Abstract
Using a chemically defined medium without l-alanine, Lactobacillus johnsonii was demonstrated to be strictly auxotrophic for that amino acid. A comparative genetic analysis showed that all known genes involved in l-alanine biosynthesis are absent from the genome of L. johnsonii. This auxotrophy was complemented by heterologous expression of the Bacillus subtilis l-alanine dehydrogenase.
Lactic acid bacteria (LAB) play important roles in food conservation because of their fermentative ability and contribute to the equilibrium of the gastrointestinal tract (GIT) microbiota. Lactobacillus johnsonii was shown to survive its passage through the GIT and to adhere to intestinal cells in vitro (25, 29). Such interactions of bacteria with the GIT may contribute to its protection against pathogenic microorganisms (28). A close association between the bacteria and their environment undoubtedly results in adaptation of their metabolic capacities. The presence of amino acids in the environment, for example, can lead to a progressive inactivation of their biosynthetic pathways (13, 14). Several genes and gene clusters in LAB have been implicated in amino acid biosynthesis (9). These include genes involved in the biosynthesis of tryptophan, histidine, and branched-chain amino acids in Lactococcus lactis (3, 11, 17) and those implicated in glutamine biosynthesis in Lactobacillus delbrueckii subsp. bulgaricus (21). Little is known, however, about l-alanine biosynthesis in LAB, as most studies have been done on Escherichia coli and Salmonella enterica serovar Typhimurium (5, 34). No strict auxotrophy for this amino acid has been reported, probably because l-alanine can be synthesized by many different metabolic pathways. Only mutants exhibiting a leaky requirement for alanine have been isolated for E. coli and S. enterica serovar Typhimurium (33).
In this work, using a chemically defined medium (CDM) without l-alanine, L. johnsonii was shown to be strictly auxotrophic for that amino acid. A comparative analysis of genes encoding enzymes that are known to be involved in l-alanine biosynthesis revealed a total absence of these genes in L. johnsonii. The l-alanine auxotrophy was complemented by heterologous expression of the l-alanine dehydrogenase gene (alaD) isolated from Bacillus subtilis (natto), a gram-positive microorganism used in the preparation of some foods.
l-Alanine auxotrophy of L. johnsonii.
L. johnsonii from the Nestlé Culture Collection (NCC533) was grown anaerobically in MRS broth (Difco, Detroit, Mich.) at 37°C for maintenance. Growth factor requirements were defined by growing L. johnsonii in a CDM (15). Optimized, this medium contains 42 components (Table 1), including amino acids, inosine, vitamins, glucose, and salts. The pH of the CDM was adjusted to 6.2. This CDM allowed rapid growth of the microorganism, with a maximum specific growth rate (μmax) of 1.6 h−1. For batch cultures, optical densities at 600 nm (OD600 values) of 2 to 3 were found.
TABLE 1.
Composition of the CDM for L. johnsoniia
| Constituent(s) | Final concn. |
|---|---|
| Sugar, salts, bases, and amino acidsb | |
| Glucose | 20 |
| Potassium acetate | 10 |
| KH2PO4 | 3.1 |
| K2HPO4 | 1.5 |
| Ammonium citrate | 2 |
| Calcium lactate, Tween 80 | 1 |
| MgSO4 · 7H2O | 0.5 |
| NaCl, MnSO4 · H2O, FeSO4 · 7H2O | 0.02 |
| Inosine | 10 |
| l-Glutamate | 0.6 |
| Glycine, l-histidine | 0.4 |
| dl-Alanine, l-lysine, l-phenylalanine, l-proline, l-serine, l-cysteine, l-arginine, l-aspartate, l-asparagine, l-leucine, l-tryptophan, l-valine | 0.2 |
| l-Threonine, dl-aminobutyrate, l-isoleucine, l-methionine, l-tyrosine | 0.1 |
| Vitaminsc | |
| Biotin | 0.05 |
| Cyanocobalamine | 0.01 |
| Folate, p-aminobenzoate | 0.1 |
| Nicotinate, calcium pentothenate, riboflavin, pyridoxine, | |
| myoinositol, l-ascorbate | 0.5 |
Final pH of CDM is 6.2.
