Abstract
Multitrophic interactions mediate the ability of fungal pathogens to cause plant disease and the ability of bacterial antagonists to suppress disease. Antibiotic production by antagonists, which contributes to disease suppression, is known to be modulated by abiotic and host plant environmental conditions. Here, we demonstrate that a pathogen metabolite functions as a negative signal for bacterial antibiotic biosynthesis, which can determine the relative importance of biological control mechanisms available to antagonists and which may also influence fungus-bacterium ecological interactions. We found that production of the polyketide antibiotic 2,4-diacetylphloroglucinol (DAPG) was the primary biocontrol mechanism of Pseudomonas fluorescens strain Q2-87 against Fusarium oxysporum f. sp. radicis-lycopersici on the tomato as determined with mutational analysis. In contrast, DAPG was not important for the less-disease-suppressive strain CHA0. This was explained by differential sensitivity of the bacteria to fusaric acid, a pathogen phyto- and mycotoxin that specifically blocked DAPG biosynthesis in strain CHA0 but not in strain Q2-87. In CHA0, hydrogen cyanide, a biocide not repressed by fusaric acid, played a more important role in disease suppression.
Molecular and biochemical analysis has demonstrated that, for many beneficial bacteria applied in agriculture as biological control agents to suppress plant diseases, production of various antimicrobial compounds is the primary mechanism of action (21, 26, 60). The polyketide antibiotic 2,4-diacetylphloroglucinol (DAPG) has received particular attention because it plays a key role in the ability of introduced Pseudomonas fluorescens strains to suppress a broad spectrum of crop diseases (29). Indigenous DAPG-producing populations have been identified as the driving force behind development of natural disease suppressiveness in certain soils under long-term monoculture (47). Conservation of phl genes for biosynthesis of DAPG among ecologically and geographically diverse antagonistic pseudomonads further supports the global importance of DAPG production in biocontrol (31, 66).
All DAPG producers so far described carry hcn genes for biosynthesis of the broad-spectrum biocide hydrogen cyanide (HCN), indicating an evolutionary linkage of the two metabolites (31, 69). Production and pathway-specific regulation of each compound, however, appear to be independent of those of the other (5, 6). Biosynthesis of antimicrobial compounds in Pseudomonas biocontrol strains is closely regulated by molecules produced by the organism itself (44, 52) and by external environmental factors, including nutritional components, soil chemical and physical properties (17, 19, 55, 56), host plant genotype (38, 43, 57), and nonpathogenic soil bacteria (45). Because these factors can determine the ability of particular strains to suppress disease, identifying them would facilitate the targeted application of strains into environments that are more favorable for effective and consistent biocontrol activity.
In biocontrol research, the focus has almost exclusively been placed on understanding how bacterial antagonists impact fungal pathogen survival and disease-causing activity. Despite the very close interaction between these bacteria and fungi, surprisingly little attention has been given to the potential impact of pathogens on biocontrol agents and their disease-suppressive activity. The best evidence to date supporting such an impact is (i) that root pathogens influence plant colonization by antagonistic pseudomonads (4, 40) and (ii) that tolerance by pathogens of particular antimicrobial metabolites diminishes the efficacy of biocontrol strains producing these compounds (12, 41). Here we report an example of signaling between a pathogen and a biocontrol agent involving genes important in disease suppression. Using promoterless lacZ fusions to the phlA and hcnA biosynthetic genes, we found that the phyto- and mycotoxin fusaric acid (32), produced by the tomato crown and root rot pathogen, Fusarium oxysporum Schlechtend.:Fr. f. sp. radicis-lycopersici Jarvis and Shoemaker, blocked biosynthesis of DAPG but not HCN in the biocontrol agent P. fluorescens CHA0 but had no major effect on DAPG production in Q2-87, another biocontrol strain of P. fluorescens. Mutational analysis confirmed that DAPG production was key to the biocontrol activity of strain Q2-87 but that it was not important for the fusaric acid-sensitive strain CHA0, which relied more on HCN production for biocontrol of this disease.
Microorganisms.
F. oxysporum f. sp. radicis-lycopersici strain 22 was isolated from tomato plants with crown and root rot disease and was obtained from C. Alabouvette, Institut National de la Recherche Agronomique, Dijon, France. The pathogen was routinely cultured on 2% malt extract (ME) agar (Oxoid Ltd., Basingstoke, Hampshire, United Kingdom) at 24°C and was stored as mycelia and conidia in ME broth with 40% glycerol at −80°C. P. fluorescens strains and plasmids used in this study are described in Table 1. P. fluorescens was routinely grown on King's B medium (KB) (34) at 27°C and was stored in KB broth with 40% glycerol at −80°C. Escherichia coli was grown in Luria-Bertani broth at 37°C (50). Plasmids were mobilized from E. coli to P. fluorescens by electroporation or by triparental matings as previously described (52).
TABLE 1.
