Abstract
Host-defense peptides inhibit bacterial growth but show little toxicity toward mammalian cells. A variety of synthetic polymers have been reported to mimic this antibacterial selectivity; however, achieving comparable selectivity for fungi is more difficult because these pathogens are eukaryotes. Here, we report nylon-3 polymers based on a novel subunit that display potent antifungal activity (MIC = 3.1 μg/mL for C. albicans) and favorable selectivity (IC10 > 400 μg/mL for 3T3 fibroblast toxicity; HC10 > 400 μg/mL for hemolysis).
Natural strategies to fend off microbial infection include production of relatively small peptides that manifest antimicrobial activity, part of the innate immune response.1 These “host-defense peptides” have diverse sequences and bioactive conformations, and their biological effects appear to arise from multiple mechanisms.2 Many host-defense peptides can adopt amphiphilic structures in which lipophilic and hydrophilic (usually cationic) side chains are segregated to distinct regions of the molecular surface.3 This global amphiphilicity is widely believed to underlie the ability of host-defense peptides to compromise bacterial membrane barrier function and thereby inhibit the growth of or kill prokaryotes.4 Numerous reports describe synthetic peptides or peptidomimetic oligomers designed to be globally amphiphilic that can serve as tools to elucidate the origins of host-defense peptide function and as candidates for therapeutic application.5 The evaluation of synthetic systems has recently expanded to include random copolymers that contain both hydrophilic and lipophilic subunits, which are much more readily prepared than are sequence-specific peptides or other oligomers.6
Antimicrobial agents have the highest potential for application when their deleterious effects are specific for microbial cells relative to human cells. Such selectivity has been achieved with a variety of compounds for bacterial growth inhibition vs. human cell destruction;6h,6m,7 the latter property is often assessed as lytic activity toward red blood cells (“hemolysis”).5e,8 Fundamental differences between prokaryotic and eukaryotic cellular membranes, including lipid composition and external surface charge density, seem to facilitate this selectivity.2,8b In contrast, it is difficult to target fungal pathogens selectively relative to human cells, because fungi are eukaryotes.9 For example, many host-defense peptides are not effective inhibitors of fungal growth at physiological ionic strength,10 and only modest antifungal vs. hemolytic selectivity has been achieved with sequence-specific oligomers.11 Here we describe a new family of nylon- 3 polymers (poly-β-peptides) that display significant and selective toxicity toward the most common fungal pathogen among humans, Candida albicans.12
Nylon-3 materials are readily prepared via ring-opening polymerization of β-lactams,13 and we have previously reported that sequence-random co-polymers that contain a lipophilic and a cationic subunit can manifest significant antibacterial activity but low hemolytic activity if the subunit identities, lipophilic-cationic subunit proportion and other parameters are optimized.6h,6m,14 The co-polymer shown in Figure 1, for example, displays a particularly favorable antibacterial activity profile.6h However, antifungal activity among previously reported nylon-3 copolymer families proved to be inseparable from hemolytic activity (unpublished). The present studies began with the preparation of a new β-lactam, NM (“no methyl”; Figure 2), which provides a cationic subunit at or below neutral pH. We were drawn to this subunit because it contains fewer saturated carbon atoms and therefore should have a lower hydrophobicity than previously examined cationic nylon-3 subunits derived from β-lactams MM and DM (“monomethyl” and “dimethyl”).6m The synthesis of NM (Figure 3) involves cycloaddition of chlorosulfonylisocyanate to an alkene, as in previous cases, but this route differs from the precedents in that the side chain nitrogen is introduced after β-lactam formation.6h,13f,15 Although the yield of the iodo-β-lactam is only modest, this potentially versatile molecule can easily be prepared on a multi-gram scale.15–16 The β-lactam bearing a Boc-protected amino group in the side chain was readily incorporated into nylon-3 co-polymers via the base-catalyzed process we have previously employed, in which the N-terminal group on each polyamide chain is specified by the choice of polymerization co-initiator.13f All polymers discussed below were prepared with 20-mer average length because previous work indicated that this size range is generally favorable in terms of maximizing antimicrobial activity and minimizing hemolytic activity.6m
Figure 1.
