Abstract
Endogenous RNA interference (endo-RNAi) pathways employ a variety of mechanisms to generate short-interfering (si) RNAs and to mediate gene silencing. In Caenorhabditis elegans, DCR-1 is essential for competing RNAi pathways - the ERI endo-RNAi pathway and the exogenous RNAi pathway. Here, we demonstrate that DCR-1 forms exclusive complexes in each pathway and further define the ERI–DCR-1 complex (ERIC). We show that the tandem-tudor protein ERI-5 potentiates ERI endo-RNAi by tethering an RNA-dependent RNA polymerase (RdRP) module to DCR-1. In the absence of ERI-5, the RdRP module is uncoupled from DCR-1. Interestingly, EKL-1, an ERI-5 paralog that specifies distinct RdRP modules in Dicer-independent endo-RNAi pathways, partially compensates for the loss of ERI-5 without interacting with DCR-1. Our results implicate tudor proteins in the recruitment of RdRP complexes to specific steps within DCR-1-dependent and DCR-1-independent endo-RNAi pathways.
Introduction
RNA interference (RNAi), discovered as a gene-silencing mechanism triggered by double-stranded (ds) RNA, was initially considered as a form of sequence-based innate immunity. It was later acknowledged that related mechanisms underlie a continuum of endogenous silencing phenomena in a broad range of organisms1.
In Caenorhabditis elegans, exogenous (exo) RNAi is triggered by foreign dsRNA, which is recognized by the type III ribonuclease DCR-1 and its co-factors the dsRNA-binding domain protein RDE-4 and the putative RNA helicase DRH-1 (ref. 2). DCR-1 processes the dsRNA into short-interfering (si) RNAs, which are specifically loaded onto the Argonaute RDE-1 (ref. 3,4) and serve as a guide, directing RDE-1 to a target mRNA through base-pair interactions. Recognition of a target by RDE-1 activates a siRNA amplification system that is dependent on the cellular RNA-dependent RNA polymerases (RdRPs) RRF-1 and EGO-1 (ref. 4–7). These so-called secondary siRNAs are synthesized de novo by RdRPs and selectively loaded onto a family of worm-specific Argonautes (WAGOs)4, which effect silencing by transcriptional8 and-or post-transcriptional means9.
Endogenous (endo) RNAi pathways refer to mechanisms whereby siRNAs are derived from, and converge back on, endogenous loci. Endo-RNAi pathways are pervasive and remarkably diverse10, but many of the underlying mechanistic details are unknown. In C. elegans, endo-RNAi pathways are required for a variety of cellular processes, including the assembly and-or maintenance of chromosome integrity11,12,13, transposon silencing and transcript surveillance14, and a gene-specific regulatory mechanism during early larval development15. Interestingly, many of these endo-RNAi pathways appear to function independently of DCR-1 (ref. 14).
The ERI endo-RNAi pathway is a DCR-1-dependent pathway that functions during sperm development as well as during embryogenesis13,16,17. In this pathway, DCR-1 is required for the biogenesis of endo-siRNAs called 26-G-RNAs, which are 26 nucleotides (nt) with a bias for a monophosphorylated 5′-guanosine (G)18,19. The 26-G-RNAs are loaded into specific Argonaute family members – ALG-3 and ALG-4 in developing sperm cells16 and ERGO-1 in the early embryo20, respectively. In the embryo, the ERI endo-RNAi mechanism activates and competes for the same RdRP-dependent siRNA amplification and WAGO system that is required for the exo-RNAi pathway13,20,21.
The molecular events that initiate ERI endo-RNAi are unknown. Our previous work identified a number of DCR-1-associated proteins that are required for ERI endo-RNAi, including the RdRP RRF-3, the Dicer-related helicase DRH-3, the Tandem-tudor domain protein ERI-5, the SAP-exonuclease ERI-1b, as well as the novel proteins ERI-3 and ERI-9 (ref. 13,17,21,22). Recent studies have also revealed that ERI endo-RNAi is dependent on the N-terminal helicase domain of DCR-1 (ref. 22).
In this study, we sought to refine the functional architecture of the exo- and endo-RNAi machineries in C. elegans. We demonstrate that two separate DCR-1 complexes are assembled to initiate the exo- and the ERI endo-RNAi pathways. We identify two distinct subunits within the ERI complex and show that an RdRP module composed of RRF-3, DRH-3 and ERI-5 is tethered to DCR-1 to potentiate ERI endo-RNAi. In addition to solving the molecular phenotype of eri-5, we demonstrate that paralogous but precisely specified RdRP modules govern DCR-1-dependent and -independent RNAi pathways.
Results
DCR-1 is in distinct exo- and endo-RNAi complexes
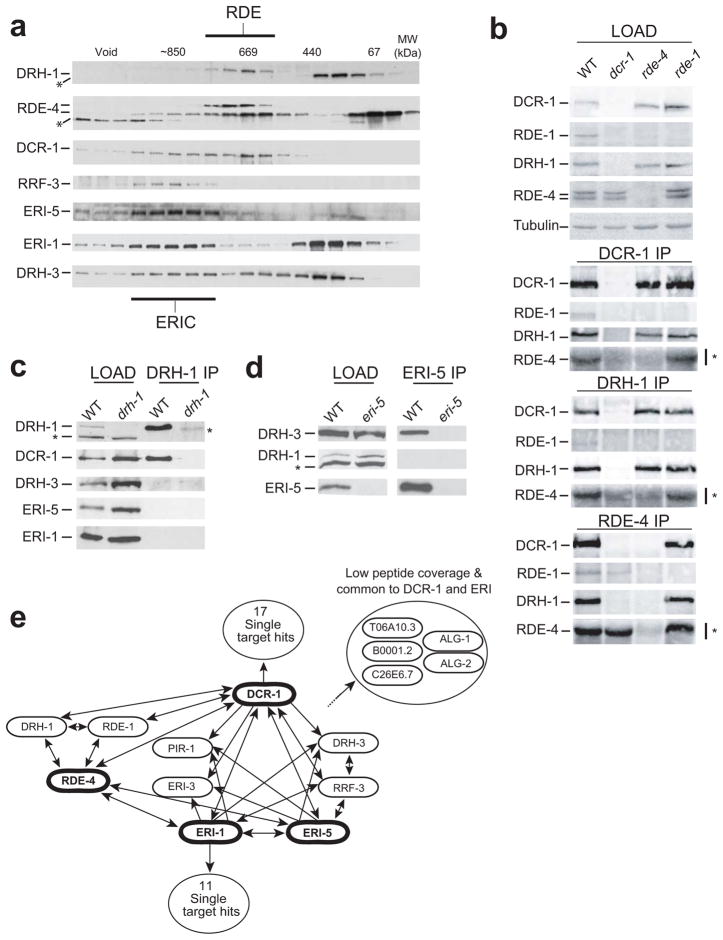
DCR-1 associates with proteins involved in microRNA-mediated silencing, exo-RNAi and endo-RNAi pathways13. To examine the distribution of DCR-1 complexes in the exo- and endo-RNAi pathways, we used gel-filtration chromatography to resolve DCR-1 complexes in an embryo extract (Fig. 1a). Western blot analyses revealed that DCR-1 was present in fractions ranging 600–950kDa in molecular weight (MW) that could be separated into two populations. The peak fractions of DCR-1 (600–700kDa) co-fractionated with factors required for exo-RNAi, including the putative RNA helicase DRH-1 and two isoforms of the dsRNA-binding protein RDE-4 (43 and 46kDa). A second complex population, representing roughly 43% of DCR-1, (750–950kDa) co-fractionated with ERI endo-RNAi proteins13, including the SAP-domain exonuclease ERI-1b, the tandem-tudor domain protein ERI-5, and the RdRP RRF-3. Strikingly, the vast majority of ERI-5 and RRF-3 was restricted to fractions corresponding to the 750–950kDa DCR-1 complex population. Finally, the Dicer-related helicase DRH-3 was present in fractions ranging from ~100kDa to 950kDa. Despite this broad distribution, DRH-3 was enriched in fractions containing ERI-1b, ERI-5, and RRF-3 (750–950kDa), but not in the 600–700kDa fractions, where the major peak of DCR-1 is found. We note in passing that a significant portion of ERI-1b, RDE-4, and DRH-3 is detected in fractions of low molecular weight (50–400kDa) and could reflect monomeric, dimeric or low molecular complexes involving these proteins. These data suggest that DCR-1 is present in distinct complexes required for exo-RNAi (RDE complex) and endo-RNAi (ERI–DCR-1 Complex, or ERIC).