P. fluorescens strains and plasmids used in this study
| Strain or plasmid | Genotype and relevant characteristicsa | Reference |
|---|---|---|
| Strains | ||
| CHA0 | Wild type from Morens, Switzerland; DAPG+ HCN+ | 63 |
| CHA5 | hcnB::Ω-Hg insertion derivative of CHA0; Hgr DAPG+ HCN− | 64 |
| CHA207 | Chromosomal hcnA′-′lacZ fusion of CHA0; DAPG+ HCN− | 7 |
| CHA630 | phlH::Tn5 single-chromosomal-insertion derivative of CHA0; Kmr DAPG− HCN+ | 52 |
| CHA631 | ΔphlA in-frame-chromosomal-deletion derivative of CHA0; DAPG− HCN+ | 52 |
| Q2-87 | Wild type from Washington State; DAPG+ HCN− | 62 |
| Q2-87::Tn5-1 | phlD::Tn5-1 chromosomal-insertion derivative of Q2-87; Kmr DAPG− HCN− | 3 |
| Plasmids | ||
| pME3013 | pVK100 with an 8-kb genomic fragment of CHA0 containing functional hcnABC genes conferring HCN biosynthesis; Tcr | 64 |
| pME6259 | pACYC177-pVS1 carrying a lacZ translational fusion to the phlA of CHA0; Tcr | 52 |
| pME6261 | pACYC177-pVS1 with a 2-kb fragment of CHA0 containing a functional phlA of CHA0 conferring DAPG biosynthesis; Tcr | 52 |
| pMON5118 | pCP13/B with a 35-kb genomic fragment of Q2-87 containing the DAPG biosynthetic locus conferring DAPG biosynthesis; Tcr | 62 |
+, positive for production; −, no production; Hgr, mecury resistant (20 μg ml−1); Kmr, kanamycin resistant (25 μg ml−1); Tcr, tetracycline resistant (125 μg ml−1 for P. fluorescens).
Effect of purified DAPG on pathogen growth.
The susceptibility of F. oxysporum f. sp. radicis-lycopersici to DAPG was tested on ME agar (pH 7.0) amended at concentrations of 0 to 300 μg ml−1. Agar plugs (4 mm in diameter) taken from 7- to 10-day-old cultures were placed inverted in the center of test plates. Hyphal radial growth was measured after 7 days of incubation at 24°C in darkness. The experiment with each treatment concentration consisted of three replicate plates and was conducted twice.
Hydroponic assay to determine the role of DAPG and HCN in biocontrol.
A soilless noncirculating hydroponic assay that simulates tomato production systems widely used worldwide (18) was used to compare the biocontrol efficacies of mutants with those of wild-type parent strains CHA0 and Q2-87. Pregerminated tomato seeds (Lycopersicum esculentum Mill.) were placed in divets of rock wool cubes (3.5 cm2 by 4 cm deep, one seed per cube; Grodania A/S, Hedehusene, Denmark), with 18 cubes per plastic tray (23.5 by 28.5 cm in diameter by 5.5 cm deep). Rock wool was saturated with nutrient solution (15) and kept at a level of approximately 1 to 2 cm deep throughout the course of experiments by addition of filter-sterilized deionized water as needed (18). Bacteria (approximately 107 CFU ml−1) and Fusarium (106 microconidia ml−1) were added to 400 ml of nutrient solution before being dispensed over the rock wool. Plants were incubated in growth chambers with 16 h of light at 22°C and 8 h of darkness at 18°C and 70% relative humidity. After 14 to 21 days, seedlings were scored for crown and root rot disease severity on a scale of 0 to 4 where 0 means no symptoms and 4 means that plants are dead or nearly so (42).
Effect of fusaric acid on bacterial gene expression.
Strains CHA0 and Q2-87 carrying a lacZ reporter gene fusion to the phlA gene of CHA0 on plasmid pME6259 were grown in 20 ml of liquid PCG medium (15, 61) in 100-ml Erlenmeyer flasks sealed with cotton plugs. Strain CHA207, carrying the chromosomal hcnA′-′lacZ fusion, was grown in OSG minimal medium (52). Culture media were inoculated with 40 μl of exponential-growth-phase cultures of the bacteria diluted to an optical density at 600 nm (OD600) of 0.05. Just prior to inoculation with bacteria, a fresh preparation of fusaric acid (5-butylpicolinic acid; molecular weight, 179.2; Sigma-Aldrich Corp., St. Louis, Mo.) dissolved in 10% (vol/vol) methanol was added to give 50 and 500 μM. Controls received the same amount of methanol. Cultures were incubated at 30°C with rotational shaking at 180 rpm. β-Galactosidase-specific activities of at least four independent cultures were determined by the method of Miller (50). After kinetics of phlA′-′lacZ reporter gene expression were obtained, acidified cultures (pH 2 with 2 N HCl) were extracted with equal volumes of ethyl acetate and the DAPG and fusaric acid concentrations were quantified by high-pressure liquid chromatography methods as previously described (15, 17). DAPG levels obtained by direct isolation were consistent with those indicated by reporter activity (data not shown), as has previously been reported (52).
Pathogen sensitivity to DAPG.
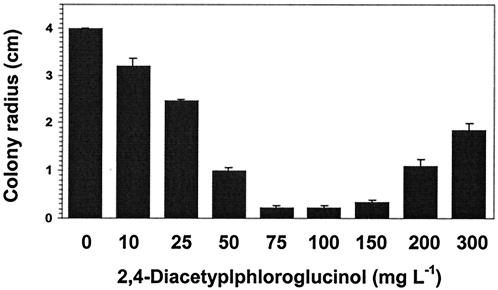
The first objective in examining molecular interactions between a tomato root pathogenic fungus and a biocontrol agent was to determine the inhibitory activity of a bacterial antimicrobial metabolite that has been identified to be primarily responsible for suppression of several other plant diseases (29, 60). We found that F. oxysporum f. sp. radicis-lycopersici was highly sensitive to pure DAPG in vitro, with considerable growth inhibition at concentrations as low as 25 μg ml−1 and nearly complete inhibition at 75 μg ml−1 (Fig. 1). This is similar to the toxicity levels that have been observed for other fungi, including other Fusarium spp (30). It also demonstrates that in situations when this antibiotic is present it is an effective growth inhibitor of this particular pathogen. The exact mechanism of action of DAPG against Fusarium is unknown. A recent report, however, indicates that DAPG at extremely low concentrations (0.4 ng ml−1) lysed zoospores and caused extreme hyphal tip disorganization in phytopathogenic Pythium ultimum (12).