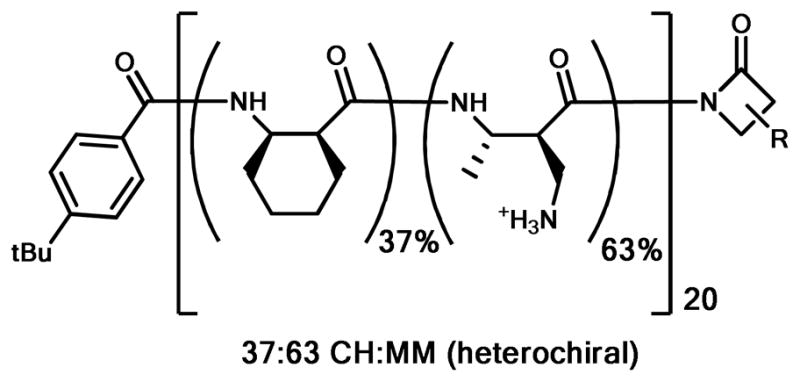
Representative sequence- and stereo-random nylon- 3 co-polymer containing subunits derived from racemic cis-cyclohexyl β-lactam (CH) and racemic monomethyl aminomethyl β-lactam (MM). R represents the side chain group for either CH or MM. This co-polymer inhibits the growth of several bacterial species at relatively low concentrations but is only weakly hemolytic.6h
Figure 2.
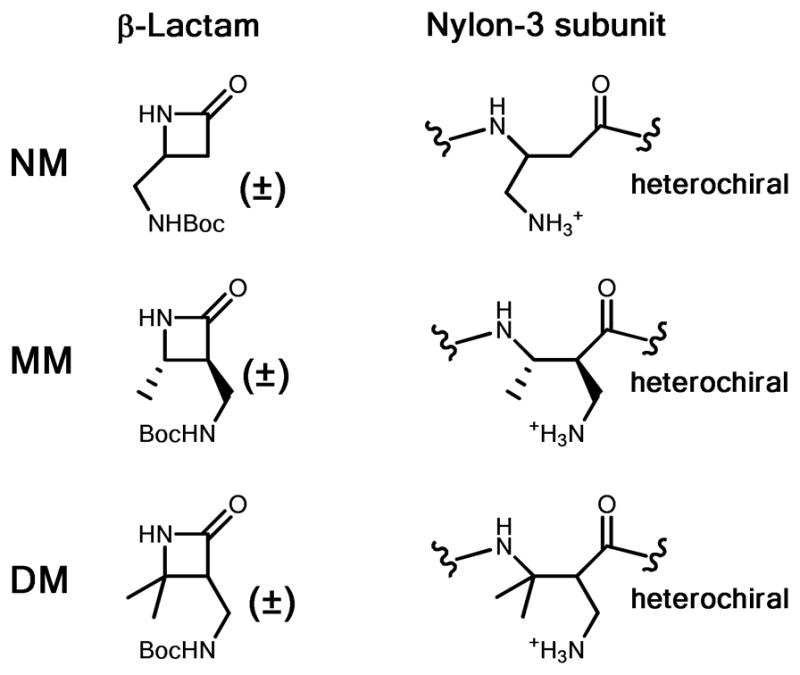
β-Lactams and corresponding hydrophilic (cationic) subunits within the nylon-3 polymer chain. All β-Lactams are racemic, and the resulting polymers are heterochiral.
Figure 3.
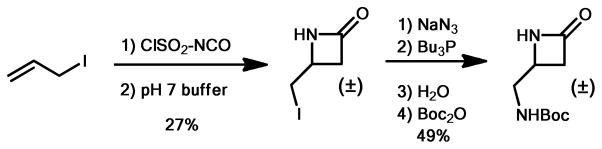
The synthesis of racemic β-lactam NM.