Concentrations are in grams per liter.
Concentrations are in milligrams per liter.
L. johnsonii was grown in the CDM in the presence and in the absence of l-alanine, and growth was indicated by increased OD readings. While growth in a complete CDM was comparable to that in MRS broth, the absence of l-alanine allowed only limited growth. This result indicates that L. johnsonii is dependent on l-alanine for growth and is strictly auxotrophic for this amino acid. When two potential precursors for l-alanine biosynthesis, l-valine and l-glutamate (see below), were also removed from the CDM, no growth was observed, indicating that this organism is auxotrophic for these two amino acids as well.
Alanine biosynthesis pathways.
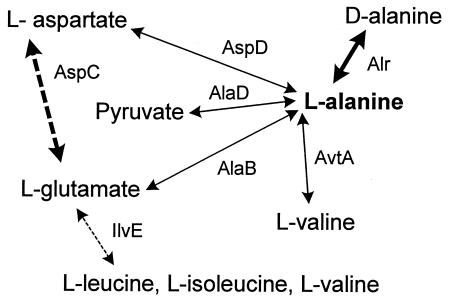
A literature survey of alanine metabolism in microorganisms identified three major biosynthetic pathways for this amino acid: reductive amination of pyruvate by alanine dehydrogenase (AlaD), decarboxylation of l-aspartate by aspartate-4-decarboxylase (AspD), and transamination from other amino acids and amines to pyruvate by aminotransferases (ATs) such as glutamate-pyruvate AT (AlaB) and valine-pyruvate AT (AvtA) (Fig. 1). A branched-chain amino acid AT (IlvE) seems to be involved in alanine metabolism under certain conditions (7, 8). In some cases, the tyrosine-repressible AT (TyrB) shows an l-alanine AT activity (33). With the exception of AlaD, all of the enzymes involved in l-alanine biosynthesis belong to the large family of pyridoxal 5′-phosphate (PLP)-dependent enzymes.
FIG. 1.
Known metabolic pathways involved in the synthesis of alanine and found in L. johnsonii (bold arrows). For enzyme abbreviations, see Table 3.
l-Alanine was shown to be essential for L. johnsonii growth. The origin of this auxotrophy was determined using the complete genome sequence of L. johnsonii. In the first step, the GenBank-deposited amino acid sequences of all enzymes known to be involved in l-alanine biosynthesis (Fig. 1) were used as a query for BLASTP analysis (1) against all predicted L. johnsonii proteins (Table 2). A similarity search performed using the default BLAST parameters revealed the absence of significant similarity (high P values) for the enzymes directly involved in l-alanine biosynthesis: AlaD, AspD, AlaB, and AvtA. However, a significant similarity (low P value) was obtained for l-alanine racemase (Alr). The LJ0272 protein shares 46% amino acid identity with the alanine racemase of Lactobacillus reuteri (32) (Table 2), an enzyme that catalyzes the conversion of l-alanine to d-alanine. The d-enantiomer is mainly used for cross-linking of adjacent peptidoglycan chains in the bacterial cell wall.
TABLE 2.