FIG. 1.
Inhibition of F. oxysporum f. sp. radicis-lycopersici by synthetic DAPG. Radial growth of hyphae on 2% ME agar was measured after 7 days of incubation at 27°C. Values (± standard errors [error bars]) represent the means of six replicates in two experiments.
Curiously though, at concentrations above 150 μg ml−1 we observed that the pathogen was again able to grow. It is unclear if fungal regrowth was from cells that escaped death from DAPG exposure or if the antibiotic had a fungistatic effect on the fungus. “Reversed toxicity” has been observed for certain fungicides when these are tested at concentrations exceeding those necessary for initial inhibition of fungal growth. A possible explanation for this is that Fusarium metabolizes DAPG, as it has been reported to metabolize related phloroglucinol compounds (65), and that the required genes are up-regulated only after exposure to high toxin concentrations. If this indeed is the case, it would be important to determine if the up-regulated genes specifically affect fungal responses to DAPG or if the efficacy of other key biocontrol compounds (e.g., pyoluteorin, phenazines, pyrrolnitrin) is also compromised. Another explanation that has recently been identified is active efflux with ABC transporter pumps to protect fungi from phenazine antibiotics produced by biocontrol bacteria (53). The ability of the pathogen to grow after exposure to hypertoxic DAPG concentrations has potentially important implications for the efficacy of biocontrol strains genetically modified to either constitutively produce or to overproduce this antibiotic (11, 51). Whether the ability to withstand high concentrations may result in selection of different fungal resistance mechanisms that confer tolerance to lower but inhibitory antibiotic doses, whether tolerance to DAPG confers cross-resistance to other antifungal bacterial metabolites, and whether these could lead to a breakdown in biocontrol should be investigated.
Role of bacterial metabolites in biocontrol of tomato crown and root rot.
We applied classical molecular analysis for determining the role of two bacterial metabolites in biocontrol of Fusarium on the tomato. Two independent sets of repeated experiments were performed. A DAPG-deficient strain Q2-87 mutant proved significantly less effective at protecting tomatoes from Fusarium attack (Fig. 2). Restoration of DAPG production to this mutant with plasmid-mobilized functional biosynthetic genes almost completely restored biocontrol activity relative to that of the wild-type strain (Fig. 2).
FIG. 2.
Biocontrol activity of wild-type P. fluorescens mutants defective for DAPG or HCN and mutants restored for DAPG or HCN production. Bacterial treatments were as follows: 1, no bacteria, 2, CHA0; 3, CHA630; 4, CHA630/pMON5118; 5, CHA5; 6, CHA5/pME3013; 7, Q2-87; 8, Q2-87::Tn5-1; 9, Q2-87::Tn5-1/pMON5118; 10, CHA631; 11, CHA631/pME6261. Biocontrol assays were conducted in a noncirculated hydroponic system (15, 18). Two sets of repeated experiments were conducted (A and B and C and D). Values (± standard errors [error bars]) represent the means of 6 (A and B) and 11 (C and D) replicates. Bars within a graph with the same letters are not significantly different according to Fisher's protected least-significant-difference test at a P of ≤0.0001.
Intriguingly, CHA0 mutants deficient for DAPG production were not significantly reduced in biocontrol efficacy. This is not to say that the CHA0 mutants were effective biocontrol agents, rather that the already relatively low level of protection provided by wild-type CHA0 was not significantly further reduced by loss of DAPG production (Fig. 2). The lack of a role for DAPG in tomato crown and root rot suppression by CHA0 was confirmed by using two different mutational events (producing CHA630 and CHA631) to abolish production of this compound. However, when HCN production was abolished in the insertion mutant CHA5, biocontrol activity was significantly impaired relative to that of the wild-type parental strain, CHA0 (Fig. 2). Restoration of HCN production with plasmid-mobilized functional hcnABC genes in CHA5/pME3013 also restored biocontrol activity (Fig. 2). This demonstrates a role for this biocidal metabolite in tomato crown and root rot biocontrol and is the first example of HCN contributing to biocontrol of Fusarium or of a plant disease in hydroponic culture. Biosynthesis of HCN is regulated by the FNR-like transcriptional regulator ANR, which up-regulates the expression of the hcnABC genes under oxygen-limited conditions (6, 27), such as those in our noncirculating hydroponic system. However, whereas these conditions may have favored HCN biosynthesis and a greater role of HCN in biocontrol, they did not preclude DAPG production. Under identical conditions, we have isolated DAPG from the rhizosphere of tomato plants inoculated with CHA0 (15).
Influence of pathogen toxin on antagonist gene expression.
