Abstract
Members of the Roseobacter lineage, an ecologically important marine clade within the class α-Proteobacteria, harbor genes for the protocatechuate branch of the β-ketoadipate pathway, a major catabolic route for lignin-related aromatic compounds. The genes of this pathway are typically clustered, although gene order varies among organisms. Here we characterize genes linked to pcaH and -G, which encode protocatechuate 3,4-dioxygenase, in eight closely related members of the Roseobacter lineage (pairwise 16S rRNA gene sequence identities, 92 to 99%). Sequence analysis of genomic fragments revealed five unique pca gene arrangements. Identical gene organization was found for isolates demonstrating species-level identity (i.e., >99% 16S rRNA gene similarity). In one isolate, six functionally related genes were clustered: pcaQ, pobA, pcaD, pcaC, pcaH, and pcaG. The remaining seven isolates lacked at least one of these genes in their clusters, although the relative order of the remaining genes was preserved. Three genes (pcaC, -H, and -G) were physically linked in all isolates. A highly conserved open reading frame (ORF) was found immediately downstream of pcaG in all eight isolates. Reverse transcription-PCR analysis of RNA from one isolate, Silicibacter pomeroyi DSS-3, provides evidence that this ORF is coexpressed with upstream pca genes. The absence of this ORF in similar bacterial pca gene clusters from diverse microbes suggests a niche-specific role for its protein product in Roseobacter group members. Collectively, these comparisons of bacterial pca gene organization illuminate a complex evolutionary history and underscore the widespread ecological importance of the encoded β-ketoadipate pathway.
The β-ketoadipate pathway provides a model system for studying mechanisms of evolution in an ecologically important catabolic pathway. This widely distributed, typically chromosomally encoded pathway plays a central role in the degradation of naturally occurring aromatic compounds that arise during the decay of lignin and other vascular plant components, as well as in that of environmental pollutants (24). The pathway is biochemically conserved, and the structural genes encoding enzymes of the pathway are similar among the phylogenetically diverse organisms that possess it (24). Despite this mechanistic conservation, studies of a limited number of soil bacteria demonstrate a remarkable diversity of this pathway in terms of gene organization, regulation, inducing metabolites, and transport systems (reviewed in reference 35). This diversification may reflect the distinctive selection pressures faced by the organisms maintaining the pathway and thus may reveal characteristic features that are specific to the bacterial group in which the pathway resides (24, 34).
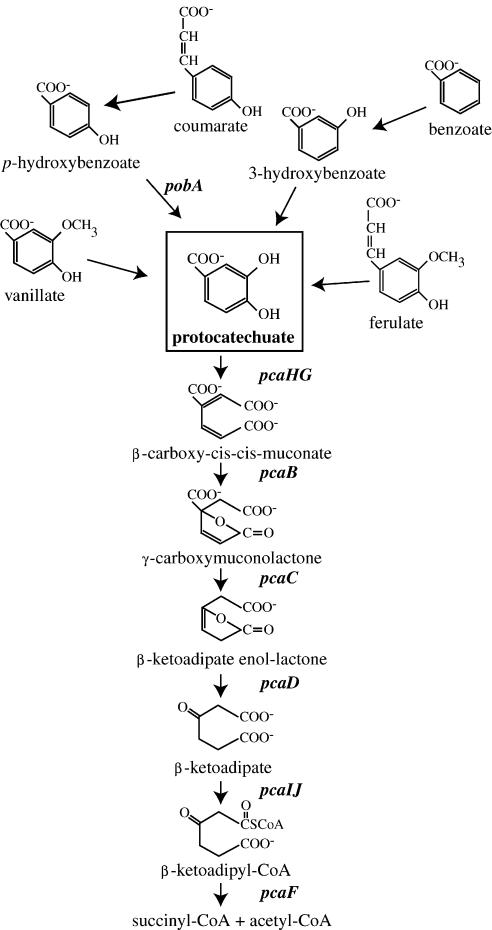
The degradation of aromatic compounds typically proceeds by conversion of the ring structure to one of several di- or trihydroxylated intermediates in preparation for enzymatic ring cleavage. In natural systems, two of the most prevalent intermediates are protocatechuate and catechol. The separate branches of the β-ketoadipate pathway are initiated as protocatechuate and catechol are cleaved between their two hydroxyl groups by protocatechuate 3,4-dioxygenase (P3,4DO) or catechol 1,2-dioxygenase, respectively. Ring fission is followed by five additional reactions leading to the generation of substrates that feed into the tricarboxylic acid cycle (Fig. 1). In organisms possessing both branches, the pathway converges to form several common intermediates including β-ketoadipate, for which the pathway is named.
FIG. 1.
Schematic of the protocatechuate branch of the β-ketoadipate pathway. Gene designations are italicized. CoA, coenzyme A.
The genes encoding the enzymes, transport proteins, and regulatory proteins of the β-ketoadipate pathway are usually present in chromosomal supraoperonic clusters (34). A remarkable example of such an assemblage is demonstrated by the protocatechuate pathway branch in Acinetobacter sp. strain ADP1, where more than 40 genes involved in the catabolism of plant-related compounds form a 56-kbp chromosomal cluster (36). However, the extent of gene clustering varies among the phylogenetically diverse microbes possessing the pathway. Pseudomonas putida demonstrates a contrasting scenario, as the genes required for the catabolism of protocatechuate are dispersed within the genome in three distinct gene clusters (34). Furthermore, gene order does not appear to be maintained within the operons except in cases where genes may coevolve because they encode subunits of a single enzyme (e.g., pcaHG and pcaIJ [see Fig. 1]) (24).
The marine Roseobacter lineage provides an ideal system for genetic studies of aromatic compound catabolism. Representatives of this lineage are abundant in coastal seawater, aerobic sediments, and decaying plant material from coastal salt marshes (9, 19, 28, 45). The presence of the protocatechuate branch of the β-ketoadipate pathway has been demonstrated in several isolates, and roseobacters have widespread capabilities for the transformation of lignin-related aromatic monomers, including p-hydroxybenzoate, vanillate, and ferulate (4, 5). Furthermore, more than half of the pcaH genes retrieved by PCR amplification from salt marsh bacterial communities could be traced to members of the Roseobacter lineage (5). To gain a better perspective on the conservation of sequence and gene arrangement in closely related organisms, we analyzed the protocatechuate branch of the β-ketoadipate pathway in eight Roseobacter group isolates.
MATERIALS AND METHODS
Bacterial isolation, growth conditions, and 16S rRNA gene analysis.
Most isolates examined in this study were cultured from seawater, sediments, or decaying salt marsh grass collected in the estuaries and coastal waters of the southeastern United States. As previously described, aromatic substrates were used in the enrichment cultures from which Sagittula stellata E-37, Sulfitobacter sp. strain EE-36, Y3F, and Y4I were isolated. For the first two strains, the high-molecular-weight (>1,000) fraction of pulp mill effluent was used (18). For the latter two isolates, a lignin-rich pulp mill by-product (Indulin) was used (4). SE45 was cultured from decaying Spartina alterniflora (smooth cord grass) by using nonselective, low-nutrient seawater plates (5). Silicibacter pomeroyi DSS-3 was derived from a marine dimethylsulfoniopropionate enrichment culture (16, 17). Finally, two isolates that were not obtained from the southeastern U.S. coast were examined: Sulfitobacter pontiacus ChLG 10, which was cultured from the Black Sea (43), and Roseovarius nubinhibens ISM, which was cultured from the Caribbean Sea (14, 16).
