Abstract
Enamel-related gene products (ERPs) are detected in non-enamel tissues such as bone. We hypothesized that, if functional, ERP expression corresponds with distinct events during osteoblast differentiation and affects bone development and mineralization. In mouse calvariae and MC3T3 cells, expression profiles of enamel-related gene products (ERPs) correlated with key events in post-natal calvarial development and MC3T3 cell mineralization. Developing skulls from both Amel- and Ambn-deficient animals were approximately 15% shorter when compared with those of wild-type controls, and their sutures remained patent for a longer period of time. Analysis of Amel- and Ambn-deficient calvariae and calvarial osteoblast cultures revealed a dramatic reduction in mineralized nodules, a significant reduction in Runx2, Sp7, Ibsp, and Msx2 expression, and a reduction in Alx4 in Amel-deficient calvariae vs. an increase in Alx4 in Ambn-deficient calvariae. Analysis of these data indicates that ERP expression follows defined developmental profiles and affects osteoblast differentiation, mineralization, and calvarial bone development. We propose that, in parallel to their role in the developing enamel matrix, ERPs have retained an evolutionary conserved function related to the biomineralization of bones.
Keywords: amelogenin, ameloblastin, osteoblast, calvaria, evolution, suture
Introduction
The mineralized tissues bone, dentin, cementum, and tooth enamel play quintessential roles in the life of organisms, acting as guardians of the brain and powerful effectors of mastication. Their formation and maintenance are tightly controlled by several phosphorylated and repeat-element-containing matrix proteins, which are encoded by a cluster of secretory phosphoprotein-encoding genes (SCPP) located on chromosome 4 (Huq et al., 2005). One half of these proteins is associated with the formation of bone and dentin, counting bone sialoprotein (BSP), osteopontin (OPN), dentin sialophosphoprotein (DSPP), dentin matrix protein (DMP), and matrix-extracellular phosphoglycoprotein (MEPE) among its members. The other half consists of the enamel-associated proteins ameloblastin (AMBN), enamelin (ENAM), amelotin (AMTN), and odontogenic ameloblast-associated protein (ODAM). Besides their affiliation with the same gene cluster, SCPP proteins contain several sequence elements, including internal repeats, acidity of select domains, phosphorylation, and negative charge, that predispose them to promote mineral nucleation through calcium ion sequestration, or to control crystal growth by adsorption onto growing crystal surfaces, or both (Addison and McKee, 2010).
The first study pointing to common gene products in enamel and other mineralized tissues was based on a monoclonal antibody generated against a hamster tooth germ (Inai et al., 1993). Subsequently, other studies have blurred the distinction between enamel-type and bone/dentin-type mineralized tissues, highlighting expression of amelogenin (Amel) in dentin, cementum, periodontal ligament, long bone, brain, and soft tissues (Deutsch et al., 2006; Li et al., 2006; Ye et al., 2006; Zeichner-David et al., 2006; Haze et al., 2007) and Ambn expression in dentin, cementum, pulp, and cranial bones (Fong et al., 1998; Spahr et al., 2002; Hao et al., 2005; Nunez et al., 2010).
The ubiquitous expression of SCPPs in multiple mineralized tissues in conjunction with similar but distinct functions suggests that SCPP proteins collaborate to control and fine-tune the growth of craniofacial mineralized tissues. Here, we have conducted studies to define the role of ERPs in craniofacial mineralized tissue development. First, ERP expression in calvarial bone and tooth enamel development was mapped and correlated with key events in calvarial mineralization. To test the role of 2 key ERPs in osteogenesis, we determined the effects of AMBN and AMEL on mineralization markers, mineral formation, and calvarial development using a diet-independent cell culture model in tandem with mutant mouse models. Together, these studies provide a re-evaluation of the roles of ERPs in mineralized tissue development.