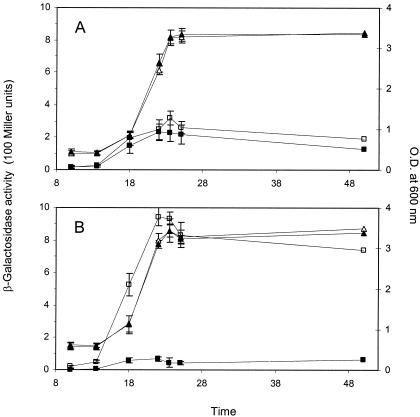
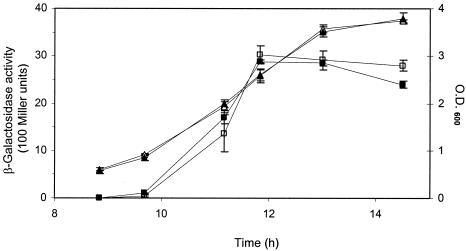
Fusaric acid completely abolished DAPG biosynthesis in strain CHA0 but had no effect in strain Q2-87, as determined by using a lacZ reporter fusion to the biosynthetic gene phlA (Fig. 3). Production by CHA0 of monoacetylphloroglucinol, a putative precursor compound (3), was also completely repressed by 50 and 500 μM fusaric acid (data not shown). Extraction of bacterial cultures and high-pressure liquid chromatography analysis found that fusaric acid was not degraded by either strain (data not shown). Fusaric acid also had no effect on HCN biosynthesis in strain CHA0, as determined by using a chromosomal hcnA′-′lacZ reporter fusion (Fig. 4). Fusaric acid can be isolated from our hydroponic tomato assays at a concentration of 0.1 μg ml−1, and even this level is sufficient to completely block DAPG production (15). These results support the conclusions drawn from our bioassays and explain why DAPG contributed to biocontrol in Q2-87 but not CHA0 and why HCN contributed to biocontrol in CHA0. Biosynthesis pathways for both metabolites are regulated via GacS and GacA (8). The fact that HCN and, in a previous study, protease (15) were not affected by fusaric acid indicates a target somewhere downstream. Recent evidence points to the DAPG pathway-specific repressor PhlF as a putative target because mutation in the phlF gene relieved sensitivity to fusaric acid in CHA0 (52). Both CHA0 and Q2-87 have phlF genes, but, as with another DAPG biosynthetic gene, phlD, there is diversity that could explain the differential responses to fusaric acid (48). Interestingly, CHA0 produces metabolites (i.e., pyoluteorin and salicylate) that also regulate DAPG biosynthesis via PhlF, indicating that the pathogen has hijacked a bacterial regulatory cascade for its self-defense purposes (14). The repressive activity of PhlF is thought to be its binding of the inverted repeated sequence phO, located downstream of the phlA transcriptional start site (1). Salicylate stabilizes the PhlF-phO complex, and this may also be the case with fusaric acid.
FIG. 3.
Kinetics of phlA′-′lacZ expression in P. fluorescens strains Q2-87 (A) and CHA0 (B) without (□) or with (▪) fusaric acid (500 μM) added to PCG liquid culture media at 27°C. OD600 values for cultures grown in the absence (▵) and presence (▴) of fusaric acid are shown. Values are means ± standard deviations from three experiments. Some of the error bars are too small to be shown.
FIG. 4.
Effect of fusaric acid on the expression of a chromosomal hcnA′-′lacZ fusion in the P. fluorescens CHA0 derivative CHA207. Specific β-galactosidase activities were determined for bacteria grown in OSG liquid culture medium at 30°C without (□) or with (▪) fusaric acid (500 μM). OD600 values for cultures grown in the absence (▵) and presence (▴) of fusaric acid are shown. Values are means ± standard deviations from three experiments. Some of the error bars are too small to be shown.
Our study provides evidence for direct signaling between a pathogen and an antagonist. Previous work identifying Pythium signals (58) found that these were likely nonspecific signals from lysed hyphae that affected bacterial trehalose genes (25). Genes in a P. fluorescens biocontrol strain which were affected by Pythium hyphal extracts appeared to contribute to long-term rhizosphere colonizing ability but were not directly involved in pathogen suppression (22). Trehalose is commonly found as a storage compound in most fungi, as well as in many bacteria and plants (23, 25, 35), and it is known to influence diverse interactions between bacteria and fungi that do not result in microbial competition (24, 37).
Implications of pathogen self-defense in biocontrol and microbial ecological interactions.
In addition to this being the first report of roles for DAPG or HCN in Fusarium suppression of tomato disease, our results have wider implications for the interpretation of studies designed to elucidate biocontrol mechanisms of action. While our study is the first example where a particular mechanism of action is biocontrol strain specific, it may not be the only case. Often, results obtained with one combination of a pathogen and biocontrol agent are generalized to conclude a role or lack thereof for a particular compound in biocontrol of a particular disease. Our results demonstrate the need to examine several strains that may have different regulatory responses to the same compounds. In situations where DAPG has been found to have no role in biocontrol (49) or in ecological fitness (9), other DAPG-producing strains may perform differently, and likewise, in the many cases where a role has been demonstrated, it cannot be assumed to be true for all DAPG-producing strains.
Our results offer a new explanation for the variation in efficacy of biocontrol agents (10, 60). Selection of strains based on possession of key biocontrol genes (46) is insufficient because strains must not only have the genes but also be able to express these genes in target environments. Previous studies identified nutritional factors, soil properties, and host plant effects on gene expression (17, 43). Here we further confirm that pathogen signals or pathogen self-defense compounds modulate gene expression in biocontrol agents (14). Moreover, in our case both CHA0 and Q2-87 possess DAPG biosynthetic genes but only Q2-87 is able to express these in the presence of a fusaric acid-producing pathogen. Preliminary data testing a collection of 42 P. fluorescens strains (16) and the findings presented here suggest that sensitivity to fusaric acid partly explains the variation in biocontrol efficacy observed with DAPG-producing pseudomonads (54). In terms of strain selection, our results provide the framework for screening DAPG-producing pseudomonads for targeted application to control Fusarium, with fusaric acid-insensitive strains likely to provide more reliable protection. A new approach may also be to develop strain mixtures by combining sensitive strains with insensitive strains and/or strains that degrade pathogen toxins (28, 59, 61). Trichoderma viridae can down-regulate genes for Fusarium mycotoxin biosynthesis (68), and mixtures of bacterial and fungal agents have already been found to improve biocontrol (20). Furthermore, selection of antagonists to control mycotoxigenic fungi (33) must consider the potential gene-regulating activity of these compounds.