Figure 1. Distinct DCR-1 complexes initiate endo- and exo-RNAi.
(a) Gel filtration on wild-type embryonic extract. DRH-1, RDE-4, DCR-1, RRF-3, ERI-5, ERI-1 and DRH-3 proteins were detected by western blot on fractions from a Superose S6 column. The fractionation of molecular weight standards (MW) is indicated. The asterisk (*) labels in DRH-1 (in the low MW fractions) and RDE-4 filtration panels indicate non-specific bands. (b) Immunoprecipitation (IP) of DCR-1, DRH-1 and RDE-4 from wild-type (WT), dcr-1, rde-4 or rde-1 mutant embryos. DCR-1, RDE-1, DRH-1 and RDE-4 proteins were detected in total lysate (LOAD) and IP by western blot. Tubulin was used as a loading control. The asterisk (*) to the right of the RDE-4 panels indicates background signal from the IgG heavy chains used for immunoprecipitation, and co-migrate with RDE-4 around 50kDa. (c) IP of DRH-1 in WT and drh-1 mutant embryos. DRH-1, DCR-1, DRH-3, ERI-5 and ERI-1 were detected by western blot. The asterisk (*) to the right and left of the DRH-1 panel indicate non-specific bands in the loading, and DRH-1 IP lanes, respectively. (d) IP of ERI-5 in WT and eri-5 mutant embryos. DRH-3, DRH-1 and ERI-5 proteins were detected by western blot. The asterisk (*) indicates the non-specific band detected in the input lanes (LOAD) of the DRH-1 blot as in panel c. (e) Interaction map of the proteins detected by MuDPIT analyses in WT embryonic extracts. Proteins in bold (DCR-1, ERI-5, ERI-1 and RDE-4) represent IP targets. See Methods for details on the epitope targeted. Arrowheads indicate interactions detected. The interactions of ERI-5 and ERI-1 in RDE-4 IP included in the diagram were only detected by western. The number of interactions detected exclusively in DCR-1 or ERI-1 MuDPIT experiments are indicated (‘17 or 11 single target hits’ circles) and may reflect divergent functions for these proteins.
Previous work strongly suggested the existence of an RDE complex comprising DCR-1, RDE-4, DRH-1 and the Argonaute RDE-1 (ref. 2). Indeed, all of these proteins were detected in immunoprecipitates of endogenous DCR-1, RDE-4, and DRH-1 (Fig. 1b). Interestingly, many of these proteins also rely on each other for their accumulation as well as for their interaction with DCR-1 (See Supplementary Results for a detailed description). Furthermore, DCR-1 and DRH-1 were detected after sequential immunoprecipitation of RDE-4 and RDE-1 (Supplementary Fig. 1b). In contrast, components of ERIC, including ERI-5, DRH-3 and ERI-1b, did not co-immunoprecipitate with DRH-1 (Fig. 1c), which was restricted to the RDE complex in our gel-filtration experiments. Conversely, DRH-1 did not co-immunoprecipitate with ERI-5, which was restricted to ERIC (Fig. 1d), while DRH-3 was detected in the ERI-5 immunoprecipitates (Fig. 1d). Finally, DRH-1 was detected with the highest peptide coverage in DCR-1 Multidimensional Protein Identification Technology (MuDPIT)13,23 analyses or when conducting MuDPIT analysis of a somatically expressed RDE-4::FLAG fusion, while DRH-1 was never detected in ERI-5 or ERI-1 MuDPIT (Fig. 1e, below). Taken together, the data indicate that the RDE complex and ERIC are biochemically distinct.
Comparative Proteomics identify ERIC components
We reported that the eri gene products ERI-1b, ERI-3, ERI-5, RRF-3 and the Dicer-related helicase DRH-3 could be detected in MuDPIT analyses on DCR-1 and ERI-1 proteins13. While both DCR-1 and ERI-1b fractionate in multiple complex populations in gel-filtration, the profile of ERI-5 suggests that it is dedicated to ERIC. Therefore, to further define ERIC components, we used MuDPIT to identify proteins that co-immunoprecipitate with ERI-5 and compared these data to previous MuDPIT analyses of DCR-1 and ERI-1 immunoprecipitates13. The interactions that could be detected from at least 2 independent sets of MuDPIT samples, and never detected in any of the controls, are depicted in Figure 1e and Table 1. DCR-1, ERI-1, ERI-5, ERI-3, DRH-3, RRF-3, and RDE-4 were the only proteins consistently detected for each of the 3 targets. The PIR-1 putative RNA triphosphatase, previously identified as a DCR-1 interaction13, was detected in some ERI-5 MuDPIT samples but with a lower peptide coverage. The sum of the masses of these 8 proteins (~810kDa) is roughly in accord with the ~850kDa mass of ERIC estimated by gel-filtration (Fig. 1a). This analysis further supports the idea that ERI-5 has only limited or no association outside the ERIC. Finally, additional proteins were identified in ERI-1 and DCR-1 immunoprecipitates but not in ERI-5 samples (Fig. 1e, and Table 1), including ALG-1, ALG-2, T06A10.3, B0001.2 and ERI-9. These proteins may be transient, indirect, or less stable components of ERIC.
Table 1.
Comparative proteomic of ERI components
| Gene | Protein | Structural Description | # Independent Detection (% Peptide Coverage) | ||
|---|---|---|---|---|---|
|
| |||||
| Common DCR-1, ERI-1 and ERI-5 Interactions | ERI-51 | ERI-1(FLAG)2 | DCR-1(HA)2 | ||
| Y38F2AR.1 | ERI-5 (61.6) | Tudor domain | 5/5 (19) | 2/2 (9) | 3/3 (24) |
| F10B5.7 | RRF-3 (201.4) | RdRP | 5/5 (12) | 2/2 (15) | 3/3 (24) |
| D2005.5 | DRH-3 (129.1) | DEAH/D box RNA helicase | 4/5 (18) | 2/2 (19) | 3/3 (35) |
| K12H4.8 | DCR-1 (210.9) | DExH box RNA helicase/RNaseIII | 4/5 (2) | 2/2 (13) | 3/3 (41) |
| T20G5.11 | RDE-4 (43.4) | dsRBD | 1/5 (6) | 2/2 (11) | 3/3 (44) |
| W09B6.3 | ERI-3 (66.4) | Novel | 1/5 (4) | 2/2 (12) | 3/3 (33) |
| T07A9.5b | ERI-1b (67.2) | SAP domain, exonuclease | 1/5 (2) | 2/2 (31) | 2/3 (13) |
| T23G7.5 | PIR-1 (30.1) | RNA phosphatase | 1/5 (7) | 1/2 (8) | 3/3 (47) |
|
| |||||
| Common DCR-1 and ERI-1 Interactions | |||||
|
| |||||
| C26E6.7 | ERI-9 (73.2) | Novel | nd | 2/2 (7) | 3/3 (7) |
| T06A10.3 | (19.9) | Novel | nd | 2/2 (11) | 2/3 (8) |
| B0001.2 | (105.1) | Novel | nd | 2/2 (7) | 2/3 (6) |
| T07D3.7 | ALG-2 (101.6) | Piwi/PAZ domain | nd | 2/2 (5) | 3 W |
| F48F7.1 | ALG-1 (110.9) | Piwi/PAZ domain | nd | 2/2 (4) | 3 W |
Immunopurifications of ERI-5 were conducted using a covalently coupled polyclonal matrix. Since peptide coverage varied between independent samples, peptide coverage for the best ERI-5 recovery is used here.