Materials & Methods
Animals and Cell Culture
Post-natal CD1 mice age 3, 10, 20, and 35 days (n = 48) (Charles River Laboratories, Wilmington, MA, USA), as well as B6;129 wild-type, Amelx- (Gibson et al., 2001), and Ambn- (n = 48) (Fukumoto et al., 2004) stage-matched mice were handled in accordance with the University of Illinois at Chicago Use of Animals in Research protocol. Mouse calvaria-derived MC3T3-E1 cells (CRL-2593) subclone 4 were maintained in α-minimum essential medium (Gibco BRL, Gaithersburg, MD, USA). Primary calvarial osteoblasts were obtained from calvariae of P1 mice (Amelx-/-, Ambn-/-, wild-type). For osteoinduction, primary calvarial cells were cultured on 6-well culture plates containing Dulbecco’s modified Eagle’s medium (Gibco BRL) supplemented with 10% fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA, USA), 50 μg/mL of ascorbic acid, and 3 μM of beta-glycerophosphate.
Alcian Blue/Alizarin Red Staining
Three CD1 mice per time-point (P3-35), and P2 B6;129 and ERP mutant mice were fixed and dehydrated with ethanol and acetone, stained with alcian blue and alizarin red S for 2 days, immersed in 0.5% potassium hydroxide solution, and stored in 80% glycerol.
RT-real-time PCR
Total mRNA was extracted from calvariae and molars of CD 1 mice at pre-determined developmental stages or from MC3T3 cells after 7, 14, 28, and 42 days’ culture in mineralization medium. To quantify mRNA expression levels, we performed RT-real-ime PCR using sequence-specific SYBR Green primers (Appendix Table 1) and the ABI Prism 7000 detection system (Applied Biosystems, Carlsbad, CA, USA). Samples were normalized with β-actin. Relative expression levels were calculated by the 2–ΔΔCt method (Livak and Schmittgen, 2001). For mRNA expression studies, day 0 in culture and P3 in calvariae were used as baseline (= 1), and β-actin was used as an endogenous control. All PCR products were sequenced at the UIC DNA facility for sequence verification.
Immunohistochemistry
Calvarial vaults from CD1 mouse heads at age P6 were fixed, decalcified, paraffin-embedded, and sectioned. Sections were processed and incubated with ERP protein primary antibodies (Appendix Table 2) at a 1:50 dilution as reported (Lu et al., 2013).
Western Blot Analysis
Calvariae from CD1 mice aged P2, P10, and P20 were ground under liquid N2. Aliquots of calvarial protein extracts were subjected to Western blotting (primary antibody dilutions listed in Appendix Table 2). Tooth proteins from extracted maxillary molars of P3 mice were used as positive controls.
In vitro Mineralization Assay
In vitro mineralization assays (alizarin red) were performed on MC3T3 cultures just before the medium was switched to mineralization medium (wk 0) and at each subsequent interval (wks 1, 2, 4, and 6).
Statistical Analyses
Analyses were performed in triplicate for 3 independent experiments. Kruskal-Wallis analysis of variance was used to determine the mean difference, and independent t tests were used to determine the pair-difference at p = .05.
Results
Enamel-related Gene Products’ (ERPs) Expression in Mouse Calvariae
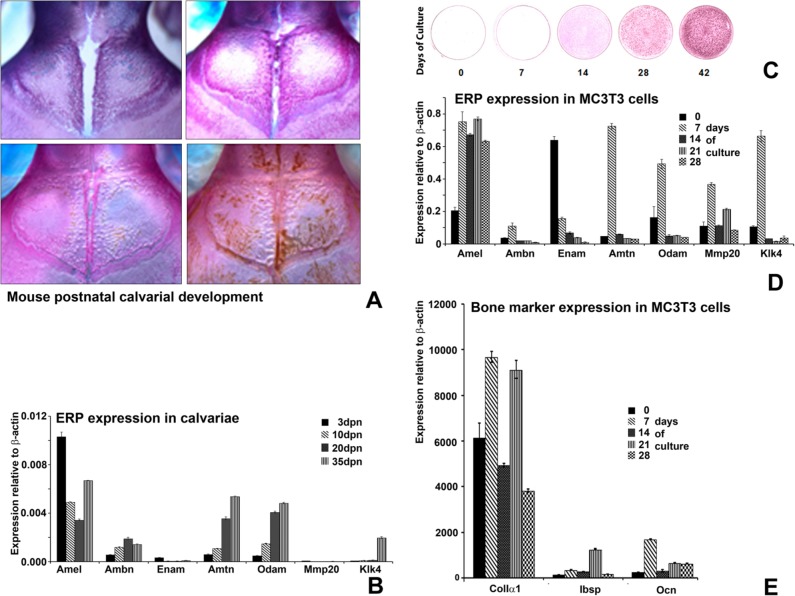
To determine whether occurrences of ERPs in tissues outside of enamel follow concise developmental patterns, we mapped longitudinal ERP expression levels in correlation with calvarial development stages from days P3-P35 (Fig. 1A). The bony bridge of the posterior frontal suture began to form in P20 animals and closed completely on day 35 (Fig. 1A). mRNA expression levels of ERPs, Amel, Ambn, Enam, Amtn, Odam, Mmp20, and Klk4 were detected in calvarial tissues at all stages of development. During calvarial development, ERP profiles changed substantially (Fig. 1B), and expression of ERPs gradually decreased from P3 to P35, with the exception of Ambn and MMP20, which peaked at P20 and then decreased gradually.