Targeted inhibition of antagonist genes by a pathogen metabolite adds a new dimension to microbial ecological interactions. This is further evidence of the ecological importance of interspecies and interphylum signaling that is only now being uncovered. Mycotoxins produced by Fusarium spp., including fusaric acid, typically have broad-spectrum antibiotic activity affecting bacteria, fungi, nematodes, insects, and mammals (39, 67), and their activity can be synergistic (13). An unexpectedly diverse range of Fusarium species have been found to produce fusaric acid and other mycotoxins in environments that require aggressive saprophytic competition with other organisms (i.e., grain [2] and crop residues [36]). The potential ecological importance of mycotoxins with respect to the saprophytic life cycles of producing fungi is largely unknown, although inhibition of bacteria and other microbial competitors or animal consumers of crop residues may enhance the saprophytic survival of plant-infectious mycotoxigenic Fusarium in crops. Understanding the genetic basis for differential responses of two closely related Pseudomonas strains carrying similar genes for biosynthesis of DAPG may help us understand the evolution of bacterial genotypes and may reveal a novel type of gene-for-gene interaction among microorganisms.
Acknowledgments
We thank Jacques Fuchs, Monika Maurhofer, Regina Notz, and Marcello Zala for technical advice; Caroline Blumer, Ursula Schnider-Keel, and Linda Thomashow for providing strains and plasmids; and Ulrich Burger for providing synthetic DAPG.
Financial support was provided by the Swiss National Foundation of Science (grant no. 3100-50522.97 to G.D. and 3100-061360.00 to C.K.) and European COST Action 830, Microbial Inoculants in Agriculture, to B.D. and G.D.
REFERENCES
- 1.Abbas, A., J. P. Morrissey, P. C. Marquez, M. M. Sheehan, I. R. Delany, and F. O'Gara. 2002. Characterization of interactions between the transcriptional repressor PhlF and its binding site at the phlA promoter in Pseudomonas fluorescens F113. J. Bacteriol. 184:3008-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bacon, C. W., J. K. Porter, W. P. Norred, and J. F. Leslie. 1996. Production of fusaric acid by Fusarium species. Appl. Environ. Microbiol. 62:4039-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bangera, M. G., and L. S. Thomashow. 1999. Identification and characterization of a gene cluster for synthesis of the polyketide antibiotic 2,4-diacetylphloroglucinol from Pseudomonas fluorescens Q2-87. J. Bacteriol. 181:3155-3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnett, S. J., I. Singleton, and M. Ryder. 1999. Spatial variation in populations of Pseudomonas corrugata 2140 and pseudomonads on take-all diseased and healthy root systems of wheat. Soil Biol. Biochem. 31:633-636. [Google Scholar]
- 5.Bender, C. L., V. Rangaswamy, and J. Loper. 1999. Polyketide production by plant-associated pseudomonads. Annu. Rev. Phytopathol. 37:175-196. [DOI] [PubMed] [Google Scholar]
- 6.Blumer, C., and D. Haas. 2000. Mechanism, regulation, and ecological role of bacterial cyanide biosynthesis. Arch. Microbiol. 173:170-177. [DOI] [PubMed] [Google Scholar]
- 7.Blumer, C., S. Heeb, G. Pessi, and D. Haas. 1999. Global GacA-steered control of cyanide and exoprotease production in Pseudomonas fluorescens involves specific ribosome binding sites. Proc. Natl. Acad. Sci. USA 96:14073-14078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bull, C. T., B. Duffy, C. Voisard, G. Défago, C. Keel, and D. Haas. 2001. Characterization of spontaneous gacS and gacA regulatory mutants of Pseudomonas fluorescens biocontrol strain CHA0. Antonie Leeuwenhoek 79:327-336. [DOI] [PubMed] [Google Scholar]
- 9.Carrol, H., Y. Moënne-Loccoz, D. N. Dowling, and F. O'Gara. 1995. Mutational disruption of the biosynthesis genes coding for the antifungal metabolite 2,4-diacetylphloroglucinol does not influence the ecological fitness of Pseudomonas fluorescens F113 in the rhizosphere of sugar beets. Appl. Environ. Microbiol. 61:3002-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cook, R. J. 1993. Making greater use of introduced mircroorganisms for biological control of plant pathogens. Annu. Rev. Phytopathol. 31:53-80. [DOI] [PubMed] [Google Scholar]
- 11.Delany, I. R., U. F. Walsh, I. Ross, A. M. Fenton, D. M. Corkery, and F. O'Gara. 2001. Enhancing the biocontrol efficacy of Pseudomonas fluorescens F113 by altering the regulation and production of 2,4-diacetylphloroglucinol-improved Pseudomonas biocontrol inoculants. Plant Soil 232:195-205. [Google Scholar]
- 12.de Souza, J. T., C. Arnould, C. Deulvot, P. Lemanceau, V. Gianinazzi-Pearson, and J. M. Raaijmakers. 2003. Effect of 2,4-diacetylphloroglucinol on Pythium: cellular responses and variation in sensitivity among propagules and species. Phytopathology 93:966-975. [DOI] [PubMed] [Google Scholar]