Growth capabilities were tested by using aromatic compounds, each provided at a 2 mM concentration, as the sole substrate in marine basal medium (MBM) with vitamins as previously described (18). Plates were incubated at 28 to 30°C in the dark.
16SrRNA sequences have been reported previously for all isolates (accession numbers are given in parentheses): S. pomeroyi DSS-3 (AF098491), Sulfitobacter sp. strain EE-36 (AF007254), R. nubinhibens ISM (AF098495), Y3F (AF253467), Y4I (AF388307), SE45 (AF388308), S. stellata E-37 (U58356), and S. pontiacus ChLG 10 (Y13155). Species-level similarity was observed between Y4I and Y3F and between Sulfitobacter sp. strain EE-36 and S. pontiacus ChLG 10. These pairs, respectively, have 100 or 99.8% identity in regions corresponding to positions 48 to 1484 or 50 to 1506 of the 16S rRNA gene in Escherichia coli.
Detection and isolation of catabolic genes from roseobacters.
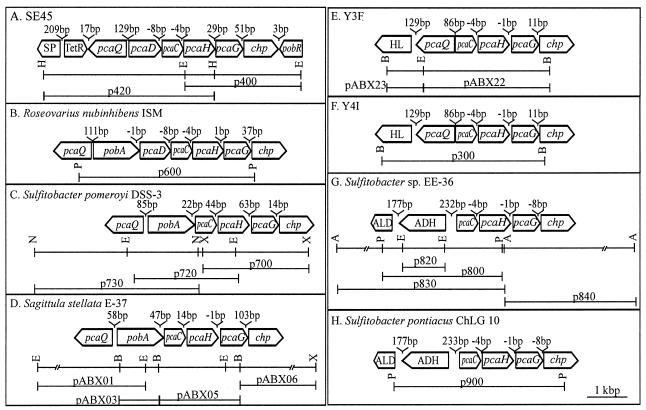
The catabolic gene clusters were initially identified in isolates by targeting a region of pcaH. A degenerate PCR primer pair designed from conserved PcaH regions (5) was used to amplify a 212-bp product from all isolates. This product was gel excised, labeled with digoxigenin (DIG) by a random priming reaction (Genius system; Roche Molecular Biochemicals, Indianapolis, Ind.), and used in Southern hybridizations with isolate genomic digestions. DNA fragments of the sizes corresponding to bands giving positive hybridization signals in Southern blot analysis were gel excised and ligated into the pZERO vector (Invitrogen Corp., Carlsbad, Calif.). Colony hybridization of genomic libraries with DIG-labeled DNA probes identified positive clones. In cases where the initial fragment did not contain all functionally related genes in the proximity of pcaHG (S. pomeroyi DSS-3, Sulfitobacter sp. strain EE-36, and SE45), adjacent fragments were identified by Southern hybridization analysis using DIG-labeled probes generated from distal ends of the primary fragment (see Fig. 2).
FIG. 2.
Restriction maps of genomic fragments from isolate SE45 (A), R. nubinhibens ISM (B), S. pomeroyi DSS-3 (C), S. stellata E-37(D), isolate Y3F (E), isolate Y4I (F), Sulfitobacter sp. strain EE-36 (G), and S. pontiacus ChLG 10 (H). The locations of the genes and their transcriptional directions are shown relative to selected restriction endonuclease recognition sites. A, ApaI; B, BamHI; E, EcoRI; N, NsiI; P, PstI; X, XhoI. Horizontal lines indicate DNA regions contained on recombinant plasmids (designations given above the lines). Genes are characterized in Table 2 and in the text.
Sequence determination of the genomic fragments was facilitated by using the GeneJumper Primer Insertion Kit for Sequencing (Invitrogen Corp.), which randomly inserted a minimal version of the Mu transposon containing two primer binding sites and a selectable marker into the target DNA.
Expression of Roseobacter group isolate DNA in E. coli.
To express the pcaHG genes from isolates SE45 and R. nubinhibens ISM under the control of the lac promoter in E. coli, p4NK and p6NB were constructed. PCR primers were used to introduce restriction sites into amplified pcaHG genes for optimal positioning in the pCYB1 expression vector (New England Biolabs, Beverly, Mass.). An NdeI cleavage site was introduced just before the ATG start codon of the pcaH gene, and either a KpnI (SE45) or a BamHI (R. nubinhibens ISM) cleavage site was introduced downstream of the pcaG stop codon by PCR amplification using the high-fidelity Pfu DNA polymerase (Stratagene, La Jolla, Calif.). The 1.3-kbp fragments were then ligated into the corresponding sites on the pCYB1 vector. The sequences of the resultant recombination plasmids were confirmed to be correct. Luria-Bertani broth cultures (100 ml) of E. coli Top10F′ cells (Invitrogen Corp.) carrying either p4NK or p6NB were grown at 30 or 25°C for 12 h. At the time of inoculation, 100 μM isopropyl-β-d-thiogalactopyranoside (IPTG) was added to the cultures. The induced cells were harvested by centrifugation, washed once with sterile Tris buffer (50 mM; pH 7.5), and stored at −20°C. Reported specific activities are averages from three independently grown and assayed cultures. No activity was detected in E. coli with the cloning vector or with recombinant plasmids in the absence of IPTG.
P3,4DO enzyme assays.
Cell pellets were suspended in 200 μl of breaking buffer [50 mM Tris-HCl, 10% glycerol, 5 mM (NH4)2SO4, 2.5 mM EDTA, 1 mM dithiothreitol (pH 7.5)]. Cell extracts were prepared as described previously (42), and P3,4DO activity was determined spectrophotometrically by measuring the decrease in absorbance at 290 nm (44). Protein concentrations were determined by the method of Bradford (2).
mRNA isolation and RT-PCR.
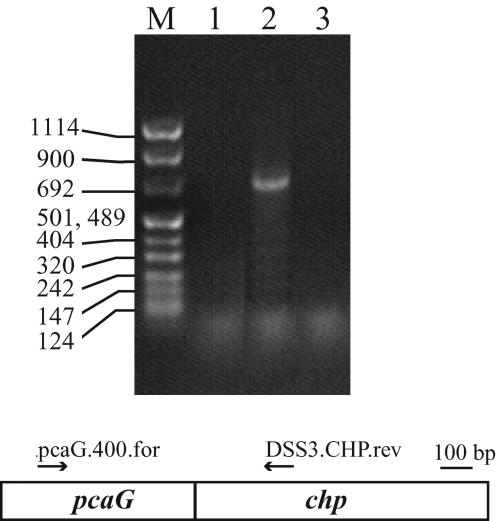
Total RNA was harvested from early-log-phase cultures of S. pomeroyi DSS-3 grown with p-hydroxybenzoate (3 mM) or succinate (10 mM) as the sole carbon source by using the RNAqueous kit (Ambion, Inc., Austin, Tex.). RNA preparations were treated with DNase (DNA-Free; Ambion, Inc.) to remove contaminating DNA. Reverse transcription reactions were carried out with a primer binding to a region within the conserved open reading frame (ORF) (primer DSS.CHP.rev, 5′ TTG ACG GTC AGT TGA TTG 3′) by using Omniscript RT (Qiagen, Valencia, Calif.) according to the manufacturer's protocols. cDNA generated in the reverse transcription step was used in subsequent PCR amplifications using primers pcaG.400.for (5′ GAG TYC TGG CAG GCC 3′) and DSS.CHP.rev. Because primer pcaG.400 is complementary to a region within pcaG, the pcaG.400-DSS.CHP.rev primer set was used to determine if the ORF and pcaG were cotranscribed. PCR thermal cycling conditions consisted of 30 cycles of 95°C for 45 s and 55°C for 45 s, followed by 72°C for 1 min. Control samples with equivalent concentrations of RNA, but lacking reverse transcriptase, were included in all reverse transcription-PCRs (RT-PCRs).