Figure 1.
Expression profiles of enamel-related gene products (ERPs) correlated with post-natal calvarial development and MC3T3 cell mineralization. (A) Mouse calvariae visualized by Alcian blue/Alizarin Red S staining. The red staining in calvarial preparations represents developing calvarial mineralized tissues. ERP mRNA expression relative to endogenous control (β-actin) mRNA was detected at all stages of development and levels of most ERPs expression, with the exception of Ambn, gradually decreased with animal age (B). (C) Alizarin red in vitro mineralization assay illustrating stages of mineralization during MC3T3-E1 cell culture over a period of 42 days. (D) Time-course of ERP mRNA expression levels relative to β-actin in MC3T3-E1 cells as revealed by real-time RT-PCR. (E) Matched real-time RT-PCR analysis of bone marker mRNA expression levels relative to β-actin Amel- (A) and Ambn- (B) deficient mice in MC3T3-E1 cells.
MC3T3 pre-osteoblasts proliferate, differentiate into osteoblasts, and exhibit signs of mineralization in long-term cell culture (Quarles et al., 1992). Here we used the MC3T3 long-term cell culture model to investigate whether ERPs display specific expression profiles during osteoblast differentiation and mineralization. RT-real-time PCR analysis indicated that all studied ERPs (Amel, Ambn, Enam, Amtn, Odam), as well as the enzymes Mmp20 and Klk4, were expressed during the entire 42 days of culture. To compare ERP expression profiles with key stages of MC3T3-related mineral formation and osteoblast differentiation in culture, we assessed mineralization levels using an in vitro mineralization assay (Fig. 1C) and identified benchmarks of osteoblast differentiation using Col1A1, Ibsp, and Ocn mRNA expression levels as reference points. ERPs exhibited distinct expression patterns and profile changes during key stages of matrix mineralization and differentiation of MC3T3 cells (Fig. 1D). Specifically, Amel and Mmp20 expression profiles revealed a bi-phasic pattern (Fig. 1D), corresponding to the onset of mineralization and the peak expression of Col1A1 and Ibsp (28 days of culture, Fig. 1E). In contrast, Ambn, Amtn, Odam, and Klk4 expression levels peaked at day 7 (Fig. 1D), corresponding to the early differentiation of MC3T3 cells (Fig. 1C), and then gradually decreased. Data are from 3 independent studies, which yielded essentially identical findings.