- 13.D'Mello, J. P. F., C. M. Placinta, and A. M. C. Macdonald. 1999. Fusarium mycotoxins: a review of global implications for animal health, welfare and productivity. Anim. Feed Sci. Technol. 80:183-205. [Google Scholar]
- 14.Duffy, B., A. Schouten, and J. M. Raaijmakers. 2003. Pathogen self-defense: mechanisms to counteract microbial antagonism. Annu. Rev. Phytopathol. 41:501-538. [DOI] [PubMed] [Google Scholar]
- 15.Duffy, B. K., and G. Défago. 1997. Zinc improves biocontrol of Fusarium crown and root rot of tomato by Pseudomonas fluorescens and represses the production of pathogen metabolites inhibitory to bacterial antibiotic biosynthesis. Phytopathology 87:1250-1257. [DOI] [PubMed] [Google Scholar]
- 16.Duffy, B. K., and G. Défago. 1997. A Fusarium pathogenicity factor blocks antibiotic biosynthesis by antagonistic pseudomonads. Phytopathology 87:S26. [DOI] [PubMed] [Google Scholar]
- 17.Duffy, B. K., and G. Défago. 1999. Environmental factors modulating antibiotic and siderophore biosynthesis by Pseudomonas fluorescens biocontrol strains. Appl. Environ. Microbiol. 65:2429-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duffy, B. K., and G. Défago. 1999. Macro- and microelement fertilizers influence the severity of Fusarium crown and root rot of tomato in a soilless production system. Hortscience 34:287-291. [Google Scholar]
- 19.Duffy, B. K., and G. Défago. 2000. Controlling instability in gacS-gacA regulatory genes during inoculant production of Pseudomonas fluorescens biocontrol strains. Appl. Environ. Microbiol. 66:3142-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duffy, B. K., A. Simon, and D. M. Weller. 1996. Combination of Trichoderma koningii with fluorescent pseudomonads for control of take-all on wheat. Phytopathology 86:188-194. [Google Scholar]
- 21.Emmert, E. A. B., and J. Handelsman. 1999. Biocontrol of plant disease: a (gram-) positive perspective. FEMS Microbiol. Lett. 171:1-9. [DOI] [PubMed] [Google Scholar]
- 22.Fedi, S., E. Tola, Y. Moënne-Loccoz, D. N. Dowling, L. M. Smith, and F. O'Gara. 1997. Evidence for signaling between the phytopathogenic fungus Pythium ultimum and Pseudomonas fluorescens F113: P. ultimum represses the expression of genes in P. fluorescens F113, resulting in altered ecological fitness. Appl. Environ. Microbiol. 63:4261-4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feofilova, E. P. 2001. The kingdom fungi: heterogeneity of physiological and biochemical properties and relationships with plants, animals, and prokaryotes. Appl. Biochem. Microbiol. 37:124-137. [Google Scholar]
- 24.Frey, P., P. Frey-Klett, J. Garbaye, O. Berge, and T. Heulin. 1997. Metabolic and genotypic fingerprinting of fluorescent pseudomonads associated with Douglas fir Laccaria bicolor mycorrhizosphere. Appl. Environ. Microbiol. 63:1852-1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gaballa, A., P. D. Abeysinghe, G. Urich, S. Matthijs, H. DeGreve, P. Cornelis, and N. Koedam. 1997. Trehalose induces antagonism towards Pythium debaryanum in Pseudomonas fluorescens ATCC 17400. Appl. Environ. Microbiol. 63:4340-4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haas, D., C. Blumer, and C. Keel. 2000. Biocontrol ability of fluorescent pseudomonads genetically dissected: importance of positive feedback regulation. Curr. Opin. Biotechnol. 11:290-297. [DOI] [PubMed] [Google Scholar]
- 27.Højberg, O., U. Schnider, H. V. Winteler, J. Sørensen, and D. Haas. 1999. Oxygen-sensing reporter strain of Pseudomonas fluorescens for monitoring the distribution of low-oxygen habitats in soil. Appl. Environ. Microbiol. 65:4085-4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karlovsky, P. 1999. Biological detoxification of fungal toxins and its use in plant breeding, feed and food production. Nat. Toxins 7:1-23. [DOI] [PubMed] [Google Scholar]
- 29.Keel, C., and G. Défago. 1997. Interactions between beneficial soil bacteria and root pathogens: mechanisms and ecological impact, p. 27-46. In A. C. Gange and V. K. Brown (ed.), Multitrophic interactions in terrestrial systems. Blackwell Scientific Publishers, London, United Kingdom.
- 30.Keel, C., U. Schnider, M. Maurhofer, C. Voisard, J. Laville, U. Burger, P. Wirthner, D. Haas, and G. Défago. 1992. Suppression of root diseases by Pseudomonas fluorescens CHA0: importance of the bacterial secondary metabolite 2,4-diacetylphloroglucinol. Mol. Plant-Microbe Interact. 1:4-13. [Google Scholar]
- 31.Keel, C., D. M. Weller, A. Natsch, G. Défago, R. J. Cook, and L. S. Thomashow. 1996. Conservation of the 2,4-diacetylphloroglucinol biosynthesis locus among fluorescent Pseudomonas strains from diverse geographic locations. Appl. Environ. Microbiol. 62:552-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kern, H. 1972. Phytotoxins produced by fusaria, p. 35-44. In R. K. S. Wood, A. Ballio, and A. Graniti (ed.), Phytotoxins in plant diseases. Academic Press, New York, N.Y.