Immunoprecipitations conducted on tagged proteins (Flag and HA, as indicated), expressed from null allele-rescuing transgenes(See Duchaine et al. 2006).
These two proteins were not detected in embryonic DCR-1 proteomic analyses, but were detected in adult purifications and with embryonic DCR-1 by western blotting (W).
ERI-5 recruits a RdRP module on DCR-1
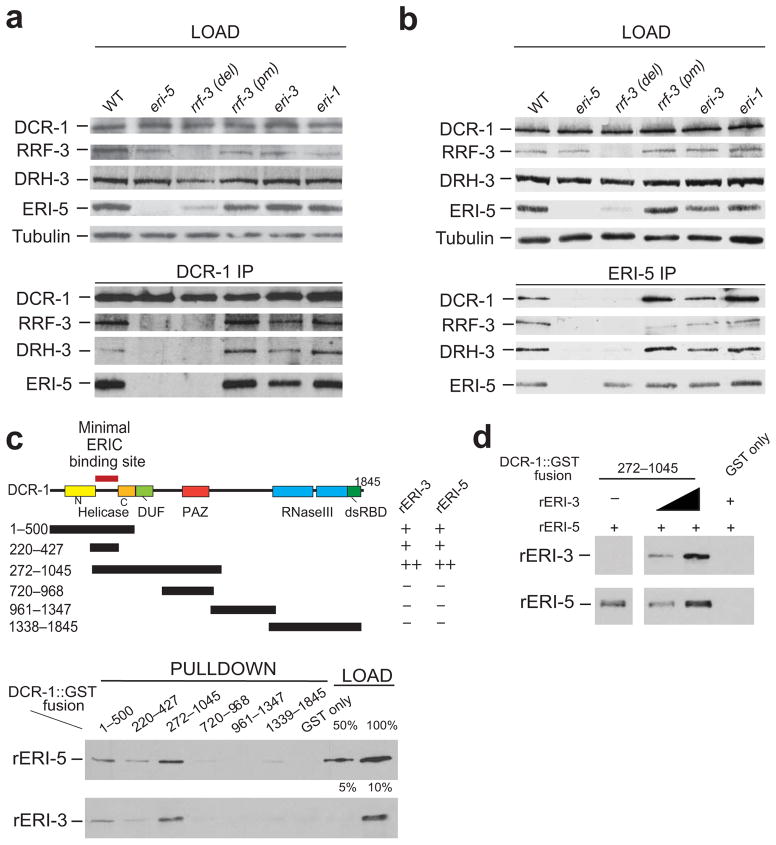
RRF-3, DRH-3 and DCR-1 were consistently the top ERI-5 interactors, suggesting that ERI-5 is most intimately associated with these components of ERIC. This idea is supported by recent work, demonstrating that EKL-1, a paralog of ERI-5, interacts with DRH-3 (ref. 14). Interestingly, the steady-state level of ERI-5 protein was dramatically reduced in the rrf-3(pk1426) deletion mutant, but was expressed at wild-type levels in the rrf-3(mg373) missense mutant that alters a conserved catalytic residue of RRF-3 (ref. 17) without altering RRF-3 expression (Fig. 2a, b, top panels, rrf-3(del) and rrf-3(pm) lanes). Quantitative RT-PCR (qRT-PCR) analysis revealed that the eri-5 mRNA level was unchanged in the rrf-3(pk1426) mutant (Supplementary Fig. 2a). Together, these results suggest that the stable expression of ERI-5 protein is dependent on RRF-3 protein and indicate a close biochemical relationship between these two proteins. The coupled expression observed for both the RDE complex and ERIC is reminiscent of similar phenomena described in the human Drosha–DGCR8, and fly Dicer2–R2D2 complexes24,25.
Figure 2. ERI-5 promotes the association of an RdRP module to DCR-1 N-terminus.
(a, b) IP of DCR-1 and ERI-5 in WT, eri-5, rrf-3 del (deletion mutant, pk1426), rrf-3 pm (point mutant, mg373), eri-3 and eri-1 mutant embryos. DCR-1, RRF-3, DRH-3 and ERI-5 were detected by western blot. Tubulin was used as a loading control. (c) (top) Map of the DCR-1-GST constructs used for the GST pull-down of recombinant (r) ERI-5 or ERI-3. The ability of each DCR-1-GST fusion to interact with rERI-5 or rERI-3 was assessed by western blot (bottom panel) to detect recombinant rERI-5-CBP or rERI-3-FLAG. The results are summarized to the right of the DCR-1 map; “−” denotes weak or no interaction, “+” denotes an interaction. (see Supplementary Fig. 2c for Coomassie Blue gel staining). Percentage (%) of the loading (bottom panel) represents the fraction of rERI-5 and rERI-3 used in the GST pull-down. (d) ERI-3 and ERI-5 bind to DCR-1(272–1045) simultaneously. An increasing amount of rERI-3 was pre-incubated with DCR-1(272–1045) prior to addition of rERI-5 and pull-down of the DCR-1 fragment.
DCR-1, ERI-5, RRF-3, and DRH-3 co-immunoprecipitated with DCR-1 and ERI-5 and these interactions were dependent on ERI-5 and RRF-3 protein (Fig. 2a, b). For example, RRF-3 and DRH-3 proteins were not detected in DCR-1 immunoprecipitates from eri-5(tm2528) or rrf-3(pk1426) mutant lysates (Fig. 2a, eri-5 and rrf-3(del) lanes). In contrast, the interactions between DCR-1, ERI-5, RRF-3 and DRH-3 remained intact in the rrf-3(mg373) catalytic mutant, as well as in eri-1(mg366) and eri-3(tm1361) mutants (Fig. 2a, rrf-3(pm), eri-1 and eri-3 lanes). Despite the reduced level of ERI-5 in the rrf-3(pk1426) mutant, a significant amount of ERI-5 was still recovered by ERI-5 immunoprecipitation, but DCR-1 was not detected and DRH-3 was weakly detected. Finally, DCR-1 was not detected in RRF-3 immunoprecipitates in the absence of ERI-5 (see below). Together, these results suggest that ERI-5, RRF-3 and DRH-3 form an inter-dependent RdRP module and indicate that the tandem-tudor domain protein ERI-5 has an important contribution in the association of this RdRP module to DCR-1. However, these results do not rule out the possibility of additional interactions between RRF-3 and DCR-1 within ERIC. It also indicates that, in stark contrast to its paralog DRH-1, DRH-3 interacts with DCR-1 strictly through RRF-3 and ERI-5.
ERI proteins interact with DCR-1 via two N-terminus sites
Previous work indicated that ERI-3 tethers ERI-1b to DCR-1 (ref. 13). The fact that the interaction between the RdRP module and DCR-1 is independent of eri-3 and eri-1 (Fig. 2a, b) suggests that ERI-5 and ERI-3 recognize distinct sites on DCR-1. To identify the direct binding site(s) for the ERI machinery on DCR-1, we expressed partially-overlapping, recombinant DCR-1 fragments fused to a GST epitope (Fig. 2c, upper panel; see Supplementary Fig. 2b for Coomassie) and used these DCR-1 fragments as bait to capture recombinant ERI-3 (rERI-3) or ERI-5 (rERI-5) proteins. rERI-5 was pulled down with fragments spanning DCR-1 residues 1–500 and 272–1045. No appreciable interaction could be detected with constructs encoding fusions of residues 720–968, 961–1347, 1338–1845, or GST alone (Fig. 2c, rERI-5 panel). Maximal binding was observed with a DCR-1 fragment that encodes residues 272–1045. A DCR-1 fragment spanning residues 220–427 still interacted with rERI-5, although slightly less efficiently. rERI-3 was captured by the same DCR-1 fragments that interact with rERI-5, but rERI-3 interacted overall more weakly than rERI-5 (Fig. 2c, rERI-3 panel, compare loading).