Immunohistochemistry and Western Blotting of ERPs in Developing Calvariae
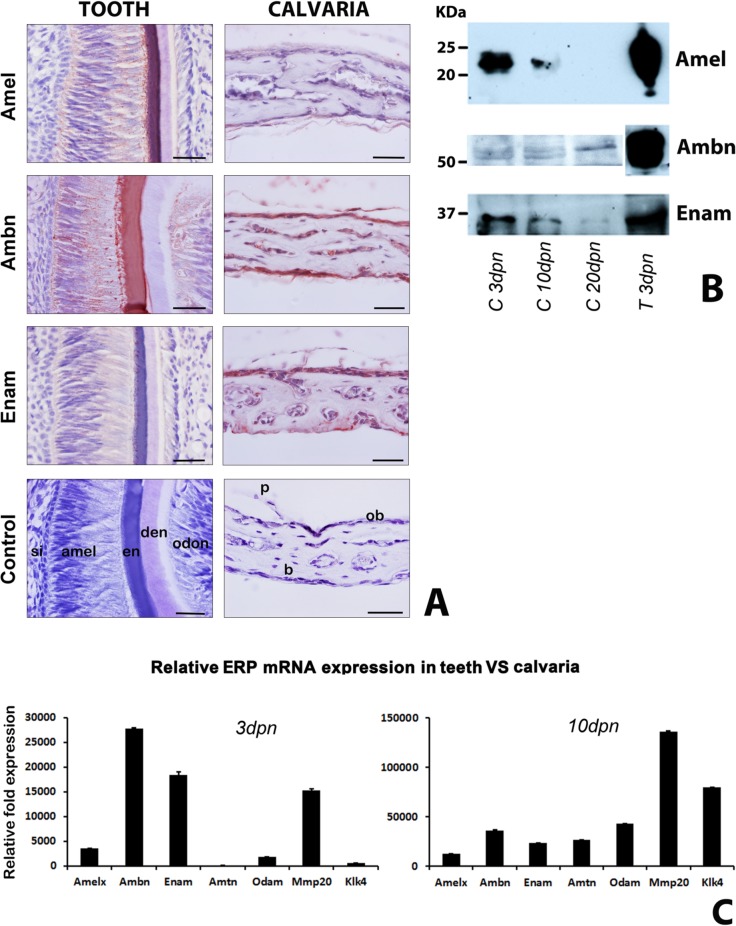
Amelogenins constitute 90% of the enamel protein matrix, whereas collagen 1a1 contributes 90% of the bone protein matrix. To determine whether ERPs contribute significantly to the non-collagenous calvarial bone protein matrix, we compared ERP protein and mRNA expression levels in developing mouse calvariae and teeth. First, Amel, Ambn, and Enam were immunohistochemically detected in calvarial tissues from P6, mainly in the bone matrix and periosteum (Fig. 2A, right panel). Weak levels of reactivity were also documented in osteoblasts, osteocytes, and periosteal cells (Fig. 2A, right panel). Amel, Ambn, and Enam were localized in ameloblasts and enamel matrices (Fig 2A, left panel). Using Western blotting, we detected all 3 ERPs (Amel, Ambn, Enam) in P3, P10, and P20 calvariae as well as in T3 teeth, with the exception of Amel in P20 calvariae (Fig. 2B). Protein levels for all 3, Amel, Ambn, and Enam, were higher in P3 calvariae than in calvariae from later stages of post-natal development, while ERP levels in T3 teeth exceeded those found in P3 calvariae by approximately 60- to 1,000-fold (calculated after adjustment for the 1:20-1:40 pre-dilution of enamel proteins in this Western blot; Fig. 2B). RT-real-time PCR teeth/calvariae expression comparisons revealed high ERP levels in 3dpn teeth when compared with calvariae, with higher Amtn, Klk4, Odam, and Amel expression levels in calvariae than other ERPs (Fig. 2C).
Figure 2.
ERP expression comparison between post-natal developing tooth organs and calvariae. (A) Positive immunoreaction (red) for amelogenin (Amel), ameloblastin (Ambn), and enamelin (Enam) in ameloblasts, enamel matrix, bone matrix, and periosteum. Scale = 100 μm. (B) Western blot detection of amelogenin (Amel), ameloblastin (Ambn), and enamelin (Enam) in calvarial bone extracts at post-natal days 3, 10, and 20 and in developing tooth organs at post-natal day 3. Note that, in this Fig., tooth extracts were diluted at a 1:20 ratio (Ambn and Enam) or a 1:40 ratio (Amel) dilution relative to calvarial extracts, for visualization of expression levels on the same blot. (C) Relative ERP mRNA expression levels between developing calvariae and teeth at post-natal days 3 and 10. ΔCt were normalized to β-actin, and ΔΔCt was performed to calculate fold changes in expression values relative to wild-type calvaria. si = stratum intermedium, amel = ameloblasts, en = enamel, de = dentin, od = odontoblasts, p = periosteum, b = bone, ob = osteoblasts.