- 33.Khan, N. I., D. A. Schisler, M. J. Boehm, P. J. Slininger, and R. J. Bothast. 2001. Selection and evaluation of microorganisms for biocontrol of Fusarium head blight of wheat incited by Gibberella zeae. Plant Dis. 85:1253-1258. [DOI] [PubMed] [Google Scholar]
- 34.King, E. O., M. K. Ward, and D. E. Raney. 1954. Two simple media for demonstration of pyocyanin and fluorescein. J. Lab. Clin. Med. 44:301-307. [PubMed] [Google Scholar]
- 35.Leyman, B., P. van Dijck, and J. M. Thevelein. 2001. An unexpected plethora of trehalose biosynthesis genes in Arabidopsis thaliana. Trends Plant Sci. 6:510-513. [DOI] [PubMed] [Google Scholar]
- 36.Lutz, M. P., G. Feichtinger, G. Défago, and B. Duffy. 2003. Mycotoxigenic Fusarium and deoxynivalenol production repress chitinase gene expression in the biocontrol agent Trichoderma atroviride P1. Appl. Environ. Microbiol. 69:3077-3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marschner, P., and D. E. Crowley. 1996. Physiological activity of a bioluminescent Pseudomonas fluorescens (strain 2-79) in the rhizosphere of mycorrhizal and non-mycorrhizal pepper (Capsicum annuum L.). Soil Biol. Biochem. 28:869-876. [Google Scholar]
- 38.Maurhofer, M., C. Keel, D. Haas, and G. Défago. 1995. Influence of plant species on disease suppression by Pseudomonas fluorescens strain CHA0 with enhanced antibiotic production. Plant Pathol. 44:40-50. [Google Scholar]
- 39.May, H. D., Q. Z. Wu, and C. K. Blake. 2000. Effects of Fusarium spp. mycotoxins fusaric acid and deoxynivalenol on the growth of Ruminococcus albus and Methanobrevibacter ruminatium. Can. J. Microbiol. 46:692-699. [DOI] [PubMed] [Google Scholar]
- 40.Mazzola, M., and R. J. Cook. 1991. Effects of fungal root pathogens on the population dynamics of biocontrol strains of fluorescent pseudomonads in the wheat rhizosphere. Appl. Environ. Microbiol. 57:2171-2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mazzola, M., D. K. Fujimoto, L. S. Thomashow, and R. J. Cook. 1995. Variation in sensitivity of Gaeumannomyces graminis to antibiotics produced by fluorescent Pseudomonas spp. and effect on biological control of take-all of wheat. Appl. Environ. Microbiol. 61:2554-2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mihuta-Grimm, L., W. A. Erb, and R. C. Rowe. 1990. Fusarium crown and root rot of tomato in greenhouse rock wool systems: sources of inoculum and disease management with benomyl. Plant Dis. 74:996-1002. [Google Scholar]
- 43.Notz, R., M. Maurhofer, U. Schnider-Keel, B. Duffy, D. Haas, and G. Défago. 2001. Biotic factors affecting expression of the 2,4-diacetylphloroglucinol biosynthesis gene phlA in Pseudomonas fluorescens biocontrol strain CHA0 in the rhizosphere. Phytopathology 91:873-881. [DOI] [PubMed] [Google Scholar]
- 44.Pierson, E. A., D. W. Wood, J. A. Cannon, F. M. Blachere, and L. S. Pierson. 1998. Interpopulation signaling via N-acyl-homoserine lactones among bacteria in the wheat rhizosphere. Mol. Plant-Microbe Interact. 11:1078-1084. [Google Scholar]
- 45.Pierson, L. S., D. W. Wood, and E. A. Pierson. 1998. Homoserine lactone-mediated gene regulation in plant-associated bacteria. Annu. Rev. Phytopathol. 36:207-225. [DOI] [PubMed] [Google Scholar]
- 46.Raaijmakers, J. M., D. M. Weller, and L. S. Thomashow. 1997. Frequency of antibiotic-producing Pseudomonas spp. in natural environments. Appl. Environ. Microbiol. 63:881-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raaijmakers, J. M., and D. M. Weller. 1998. Natural plant protection by 2,4-diacetylphloroglucinol-producing Pseudomonas spp. in take-all decline soil. Mol. Plant-Microbe Interact. 11:144-152. [Google Scholar]
- 48.Ramette, A., Y. Moënne-Loccoz, and G. Défago. 2001. Polymorphism of the polyketide synthase gene phlD in biocontrol fluorescent pseudomonads producing 2,4-diacetylphloroglucinol and comparison of PhlD with plant polyketide synthases. Mol. Plant-Microbe Interact. 14:639-652. [DOI] [PubMed] [Google Scholar]
- 49.Resca, R., M. Basaglia, S. Poggiolini, P. Vian, S. Bardin, U. F. Walsh, C. M. E. Barreiros, F. O'Gara, M. P. Nuti, S. Casella, and U. Peruch. 2001. An integrated approach for the evaluation of biological control of the complex Polymyxa betae beet necrotic yellow vein virus, by means of seed inoculants. Plant Soil 232:215-226. [Google Scholar]