Sequence determination and analysis.
DNA sequences were determined with double-stranded templates and primers that recognized the cloning vector or the GeneJumper transposon. When necessary, new oligonucleotide primers were made based on previously sequenced regions. Either an ABI3700 or an ABI310 automated DNA sequencer (Applied Biosystems, Foster City, Calif.) was used.
Homology searches (BLAST) were carried out at the network server of the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov). Sequences were analyzed using the Wisconsin Package 10.1 (Accelrys, Burlington, Mass.). Phylogenetic trees were constructed for sequences with the PHYLIP package (13) by using evolutionary distances (Jukes Cantor or Kimura) and tree-building algorithms (Neighbor or Fitch) or a parsimony method (ProtPars). The pcaC, -H, and -G gene sequences were tested for intragenic recombination by using Partial Likelihoods Assessed through Optimisation (PLATO; available at http://evolve.zoo.ox.ac.uk/program). The conserved hypothetical proteins were analyzed for the presence of signature sequence motifs by using the PROSITE (http://ca.expasy.ch/prosite/), PRINTS (http://www.bioinf.man.ac.uk/dbbrowser/PRINTS/), and BLOCKS (www.blocks.fhcrc.org/) programs. The conserved hypothetical protein was also searched for putative membrane-spanning regions by using the MEMSAT2 program (http://bioinf.cs.ucl.ac.uk/psipred).
Nucleotide sequence accession numbers.
Sequences reported here were submitted to the GenBank database under the following accession numbers: AF253465, AF253466, and AY457916 to AY457921.
RESULTS
Choice of Roseobacter group isolates. Studies of salt marsh bacterial communities indicate that the ability to cleave protocatechuate is a common and environmentally significant trait of the Roseobacter lineage (5). In agreement with this conclusion, pcaH sequences were obtained from 16 of 19 Roseobacter group members screened by PCR with a degenerate primer set (5). To characterize the genetic regions associated with this trait, isolates were selected from a roseobacter collection of salt marsh and coastal ocean isolates (5, 19, 20). Five strains (S. stellata E-37, S. pontiacus ChLG 10, R. nubinhibens ISM, and isolates Y3F and SE45) were chosen for this study because their pcaH sequences were identical to a 159-bp region of pcaH retrieved from bacterial communities associated with decaying plant material (5). Two additional isolates, Y4I and Sulfitobacter sp. strain EE-36, were selected because 16S rRNA analysis demonstrated species-level similarity to other chosen isolates, Y3F and S. pontiacus ChLG 10, respectively (see Materials and Methods). Finally, S. pomeroyi DSS-3 was selected because it is a well-characterized representative of a large, distinct subgroup within the Roseobacter lineage (16). The marine Roseobacter lineage forms a monophyletic clade within the α-3 subclass of the Proteobacteria (19). Pairwise sequence identities of 16S rRNA genes among the eight roseobacters of this study ranged from 92 to 100%.
Growth on aromatic compounds.
To ensure that the genes under investigation have physiological relevance, growth was tested on aromatic compounds degraded via protocatechuate by other bacteria (Table 1). Although only SE45 grew on all eight substrates, every Roseobacter isolate grew on at least four of these compounds. Furthermore, all grew well on p-hydroxybenzoate, a feature common to organisms possessing the protocatechuate branch of the β-ketoadipate pathway (24). Interestingly, the two isolate pairs with species-level similarity (Y3F and Y4I; Sulfitobacter sp. strain EE-36 and S. pontiacus ChLG 10) had different substrate utilization profiles (Table 1). Collectively, these growth capabilities support the involvement of the pca genes in protocatechuate catabolism by these bacteria.
TABLE 1.
Growth of Roseobacter group isolates on aromatic compounds
| Roseobacter group isolate | Growtha of isolate on the following aromatic compoundb:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Benzoate | Chlorogenate | 4-CB | Coumarate | Ferulate | POB | PCA | Vanillate | |
| R. nubinhibens ISM | − | − | − | +/− | + | + | + | − |
| SE45 | + | + | + | + | + | + | + | + |
| S. stellata E-37 | + | + | − | + | + | + | − | + |
| S. pomeroyi DSS-3 | + | − | + | + | + | + | + | − |
| Y3F | + | + | + | + | − | + | + | + |
| Y4I | +/− | + | − | +/− | − | + | − | + |
| S. pontiacus ChLG 10 | − | +/− | − | +/− | − | + | + | − |
| Sulfitobacter sp. strain EE-36 | +/− | +/− | − | + | + | + | + | − |
Growth was determined on MBM plates containing 2 mM growth substrate plus 0.1% vitamins. +, growth within 4 days; +/−, growth within 14 days; −, no growth within 14 days.
4-CB, 4-chlorobenzoate; POB, p-hydroxybenzoate; PCA, protocatechuate.
Identification of catabolic genes in roseobacters.
Genomic fragments hybridizing to pcaH were identified and isolated from S. pontiacus ChLG 10, Sulfitobacter sp. strain EE-36, S. pomeroyi DSS-3, R. nubinhibens ISM, Y4I, and SE45. DNA sequence and homology analysis indicated that these fragments carried multiple genes of the protocatechuate branch of the β-ketoadipate pathway. Genes predicted to encode γ-carboxymuconolactone decarboxylase (PcaC), and P3,4DO (PcaHG) (Fig. 1) were identified in these six roseobacters as well as in two previously characterized Roseobacter group members, S. stellata E-37 and isolate Y3F (4). A comparison of the deduced genomic arrangements in all eight strains is shown in Fig. 2. In some isolates, neighboring sequences were predicted to encode an enol-lactone hydrolase (PcaD), p-hydroxybenzoate hydroxylase (PobA), and/or a LysR-type transcriptional regulator (PcaQ). In strains carrying pcaQ, its upstream and divergent location relative to the adjacent pca gene cluster was consistent with the predicted regulatory function (41). No ORFs showing similarity to transcriptional regulators were evident in the immediate vicinity of the pca genes in either Sulfitobacter strain. The orientation of the pca genes and their proximity to one another suggest that they are transcribed as a single unit (Fig. 2), as has been demonstrated previously for some soil bacteria (8).
G+C contents have been determined to be 65.0, 66.8, and 66.0% for isolates S. stellata E-37, S. pomeroyi DSS-3, and R. nubinhibens ISM, respectively (16, 20). The G+C contents of the pca genes (Table 2) are consistent with those of the genomes of these organisms. The lengths of the pobA and pca genes as well as the molecular masses, calculated from the deduced amino acids (Table 2), are consistent with those found for their soil counterparts. Additional ORFs located in the vicinity of the pca genes are described in Table 3.
TABLE 2.