Sequence similarities to l-alanine biosynthesis-related enzymes encoded in the L. johnsonii genome
| Enzyme [name/EC no./Pfam motif(s)] | Organism (SwissProt ID or NCBI identification no.) | Best BLAST hits in L. johnsonii genome (P value, % identity) | L. johnsonii proteina |
|---|---|---|---|
| l-Alanine dehydrogenase (AlaD/EC 1.4.1.1/AlaDh_PNT) | Mycobacterium tuberculosis (DHA_MYCTU) | NSb | |
| Listeria innocua (NP_465104) | NS | ||
| Aspartate 4-decarboxylase (AspD/EC 4.1.1.12/aminotran_1_2) | Alcaligenes faecalis subsp. faecalis (GI 14279103) | 2 × 10−10 | LJ1390 |
| Glutamate-pyruvate AT (AlaB/EC:2.6.1.2/aminotran_1_2) | Saccharomyces cerevisiae (GI 6320317) | 2 × 10−10 1 × 10−8 | LJ1390 LJ0915 |
| Valine-pyruvate AT (AvtA/EC:2.6.1.66/aminotran_1_2) | Yersinia pestis avtA (GI 15981974) | 6 × 10−5 2 × 10−4 | LJ1390 LJ0915 |
| Alanine racemase (Alr/EC:5.1.1./Ala_racemase) | B. subtilis (ALR1_BACSU) | 5 × 10−72, 40 | LJ0272 |
| L. reuteri (ALR1_LACRE) | 3 × 10−90, 46 | LJ0272 | |
| Branched-chain amino acid AT (IIvE/EC:2.6.1.42/aminotran_4) | E. coli (IIVE_ECOLI) | NS | |
| Lactoccocus lactis (GI 12724265) | NS | ||
| Tyrosine AT (TyrB/EC:2.6.1.5/aminotran_1_2) | E. coli (GI 16131880) | NS | |
| B. subtilis (HIS8_BACSU) | NS | ||
| Aspartate AT (AspC/EC:2.6.1.1/aminotran_1_2, Cys_Met_Meta_PP, DegT_DnrJ_EryC1) | Streptococcus pyogenes (GI 15674718) Lactococcus lactis subsp. lactis aspC (GI 12723010) E. coli (YFBQ_ECOLI) | 1 × 10−52, 34 8 × 10−9, 19 3 × 10−21 25 3 × 10−10, 23 6× 10−20, 26 1× 10−8, 21 | LJ1390 LJ0915 LJ1390 LJ0915 LJ1390 LJ0915 |
Among the enzymes indirectly involved in l-alanine biosynthesis, IlvE and TyrB showed no significant similarity to L. johnsonii proteins; however, two proteins, LJ0915 and LJ1390, were found to exhibit 23 and 25% amino acid identity, respectively, with AspC of Lactococcus lactis (Table 2). So far, additional known ATs potentially implicated in l-alanine metabolism (EC 2.6.1.15, 2.6.1.21, 2.6.1.44, 2.6.1.51, 2.6.1.71, and 2.6.1.22) have not been found to be encoded in prokaryotic genomes.
To further refine the searches, a TBLASTN similarity search was performed to compare the query enzyme sequences (Table 2) with the genomic L. johnsonii DNA sequence translated in the six frames. This search confirmed the absence of all four enzymes directly involved in l-alanine biosynthesis. Then, protein motif searches on protein profiles including the Pfam (4) alanine racemase motif, the AT class I, II, IV, and V motifs, and the DegT/DnrJ/EryC1/StrS aminotransferase family motifs (Table 3) were performed using the DNA search algorithm Wise2 (http://www.ebi.ac.uk/Wise2/). Using the alanine racemase motif, one hit within the L. johnsonii genome matched the previously identified putative racemase LJ0272. Using AT class I and II motifs, two hits matched the two previously identified PLP-dependent ATs, LJ0915 and LJ1390 (Table 2). LJ0915 showed 50% sequence identity to a recently described L. delbrueckii cystathionase (2) that catalyzes the cleavage of l-cystathionine to homocysteine, pyruvate, and ammonia via an α/β-elimination reaction (10). LJ1390 showed 43% sequence identity to the Lactococcus lactis aromatic amino acid-specific AT (27), which plays an important role in the biosynthesis of amino acids and in the conversion of amino acids to aromatic compounds. Both enzymes are putative PLP-dependent ATs, but neither is involved in l-alanine biosynthesis. In addition, the AT class V motif allowed identification of three more PLP-dependent ATs: LJ0953, LJ0984, and LJ1140 (Table 3). The first two ATs are putative cysteine desulfurases (NifS-like proteins [24]), a group of enzymes mediating chemical reactions common to diverse metabolic pathways and exhibiting sequence homology to PLP-dependent ATs. This type of enzyme is not involved in l-alanine metabolism. The last AT may be involved in the production of l-alanine from selenocysteine. Members of the large family of PLP-dependent enzymes exhibit several different enzymatic activities, among them the aminotransferase activity that is found in a wide range of enzymes having the same function of amino group transfer but with many different specificities for the donors and acceptors. These specificities were evolutionarily optimized to accelerate the required reactions, so that the turnover of all other alternative reactions was several orders of magnitude lower (22). Many PLP-dependent ATs are found in other bacteria; for example, Listeria innocua has 16 and Lactococcus lactis has 14. The small number of ATs found in L. johnsonii may explain why this bacterium cannot synthesize l-alanine.