The antifungal activity of new NM-containing co-polymers (Figure 4) was evaluated with a clinically isolated strain of C. albicans (K1).17 The minimum inhibitory concentration (MIC) was measured using a protocol suggested by the Clinical and Laboratory Standard Institute (previously known as the National Committee for Clinical Laboratory Standard)18. In order to assess the effects of the new polymers on mammalian cells, we determined the concentration necessary for 10% lysis of human red blood cells (HC10), and the concentration necessary to induce 10% cell death in NIH 3T3 fibroblasts (IC10). Previously we have used the minimum hemolytic concentration (MHC) as a metric of red blood cell disruption, but we shifted to HC10 for the present studies because it was sometimes difficult to identify the lowest polymer concentration that displayed a non-zero extent of hemolysis.6h,6m The fibroblast assays provide an alternative measure, relative to hemolysis, of toxicity toward mammalian cells. Amphotericin B (AmpB), which is used clinically for C. albicans infections but associated with high toxicity toward mammalian cells, served as a positive control in these studies.19 Results are summarized in Table 1.
Figure 4.
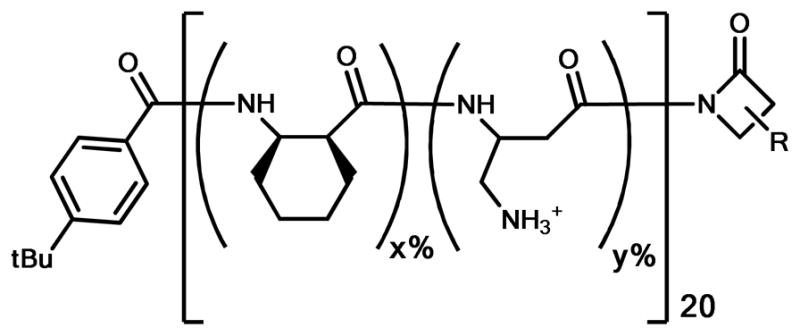
The structure of CH:NM co-polymers. All copolymers are heterochiral and sequence-random. x + y = 100, y = 40, 50, 60, 70, 80, or 90. R represents the side chain group of either CH or NM.
Table 1.
Physical and biological properties of nylon-3 polymers
| polymer composition | DPa | PDIb | MIC, μg/mLc | IC10, μg/mLd | HC10, μg/mLe |
|---|---|---|---|---|---|
| 60:40 CH:NM | 23 | 1.29 | 100 | > 400 | 100–200 |
| 50:50 CH:NM | 23 | 1.29 | 50 | > 400 | 200 |
| 40:60 CH:NM | 21 | 1.29 | 13 | > 400 | > 400 |
| 30:70 CH:NM | 20 | 1.26 | 6.3 | > 400 | > 400 |
| 20:80 CH:NM | 22 | 1.33 | 3.1 | 100–200 | > 400 |
| 10:90 CH:NM | 17 | 1.24 | 3.1 | > 400 | > 400 |
| NM | 20 | 1.13 | 3.1 | > 400 | > 400 |
| MM | 22 | 1.03 | 200 | > 400 | > 400 |
| DM | 18 | 1.13 | 6.3 | 50 | 3.1 |
| Ampho. Bf | N/A | N/A | 0.78 | < 1.5 | ND |
DP (degree of polymerization) indicates average polymer length (number of subunits).
PDI is polydispersity index.
MIC indicates the minimum inhibitory concentration for fungal growth as determined for C. albicans in the planktonic form.
IC10 indicates the concentration necessary to induce 10% cell death in NIH 3T3 fibroblasts.
HC10 indicates the concentration necessary for 10% lysis of human red blood cell.
Amphotericin B was dissolved in 1:1 DMSO:water as the stock solution for bioassay. ND indicates hemolysis data were not obtained. All polymers have an N-terminal p-tbutylbenzoyl group.