Characterization of β-ketoadipate pathway genes identified in Roseobacter group isolates
| Roseobacter group isolate |
pcaQ
|
pobA
|
pcaD
|
pcaC
|
pcaH
|
pcaG
|
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % G+C | Length (bp) | Stop codona | MMb (kDa) | % G+C | Length (bp) | Stop codon | MM (kDa) | % G+C | Length (bp) | Stop codon | MM (kDa) | % G+C | Length (bp) | Stop codon | MM (kDa) | % G+C | Length (bp) | Stop codon | MM (kDa) | % G+C | Length (bp) | Stop codon | MM (kDa) | |
| R. nubinhibens ISM | 62.1 | 227c | 63.2 | 1,170 | TGA | 43.4 | 71.9 | 789 | TGA | 27.8 | 66.7 | 411 | TGA | 15 | 62.7 | 729 | TGA | 27.3 | 65.5 | 621 | TAG | 22.6 | ||
| SE45 | 68.5 | 930 | TGA | 33.7 | 68.7 | 789 | TGA | 27.6 | 66.7 | 387 | TGA | 14.1 | 63.5 | 726 | TGA | 27.1 | 66.8 | 693 | TGA | 25.3 | ||||
| S. stellata E-37 | 65.7 | 960 | TAA | 34.4 | 63.7 | 1,181 | TGA | 44.1 | 64.6 | 393 | TGA | 14.4 | 66.7 | 723 | TGA | 26.8 | 65.1 | 602 | TGA | 21.9 | ||||
| S. pomeroyi DSS-3 | 66.8 | 918 | TGA | 33.1 | 63.6 | 1,170 | TAG | 43.8 | 64.8 | 378 | TGA | 13.8 | 63.0 | 729 | TGA | 27.3 | 66.0 | 621 | TGA | 22.6 | ||||
| Y3F | 69.1 | 929 | TAG | 32.9 | 67.3 | 395 | TGA | 14.3 | 63.7 | 738 | TGA | 27.7 | 64.2 | 618 | TGA | 22.9 | ||||||||
| Y4I | 69.2 | 932 | TAG | 33.1 | 69.1 | 396 | TGA | 14.3 | 64.8 | 738 | TGA | 27.7 | 64.4 | 621 | TGA | 22.8 | ||||||||
| S. pontiacus ChLG 10 | 62.7 | 381 | TGA | 13.7 | 60.1 | 732 | TGA | 27.1 | 59.4 | 606 | TGA | 22.1 | ||||||||||||
| Sulfitobacter sp. strain EE-36 | 61.9 | 381 | TGA | 13.7 | 60.7 | 732 | TGA | 27 | 58.9 | 606 | TGA | 22.2 | ||||||||||||
Stop codon for deduced gene product.
Molecular mass of deduced gene product.
Partial gene sequence was obtained.
TABLE 3.
Additional ORFs in the vicinity of the pca genes
| Strain(s) and gene designationa | Putative function or role | Most similar gene and/or bacterium
|
||
|---|---|---|---|---|
| Designation | % Amino acid similarity (identity) | SwissProt accession no. | ||
| SE45 | ||||
| SP | Shikimate dehydrogenase | aroE2, Ralstonia solanacearum | 65 (57) | NP_522957 |
| TetR | TetR-like transcriptional regulator | Agrobacterium tumefaciens | 59 (50) | NP_356128 |
| pobR | AraC-like transcriptional regulator | pobR, Rhizobium leguminosarum | 42 (32) | AAA83008 |
| Y3F and Y4I, HL | Zn-dependent hydrolase | soxG, Paracoccus denitrificans | 60 (51) | X79242 |
| Sufitobacter strains | ||||
| ALD | Aldehyde dehydrogenase, subunit III | Mesorhizobium loti | 70 (58) | NP_104855 |
| ADH | Glutathione-dependent alcohol dehydrogenase | adhI, Rhodobacter sphaeroides | 87 (80) | P72324 |
As shown in Fig. 2.
Sequence analysis of the pcaC, -H, and -G genes from roseobacters.
The three genes present in all eight isolates encode two distinct enzymes catalyzing reactions in the degradation of protocatechuate (pcaC, pcaH, and pcaG [Fig. 1]). Among the Roseobacter group isolates, pairwise comparisons of the deduced amino acid sequences for each comparable protein (PcaC, PcaH, and PcaG) ranged from approximately 55 to 76% identity. Comparisons were also carried out with the corresponding sequences from the soil bacteria Acinetobacter sp. strain ADP1, Pseudomonas aeruginosa PAO1, P. putida ATCC 23975, Sinorhizobium meliloti 1021, Mesorhizobium loti MAFF303099, Caulobacter crescentus CB15, Agrobacterium tumefaciens C58, and Rhodococcous opacus 1CP. The identities of the P. aeruginosa PAO1, S. meliloti 1021, M. loti MAFF303099, and C. crescentus CB15 genes were inferred from genomic sequence data. In these comparisons, the identities between corresponding deduced protein sequences ranged from approximately 28 to 39%.
Residues known to be important for catalytic function were well conserved within all of the Roseobacter group PcaHG sequences. These included residues involved in coordinating the nonheme Fe3+ (Tyr408, Tyr447, His460, and His462) as well as critical active-site residues (e.g., Pro15, Arg133, Trp400, Trp449, and Ile491) (30, 48). In all, 85 of the ∼450 residues were completely conserved in the 16 PcaHG sequences examined. Sequence analysis revealed a 72-bp insertion in SE45 pcaG that was confirmed by PCR amplification and sequencing of the region from genomic DNA preparations (data not shown). The position of the insertion in SE45 corresponds to the region near residue 125 of PcaG from Acinetobacter sp strain ADP1. Based on the crystal structures of PcaHG from P. putida ATCC 23975 and Acinetobacter sp. strain ADP1 (30, 48), the insertion in SE45 is likely to reside in a loop on the outside face of the enzyme.
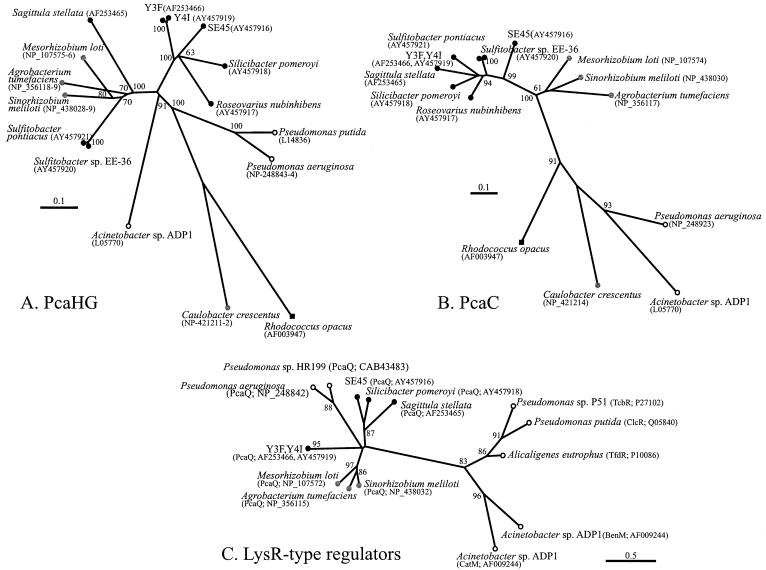
The Roseobacter group PcaHG sequences do not form a cohesive lineage, as indicated by phylogenetic trees (Fig. 3A). The overall tree topology was identical for trees constructed for individual and combined PcaG and PcaH subunits. Sequences from S. stellata E-37 and the two Sulfitobacter strains cluster with α-proteobacterial sequences but not with the other Roseobacter group isolates (Fig. 3A). However, the deep branches are difficult to resolve, and thus the phylogenetic relationship of these proteins within the context of other α-proteobacteria is difficult to interpret with the sequences on hand. In contrast, the phylogenetic relationships among the PcaC proteins are similar to those determined from 16S rRNA gene analysis (Fig. 3B and 4). Branch order was maintained when PcaC and PcaHG trees were constructed by using alternative tree-building algorithms (Fitch) and a parsimony method (ProtPars) (data not shown).
FIG. 3.