TABLE 3.
L. johnsonii proteins presenting Pfam motifs of enzymes potentially involved in the metabolism of l-alanine and with best hit in GenBank
| L. johnsonii protein | Description | Pfam motif(s)a of L. johnsonii protein | Best hit in GenBank (NCBI identification no.) | P value and % identity |
|---|---|---|---|---|
| LJ0915 | Cystathionase PLP-dependent AT | aminotran_1_2 | L. delbrueckii cystathionase (GI 20385889) | 10 × 10−100, 50 |
| LJ1390 | PLP-dependent AT | aminotran_1_2, OKR_DC_1, DegT_DnrJ_EryC1 | Lactococcus lactis aromatic-amino acid specific AT (GI 15672039) | 6 × 10−79, 43 |
| LJ0953 | Cysteine desulfurase (NifS-like protein) | aminotran_5, pyridoxal_deC | L. delbrueckii nifS-like gene (GI 43985) | 10−131, 69 |
| LJ0984 | Cysteine desulfurase (NifS-like protein) | aminotran_5 | B. subtilis nifS-like gene (GI 16080011) | 10−78, 46 |
| LJ1140 | Selenocysteine lyase (CsdB; EC 4.4.1.16) | aminotran1_2 and_5, DegT_DnrJ_EryC1, pyridoxal_deC | Streptococcus pyogenes putative AT (GI 19745416) | 10−129, 57 |
Pfam motifs: AlaDh_PNT, PF01262, alanine dehydrogenase/pyridine nucleotide transhydrogenase; aminotran_1_2, PF00155; aminotran_4, PF01063; aminotran_5, PF00266, aminotransferase class I, II, IV, and V; Ala_racemase, PF00842, alanine racemase; DegT_DnrJ_EryC1, PF01041; DegT/DnrJ/EryC1/StrS, PF01041, aminotransferase family; OKR_DC_1, PF01276, Orn/Lys/Arg decarboxylase; pyridoxal_deC, PF00282, PLP-dependent decarboxylase; Cys_Met_Meta_PP, PF01053, Cys/Met metabolism PLP-dependent enzyme.
Expression of the B. subtilis alanine dehydrogenase in L. johnsonii.
In order to confirm that l-alanine auxotrophy in L. johnsonii is due to the absence of an enzyme involved in l-alanine biosynthesis, alanine dehydrogenase (AlaD) was expressed heterologously. AlaD is responsible for the biosynthesis of l-alanine from pyruvate and ammonia (31). This enzyme was shown to convert Lactococcus lactis from a homolactic acid fermenter to a homoalanine fermenter through heterologous expression of AlaD after inactivation of lactate dehydrogenase (20). The alaD gene was isolated from B. subtilis (natto) (NCC156) by DNA amplification using a forward primer, A722 (5′ GATGTTTAGACCATGATCATAGGGGTTCC), and a reverse primer containing a BamHI site (in italics), A723 (5′ TTGGGATCCCGCCATATTGCTGAACAGCC) (Microsynth, Balgach, Switzerland). The ldh promoter of Streptococcus thermophilus (NCC9019) was amplified using a forward primer containing an EcoRI site (in italics), A720 (5′ TGTGAATTCTAGATAGATGAGTC), and a reverse primer, A721 (5′ ACCCCTATGATCATGGTCTAAACATCTCC). Both amplification products were fused by DNA amplification using primers A720 and A723. The final product was cleaved with EcoRI and BamHI and cloned into pNZ124 (26). The resulting plasmid, pHK15, was transformed into Lactococcus lactis MG1363 (16) by electroporation (19). Plasmid DNA was purified using a minipreparation kit (Gibco BRL) and transformed into L. johnsonii. An 8-h starter culture of L. johnsonii was diluted 1/100 in MRS broth containing 0.5 M sucrose and grown overnight at 37°C under anaerobic conditions. After being diluted again under the same conditions, the culture was grown to an OD600 of 0.6. Cells were washed twice in an ice-cold solution comprising 1 M sucrose and 2.5 mM CaCl2 and resuspended at 1/50 of the culture volume in the same solution containing 10% glycerol for storage at −80°C. Cell suspension (40 μl) was mixed with 10 to 100 ng of plasmid DNA, electroporated (2.0 kV, 25 μF, 200 W) using a GenePulser apparatus (Bio-Rad Laboratories, Hercules, Calif.), mixed with 1 ml of MRS broth containing 20 mM MgCl2 and 2 mM CaCl2, and incubated for 2 to 3 h at 37°C before being plated on MRS agar containing 5 μg of chloramphenicol/ml. Plates were incubated at 37°C for 2 days under anaerobic conditions. The observed transformation efficiency was 104 transformants/μg of pHK15 DNA.