We began by examining random co-polymers (Figure 4) formed from new β-lactam NM and cyclohexyl β-lactam CH, because the latter had given rise to selective antibacterial copolymers when paired with the cationic subunit derived from MM (Figure 1).6h All of the new polymers bore a p-t-butylbenzoyl group at the N-terminus, as in previous antibacterial examples. The maximum proportion of the cyclohexyl subunit that could be used without compromising aqueous solubility, 60:40 CH:NM, led to weak antifungal activity and weak hemolytic activity (MIC and HC10 ~ 100 μg/mL). Antifungal activity steadily increased (i.e., MIC decreased) as the proportion of the lipophilic subunit declined, and no co-polymer containing > 50% of the cationic subunit manifested detectable hemolytic activity. Members of this polymer family were generally not toxic toward mouse fibroblasts. The activity levels observed for CH:NM co-polymers with ≥ 80% of the cationic subunit, on a μg/mL basis, approached that of AmpB, but were accompanied by substantially less fibroblast cytotoxicity than AmpB. Replacing the p-t-butylbenzyol end-group with an acetyl end-group did not alter the biological activity of poly-NM. The NM homopolymer displayed antifungal activity comparable to that of the most active CH:NM copolymers. Follow-up studies showed that poly-NM is fungicidal at the MIC, rather than merely inhibitory toward fungal growth.20
The excellent activity profile observed for poly-NM contrasts with the behavior observed for two other cationic nylon-3 homopolymers, poly-MM and poly-DM (Table 1). Poly-MM shows very little antifungal activity, and this homopolymer is also not hemolytic or toxic toward 3T3 fibroblasts. Poly-DM, on the other hand, approximately matches poly-NM in activity against C. albicans, but poly- DM is hemolytic and moderately toxic toward 3T3 fibroblasts.
Poly-NM was evaluated for antibacterial activity against a panel of four species that we have previously used to assess poly-MM and poly-DM as well as cationic-hydrophobic copolymers (Table 2).6m The antibacterial effects of poly-NM were generally comparable to those of the other two cationic nylon-3 homopolymers: significant activity was observed for Bacillus subtilis, which seems to be highly susceptible to a wide array of peptides and peptidomimetic oligomers and polymers, but all three homopolymers were considerably less active against Escherichia coli, Enterococcus faecium and Staphylococcus aureus. The generally low antibacterial activity of poly-MM and poly-DM has previously been rationalized in terms of their lack of hydrophobic subunits (e.g., the subunit derived from CH), which may limit their ability to disrupt bacterial membranes.6m,14 From this perspective, the relatively low antibacterial activity of poly- NM is not surprising. The potent antifungal activity of poly- NM is noteworthy in the context of this limited antibacterial activity.
Table 2.
Antibacterial activities of cationic nylon-3 homopolymers
| MIC,a μg/mL | ||||
|---|---|---|---|---|
| polymer | E. coli | B. subtilis | E. faecium | S. aureus |
| NM | 50 | 6.3 | > 200 | 100 |
| MM | > 200 | 6.3 | > 200 | 100 |
| DM | 100 | 3.1 | 100 | 50 |
MIC is the minimum inhibitory concentration for bacterial growth.
The data we have presented show that nylon-3 polymers containing subunits derived from the new β-lactam NM display potent antifungal activity without a strong tendency to disrupt human red blood cell membranes or strong toxicity toward 3T3 fibroblasts. It is particularly intriguing that poly-NM displays such profound differences in biological activity relative to the structurally similar cationic nylon-3 homopolymers poly-MM and poly-DM. There are several differences among the subunits of these three polymers: (1) the added side-chain carbons in poly-MM and poly-DM relative to poly-NM cause a modest increase in hydrophobicity;20 (2) the added carbons alter backbone flexibility; (3) the point of attachment of the aminomethyl side chain in NM differs from that in MM and DM (α-carbon vs. β-carbon). Further studies will be necessary to determine the mechanism by which these seemingly subtle molecularlevel changes exert such a substantial influence on biological activity. We have previously proposed that nylon-3 copolymers exert antibacterial effects via disruption of prokaryotic cell membranes, and this hypothesis has been supported by studies of the 40:60 CH:MM co-polymer (Figure 1) with synthetic vesicles of varying lipid composition.14 However, our finding that maximal antifungal activity is manifested by poly-NM, the least hydrophobic nylon-3 polymer we have examined to date, raises the possibility that NM-containing polymers act via a mechanism that does not involve disturbance of lipid bilayers. The surprising biological activity profile discovered for NM-based nylon-3 suggests that antifungal applications of these new materials be pursued.
Supplementary Material
Acknowledgments
This research was supported by the NIH (R21EB013259 and R01GM093265). X. C. and Z. H. were supported in part by the Nanoscale Science and Engineering Center at UWMadison (DMR-0425880). In addition, Z. H. was supported in part by a Fulbright Fellowship.
Footnotes
B.W. and S.H.G. are co-inventors on a patent application that covers the polymers described here.