Thus, the minimal fragment of DCR-1 bound by rERI-5 and rERI-3 (220–427) correspond to the non-conserved linker between the conserved N- and C-terminal portions of the helicase domain (Fig. 2c). However, optimal binding may require additional residues, or a contribution by surrounding polypeptide sequences for the folding of this region. These two interactions may occur simultaneously within ERIC, as pre-binding increasing amounts of rERI-3 did not preclude, but slightly improved, rERI-5 binding to DCR-1 (Fig. 2d). Together with the immunoprecipitation data described above (Fig. 2a and b), these data suggest that the ERI proteins directly interact with different, and possibly neighboring, sites near the N-terminus of DCR-1 (Fig. 2c).
ERI-5 potentiates small RNA biogenesis in ERI endo-RNAi
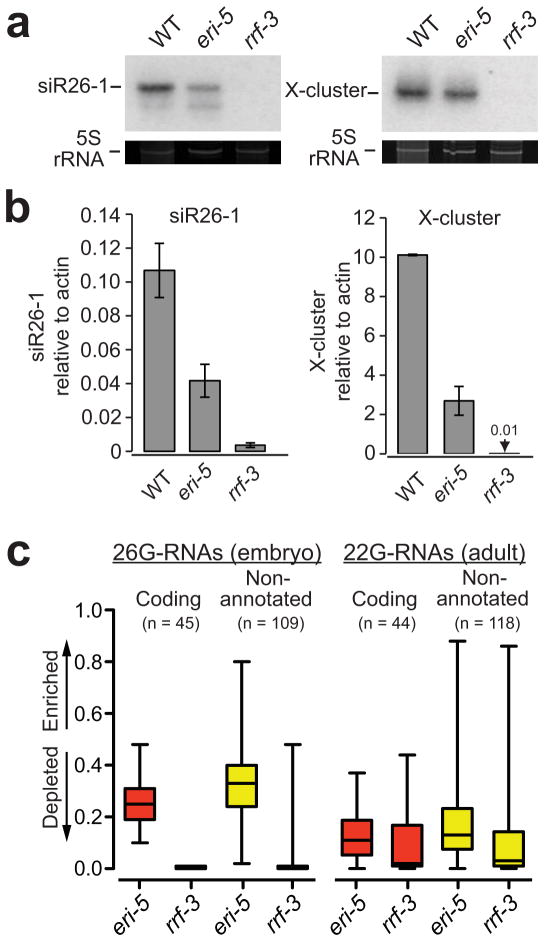
DCR-1, ERI-1, ERI-3, RRF-3 and DRH-3 are absolutely required for the biogenesis of 26-G-RNAs and the downstream 22G-RNAs13,20–22,26. Interestingly, these small RNA species were only partially reduced in eri-5 point mutants13. The eri-5(tm2528) deletion allele used in this study is likely to be a null allele that completely removes the first tudor domain and alters the reading frame of the remaining eri-5 locus (Supplementary Fig. 2c). Northern blot analyses of small RNAs expressed in embryos revealed that siR26–1, an ERI-dependent 26-G-RNA derived from the C40A11.10 ERI target, and 22G-RNAs from the X-cluster locus were only partially reduced in eri-5(tm2528) mutants (Fig. 3a). These small RNAs were not detected in rrf-3(pk1425) mutants (Fig. 3a). Using qRT-PCR to measure the level of these small RNAs, we estimated that the expression of siR26–1 and the X-cluster 22G-RNA in eri-5(tm2528) is one-third to one-quarter of the wild-type level (Fig. 3b).
Figure 3. ERI-5 potentiates ERI endo-RNAi small RNA biogenesis.
(a) Northern and (b) qRT-PCR analysis of C40A11.10 26-G-RNAs siRNA species (siR26–1) as indicated in WT, eri-5 and rrf-3 (pk1426) mutant embryos. The C40A11.10 probe detected both 26-G- and 22G-RNAs. 5S ribosomal RNA (rRNA) ethidium bromide staining is shown as a loading control in a. The mean of at least three independent experiments is depicted as the ratio of siR26–1 or X-cluster relative to actin. Error bars indicate s.d. (c) Box and whisker plots show the enrichment or depletion of small RNAs targeting 26-G-RNA coding genes (red) and non-annotated 26-G-RNA clusters (yellow) in the indicated mutant. The left panel is an analysis of 26nt antisense reads from embryo small RNA libraries that target the 26-G-RNA loci. The right panel is an analysis of all antisense reads from adult small RNA libraries that target the 26-G-RNA loci. The majority of reads in the adult samples are 22G-RNAs. Values approaching 1 indicate enrichment of small RNA; values approaching 0 indicate depletion. Relative enrichment was calculated as the ratio of mutant per (mutant + wild-type). The top and bottom of each box represent the 75th and 25th percentiles, respectively. The horizontal line within each box represents the median value. The number of loci used to generate box and whisker plots is indicated above each plot and the data are provided in Supplementary Data 1 and 2.
The reduced level of the ERI-dependent endo-siRNAs suggested that ERI-5 could play a role in either the specificity or the efficiency of 26-G-RNA biogenesis. To measure the overall level of 26-G-RNAs in the eri-5 mutant, we cloned and deep-sequenced small RNAs from eri-5, rrf-3 and wild-type embryos using a method that is compatible with small RNAs bearing a 5′-monophosphate, including 26-G-RNAs, miRNAs and 21U-RNAs18,19. As expected, 26-G-RNAs were completely abolished in rrf-3 mutant embryos (Fig. 3c). Of the few 26 nt sequences cloned from the rrf-3 sample, most were reads with a 5′A and primarily corresponded to miRNA precursors or to mRNA degradation products. The overall 26-G-RNA level was reduced by ~50% in eri-5 embryos relative to wild-type (Supplementary Fig. 3a,b), consistent with northern blot and qRT-PCR analyses of siR26–1. The 26-G-RNAs that were cloned from eri-5 mutant embryos map to the previously defined 26-G-RNA loci and are not derived from novel loci (Fig. 3c and not shown)20, indicating that the specificity of ERI endo-RNAi remains unchanged in the eri-5 mutant. The 26-G-RNA reads targeting more than 75% of genes and 50% of non-annotated loci were depleted at least 2-fold in the eri-5 mutant embryos (Fig. 3c, left panel), with a median 4-fold depletion of 26-G-RNAs that target genes and 2-fold for those that target non-annotated loci. Together, these results suggest that ERI-5 promotes the efficiency of 26-G-RNA biogenesis by coupling the activities of RRF-3 and DCR-1.
To analyze 22G-RNAs in eri-5(tm2528), we cloned small RNAs from adult animals using a method compatible with small RNAs bearing a 5′ tri-phosphate14. Consistent with the X-cluster northern blot analysis (Fig. 3a), 22G-RNAs targeting 26-G-RNA loci were substantially reduced in the eri-5 mutant, but less severely than in the rrf-3 mutant (Fig. 3c, right panel). The median depletion of 22G-RNAs targeting 26-G-RNA loci was 9-fold in the eri-5 mutant and 50-fold in the rrf-3 mutant (Fig. 3c). As previously observed for ergo-1 and rrf-3 mutants20, the ERI-independent 22G-RNA populations that are important for genome surveillance and chromosome segregation appear to be unaffected in the eri-5 mutant (Supplementary Fig. 3c,d). These findings suggest that efficient 26-G-RNA biogenesis is important for the amplification and accumulation of the downstream 22G-RNAs in the ERI pathway.