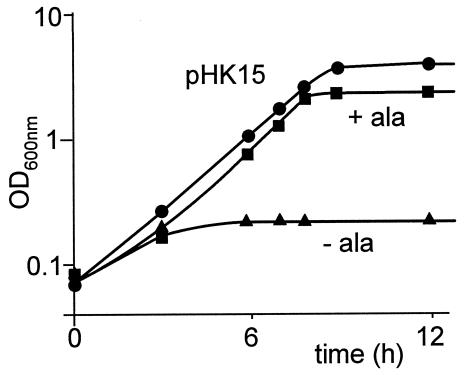
The recombinant L. johnsonii harboring the pHK15 plasmid was grown at 37°C in CDM containing chloramphenicol (5 μg/ml) but no l-alanine. This medium allowed rapid development of the recombinant microorganism (Fig. 2), with a maximum specific growth rate of 1.4 h−1, comparable to that of the wild-type L. johnsonii (1.6 h−1) grown in a complete CDM. In the absence of l-alanine, only residual growth was observed for the wild-type L. johnsonii (Fig. 2). This residual growth could be due to internal pools of l-alanine. The introduction of pHK15 into L. johnsonii conferred l-alanine prototrophy to the microorganism, indicating a successful heterologous expression of the B. subtilis alaD gene and an efficient activity of the alanine dehydrogenase.
FIG. 2.
Growth of wild-type L. johnsonii in CMD in the presence (+ala) and absence (-ala) of alanine and after transformation with pHK15 expressing B. subtilis AlaD in the absence of alanine (pHK15). Growth measurements were repeated three times with no significant variations.
Conclusion.
L. johnsonii is the first bacterium found to be strictly auxotrophic for l-alanine. This auxotrophy was complemented by overexpression of the B. subtilis alanine dehydrogenase gene (alaD). Amino acid auxotrophy has been reported to occur via inactivation of genes involved in their synthesis because of point mutations (9). In L. johnsonii, genes involved in l-alanine biosynthesis not only were inactivated by point mutations but were apparently deleted during evolution in an environment rich in l-alanine. It has also been shown that bacterial genomes are subjected to a high genomic deletion rate that has resulted in reduction of their genome sizes and deletion of unnecessary genes (23). Once genes essential for the synthesis of a particular amino acid have been deleted, the bacterium becomes dependent on the supply of the amino acid. Specific Lactobacillus strains are known to be residents of the nutrient-rich small intestine, where they may contribute to protection against pathogens and toxins, known as a probiotic property (18, 28). As a consequence, the deletion of genes encoding enzymes essential for amino acids synthesis, like in L. johnsonii, has led to a commensal relationship between the bacterium and the human host. More extensive gene deletions are found in Buchnera sp., which has established a symbiotic relationship with aphids living inside specialized cells called bacteriocytes; the size of its genome was reduced over time to 0.64 MB (30). Such a tendency for genome reduction through adaptation reminds us that a bacterium and a primitive eukaryote may have once established a symbiosis based on respiration for the evolution of mitochondria (12). For most lactobacilli that are adapted to grow in nutrient-rich environments, genome sizes are still around 2 MB (1.99 MB for L. johnsonii). In contrast, a larger coding capacity, such as that of E. coli, with a genome size of 4.6 MB, indicates the maintenance of higher metabolic diversity and, thus, the potential to overcome nutrient-poor environments (6). This kind of ecogenomic approach, involving the analysis of complete bacterial genomes, will eventually lead to identification of the complete metabolic potential, which will help to define the natural ecological environment of microorganisms.