Experimental details for synthesis and characterization of nylon-3 polymers, antifungal and antibacterial assays, cytotoxicity on 3T3 fibroblasts and hemolysis on human RBCs. This information is available free of charge via the Internet at http://pubs.acs.org/.
Contributor Information
Kristyn S. Masters, Email: kmasters@wisc.edu.
Samuel H. Gellman, Email: gellman@chem.wisc.edu.
References
- 1.(a) Zasloff M. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]; (b) Boman HG. J Intern Med. 2003;254:197–215. doi: 10.1046/j.1365-2796.2003.01228.x. [DOI] [PubMed] [Google Scholar]; (c) Hancock REW, Sahl HG. Nat Biotechnol. 2006;24:1551–1557. doi: 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]; (d) Steinstraesser L, Kraneburg UM, Hirsch T, Kesting M, Steinau HU, Jacobsen F, Al-Benna S. Int J Mol Sci. 2009;10:3951–3970. doi: 10.3390/ijms10093951. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Diamond G, Beckloff N, Weinberg A, Kisich KO. Curr Pharm Design. 2009;15:2377–2392. doi: 10.2174/138161209788682325. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Yeung ATY, Gellatly SL, Hancock REW. Cell Mol Life Sci. 2011;68:2161–2176. doi: 10.1007/s00018-011-0710-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yeaman MR, Yount NY. Pharmacol Rev. 2003;55:27–55. doi: 10.1124/pr.55.1.2. [DOI] [PubMed] [Google Scholar]
- 3.(a) van’t Hof W, Veerman ECI, Helmerhorst EJ, Amerongen AVN. Biol Chem. 2001;382:597–619. doi: 10.1515/BC.2001.072. [DOI] [PubMed] [Google Scholar]; (b) Sitaram N, Nagaraj R. Curr Pharm Design. 2002;8:727–742. doi: 10.2174/1381612023395358. [DOI] [PubMed] [Google Scholar]
- 4.Tossi A, Sandri L, Giangaspero A. Biopolymers. 2000;55:4–30. doi: 10.1002/1097-0282(2000)55:1<4::AID-BIP30>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 5.(a) Wade D, Boman A, Wahlin B, Drain CM, Andreu D, Boman HG, Merrifield RB. P Natl Acad Sci USA. 1990;87:4761–4765. doi: 10.1073/pnas.87.12.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Maloy WL, Kari UP. Biopolymers. 1995;37:105–122. doi: 10.1002/bip.360370206. [DOI] [PubMed] [Google Scholar]; (c) Dathe M, Schumann M, Wieprecht T, Winkler A, Beyermann M, Krause E, Matsuzaki K, Murase O, Bienert M. Biochemistry. 1996;35:12612–12622. doi: 10.1021/bi960835f. [DOI] [PubMed] [Google Scholar]; (d) Hamuro Y, Schneider JP, DeGrado WF. J Am Chem Soc. 1999;121:12200–12201. [Google Scholar]; (e) Porter EA, Wang XF, Lee HS, Weisblum B, Gellman SH. Nature. 2000;404:565–565. doi: 10.1038/35007145. [DOI] [PubMed] [Google Scholar]; (f) Liu DH, DeGrado WF. J Am Chem Soc. 2001;123:7553–7559. doi: 10.1021/ja0107475. [DOI] [PubMed] [Google Scholar]; (g) Tew GN, Liu DH, Chen B, Doerksen RJ, Kaplan J, Carroll PJ, Klein ML, DeGrado WF. P Natl Acad Sci USA. 2002;99:5110–5114. doi: 10.1073/pnas.082046199. [DOI] [PMC free article] [PubMed] [Google Scholar]; (h) Porter EA, Weisblum B, Gellman SH. J Am Chem Soc. 2002;124:7324–7330. doi: 10.1021/ja0260871. [DOI] [PubMed] [Google Scholar]; (i) Raguse TL, Porter EA, Weisblum B, Gellman SH. J Am Chem Soc. 2002;124:12774–12785. doi: 10.1021/ja0270423. [DOI] [PubMed] [Google Scholar]; (j) Oren Z, Ramesh J, Avrahami D, Suryaprakash N, Shai Y, Jelinek R. Eur J Biochem. 