Tandem-tudor domain proteins are required for ERI endo-siRNA biogenesis
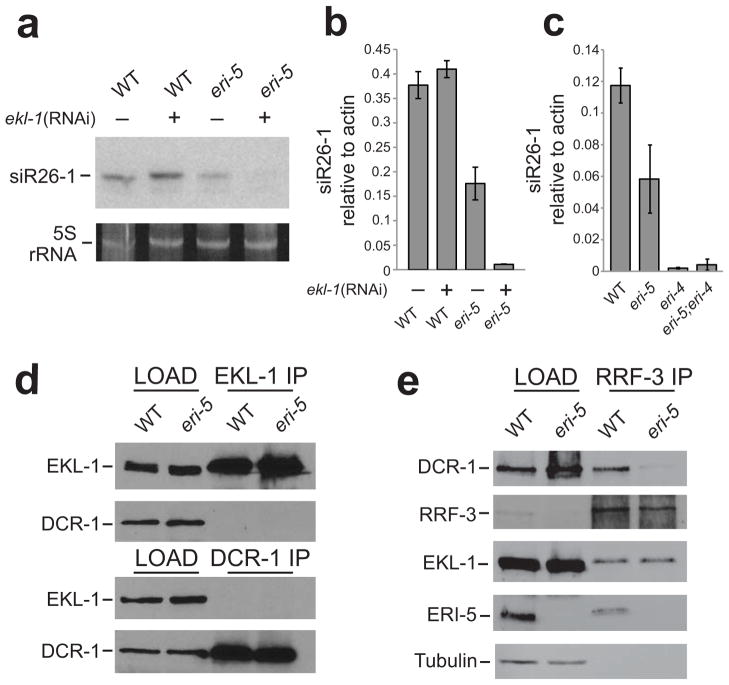
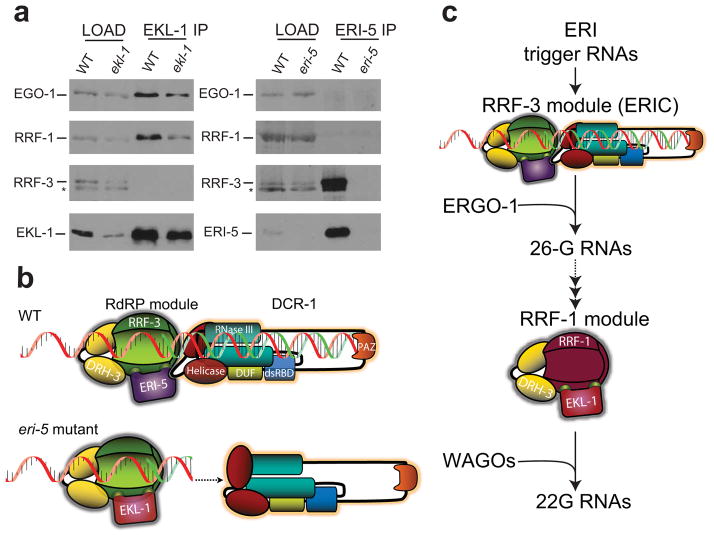
Our data indicate that a complete loss of ERI-5, which uncouples the interaction of RRF-3 with DCR-1, results in a reduction of ERI endo-siRNAs. One possible explanation for this partial loss of endo-siRNAs is that EKL-1, a paralog of ERI-5, may partially compensate for the loss of ERI-5. To test this hypothesis, we measured C40A11.10 26-G-RNA levels by qRT-PCR in the eri-5(tm2528) animals where ekl-1 was depleted by RNAi. Since a strong exo-RNAi response can mildly reduce endo-siRNA levels13, we used sel-1(RNAi) as a negative control (Fig. 4a,b, Supplementary Fig. 4). ekl-1(RNAi) alone did not affect the level of C40A11.10 26-G-RNAs, indicating that EKL-1 does not normally contribute to 26-G-RNA biogenesis. However, ekl-1(RNAi) obliterated the remaining 26-G-RNAs in the eri-5(tm2528) mutant (Fig. 4a,b), indicating that EKL-1 is not redundant with ERI-5, but can partially compensate for the loss of ERI-5 in the production of at least some of the endo-siRNAs.
Figure 4. Tandem-tudor domain proteins are required for ERI endo-siRNA biogenesis.
(a) Northern and (b) qRT-PCR analysis of C40A11.10 26-G-RNAs (siR26–1) in sel-1 (RNAi) (a negative control, marked with (−)), ekl-1(RNAi), eri-5 and eri-5; ekl-1(RNAi) embryos. The mean of at least three independent experiments is depicted as the ratio of siR26–1 relative to actin. Error bars indicate s.d. (c) qRT-PCR analysis of C40A11.10 26-G-RNAs (siR26–1) in WT, eri-5, eri-4 and double eri-5; eri-4 mutant embryos. The mean of at least three independent experiments is depicted as the ratio of siR26–1 relative to actin. Error bars indicate s.d. (d) IP of EKL-1 and DCR-1 in WT and eri-5 mutant embryos. EKL-1 and DCR-1 proteins were detected by western blot. (e) IP of RRF-3 in WT and eri-5 mutant embryos. DCR-1, RRF-3, EKL-1 and ERI-5 proteins were detected by western blot. Tubulin was used as a loading control.
Because EKL-1 is required for small RNA pathways that are independent of DCR-112,14, we addressed the possibility that molecular replacement of ERI-5 by EKL-1 may have altered the requirements for 26-G-RNA biogenesis. In particular, we asked whether the 26-G-RNAs that remain in the eri-5 mutant are dependent on DCR-1. We took advantage of an Eri allele of dcr-1, eri-4(mg375), which carries a missense mutation in the helicase domain of dcr-1 and completely abrogates ERI-dependent endo-siRNAs, including the 26-G-RNAs, while maintaining interactions between ERIC components17,22. Indeed, C40A11.10 26-G-RNAs were completely dependent on ERI-4. eri-4(mg375) completely abolished the EKL-1-dependent 26-G-RNAs that accumulate in the eri-5(tm2528) background (Fig. 4c, eri-5; eri-4). Although the 26-G-RNAs that accumulate in the absence of ERI-5 are dependent on EKL-1 and DCR-1, a stable interaction between EKL-1 and DCR-1 was not detected in reciprocal immunoprecipitation experiments (Fig. 4d). Therefore, although RRF-3 and DCR-1 do not stably interact in the eri-5 mutant, DCR-1 is nonetheless required for the production of EKL-1-dependent 26-G-RNAs.
Since EKL-1 partially compensates for the loss of ERI-5, we asked whether EKL-1 co-immunoprecipitates with RRF-3 in WT and eri-5 backgrounds. Surprisingly, a small fraction of EKL-1 co-purified with RRF-3 equally well in both WT and eri-5 backgrounds (Fig. 4e). These results indicate that a minor fraction of EKL-1 normally interacts with RRF-3, but its contribution to 26-G-RNA biogenesis is only observed in the absence of ERI-5.
Tandem-tudor proteins define paralogous RdRP modules
The molecular replacement of ERI-5 by EKL-1 in the RRF-3 RdRP module suggests that the RdRPs RRF-1 and EGO-1 may adopt the same modular organization in the other RNAi pathways. Indeed, Gu and coworkers14 recently demonstrated that EKL-1 and the RdRPs RRF-1 and EGO-1 co-immunoprecipitate with DRH-3 and function in multiple DCR-1-independent endo-RNAi pathways. Hence, at least three independent RdRP ‘modules’ may co-exist in C. elegans to specifically orchestrate different steps in RNAi pathways. To test this hypothesis, we immunoprecipitated EKL-1 and ERI-5 proteins, and identified the associated RdRPs by western blot. Both EGO-1 and RRF-1 efficiently co-immunoprecipitated with EKL-1 (Fig. 5a left panel). Although RRF-3 immunoprecipitation did reveal a weak interaction with EKL-1 (Fig. 4e), RRF-3 was not detected in the EKL-1 immunoprecipitates. Furthermore, while RRF-3 was strongly enriched by ERI-5 immunoprecipitation, EGO-1 or RRF-1 were not detected in the ERI-5 immunoprecipitates (Fig. 5a right panel). Altogether, our results indicate that tandem-tudor proteins specify RdRP modules in DCR-1-dependent and independent RNAi pathways (see Model, Fig. 5b, c, and Discussion).