(A) Phylogenetic tree of PcaHG protein sequences. The tree is based on the deduced amino acid sequence encoded by the pcaHG genes and is unrooted, with PcaHG from R. opacus 1CP as the outgroup. (B) Phylogenetic tree of PcaC protein sequences. The tree is based on the deduced amino acid sequence encoded by the pcaC genes and is unrooted. In R. opacus 1CP and C. crescentus CB15, pcaC and pcaD have fused into a single gene (pcaL) encoding a protein demonstrating both PcaC and PcaD activities (12); the PcaC-like segment of PcaL was used in this analysis. The protein sequence from R. opacus 1CP served as the outgroup. (C) Phylogenetic tree of LysR-type protein sequences. The tree is based on deduced amino acids encoded by the pcaQ, tcbR (47), clcR (6), tfdR (27), benM, and catM (7) genes and is unrooted, with CatR (40) as the outgroup. Bootstrap values of ≥50% are given at branch nodes. Bars indicate Kimura distances. Solid circles, Roseobacter group members; shaded circles, non-Roseobacter α-proteobacteria; open circles, γ-proteobacteria; solid squares, gram-positive bacteria. GenBank accession numbers are provided in parentheses.
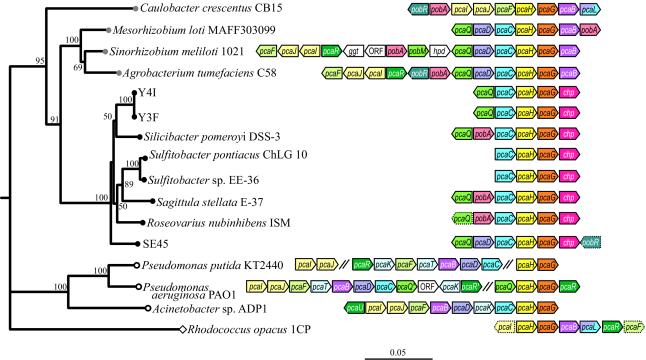
FIG. 4.
Phylogenetic tree of 16S rRNA gene sequences. The tree is based on 1,300 nucleotides (positions 111 to 1411; E. coli numbering system), with the gene from R. opacus 1CP (AB032565) as the outgroup. Bootstrap values of ≥50% are given at branch nodes. Bar indicates Jukes-Cantor distances. The pca gene organization of each of the following strains is shown: C. crescentus CB15 (SwissProt accession no. NP_421206 to -14), M. loti MAFF303099 (NP_107572 to -8), S. meliloti 1021 (NP_438027 to -41), A. tumefaciens C58 (NP_356109 to -20), isolate Y4I (GenBank accession no. AY457919), isolate Y3F (AF253466), S. pomeroyi DSS-3 (AY457918), S. pontiacus ChLG 10 (AY457921), Sulfitobacter sp. strain EE-36 (AY457920), S. stellata E-37 (AF253465), R. nubinhibens ISM (AY457917), isolate SE45 (AY457916), P. putida KT2440 (AAN69545 to -6, AAN66998 to -67004, AAN70228 to -9), P. aeruginosa PAO1 (NP_248917 to -27, NP_248842 to -5), Acinetobacter sp. strain ADP1 (L05770), and R. opacus 1CP (AF003947). pcaB, β-carboxy-cis,cis-muconate lactonizing enzyme; pcaC, γ-carboxy-muconolactone decarboxylase; pcaD, β-ketoadipate enol-lactone hydrolase; pcaHG, P3,4DO; pcaF, β-ketoadipyl coenzyme A thiolase; pcaIJ, β-ketoadipate succinyl coenzyme A transferase; pcaK and pcaT, transport proteins; pcaL, pcaDC gene fusion; pobA, p-hydroxybenzoate hydroxylase; hpd, p-hydroxyphenylpyruvate dioxygenase; ggt, γ-glutamyltranspeptidase; pcaQ, pcaQ′, pobR, pcaR′, pobM, pcaU, and pcaR, transcriptional regulators. Dark blue, AraC-type transcriptional regulator; dark green, IclR-type transcriptional regulator; light green, LysR-type transcriptional regulator. Double diagonal lines indicate that >10 kbp separates the transcriptional units. Dotted outlines indicate that only partial sequences have been obtained for these genes.
Incongruent topologies of the PcaC and PcaHG phylogenetic trees could be the result of recombination (inter- and/or intragenic) or differences in rates of evolutionary change among the corresponding genes. This was tested by using the PLATO program (21). We found no evidence of intragenic recombination within the sets of 16 pcaC, pcaH, or pcaG genes analyzed (8 from roseobacters and 8 from phylogenetically diverse soil bacteria). Furthermore, the relative rates of evolution were assessed by the method outlined by Dykhuizen and Green (11). The similarity in the percentages of divergence of pcaC, -H, and -G among the 16 genes analyzed (32.2, 34.2, and 32.7%, respectively) suggested that the genes are evolving at approximately the same rate. Significant differences were evident for all three genes relative to the 16S rRNA gene, which had significantly lower sequence divergence as determined by analysis of variance (9.6%; P < 0.001). A regression of pairwise distances of each of the PcaC, PcaH, and PcaG proteins against pairwise distances of the 16S rRNA gene provided slopes that were not significantly different from one another, likewise suggesting similar rates of evolution of the pca genes. Therefore, the different topologies seen for the PcaC and PcaHG protein trees might simply be attributed to the relatively small data set available for analysis.
Expression of the pcaHG genes in E. coli.
To investigate function, the pcaHG genes of two roseobacters, SE45 and R. nubinhibens ISM, were transformed into E. coli, a bacterium that does not encode P3,4DO. The roseobacter pcaHG genes resulted in IPTG-inducible P3,4DO activity in cell extracts of the plasmid-bearing E. coli strains (258 ± 0.053 or 379 ± 103 nmol/min/mg for the SE45 or R. nubinhibens ISM genes, respectively). Surprisingly, for the R. nubinhibens ISM genes, activity was observed when plasmid-bearing E. coli strains were grown at 25°C but not at 30°C. Although the reason for the temperature sensitivity is not evident, difficulties have been encountered previously with heterologous production of P3,4DO in E. coli (38). Possible toxicity associated with expression of pcaHG could result in a decreased plasmid copy number (10) or the sequestration of the protein in inclusion bodies. Such problems could be more pronounced when the host is grown at 30°C. Nevertheless, the activity of the PcaHG enzyme from R. nubinhibens ISM is indicative of a functional protocatechuate pathway. The broader functional significance of this pathway within the lineage is further supported by a previous report of P3,4DO activity in enzymes from two additional roseobacters, S. stellata E-37 and isolate Y3F (4).
Sequence analysis of additional pca-related genes.
The pobA gene, identified in S. stellata E-37, S. pomeroyi DSS-3, and R. nubinhibens ISM (Fig. 2), should encode a hydroxylase for the conversion of p-hydroxybenzoate to protocatechuate (Fig. 1). The deduced amino acid sequences of PobA from the roseobacters were >64% identical to comparable sequences from soil bacteria. The regions associated with flavin adenine dinucleotide and substrate binding (46) were highly conserved among all the sequences.
Pairwise comparisons of deduced amino acid sequences of Roseobacter group PcaD proteins ranged from 60 to 70%. The deduced amino acid sequence also suggests homology to the CatD protein, which catalyzes the analogous reaction in the catechol branch of the β-ketoadipate pathway (23). The conserved active-site cysteine demonstrated to be critical to hydrolysis in PcaD from Pseudomonas sp. strain B13 (37) is present in the sequences of both Roseobacter group isolates. The amino acids surrounding this residue are also fairly well conserved (GYXXXCXA).