2002;269:3869–3880. doi: 10.1046/j.1432-1033.2002.03080.x. [DOI] [PubMed] [Google Scholar]; (k) Patch JA, Barron AE. J Am Chem Soc. 2003;125:12092–12093. doi: 10.1021/ja037320d. [DOI] [PubMed] [Google Scholar]; (l) Liu DH, Choi S, Chen B, Doerksen RJ, Clements DJ, Winkler JD, Klein ML, DeGrado WF. Angew Chem Int Edit. 2004;43:1158–1162. doi: 10.1002/anie.200352791. [DOI] [PubMed] [Google Scholar]; (m) Papo N, Shai Y. Biochemistry. 2004;43:6393–6403. doi: 10.1021/bi049944h. [DOI] [PubMed] [Google Scholar]; (n) Schmitt MA, Weisblum B, Gellman SH. J Am Chem Soc. 2004;126:6848–6849. doi: 10.1021/ja048546z. [DOI] [PubMed] [Google Scholar]; (o) Li X, Li YF, Han HY, Miller DW, Wang GS. J Am Chem Soc. 2006;128:5776–5785. doi: 10.1021/ja0584875. [DOI] [PubMed] [Google Scholar]; (p) Olsen CA, Bonke G, Vedel L, Adsersen A, Witt M, Franzyk H, Jaroszewski JW. Org Lett. 2007;9:1549–1552. doi: 10.1021/ol070316c. [DOI] [PubMed] [Google Scholar]; (q) Meng H, Kumar K. J Am Chem Soc. 2007;129:15615–15622. doi: 10.1021/ja075373f. [DOI] [PubMed] [Google Scholar]; (r) Chongsiriwatana NP, Patch JA, Czyzewski AM, Dohm MT, Ivankin A, Gidalevitz D, Zuckermann RN, Barron AE. P Natl Acad Sci USA. 2008;105:2794–2799. doi: 10.1073/pnas.0708254105. [DOI] [PMC free article] [PubMed] [Google Scholar]; (s) Choi S, Isaacs A, Clements D, Liu DH, Kim H, Scott RW, Winkler JD, DeGrado WF. P Natl Acad Sci USA. 2009;106:6968–6973. doi: 10.1073/pnas.0811818106. [DOI] [PMC free article] [PubMed] [Google Scholar]; (t) Olsen CA, Ziegler HL, Nielsen HM, Frimodt-Moller N, Jaroszewski JW, Franzyk H. Chembiochem. 2010;11:1356–1360. doi: 10.1002/cbic.201000232. [DOI] [PubMed] [Google Scholar]; (u) Hu J, Chen CX, Zhang SZ, Zhao XC, Xu H, Zhao XB, Lu JR. Biomacromolecules. 2011;12:3839–3843. doi: 10.1021/bm201098j. [DOI] [PubMed] [Google Scholar]
- 6.(a) Gelman MA, Weisblum B, Lynn DM, Gellman SH. Org Lett. 2004;6:557–560. doi: 10.1021/ol036341+. [DOI] [PubMed] [Google Scholar]; (b) Senuma M, Tashiro T, Iwakura M, Kaeriyama K, Shimura Y. J Appl Polym Sci. 1989;37:2837–2843. [Google Scholar]; (c) Li GJ, Shen JR, Zhu YL. J Appl Polym Sci. 1998;67:1761–1768. [Google Scholar]; (d) Tiller JC, Liao CJ, Lewis K, Klibanov AM. P Natl Acad Sci USA. 2001;98:5981–5985. doi: 10.1073/pnas.111143098. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Ilker MF, Nusslein K, Tew GN, Coughlin EB. J Am Chem Soc. 2004;126:15870–15875. doi: 10.1021/ja045664d. [DOI] [PubMed] [Google Scholar]; (f) Kuroda K, DeGrado WF. J Am Chem Soc. 2005;127:4128–4129. doi: 10.1021/ja044205+. [DOI] [PubMed] [Google Scholar]; (g) Fuchs AD, Tiller JC. Angew Chem Int Edit. 2006;45:6759–6762. doi: 10.1002/anie.200602738. [DOI] [PubMed] [Google Scholar]; (h) Mowery BP, Lee SE, Kissounko DA, Epand RF, Epand RM, Weisblum B, Stahl SS, Gellman SH. J Am Chem Soc. 2007;129:15474–15476. doi: 10.1021/ja077288d. [DOI] [PubMed] [Google Scholar]; (i) Sellenet PH, Allison B, Applegate BM, Youngblood JP. Biomacromolecules. 