- 50.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 51.Schnider, U., C. Keel, C. Blumer, J. Troxler, G. Défago, and D. Haas. 1995. Amplification of the house-keeping sigma factor in Pseudomonas fluorescens CHA0 enhances antibiotic production and improves biocontrol abilities. J. Bacteriol. 177:5387-5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schnider-Keel, U., A. Seematter, M. Maurhofer, C. Blumer, B. Duffy, C. Reimmann, R. Notz, G. Défago, D. Haas, and C. Keel. 2000. Autoinduction of 2,4-diacetylphloroglucinol biosynthesis in the biocontrol agent Pseudomonas fluorescens CHA0 and repression by the bacterial metabolites salicylate and pyoluteorin. J. Bacteriol. 182:1215-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schoonbeek, H., J. M. Raaijmakers, and M. A. De Waard. 2002. Fungal ABC transporters and microbial interactions in natural environments. Mol. Plant-Microbe Interact. 15:1165-1172. [DOI] [PubMed] [Google Scholar]
- 54.Sharifi-Tehrani, A., M. Zala, A. Natsch, Y. Moënne-Loccoz, and G. Défago. 1998. Biocontrol of soil-borne fungal plant disease by 2,4-diacetylphloroglucinol-producing fluorescent pseudomonads with different restriction profiles of amplified 16S rDNA. Eur. J. Plant Pathol. 104:631-643. [Google Scholar]
- 55.Slininger, P. J., and M. A. Jackson. 1992. Nutritional factors regulating growth and accumulation of phenazine 1-carboxylic acid by Pseudomonas fluorescens 2-79. Appl. Microbiol. Biotechnol. 37:388-392. [Google Scholar]
- 56.Slininger, P. J., and M. A. Shea-Wilbur. 1995. Liquid-culture pH, temperature, and carbon (not nitrogen) source regulate phenazine productivity of the take-all biocontrol agent Pseudomonas fluorescens 2-79. Appl. Microbiol. Biotechnol. 43:794-800. [DOI] [PubMed] [Google Scholar]
- 57.Smith, K. P., and R. M. Goodman. 1999. Host variation for interactions with beneficial plant-associated microbes. Annu. Rev. Phytopathol. 37:473-491. [DOI] [PubMed] [Google Scholar]
- 58.Smith, L. M., E. Tola, P. deBoer, and F. O'Gara. 1999. Signalling by the fungus Pythium ultimum represses expression of two ribosomal RNA operons with key roles in the rhizosphere ecology of Pseudomonas fluorescens F113. Environ. Microbiol. 1:495-502. [DOI] [PubMed] [Google Scholar]
- 59.Thangavelu, R., A. Palaniswami, G. Ramakrishnan, S. Doraiswamy, S. Muthukrishnan, and R. Velazhahan. 2001. Involvement of fusaric acid detoxification by Pseudomonas fluorescens strain Pf10 in the biological control of Fusarium wilt of banana caused by Fusarium oxysporum f. sp. cubense. J. Plant Dis. Prot. 108:433-445. [Google Scholar]
- 60.Thomashow, L. S., and D. M. Weller. 1996. Current concepts in the use of introduced bacteria for biological disease control: mechanisms and antifungal metabolites, p. 187-235. In G. Stacey and N. T. Keen (ed.), Plant-microbe interactions, vol. 1. Chapman & Hall, New York, N.Y.
- 61.Toyoda, H., H. Hashimoto, R. Utsumi, H. Kobayashi, and S. Ouchi. 1988. Detoxification of fusaric acid by a fusaric acid-resistant mutant of Pseudomonas solanacearum and its application to biological control of Fusarium wilt of tomato. Phytopathology 78:1307-1311. [Google Scholar]
- 62.Vincent, M. N., L. A. Harrison, J. M. Brackin, P. A. Kovacevich, P. Mukerji, D. M. Weller, and E. A. Pierson. 1991. Genetic analysis of the antifungal activity of a soil-borne Pseudomonas aureofaciens strain. Appl. Environ. Microbiol. 57:2928-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Voisard, C., C. T. Bull, C. Keel, J. Laville, M. Maurhofer, U. Schnider, G. Défago, and D. Haas. 1994. Biocontrol of root diseases by Pseudomonas fluorescens CHA0: current concepts and experimental approaches, p. 67-89. In F. O'Gara, D. Dowling, and B. Boesten (ed.), Molecular ecology of rhizosphere microorganisms. VCH Publishers, Weinheim, Germany.
- 64.Voisard, C., C. Keel, D. Haas, and G. Défago. 1989. Cyanide production by Pseudomonas fluorescens helps suppress black root rot of tobacco under gnotobiotic conditions. EMBO J. 8351-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Walker, J. R. L., and B. G. Taylor. 1983. Metabolism of phloroglucinol by Fusarium solani. Arch. Microbiol. 134:123-126. [Google Scholar]
- 66.Wang, C. X., A. Ramette, P. Punjasamarnwong, M. Zala, A. Natsch, Y. Moënne-Loccoz, and G. Défago. 2001. Cosmopolitan distribution of phlD-containing dicotyledonous crop-associated biocontrol pseudomonads of worldwide origin. FEMS Microbiol. Ecol. 37:105-116. [Google Scholar]
- 67.Wang, H. X., and T. B. Ng. 1999. Pharmacological activities of fusaric acid (5-butylpicolinic acid). Life Sci. 65:849-856. [DOI] [PubMed] [Google Scholar]
- 68.Yates, I. E., F. Meredith, W. Smart, C. W. Bacon, and A. J. Jaworski. 1999. Trichoderma viridae suppresses fumonisin B-1 production by Fusarium moniliforme. J. Food Prot. 62:1326-1332. [DOI] [PubMed] [Google Scholar]
- 69.Zala, M., C. Gyawali, B. Duffy, C. Keel, and G. Défago. 1999. Application of phloroglucinol (PHL) production as a phenotypic marker for selecting better biocontrol bacteria. Phytopathology 89:S88-S89. [Google Scholar]