The Roseobacter group PcaQ proteins, putative LysR-type regulators, were found to be 39 to 76% identical to PcaQ proteins from other α-proteobacteria (A. tumefaciens C58, S. meliloti 1021, and M. loti MAFF303099). Sequence similarity to several LysR-type regulators of the catechol branch of the β-ketoadipate pathway and of the modified ortho-cleavage chlorocatechol pathway was also observed (Fig. 3C). Pairwise PcaQ comparisons showed 45.0 to 98.7% deduced amino acid similarity among the roseobacters. The region of highest similarity among the PcaQ proteins was in the amino terminus, an area presumed to comprise a helix-turn-helix motif for DNA binding (41). A 55-residue stretch of amino acids in the central region of the protein proposed to be involved in inducer binding (7) appears more highly conserved among the PcaQ proteins than among the other LysR-type proteins. By analogy to PcaQ of A. tumefaciens A348 and other LysR-type regulators, the Roseobacter group PcaQ proteins might be expected to negatively autoregulate their own synthesis and also to activate the expression of genes downstream of, and divergently transcribed from, pcaQ (33).
A conserved hypothetical protein present in all roseobacters.
A highly conserved ORF, designated a conserved hypothetical protein gene (chp), was found immediately downstream of, and oriented in the same direction as, pcaG in all eight roseobacters (Fig. 2). The complete gene was retrieved from S. stellata E-37 and isolate SE45 and was found to be 828 or 837 bp long, respectively. The two genes share 73.9% identity at the nucleotide level. Partial sequences of the chp gene were obtained for the remaining six roseobacters. Pairwise comparisons of S. stellata E-37, SE45, S. pontiacus ChLG 10, and S. pomeroyi DSS-3 show ≥71.8% nucleotide identity for the chp gene.
The location of the chp gene suggests that it may be cotranscribed with the adjacent pca genes (Fig. 2). This suggestion was supported by RT-PCR analysis of RNA isolated from cultures of S. pomeroyi DSS-3 grown on succinate (uninduced) or p-hydroxybenzoate (induced), a metabolic precursor of protocatechuate (Fig. 5). Amplification of cDNA containing pcaG and chp was achieved in induced cultures only. However, the CHP function remains unknown. Protein motifs or signature sequences that might reveal catalytic or functional properties of this hypothetical protein (see Materials and Methods) were not apparent. The absence of transmembrane helices suggests that the putative protein is cytoplasmic. Sequence similarity places the CHP within the Pfam protein family DUF849 (www.sanger.ac.uk/Software/Pfam). Not including the Roseobacter group CHP sequences described here, the family currently consists of 24 prokaryotic sequences. All are hypothetical proteins with no known function. With the exception of the Roseobacter group chp genes, none of the genes encoding Pfam DUF849 members are clustered with genes for aromatic compound catabolism.
FIG. 5.
RT-PCR analysis of chp in S. pomeroyi DSS-3 cultures. RNAs extracted from S. pomeroyi DSS-3 cultures grown on p-hydroxybenzoate (lane 2) or succinate (lane 3) served as the template in RT-PCRs. The orientations and locations of the primers used in amplification reactions are shown beneath the gel. Lane 1, a reaction lacking template; lane M, DNA molecular weight markers. Sizes of the standard (in base pairs) are shown to the left of the gel.
Species-level comparisons.
Sequence similarity in pca gene-containing regions was determined for the two pairs of isolates demonstrating species-level similarity of the 16S rRNA gene. Y3F and Y4I share 98.2% sequence identity over 3,354 bp, and Sulfitobacter sp. strain EE-36 and S. pontaicus ChLG 10 share 97.6% identity over 3,597 bp.
DISCUSSION
The diverse nature of the organization of genes involved in the catabolism of protocatechuate among closely related organisms is evident from the sequence analysis of eight roseobacters. Five unique gene arrangements were identified, with identical gene organization in each of the two species pairs (Fig. 2). Identical pca gene organization was also evident for S. stellata E-37 and S. pomeroyi DSS-3, which share 93.1% sequence identity of the 16S rRNA gene and 64.1% sequence similarity over a 4.97-kbp genomic region containing the pobA and pca genes.
In the Sulfitobacter isolates, the absence of a regulatory gene in the immediate vicinity of pcaCHG differs from the genetic arrangement in the other Roseobacter group isolates. Since it is common for transcriptional regulators to control the expression of distal genes, this arrangement does not imply anything significant about whether pca gene expression is constitutive or regulated in the Sulfitobacter strains. For example, the PcaR regulator in P. putida is encoded in a pca region that is distant from the pcaHG genes that it also regulates (32) (Fig. 4). However, these variations in genetic arrangement necessarily affect which genes will be coordinately transcribed within operons and regulons. Thus, the eight roseobacters in this study appear to represent at least five alternative regulatory schemes for the catabolism of p-hydroxybenzoate and/or protocatechuate (Fig. 2).
The presence of the chp homologs immediately downstream of pcaG in all eight isolates is intriguing. The proximity and orientation of this putative gene suggest that it is cotranscribed with the pca genes, as demonstrated for one of the roseobacters (Fig. 5). Therefore, it might be expected to play a role in the degradation of protocatechuate or related aromatic compounds. Since this gene is uniquely associated with the pca genes of Roseobacter group isolates, its function may be related to a substrate or environmental feature that characterizes the ecological niche of this bacterial group. Further genetic and biochemical investigations are necessary to elucidate the function of CHP.
Six enzymatic steps encoded by eight distinct genes (pcaGHBCDIJF) complete the conversion of protocatechuate to tricarboxylic acid cycle intermediates (see Fig. 1). Clustering of all eight genes within a single operon can be found in some bacteria. However, it appears that distribution of these functionally related genes in two or more distinct genetic loci is the norm (Fig. 4). A preliminary genome sequence has recently become available for the Roseobacter group member S. pomeroyi DSS-3 (www.tigr.org); it indicates that the pca genes are found in four distinct regions of the genome, grouped as pcaCHG, pcaBD, pcaIJ, and pcaF. A defining characteristic of the Roseobacter group pcaCHG clusters appears to be the absence of the gene encoding the enzyme mediating the step immediately following ring cleavage, pcaB. In other organisms examined to date, pcaB is physically linked with the genes encoding the two subsequent reactions (pcaCD or pcaL) (Fig. 4), in agreement with the notion that coordinated expression of genes is an economical strategy for maintaining the fluidity of sequential enzymatic steps. This coordination may be particularly relevant for the gene encoding PcaB, because its substrate, β-carboxymuconate, is known to be a toxic intermediary metabolite (22). Examination of the regulatory strategies invoked by members of the Roseobacter lineage may reveal unique mechanisms to ensure stringent coordination of genes involved in the formation and degradation of β-carboxymuconate.
Whether or not both branches of the β-ketoadipate pathway are present within a single bacterium may be pivotal in dictating the genetic organization of genes in the pathway. Both the catechol and protocatechuate branches of the pathway have been found in phylogenetically diverse soil microorganisms including Acinetobacter sp. strain ADP1, P. putida KT2440, P. aeruginosa PAO1, and R. opacus 1CP. In contrast, there is no evidence of the catechol branch of the pathway in the Roseobacter group isolates (4; www.tigr.org) or in other pca-containing α-proteobacteria, including A. tumefaciens C58, S. meliloti 1021, M. loti MAFF303099, or C. crescentus CB15, based on genome sequence analyses. Organisms possessing both branches of the pathway may require additional regulatory mechanisms for dictating a preferential hierarchy in substrate uptake when presented with the mixture of aromatic compounds found in natural environments (1, 15, 29). This cross-regulation between the two branches of the pathway might serve to prevent mismatched interactions between the structurally similar enzymes and substrates of the pathway (3). In addition, the presence of both branches of the pathway may lead to a greater selective pressure to maintain the genes of each branch within a minimum number of transcriptional units.