2007;8:19–23. doi: 10.1021/bm0605513. [DOI] [PubMed] [Google Scholar]; (j) Waschinski CJ, Zimmermann J, Salz U, Hutzler R, Sadowski G, Tiller JC. Adv Mater. 2008;20:104. [Google Scholar]; (k) Sambhy V, Peterson BR, Sen A. Angew Chem Int Edit. 2008;47:1250–1254. doi: 10.1002/anie.200702287. [DOI] [PubMed] [Google Scholar]; (l) Palermo EF, Sovadinova I, Kuroda K. Biomacromolecules. 2009;10:3098–3107. doi: 10.1021/bm900784x. [DOI] [PubMed] [Google Scholar]; (m) Mowery BP, Lindner AH, Weisblum B, Stahl SS, Gellman SH. J Am Chem Soc. 2009;131:9735–9745. doi: 10.1021/ja901613g. [DOI] [PubMed] [Google Scholar]; (n) Findlay B, Zhanel GG, Schweizer F. Antimicrob Agents Ch. 2010;54:4049–4058. doi: 10.1128/AAC.00530-10. [DOI] [PMC free article] [PubMed] [Google Scholar]; (o) Stratton TR, Howarter JA, Allison BC, Applegate BM, Youngblood JP. Biomacromolecules. 2010;11:1286–1290. doi: 10.1021/bm1000839. [DOI] [PubMed] [Google Scholar]; (p) Tew GN, Scott RW, Klein ML, Degrado WF. Accounts Chem Res. 2010;43:30–39. doi: 10.1021/ar900036b. [DOI] [PMC free article] [PubMed] [Google Scholar]; (q) Nederberg F, Zhang Y, Tan JPK, Xu KJ, Wang HY, Yang C, Gao SJ, Guo XD, Fukushima K, Li LJ, Hedrick JL, Yang YY. Nat Chem. 2011;3:409–414. doi: 10.1038/nchem.1012. [DOI] [PubMed] [Google Scholar]; (r) Li P, Poon YF, Li WF, Zhu HY, Yeap SH, Cao Y, Qi XB, Zhou CC, Lamrani M, Beuerman RW, Kang ET, Mu YG, Li CM, Chang MW, Leong SSJ, Chan-Park MB. Nat Mater. 2011;10:149–156. doi: 10.1038/nmat2915. [DOI] [PubMed] [Google Scholar]; (s) Wang YQ, Xu JJ, Zhang YH, Yan HS, Liu KL. Macromol Biosci. 2011;11:1499–1504. doi: 10.1002/mabi.201100196. [DOI] [PubMed] [Google Scholar]; (t) Timofeeva L, Kleshcheva N. Appl Microbiol Biot. 2011;89:475–492. doi: 10.1007/s00253-010-2920-9. [DOI] [PubMed] [Google Scholar]
- 7.Lienkamp K, Madkour AE, Musante A, Nelson CF, Nusslein K, Tew GN. J Am Chem Soc. 2008;130:9836–9843. doi: 10.1021/ja801662y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.(a) Haberman E. Science. 1972;177:314–322. doi: 10.1126/science.177.4046.314. [DOI] [PubMed] [Google Scholar]; (b) Shai Y. Bba- Biomembranes. 1999;1462:55–70. doi: 10.1016/s0005-2736(99)00200-x. [DOI] [PubMed] [Google Scholar]
- 9.Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG, White TC. Sci Transl Med. 2012;4:165rv113. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 10.Helmerhorst EJ, Reijnders IM, van’t Hof W, Veerman ECI, Amerongen AVN. Febs Lett. 1999;449:105–110. doi: 10.1016/s0014-5793(99)00411-1. [DOI] [PubMed] [Google Scholar]