Figure 5. Roles and paralog organization of RdRP modules in ERI endo-RNAi.
(a) IP of EKL-1 in WT and ekl-1(RNAi) (ekl-1 lanes) embryos, and IP of ERI-5 in WT and eri-5 mutant embryos. The RdRPs EGO-1, RRF-1, RRF-3, and the tudor domain EKL-1 and ERI-5 proteins were detected by western blot. Asterisk (*) indicates a non-specific band. (b) Model of the molecular compensation of ERI-5 by EKL-1. Interactions between the RdRP module and the N-terminal helicase domain of DCR-1 couple the generation of dsRNA by RRF-3 with processive DCR-1 activity. In the eri-5 mutant, this coupling is lost and the auto-inhibitory function of the helicase domain predominates, resulting in inefficient 26-G-RNA production. (c) Paralogous RdRP modules function sequentially in ERI endo-RNAi. An RdRP module comprised of RRF-3, DRH-3 and ERI-5 together with DCR-1 function at the initial step to generate 26-G-RNAs, the primary siRNAs of the ERI pathway that program ERGO-1. A paralogous RdRP module comprised of RRF-1, DRH-3 and EKL-1 is responsible for secondary siRNA generation that is independent of DCR-1. This abundant pool of small RNAs programs the WAGO Argonautes to effect endo-RNAi silencing. Paralogous EGO-1 complexes may be involved in this and other RNAi pathways. Some of the ERIC components were omitted from the model for clarity.
Discussion
Using a combination of molecular and proteomic approaches, we demonstrate that DCR-1 forms distinct complexes that mediate exo-RNAi (RDE complex) and ERI endo-RNAi (ERIC). Our analysis of ERIC indicates that an RdRP module comprised of RRF-3, DRH-3 and ERI-5 is recruited to DCR-1 by the tandem-tudor domain protein ERI-5. Furthermore, this report reveals the importance of interactions between RdRPs, tandem-tudor domain proteins and DCR-1 and provides a comprehensive interaction framework for the functional organization of RdRPs in C. elegans.
The significance of DCR-1 interaction for ERI endo-RNAi
ERIC is composed of RRF-3, ERI-5, and DRH-3 (an RdRP ‘module’), ERI-1 and ERI-3 (together), RDE-4, and additional proteins that possibly interact in a transient, or unstable manner. The physical coupling between DCR-1 and an RdRP presented here is reminiscent of the molecular organization of Dicer-RdRP complexes described in Schizosaccharomyces pombe and Tetrahymena thermophila27,28. Intriguingly, in S. pombe the C-terminal dsRNA binding domain of Dcr1 both physically and functionally couples the RdRP complex to Dcr1 (ref. 27). In contrast, within ERIC, both the RdRP module and the ERI-1-ERI-3 module bind to the N-terminus of DCR-1. The configuration of each complex is essential to promote efficient siRNA synthesis. The interaction between the RdRP module and the N-terminus of DCR-1 likely bears additional significance in light of the properties recently attributed to this region. The ATPase activity of the helicase domain of DCR-1 is essential for 26-G-RNA accumulation and ERI endo-RNAi22. Furthermore, the ATPase activity is required for recognition and processing of blunt-ended dsRNA substrates into 26nt siRNAs in vitro29. Interestingly, the N-terminus of human Dicer exerts an auto-inhibitory function on its RNaseIII activity30. How can DCR-1’s known N-terminus functions be linked with its scaffolding of the ERI endo-RNAi machinery? The interaction between ERI proteins and the helicase domain of DCR-1 effectively couple the generation of dsRNA by RRF-3 to the processive ‘mode’ of DCR-1 responsible for 26-G-RNA generation29. The interactions within ERIC may mediate conformational changes that stimulate DCR-1 activity (Fig. 5b, WT panel). In the absence of these interactions (e.g. in the eri-5 mutant), the auto-inhibitory function of the helicase domain of DCR-1 predominates, resulting in inefficient 26-G-RNA production (Fig. 5b, eri-5 mutant).
Although this is an attractive model, a number of questions on the biogenesis of 26-G-siRNAs in ERI endo-RNAi remain, including the asymmetric accumulation of antisense 26-G-siRNAs, and the strong bias for the 5′ G nucleotide. Answers to these lingering conundrums will require further characterization of each module, recapitulation of the ERIC using recombinant proteins, and additional structural details regarding its dynamic organization.
Functional organization of paralog RdRP modules in RNAi
A number of previous publications have linked EKL-1 and DRH-3 to various endo- and exo-RNAi functions12,13,15,31,32. Of particular relevance to this work, Gu et al. found that EKL-1 and the RdRPs RRF-1 and EGO-1 were present in DRH-3 immunoprecipitates and postulated that these proteins were assembled as a ‘core’ complex to mediate RdRP functions in multiple endo-RNAi pathways14. The results presented here refine and extend this view. Indeed, our data reveal that RRF-3, EGO-1, and RRF-1 are assembled into analogous modules comprised of an RdRP, a tandem-tudor domain protein and the DRH-3 helicase. However, these modules participate in strikingly different mechanisms of small RNA biogenesis. While the EGO-1 and RRF-1 modules are capable of generating small RNAs in a DCR-1-independent manner7, the RRF-3 module requires DCR-1 and several other factors including ERI-1 and ERI-3 for efficient generation of the 26-G-RNAs. This surprisingly mechanistic diversification of paralogous RdRP modules is emphasized by sequential activity of distinct RdRP modules in the ERI endo-RNAi pathway (Fig. 5c)20,21. Thus, our findings indicate that the specificity of paralogous RdRP modules is imparted by the tandem-tudor domain proteins ERI-5 and EKL-1.
The roles of tandem-tudor domain proteins in RNAi
Our data indicate that the tandem-tudor domain proteins ERI-5 and EKL-1 carry out at least two distinct functions in ERIC. First, they are essential co-factors for RRF-3 activity, and second, the interaction with ERI-5 is essential for the stable recruitment of RRF-3 to DCR-1. Although EKL-1 palliates, at least partially, the 26-G-RNA defects in the eri-5 mutant, it cannot compensate for the defect in DCR-1 recruitment, resulting in attenuated ERI endo-RNAi. The partial molecular compensation by EKL-1 indicates that a stable association with DCR-1 potentiates, but is not essential for, 26-G-RNA biogenesis.
In the absence of ERI-5, EKL-1 does not appear to alter the target specificity of ERI endo-RNAi. Furthermore, DRH-3 is a common component of RdRP modules in C. elegans. Hence, it is unlikely that DRH-3 or the tandem-tudor domain proteins act as determinants that recognize the ERI endo-RNAi triggers. Possible determinants may lie within the N-terminus of RRF-3, a portion that has significantly diverged from the other RdRPs in C. elegans. Alternatively, another ERIC component may act as a trans-acting factor(s) that recognizes the RNA template.