The importance of transport proteins in defining the biological individuality of organisms harboring the β-ketoadipate pathway is becoming increasingly evident (35, 49). Acinetobacter sp. strain ADP1, P. putida KT2440, and P. aeruginosa PAO1, members of the γ-Proteobacteria, have catabolic pca regions that include pcaK, a gene that encodes a transport protein in the major facilitator superfamily (31). Similar genetic arrangements have also been identified in some α-proteobacteria. For example, C. crescentus CB15 has a pcaK-like gene immediately downstream of pcaL (NP_421214). However, in some α-proteobacteria the pca regions include genes that may encode proteins of the ABC transport family. Examples of this type of arrangement are seen in A. tumefaciens C58 (NP_356121) and M. loti MAFF303099 (NP_107571). In other cases, such as S. meliloti 1021, no genes encoding transport proteins are evident in the immediate vicinity of the pca genes. As depicted in Fig. 2, the pcaHG regions from the Roseobacter isolates did not include transport genes. In these isolates, transport genes could be associated with pca genes, such as pcaB, that are not linked to the regions characterized. In fact, S. pomeroyi DSS=3 has a gene adjacent to pcaBD that may encode a major facilitator-type transporter (www.tigr.org). The location of transport genes may further reflect the specific compound(s) that serves as a natural substrate(s) and inducer(s) of the pathway. Roseobacters can degrade diverse compounds via the protocatechuate branch of the pathway, yet regulation remains to be investigated in depth (4, 5).
Although the organization of genes within operons provides coordinated and potentially economic genetic regulation, operon organization does not tend to be conserved. Operon rearrangement appears to be selectively neutral in long-term evolution (25). However, despite extensive changes in individual operons (Fig. 4), it is remarkable that multiple transcriptional units with related function remain clustered in diverse and distantly related bacteria. Various factors have been proposed to account for the selective advantage conferred by this type of supraoperonic clustering during the course of evolution. This genetic arrangement facilitates the horizontal gene transfer of complete functional units (26) and provides the potential for multiple functionally related genes to be coamplified (39). A related topic that has received far less attention because it is difficult to address experimentally is whether the subtle variations in genetic organization are ecologically significant. By characterizing the specific arrangements of functionally related pca genes in different bacteria, these studies provide a first step in assessing the broader ecological implications of genetic variation for niche partitioning.
Acknowledgments
We are grateful to William Whitman for helpful discussions.
This work was supported by NSF grants MCB-0084164 to the Sapelo Island Microbial Observatory (M.A.M.), MCB-0135210 for the genome sequencing of S. pomeroyi DSS-3 (M.A.M.), and MCB-0212604 (to E.L.N.).
REFERENCES
- 1.Ampe, F., and N. D. Lindley. 1995. Acetate utilization is inhibited by benzoate in Alcaligenes eutrophus: evidence for transcriptional control of the expression of acoE coding for acetyl coenzyme A synthetase. J. Bacteriol. 177:5826-5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 3.Brzostowicz, P. C., A. B. Reams, T. J. Clark, and E. L. Neidle. 2003. Transcriptional cross-regulation of the catechol and protocatechuate branches of the β-ketoadipate pathway contributes to carbon source-dependent expression of the Acinetobacter sp. strain ADP1 pobA gene. Appl. Environ. Microbiol. 69:1598-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buchan, A., L. S. Collier, E. L. Neidle, and M. A. Moran. 2000. Key aromatic-ring-cleaving enzyme, protocatechuate 3,4-dioxygenase, in the ecologically important marine Roseobacter lineage. Appl. Environ. Microbiol. 66:4662-4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchan, A., E. L. Neidle, and M. A. Moran. 2001. Diversity of the ring-cleaving dioxygenase gene pcaH in a salt marsh bacterial community. Appl. Environ. Microbiol. 67:5801-5809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coco, W. M., R. K. Rothmel, S. Henikoff, and A. M. Chakrabarty. 1993. Nucleotide sequence and initial functional characterization of the clcR gene encoding a LysR family activator of the clcABD chlorocatechol operon in Pseudomonas putida. J. Bacteriol. 175:417-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collier, L. S., G. L. Gaines, and E. L. Neidle. 1998. Regulation of benzoate degradation in Acinetobacter sp. strain ADP1 by BenM, a LysR-type transcriptional activator. J. Bacteriol. 180:2493-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dal, S., I. Steiner, and U. Gerischer. 2002. Multiple operons connected with catabolism of aromatic compounds in Acinetobacter sp. strain ADP1 are under carbon catabolite repression. J. Mol. Microbiol. Biotechnol. 4:389-404. [PubMed] [Google Scholar]
- 9.Dang, H. Y., and C. R. Lovell. 2000. Bacterial primary colonization and early succession on surfaces in marine waters as determined by amplified rRNA gene restriction analysis and sequence analysis of 16S rRNA genes. Appl. Environ. Microbiol. 66:467-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D'Argenio, D. A., M. W. Vetting, D. H. Ohlendorf, and L. N. Ornston. 1999. Substitution, insertion, deletion, suppression, and altered substrate specificity in functional protocatechuate 3,4-dioxygenases. J. Bacteriol. 181:6478-6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dykhuizen, D. E., and L. Green. 1991. Recombination in Escherichia coli and the definition of biological species. J. Bacteriol. 173:7257-7268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eulberg, D., S. Lakner, L. A. Golovleva, and M. Schlömann. 1998. Characterization of a protocatechuate catabolic gene cluster from Rhodococcus opacus 1CP: evidence for a merged enzyme with 4-carboxymuconolactone-decarboxylating and 3-oxoadipate enol-lactone-hydrolyzing activity. J. Bacteriol. 180:1072-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Felsenstein, J. 1989. PHYLIP—Phylogeny Inference Package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 14.Fuhrman, J. A., S. H. Lee, Y. Masuchi, A. A. Davis, and R. M. Wilcox. 1994. Characterization of marine prokaryotic communities via DNA and RNA. Microb. Ecol. 28:133-145. [DOI] [PubMed] [Google Scholar]
- 15.Gaines, G. L., L. Smith, and E. L. Neidle. 1996. Novel nuclear magnetic resonance spectroscopy methods demonstrate preferential carbon utilization by Acinetobacter calcoaceticus. J. Bacteriol. 178:6833-6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.González, J. M., J. S. Covert, W. B. Whitman, J. Henriksen, F. Mayer, B. Scharf, R. Schmitt. A. Buchan, J. A. Fuhrman, R. P. Kiene, and M. A. Moran. 2003. Silicibacter pomeroyi sp. nov. and Roseovarius nubinhibens sp. nov., two dimethylsulfoniopropionate-demethylating bacteria from marine environments. Int. J. Syst. E vol. Microbiol. 53:1261-1269. [DOI] [PubMed] [Google Scholar]
- 17.González, J. M., R. P. Kiene, and M. A. Moran. 1999. Transformations of sulfur compounds by an abundant lineage of marine bacteria in the α-subclass of the class Proteobacteria. Appl. Environ. Microbiol. 65:3810-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.González, J. M., F. Mayer, M. A. Moran, R. E. Hodson, and W. B. Whitman. 1997. Sagittula stellata gen. nov., sp. nov., a lignin-transforming bacterium from a coastal environment. Int. J. Syst. Bacteriol. 47:773-780. [DOI] [PubMed] [Google Scholar]