Amel and Ambn Affect Developing Skull Size and Suture Closure
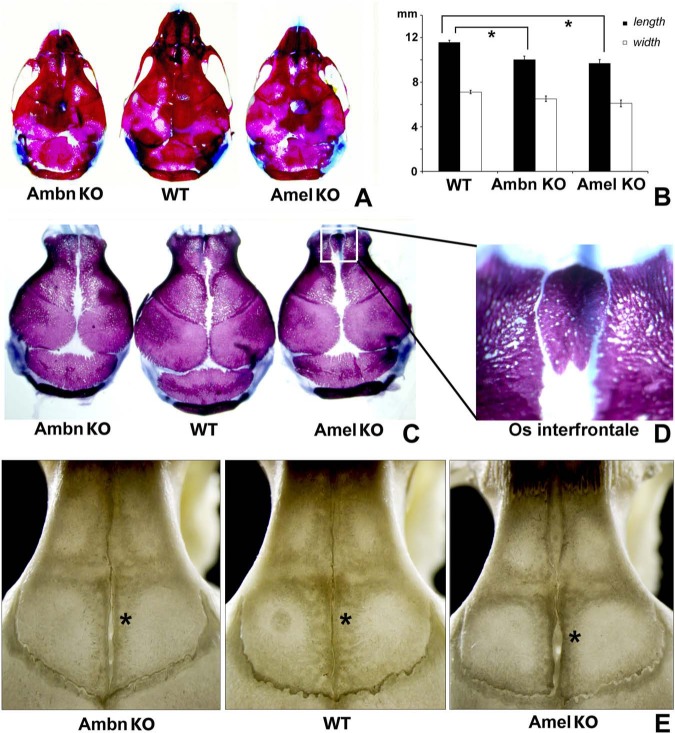
In developing calvarial bone matrices and cultured MC3T3 cells, ERP levels were well beyond detection threshold, stage-specific, and time-dependent. These findings prompted us to determine whether the loss of key ERPs in calvarial cells would affect skull growth (Fig. 3). Phenotype penetrance was 100%, and an interfrontal bone in Amel-deficient mice was present in 40% of the animals. Both skull length and width of P2 Ambn- and the Amel-deficient mice were significantly reduced compared with those of wild-type controls (16.2% Ambn-/- and 13.4% Amel-/- length reduction, p < .05; Figs. 3A, 3B). Moreover, suture closure was delayed in Ambn- and Amel-deficient animals (Fig. 3C), and an interfrontal bone was frequently found between the frontal bones of Amel-deficient mice (Fig. 3D). The interfrontal suture remained patent in 60 dpn in Amel-/- and Ambn-/- mice, and frontal bone length in Amel-/- mice was substantially reduced (Fig. 3E).
Figure 3.
Comparison of skull dimensions and features among Ambn-deficient (Ambn KO), wild-type (WT), and amelogenin-deficient (Amel KO) mice. (A) Whole-skull comparison among representative skulls; (B) length and width comparison among Ambn KO, Amel KO, and WT mice; (C) calvarial vaults after removal of the cranial base; (D) pronounced interfrontal bone (Os frontale) in Amel-deficient mice; and (E) frontal suture patency in Amel KO, Ambn KO, and WT mice. *p < .05.