Tudor domains adopt a fold similar to the chromo domains and bind to methylated derivatives of arginine or lysine33,34. Our evidence suggests that ERI-5 may recognize such modified amino acid(s) within ERIC. The essential role of ERI-5 in recruiting RRF-3 to DCR-1, the high peptide coverage for RRF-3 in ERI-5 MuDPIT experiments, and the coupled stability of ERI-5 protein with RRF-3 indicate a robust, and likely direct, interaction between ERI-5 and RRF-3. Yet, recombinant RRF-3 (which may not be methylated in vitro) did not interact directly with recombinant ERI-5 in vitro (not shown), while ERI-5 strongly binds DCR-1 under the same conditions. Therefore, we speculate that in vivo modification of RRF-3 (although we do not rule out DRH-3) generates a tudor-binding site(s) for ERI-5 and-or EKL-1 (Fig. 5b, c). In light of the role of EKL-1 in paralogous RdRP modules as well as its compensatory function in eri-5 mutants, a similar interaction between EKL-1 and the RdRPs RRF-1, EGO-1 or RRF-3 could occur. Hence, the identification of the modified amino acid(s) within RRF-3, and the other RdRPs, represents an important next research avenue. We note that this hypothesis for the function of the tandem-tudor domains of ERI-5 and EKL-1 is quite distinct from, but not mutually exclusive of, the RITS-like interaction proposed for EKL-1 with chromatin marks in the CSR pathway12.
ONLINE METHODS
C. elegans strains and RNAi
The Bristol strain N2 was used as the standard wild-type (WT) strain. Alleles used are listed by chromosome as follows: LGII: rrf-3(pk1426), rrf-3(mg373), eri-3(tm1361); LGIII: dcr-1(ok247), eri-4(mg375), rde-4(ne337); LGIV: eri-1(mg366), eri-5(tm2528), drh-1(tm1329); LGV: rde-1(ne300). C. elegans were cultured as previously described 35. RNAi was carried out as in 36.
C. elegans Preparations
C. elegans embryonic pellets were prepared as described in 13. Samples were suspended on ice in 3 volumes of lysis buffer (25 mM HEPES-KOH pH7.5, 10 mM KOAc, 2 mM Mg(OAc)2, 100 mM KCl, 1 % (v/v) Triton X-100 and 1 mM DTT) with a 4× concentration of Complete protease inhibitors (Roche) and homogenized in a stainless steel Dounce homogenizer for 30–40 strokes on ice. The resulting slurry was clarified twice at 17,000 × g for 10 min at 4 °C.
Gel Filtration Analysis
Approximately 4 mg of WT C. elegans embryonic extract were loaded onto an equilibrated Superose 6 gel filtration column (GE Healthcare) according to the supplier’s instructions. The protein fractions were collected, precipitated with acetone, and 25 μg were loaded on a 4–15 % (v/v) gel (BioRad) for western analysis.
Multidimensional Protein Identification (MuDPIT)
Samples preparation and analysis were performed as previously described in 13. For the analyses presented, previously described complex array expressing DCR-1::8xHA, and internal ERI-1::3xFLAG fusions were utilized. For ERI-5 samples, affinity-purified polyclonal antibodies directed against the FL ERI-5 (below) were used as an affinity matrix, while for RDE-4 a FLAG fusion expressed from a simple array was utilized.
Antibodies
Polyclonal antibodies against RRF-3, ERI-5 and EKL-1 were raised in rabbits or mice. Anti-RRF-3 antibodies were raised against a synthetic C-terminal peptide IANNVVPNEVRDEFL, conjugated to KLH (Capralogics). Anti-ERI-5 and EKL-1 antibodies were raised against full-length eri-5 and ekl-1 cDNA in both rabbits (Capralogics) and mice in the case of ERI-5, and in mice only for EKL-1.
Immunoprecipitations
DRH-1, RDE-4, DCR-1, ERI-5, RRF-3 and EKL-1 affinity purified antibodies or antiserums, were incubated with 1–5 mg of C. elegans embryo lysate at 4 °C for 1 hr. Immune complexes were precipitated with Protein A/G Sepharose beads (GE Healthcare) or Protein A Dynabeads (Invitrogen) and washed with cold lysis buffer.
Western Blot Analysis
Proteins immobilized on Hybond-C Extra membrane (GE Healthcare) were probed with the following antibodies: anti-DCR-1 (1:4000), anti-RDE-1 (1:200), anti-DRH-1 (1:1000), anti-RDE-4 (1:1000), anti-DRH-3 (1:300), anti-ERI-5 (1:200), anti-ERI-1 (1:1000), anti-RRF-3 (1:2000), anti-RRF-1 (1:1000), anti-EGO-1 (1:1000), anti-EKL-1 (1:1000) and anti-tubulin (1:5000) (abcam) in PBST (137 mM NaCl, 2.7 mM KCl, 10 mM Phosphate (pH 7.4), 0.1 % (v/v) Tween-20) with 5 % (w/v) non fat dried milk. Rabbit and mouse true blot HRP- conjugated secondary antibodies (eBiosciences) were used at 1:1000.
Recombinant Proteins
The cDNA for eri-5, eri-3, ekl-1 and dcr-1 were obtained by RT-PCR, sequenced and cloned into pCal-KC and-or pET vectors. The plasmids were transformed into ArcticExpress (DE3) (Stratagene) or BL-21 (DE3) pLysS (Promega) bacterial cells and expression was induced with IPTG according to the supplier’s instructions.
GST Pull-Down
Approximately 3 μg of purified DCR-1-GST fusions were incubated with Glutathione-Sepharose beads (GE Healthcare) in STE buffer (10 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA). The bead-bound fusions were incubated 1 h with 1 μg of purified ERI-5-CBP or ERI-3-FLAG recombinant proteins in bead binding buffer (50 mM HEPES-KOH pH 7.5, 150 mM KCl, 1 mM Mg(OAc)2, 10 % (v/v) glycerol, 1 % (v/v) Triton X-100). GST pull-downs were washed with bead binding buffer containing 500 mM KCl and analyzed by western blot with anti-CBP (1:1000) (GenScript) or anti-FLAG (1:2500) (Sigma) antibodies. For the simultaneous interaction pull down (Figure 2d), 0, 0.3 and 3 μg of ERI-3-FLAG were pre-bound to the 272–1045 fragment of DCR-1 fused to GST and bound to beads. The beads were washed, and 1 μg of ERI-5-CBP was used to assay for the second interaction, as indicated above.
RNA Preparation and Northern Analysis
Total RNA from C. elegans embryos was prepared using the TRIZOL method (Invitrogen). Small-RNA species were enriched using the mirVana kit (Ambion). 5 to 10 μg of enriched small RNA were resolved per lane on a 15 % (v/v) UREA-TBE gel. Transfer, hybridization and blot analysis were conducted as previously described 37.
Real-Time PCR
For X-cluster small RNAs and 26-G-RNA (C40A11.10) analyses, real-time PCR was performed as previously described38. Primers used for this assay are listed in the Supplementary Methods.
Small RNA Cloning, Deep Sequencing and Data Analyses
Small RNAs isolated fromembryoswereclonedusinga5′-ligation-dependentmethodasdescribed14, except that 5′- and 3′-adapters were ligated without pretreatment of small RNAs. This method favors the cloning of mono-phosphorylated, small RNAs including 26G-RNAs, miRNAsand21U-RNAs.SmallRNAsisolatedfromadultwormswerepretreated sequentially with calf-intestine phosphatase (New England Biolabs) and polynucleotide kinase (New England Biolabs), to convert the 5′ tri-phosphate of 22G-RNAs to mono-phosphate, and then cloned as described1,4. Libraries were sequenced at the University of Massachusetts Deep Sequencing Core (Worcester, MA) using an Illumina Genome AnalyzerII.Data were processed and analyzed using customPERLscripts as described14,20 Data were graphed using DeltaGraph (RedRockSoftware) or Prism (GraphPad).
