbonate secretion. Gastroenterology. 2010 Aug;139(2):620–631. doi: 10.1053/j.gastro.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 24.Schneider A, Larusch J, Sun X, Aloe A, Lamb J, Hawes R, et al. Combined Bicarbonate Conductance-Impairing Variants in CFTR and SPINK1 Variants Are Associated With Chronic Pancreatitis in Patients Without Cystic Fibrosis. Gastroenterology. 2011 Jan;140(1):162–171. doi: 10.1053/j.gastro.2010.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen JM, Ferec C. Chronic pancreatitis: genetics and pathogenesis. Annu Rev Genomics Hum Genet. 2009;10:63–87. doi: 10.1146/annurev-genom-082908-150009. [DOI] [PubMed] [Google Scholar]
- 26.Whitcomb DC. Genetic aspects of pancreatitis. Annu Rev Med. 2010;61:413–424. doi: 10.1146/annurev.med.041608.121416. [DOI] [PubMed] [Google Scholar]
- 27.Whitcomb DC. Going MAD: Development of a "Matrix Academic Division" to Facilitate Translating Research to Personalized Medicine. Academic Medicine: Journal of the Association of American Medical Colleges. 2011 Sep 26;86(11):1353–1359. doi: 10.1097/ACM.0b013e3182303d7a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yadav D, Hawes RH, Brand RE, Anderson MA, Money ME, Banks PA, et al. Alcohol consumption, cigarette smoking, and the risk of recurrent acute and chronic pancreatitis. Arch Intern Med. 2009 Jun 8;169(11):1035–1045. doi: 10.1001/archinternmed.2009.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whitcomb DC. Hereditary Pancreatitis: New insights into acute and chronic pancreatitis. Gut. 1999;45:317–322. doi: 10.1136/gut.45.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yadav D, Whitcomb DC. The role of alcohol and smoking in pancreatitis. Nat Rev Gastroenterol Hepatol. 2010 Mar;7(3):131–145. doi: 10.1038/nrgastro.2010.6. [DOI] [PubMed] [Google Scholar]
- 31.Deng X, Wang L, Elm MS, Gabazadeh D, Diorio GJ, Eagon PK, et al. Chronic alcohol consumption accelerates fibrosis in response to cerulein-induced pancreatitis in rats. Am J Pathol. 2005 Jan;166(1):93–106. doi: 10.1016/S0002-9440(10)62235-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nordback I, Pelli H, Lappalainen-Lehto R, Jarvinen S, Raty S, Sand J. The recurrence of acute alcohol-associated pancreatitis can be reduced: a randomized controlled trial. Gastroenterology. 2009 Mar;136(3):848–855. doi: 10.1053/j.gastro.2008.11.044. [DOI] [PubMed] [Google Scholar]
- 33.Talamini G, Bassi C, Falconi M, Sartori N, Vaona B, Bovo P, et al. Smoking cessation at the clinical onset of chronic pancreatitis and risk of pancreatic calcifications. Pancreas. 2007 Nov;35(4):320–326. doi: 10.1097/mpa.0b013e31812e965e. [DOI] [PubMed] [Google Scholar]
- 34.Van Goor F, Hadida S, Grootenhuis PD, Burton B, Stack JH, Straley KS, et al. Correction of the F508del-CFTR protein processing defect in vitro by the investigational drug VX-809. Proceedings of the National Academy of Sciences of the United States of America. 2011 Nov 15;108(46):18843–18848. doi: 10.1073/pnas.1105787108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morinville VD, Lowe ME, Elinoff BD, Whitcomb DC. Hereditary pancreatitis amlodipine trial: a pilot study of a calcium-channel blocker in hereditary pancreatitis. Pancreas. 2007 Nov;35(4):308–312. doi: 10.1097/mpa.0b013e318120023a. [DOI] [PubMed] [Google Scholar]
- 36.Shimosegawa T, Chari ST, Frulloni L, Kamisawa T, Kawa S, Mino-Kenudson M, et al. International consensus diagnostic criteria for autoimmune pancreatitis: guidelines of the International Association of Pancreatology. Pancreas. 2011 Apr;40(3):352–358. doi: 10.1097/MPA.0b013e3182142fd2. [DOI] [PubMed] [Google Scholar]
- 37.Mullady DK, Yadav D, Amann ST, O'Connell MR, Barmada MM, Elta GH, et al. Type of pain, pain-associated complications, quality of life, disability and resource utilisation in chronic pancreatitis: a prospective cohort study. Gut. 2011 Jan;60(1):77–84. doi: 10.1136/gut.2010.213835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clarke B, Slivka A, Tomizawa Y, Sanders M, Papachristou G, Whitcomb DC, et al. Endoscopic Therapy is Effective for Patients with Chronic Pancreatitis. Clinical gastroenterology and hepatology : The official clinical practice journal of the American Gastroenterological Association. 2012 Jan 12; doi: 10.1016/j.cgh.2011.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sutton JM, Schmulewitz N, Sussman JJ, Smith M, Kurland JE, Brunner JE, et al. Total pancreatectomy and islet cell autotransplantation as a means of treating patients with genetically linked pancreatitis. Surgery. 2010 Oct;148(4):676–685. doi: 10.1016/j.surg.2010.07.043. discussion 85-6. [DOI] [PubMed] [Google Scholar]
- 40.Bellin MD, Freeman ML, Schwarzenberg SJ, Dunn TB, Beilman GJ, Vickers SM, et al. Quality of life improves for pediatric patients after total pancreatectomy and islet autotransplant for chronic pancreatitis. Clinical Gastroenterology and Hepatology : the official clinical practice journal of the American Gastroenterological Association. 2011 Sep;9(9):793–799. doi: 10.1016/j.cgh.2011.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.LaRusch J, Barmada MM, Solomon S, Whitcomb DC. Whole Exome Sequencing Identifies Multiple, Complex Etiologies in an Idiopathic Hereditary Pancreatitis Kindred. JOP. 2012 May;13(3):1001–1006. [PMC free article] [PubMed] [Google Scholar]