- 19.González, J. M., and M. A. Moran. 1997. Numerical dominance of a group of marine bacteria in the α-subclass of the class Proteobacteria in coastal seawater. Appl. Environ. Microbiol. 63:4237-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.González, J. M., W. B. Whitman, R. E. Hodson, and M. A. Moran. 1996. Identifying numerically abundant culturable bacteria from complex communities: an example from a lignin enrichment culture. Appl. Environ. Microbiol. 62:4433-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grassly, N. C., and E. C. Holmes. 1997. A likelihood method for the detection of selection and recombination using nucleotide sequences. Mol. Biol. Evol. 14:239-247. [DOI] [PubMed] [Google Scholar]
- 22.Hartnett, G. B., B. Averhoff, and L. N. Ornston. 1990. Selection of Acinetobacter calcoaceticus mutants deficient in the p-hydroxybenzoate hydroxylase gene (pobA), a member of a supraoperonic cluster. J. Bacteriol. 172:6160-6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hartnett, G. B., and L. N. Ornston. 1994. Acquisition of apparent DNA slippage structures during extensive evolutionary divergence of pcaD and catD genes encoding identical catalytic activities in Acinetobacter calcoaceticus. Gene 142:23-29. [DOI] [PubMed] [Google Scholar]
- 24.Harwood, C. S., and R. E. Parales. 1996. The β-ketoadipate pathway and the biology of self-identity. Annu. Rev. Microbiol. 50:553-590. [DOI] [PubMed] [Google Scholar]
- 25.Itoh, T., K. Takemoto, H. Mori, and T. Gojobori. 1999. Evolutionary instability of operon structures disclosed by sequence comparisons of complete microbial genomes. Mol. Biol. Evol. 16:332-346. [DOI] [PubMed] [Google Scholar]
- 26.Lawrence, J. G., and J. R. Roth. 1996. Selfish operons: horizontal transfer may drive the evolution of gene clusters. Genetics 143:1843-1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matrubutham, U., and A. R. Harker. 1994. Analysis of duplicated gene sequences associated with tfdR and tfdS in Alicaligenes eutrophus JMP34. J. Bacteriol. 176:2348-2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mullins, T. D., T. B. Britschgi, R. L. Krest, and S. J. Giovannoni. 1995. Genetic comparisons reveal the same unknown bacterial lineages in Atlantic and Pacific bacterioplankon communities. Limnol. Oceanogr. 40:148-158. [Google Scholar]
- 29.Nichols, N. N., and C. S. Harwood. 1995. Repression of 4-hydroxybenzoate transport and degradation by benzoate: a new layer of regulatory control in the Pseudomonas putida β-ketoadipate pathway. J. Bacteriol. 177:7033-7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orville, A. M., J. D. Lipscomb, and D. H. Ohlendorf. 1997. Crystal structures of substrate and substrate analog complexes of protocatechuate 3,4-dioxygenase: endogenous Fe3+ ligand displacement in response to substrate binding. Biochemistry 36:10052-10066. [DOI] [PubMed] [Google Scholar]
- 31.Pao, S. S., I. T. Paulsen, and M. H. Saier. 1998. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 62:1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parales, R. E., and C. S. Harwood. 1993. Regulation of the pcaIJ genes for aromatic acid degradation in Pseudomonas putida. J. Bacteriol. 175:5829-5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parke, D. 1996. Characterization of PcaQ, a LysR-type transcriptional activator required for catabolism of phenolic compounds in Agrobacterium tumefaciens. J. Bacteriol. 178:266-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parke, D. 1997. Acquisition, reorganization, and merger of genes: novel management of the β-ketoadipate pathway in Agrobacterium tumefaciens. FEMS Microbiol. Lett. 146:3-12. [Google Scholar]
- 35.Parke, D., D. A. D'Argenio, and L. N. Ornston. 2000. Bacteria are not what they eat: that is why they are so diverse. J. Bacteriol. 182:257-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parke, D., and L. N. Ornston. 2003. Hydroxycinnamate (hca) catabolic genes from Acinetobacter sp. strain ADP1 are repressed by HcaR and are induced by hydroxycinnamoyl-coenzyme A thioesters. Appl. Environ. Microbiol. 69:5398-5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pathak, D., G. Ashley, and D. Ollis. 1991. Thiol protease-like active site found in the enzyme dienelactone hydrolase—localization using biochemical, genetic and structural tools. Proteins 9:267-279. [DOI] [PubMed] [Google Scholar]
- 38.Petersen, E. I., J. Zuegg, D. W. Ribbons, and H. Schwab. 1996. Molecular cloning and homology modeling of protocatechuate 3,4-dioxygenase from Pseudomonas marginata. Microbiol. Res. 151:359-370. [DOI] [PubMed] [Google Scholar]
- 39.Reams, A. B., and E. L. Neidle. 2003. Genome plasticity in Acinetobacter: new degradative capabilities acquired by spontaneous amplification of large chromosomal segments. Mol. Microbiol. 47:1291-1304. [DOI] [PubMed] [Google Scholar]
- 40.Rothmel, R. K., T. L. Aldrich, J. E. Houghton, W. M. Coco, L. N. Ornston, and A. M. Chakrabarty. 1990. Nucleotide sequencing and characterization of Pseudomonas putida catR: a postive regulator of the catBC operon is a member of the LysR family. J. Bacteriol. 172:922-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schell, M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47:597-626. [DOI] [PubMed] [Google Scholar]
- 42.Shanley, M. S., E. L. Neidle, R. E. Parales, and L. N. Ornston. 1986. Cloning and expression of Acinetobacter calcoaceticus catBCDE genes in Pseudomonas putida and Escherichia coli. J. Bacteriol. 165:557-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sorokin, D. Y., and A. M. Lysenko. 1993. Heterotrophic bacteria from the Black Sea oxidizing reduced sulfur compounds to sulfate. Mikrobiologiya 62:1018-1031. [Google Scholar]
- 44.Stanier, R. Y., and J. L. Ingraham. 1954. Protocatechuic acid oxidase. J. Biol. Chem. 210:799-808. [PubMed] [Google Scholar]
- 45.Suzuki, M. T., M. S. Rappé, Z. W. Haimberger, H. Winfield, N. Adair, J. Ströbel, and S. J. Giovannoni. 1997. Bacterial diversity among small-subunit rRNA gene clones and cellular isolates from the same seawater sample. Appl. Environ. Microbiol. 63:983-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Berkel, W., A. Westphal, K. Eschrich, M. Eppink, and A. Dekok. 1992. Substitution of Arg214 at the substrate-binding site of p-hydroxybenzoate hydroxylase from Pseudomonas fluorescens. Eur. J. Biochem. 210:411-419. [DOI] [PubMed] [Google Scholar]
- 47.van der Meer, J. R., A. C. Frijters, J. H. Leveau, R. I. Eggen, A. J. Zehnder, and W. M. de Vos. 1991. Characterization of the Pseudomonas sp. strain P51 gene tcbR, a LysR-type transcriptional activator of the tcbCDEF chlorocatechol oxidative operon, and analysis of the regulatory region. J. Bacteriol. 173:3700-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vetting, M. W., D. A. D'Argenio, L. N. Ornston, and D. H. Ohlendorf. 2000. Structure of Acinetobacter strain ADP1 protocatechuate 3,4-dioxygenase at 2.2 Å resolution: implications for the mechanism of an intradiol dioxygenase. Biochemistry 39:7943-7955. [DOI] [PubMed] [Google Scholar]
- 49.Young, D. A., D. Parke, D. A. D'Argenio, M. A. Smith, and L. N. Ornston. 2001. Evolution of a catabolic pathway. ASM News 67:362-369. [Google Scholar]