Supplementary Material
Acknowledgments
We thank Chris Rocheleau for comments on the manuscript, Noriko Uetani for conceptual and artistic contributions to the model, Shohei Mitani and his group at the Department of Physiology, Tokyo Women’s Medical University School of Medicine (Tokyo, Japan) for the generation of the eri-5(tm2528) allele introduced in this manuscript. We thank Ian MacRae and Noah Welker for discussions on the ERIC model. We also thank Allison Haggarty for her assistance in the development of some of the polyclonal antisera. This work was supported by the National Sciences and Engineering Council of Canada RGPIN 341457 (T.F.D.), the Canadian Institute of Health Research MOP 86577 (T.F.D.), the Canada Foundation for Innovation (CFI), and the Fonds de la Rercherche en Santé du Québec, Chercheur-Boursier Salary Award J2, (T.F.D.)
Footnotes
Author contributions
CT performed the experiments presented in Figures 1C, 2C,D, 3A,B, 4A,C & 5A, prepared the figures, and assisted with the preparation of the manuscript. NM performed the experiments presented in Figures 1A,D, the ERI-5 samples in 1E, 2A,B. MF performed the experiments presented in Figures 4B,D, and assisted with the model. James Wohlschlegel performed the MuDPIT analyses of IP samples. DC and JV conducted the experiments in Figure 3C. DC provided scientific advice, and assisted with the redaction of the manuscript. TFD performed the experiments in Figure 1B, wrote the manuscript, and directed the project.
References
- 1.Mello CC, Conte D., Jr Revealing the world of RNA interference. Nature. 2004;431:338–42. doi: 10.1038/nature02872. [DOI] [PubMed] [Google Scholar]
- 2.Tabara H, Yigit E, Siomi H, Mello CC. The double-stranded RNA binding protein RDE-4 interacts in vivo with RDE-1, DCR-1 and a conserved DExH-box helicase to direct RNA interference in C. elegans. Cell. 2002 doi: 10.1016/s0092-8674(02)00793-6. [DOI] [PubMed] [Google Scholar]
- 3.Tabara H, et al. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–32. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- 4.Yigit E, et al. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006;127:747–57. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]
- 5.Pak J, Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007;315:241–4. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- 6.Sijen T, Steiner FA, Thijssen KL, Plasterk RH. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science. 2007;315:244–7. doi: 10.1126/science.1136699. [DOI] [PubMed] [Google Scholar]
- 7.Aoki K, Moriguchi H, Yoshioka T, Okawa K, Tabara H. In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. EMBO J. 2007;26:5007–19. doi: 10.1038/sj.emboj.7601910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guang S, et al. Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature. 2010;465:1097–101. doi: 10.1038/nature09095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–11. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 10.Ketting RF. The many faces of RNAi. Dev Cell. 2011;20:148–61. doi: 10.1016/j.devcel.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 11.Talsky KB, Collins K. Initiation by a eukaryotic RNA-dependent RNA polymerase requires looping of the template end and is influenced by the template-tailing activity of an associated uridyltransferase. J Biol Chem. 2010;285:27614–23. doi: 10.1074/jbc.M110.142273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Claycomb JM, et al. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell. 2009;139:123–34. doi: 10.1016/j.cell.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duchaine TF, et al. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell. 2006;124:343–54. doi: 10.1016/j.cell.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 14.Gu W, et al. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol Cell. 2009;36:231–44. doi: 10.1016/j.molcel.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rocheleau CE, et al. The Caenorhabditis elegans ekl (enhancer of ksr-1 lethality) genes include putative components of a germline small RNA pathway. Genetics. 2008;178:1431–43. doi: 10.1534/genetics.107.084608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conine CC, et al. Argonautes ALG-3 and ALG-4 are required for spermatogenesis-specific 26G-RNAs and thermotolerant sperm in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2010;107:3588–93. doi: 10.1073/pnas.0911685107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pavelec DM, Lachowiec J, Duchaine TF, Smith HE, Kennedy S. Requirement for the ERI/DICER complex in endogenous RNA interference and sperm development in Caenorhabditis elegans. Genetics. 2009;183:1283–95. doi: 10.1534/genetics.109.108134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Batista PJ, et al. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol Cell. 2008;31:67–78. doi: 10.1016/j.molcel.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruby JG, et al. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 20.Vasale JJ, et al. Sequential rounds of RNA-dependent RNA transcription drive endogenous small-RNA biogenesis in the ERGO-1/Argonaute pathway. Proc Natl Acad Sci U S A. 2010;107:3582–7. doi: 10.1073/pnas.0911908107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gent JI, et al. Distinct phases of siRNA synthesis in an endogenous RNAi pathway in C. elegans soma. Mol Cell. 2010;37:679–89. doi: 10.1016/j.molcel.2010.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Welker NC, et al. Dicer’s helicase domain is required for accumulation of some, but not all, C. elegans endogenous siRNAs. Rna. 2010;16:893–903. doi: 10.1261/rna.2122010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wolters DA, Washburn MP, Yates JR., 3rd An automated multidimensional protein identification technology for shotgun proteomics. Anal Chem. 2001;73:5683–90. doi: 10.1021/ac010617e. [DOI] [PubMed] [Google Scholar]
- 24.Gu SG, et al. Distinct ribonucleoprotein reservoirs for microRNA and siRNA populations in C. elegans. Rna. 2007;13:1492–504. doi: 10.1261/rna.581907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Q, et al. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301:1921–5. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- 26.Han T, et al. 26G endo-siRNAs regulate spermatogenic and zygotic gene expression in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:18674–9. doi: 10.1073/pnas.0906378106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Colmenares SU, Buker SM, Buhler M, Dlakic M, Moazed D. Coupling of double-stranded RNA synthesis and siRNA generation in fission yeast RNAi. Mol Cell. 2007;27:449–61. doi: 10.1016/j.molcel.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 28.Lee SR, Collins K. Physical and functional coupling of RNA-dependent RNA polymerase and Dicer in the biogenesis of endogenous siRNAs. Nat Struct Mol Biol. 2007;14:604–10. doi: 10.1038/nsmb1262. [DOI] [PubMed] [Google Scholar]
- 29.Welker NC, et al. Dicer’s helicase domain discriminates dsRNA termini to promote an altered reaction mode. Mol Cell. 2011;41:589–99. doi: 10.1016/j.molcel.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim JK, Cho JH. Change of external auditory canal pH in acute otitis externa. The Annals of otology, rhinology, and laryngology. 2009;118:769–72. [PubMed] [Google Scholar]
- 31.Cho KJ, et al. Structural features of cephalosporin acylase reveal the basis of autocatalytic activation. Biochemical and biophysical research communications. 2009;390:342–8. doi: 10.1016/j.bbrc.2009.09.134. [DOI] [PubMed] [Google Scholar]
- 32.Kim EH, Park SH, Kim JK. Methyl jasmonate triggers loss of grain yield under drought stress. Plant signaling & behavior. 2009;4:348–9. doi: 10.4161/psb.4.4.8199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomson T, Lasko P. Drosophila tudor is essential for polar granule assembly and pole cell specification, but not for posterior patterning. Genesis. 2004;40:164–70. doi: 10.1002/gene.20079. [DOI] [PubMed] [Google Scholar]
- 34.Kim JK, et al. Compact all-fiber Bessel beam generator based on hollow optical fiber combined with a hybrid polymer fiber lens. Optics letters. 2009;34:2973–5. doi: 10.1364/OL.34.002973. [DOI] [PubMed] [Google Scholar]
- 35.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–12. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- 37.Wu E, et al. Pervasive and cooperative deadenylation of 3′UTRs by embryonic microRNA families. Mol Cell. 2010;40:558–70. doi: 10.1016/j.molcel.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raymond CK, Roberts BS, Garrett-Engele P, Lim LP, Johnson JM. Simple, quantitative primer-extension PCR assay for direct monitoring of microRNAs and short-interfering RNAs. RNA. 2005;11:1737–44. doi: 10.1261/rna.2148705. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.