Acknowledgments
This work was supported by a grant from EU Biotech II, contract BIO4CT960439 (CH-OFES 96.0018).
We gratefully acknowledge Maria Karmirantzou and David Pridmore for their help with the L. johnsonii genome, Anne Bauché for help with the CDM, Harald Bruessow and Annick Mercenier for critical reading of the manuscript, and Elizabeth Prior for reviewing the manuscript.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Aubel, D., J. E. Germond, C. Gilbert, and D. Atlan. 2002. Isolation of the patC gene encoding the cystathionine beta-lyase of Lactobacillus delbrueckii subsp. bulgaricus and molecular analysis of inter-strain variability in enzyme biosynthesis. Microbiology 148:2029-2036. [DOI] [PubMed] [Google Scholar]
- 3.Bardowski, J., S. D. Ehrlich, and A. Chopin. 1992. Tryptophan biosynthesis genes in Lactococcus lactis subsp. lactis. J. Bacteriol. 174:6563-6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bateman, A., E. Birney, L. Cerruti, R. Durbin, L. Etwiller, S. R. Eddy, S. Griffiths-Jones, K. L. Howe, M. Marshall, and E. L. Sonnhammer. 2002. The Pfam protein families database. Nucleic Acids Res. 30:276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berg, C. M., W. A. Whalen, and L. B. Archambault. 1983. Role of alanine-valine transaminase in Salmonella typhimurium and analysis of an avtA::Tn5 mutant. J. Bacteriol. 155:1009-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 7.Blazey, D. L., and R. O. Burns. 1980. Gene ilvY of Salmonella typhimurium. J. Bacteriol. 142:1015-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blazey, D. L., R. Kim, and R. O. Burns. 1981. Molecular cloning and expression of the ilvGEDAY genes from Salmonella typhimurium. J. Bacteriol. 147:452-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chopin, A. 1993. Organization and regulation of genes for amino acid biosynthesis in lactic acid bacteria. FEMS Microbiol. Rev. 12:21-37. [DOI] [PubMed] [Google Scholar]
- 10.Clausen, T., B. Laber, and A. Messerschmidt. 1997. Mode of action of cystathionine beta-lyase. Biol. Chem. 378:321-326. [PubMed] [Google Scholar]
- 11.Delorme, C., S. D. Ehrlich, and P. Renault. 1992. Histidine biosynthesis genes in Lactococcus lactis subsp. lactis. J. Bacteriol. 174:6571-6579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doolittle, W. F. 1998. A paradigm gets shifty. Nature 392:15-16. [DOI] [PubMed] [Google Scholar]
- 13.Driessen, A. J. 1989. Secondary transport of amino acids by membrane vesicles derived from lactic acid bacteria. Antonie Leeuwenhoek 56:139-160. [DOI] [PubMed] [Google Scholar]
- 14.Driessen, A. J., J. Kodde, S. de Jong, and W. N. Konings. 1987. Neutral amino acid transport by membrane vesicles of Streptococcus cremoris is subject to regulation by internal pH. J. Bacteriol. 169:2748-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elli, M., R. Zink, A. Rytz, R. Reniero, and L. Morelli. 2000. Iron requirement of Lactobacillus spp. in completely chemically defined growth media. J. Appl. Microbiol. 88:695-703. [DOI] [PubMed] [Google Scholar]
- 16.Gasson, M. J. 1983. Genetic transfer systems in lactic acid bacteria. Antonie Leeuwenhoek 49:275-282. [DOI] [PubMed] [Google Scholar]
- 17.Godon, J. J., M. C. Chopin, and S. D. Ehrlich. 1992. Branched-chain amino acid biosynthesis genes in Lactococcus lactis subsp. lactis. J. Bacteriol. 174:6580-6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hammes, W. P., and R. F. Vogel. 1995. The genus Lactobacillus, p. 19-54. In B. J. B. Wood and W. H. Holzappel (ed.), The genera of lactic acid bacteria. Blackie Academic and Professional, London, England.