- 11.(a) Karlsson AJ, Pomerantz WC, Weisblum B, Gellman SH, Palecek SP. J Am Chem Soc. 2006;128:12630–12631. doi: 10.1021/ja064630y. [DOI] [PubMed] [Google Scholar]; (b) Karlsson AJ, Pomerantz WC, Neilsen KJ, Gellman SH, Palecek SP. Acs Chem Biol. 2009;4:567–579. doi: 10.1021/cb900093r. [DOI] [PubMed] [Google Scholar]; (c) Makovitzki A, Avrahami D, Shai Y. P Natl Acad Sci USA. 2006;103:15997–16002. doi: 10.1073/pnas.0606129103. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Chongsiriwatana NP, Miller TM, Wetzler M, Vakulenko S, Karlsson AJ, Palecek SP, Mobashery S, Barron AE. Antimicrob Agents Ch. 2011;55:417–420. doi: 10.1128/AAC.01080-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. Clin Infect Dis. 2004;39:309–317. doi: 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- 13.(a) Králíček J, Šebenda J. Journal of Polymer Science. 1958;30:493–499. [Google Scholar]; (b) Hall HK. J Am Chem Soc. 1958;80:6404–6409. [Google Scholar]; (c) Graf R, Lohaus G, Böner K, Schmidt E, Bestian H. Angewandte Chemie International Edition in English. 1962;1:481–488. [Google Scholar]; (d) de Ilarduya AM, Aleman C, Garcia-Alvarez M, Lopez-Carrasquero F, Munoz-Guerra S. Macromolecules. 1999;32:3257–3263. [Google Scholar]; (e) Hashimoto K. Prog Polym Sci. 2000;25:1411–1462. [Google Scholar]; (f) Zhang JH, Kissounko DA, Lee SE, Gellman SH, Stahl SS. J Am Chem Soc. 2009;131:1589–1597. doi: 10.1021/ja8069192. [DOI] [PubMed] [Google Scholar]; (g) Lee MR, Stahl SS, Gellman SH, Masters KS. J Am Chem Soc. 2009;131:16779–16789. doi: 10.1021/ja9050636. [DOI] [PMC free article] [PubMed] [Google Scholar]; (h) Dane EL, Grinstaff MW. J Am Chem Soc. 2012;134:16255–16264. doi: 10.1021/ja305900r. [DOI] [PMC free article] [PubMed] [Google Scholar]; (i) Liu RH, Masters KS, Gellman SH. Biomacromolecules. 2012;13:1100–1105. doi: 10.1021/bm201847n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Epand RM, Epand RF, Mowery BP, Lee SE, Stahl SS, Lehrer RI, Gellman SH. J Mol Biol. 2008;379:38–50. doi: 10.1016/j.jmb.2008.03.047. [DOI] [PubMed] [Google Scholar]
- 15.Tanaka T, Miyadera T. Heterocycles. 1982;19:1497–1500. [Google Scholar]
- 16.(a) Salzmann TN, Ratcliffe RW, Christensen BG, Bouffard FA. J Am Chem Soc. 1980;102:6161–6163. doi: 10.1098/rstb.1980.0037. [DOI] [PubMed] [Google Scholar]; (b) Brennan J, Richardson G, Stoodley RJJ. Chem Soc-Chem Commun. 1980:49–49. [Google Scholar]
- 17.Andes D, Lepak A, Nett J, Lincoln L, Marchillo K. Antimicrob Agents Ch. 2006;50:2384–2394. doi: 10.1128/AAC.01305-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.NCCLS document M27- A2. 2. National Committee for Clinical Laboratory Standards; Wayne, PA: 2002. Reference method for broth dilution antifungal susceptibility testing of yeast: Approved standard. [Google Scholar]
- 19.(a) Chu P, Sadullah S. Curr Med Res Opin. 2009;25:3011–3020. doi: 10.1185/03007990903379077. [DOI] [PubMed] [Google Scholar]; (b) Kagan S, Ickowicz D, Shmuel M, Altschuler Y, Sionov E, Pitusi M, Weiss A, Farber S, Domb AJ, Polacheck I. Antimicrob Agents Ch. 2012;56:5603–5611. doi: 10.1128/AAC.00612-12. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Laniado-Laborin R, Cabrales-Vargas MN. Rev Iberoam Micol. 2009;26:223–227. doi: 10.1016/j.riam.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 20.Please see the supporting information.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