- 19.Holo, H., and I. F. Nes. 1989. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl. Environ. Microbiol. 55:3119-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hols, P., M. Kleerebezem, A. N. Schanck, T. Ferain, J. Hugenholtz, J. Delcour, and W. M. de Vos. 1999. Conversion of Lactococcus lactis from homolactic to homoalanine fermentation through metabolic engineering. Nat. Biotechnol. 17:588-592. [DOI] [PubMed] [Google Scholar]
- 21.Ishino, Y., P. Morgenthaler, H. Hottinger, and D. Soll. 1992. Organization and nucleotide sequence of the glutamine synthetase (glnA) gene from Lactobacillus delbrueckii subsp. bulgaricus. Appl. Environ. Microbiol. 58:3165-3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.John, R. A. 1995. Pyridoxal phosphate-dependent enzymes. Biochim. Biophys. Acta 1248:81-96. [DOI] [PubMed] [Google Scholar]
- 23.Lawrence, J. G., R. W. Hendrix, and S. Casjens. 2001. Where are the pseudogenes in bacterial genomes? Trends Microbiol. 9:535-540. [DOI] [PubMed] [Google Scholar]
- 24.Leong-Morgenthaler, P., S. G. Oliver, H. Hottinger, and D. Soll. 1994. A Lactobacillus nifS-like gene suppresses an Escherichia coli transaminase B mutation. Biochimie 76:45-49. [DOI] [PubMed] [Google Scholar]
- 25.Link-Amster, H., F. Rochat, K. Y. Saudan, O. Mignot, and J. M. Aeschlimann. 1994. Modulation of a specific humoral immune response and changes in intestinal flora mediated through fermented milk intake. FEMS Immunol. Med. Microbiol. 10:55-63. [DOI] [PubMed] [Google Scholar]
- 26.Platteeuw, C., G. Simons, and W. M. de Vos. 1994. Use of the Escherichia coli β-glucuronidase (gusA) gene as a reporter gene for analyzing promoters in lactic acid bacteria. Appl. Environ. Microbiol. 60:587-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rijnen, L., S. Bonneau, and M. Yvon. 1999. Genetic characterization of the major lactococcal aromatic aminotransferase and its involvement in conversion of amino acids to aroma compounds. Appl. Environ. Microbiol. 65:4873-4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salminen, S., E. Isolauri, and E. Salminen. 1996. Probiotics and stabilisation of the gut mucosal barrier. Asia Pac. J. Clin. Nutr. 5:53-56. [PubMed] [Google Scholar]
- 29.Schiffrin, E. J., D. Brassart, A. L. Servin, F. Rochat, and A. Donnet-Hughes. 1997. Immune modulation of blood leukocytes in humans by lactic acid bacteria: criteria for strain selection. Am. J. Clin. Nutr. 66:515S-520S. [DOI] [PubMed] [Google Scholar]
- 30.Shigenobu, S., H. Watanabe, M. Hattori, Y. Sakaki, and H. Ishikawa. 2000. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407:81-86. [DOI] [PubMed] [Google Scholar]
- 31.Siranosian, K. J., K. Ireton, and A. D. Grossman. 1993. Alanine dehydrogenase (ald) is required for normal sporulation in Bacillus subtilis. J. Bacteriol. 175:6789-6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson, A., H. Griffin, and M. J. Gasson. 2002. Characterization of an alanine racemase gene from Lactobacillus reuteri. Curr. Microbiol. 44:246-250. [DOI] [PubMed] [Google Scholar]
- 33.Wang, M. D., L. Buckley, and C. M. Berg. 1987. Cloning of genes that suppress an Escherichia coli K-12 alanine auxotroph when present in multicopy plasmids. J. Bacteriol. 169:5610-5614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whalen, W. A., and C. M. Berg. 1984. Gratuitous repression of avtA in Escherichia coli and Salmonella typhimurium. J. Bacteriol. 158:571-574. [DOI] [PMC free article] [PubMed] [Google Scholar]