Effects of Amel and Ambn on Osteoblast Differentiation
To investigate whether changes in skull size and mineralization were directly affected by ERP function and were independent of feeding behavior in ERP mutant mice, we assayed osteoblast mineralization and key mineralization marker gene expression. There was a significant reduction of Ibsp mRNA expression in Amel-deficient calvariae, while Sp7 was reduced in Ambn-deficient calvariae when compared with calvarial mRNA of wild-type littermates (p < .05, Figs. 4A, 4B). The suture-patency-related transcription factor Msx2 was reduced in calvariae of both Amel- and Ambn-deficient mice, while Alx4 was reduced in Amel-deficient mice and increased in Ambn-deficient mice. Twist1 was reduced only in Ambn-deficient mice, while Fgfr2 was increased in Amel-deficient mice and not in Ambn-deficient mice (Figs. 4C, 4D). There was a highly significant 5- to 10-fold reduction in Runx2, Sp7, and Ibsp bone marker mRNA expression levels in Amel-deficient calvarial osteoblasts (p < .05, Fig. 4E) and a highly significant approximately 50% reduction in Runx2, Sp7, and Ibsp expression in Ambn-deficient calvarial osteoblasts (p < .05, Fig. 4F). Mineral nodule formation in Amel- and Ambn-deficient calvarial osteoblasts after 4-week culture was severely reduced (Fig. 4G).
Figure 4.
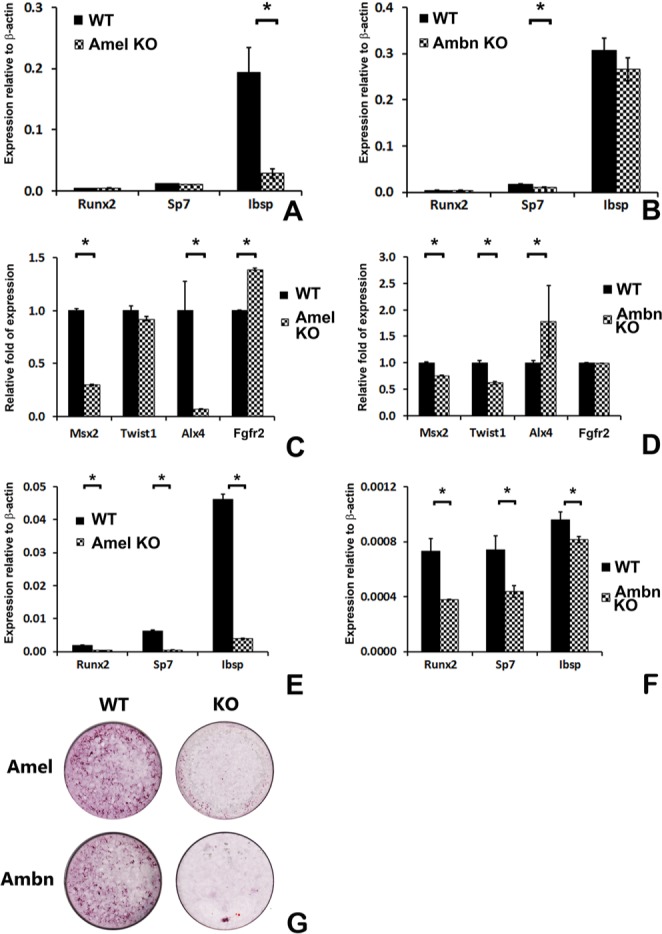
Bone transcription factor and marker expression and mineralization levels in calvariae and calvarial osteoblasts from ERP mutant mice. Bone marker RT-real-time PCR of calvariae (A,B) from Amel- and Ambn-deficient mice, suture-patency-related gene (C,D), and calvarial osteoblasts from Amel- (E) and Ambn- (F) deficient mice. Msx2 expression was reduced, while Alx4 expression showed different expression levels in calvariae from Amel- (C) and Ambn- (D) deficient mice. Note the significant reduction in bone marker and transcription factor gene expression levels, e.g., Runx2, Sp7, and Ibsp, in calvarial osteoblasts after loss of Amel and Ambn (E,F). (G) In vitro mineralization assay of primary calvarial osteoblasts from Amel- and Ambn-deficient mice compared with those of wild-types. *p < .05.
Discussion
The present study was designed to address 3 aspects relative to the presence of ERPs in tissues outside of enamel: (i) ERP expression levels in bone, (ii) correlation of ERPs in developing calvarial bone and in differentiating pre-osteoblasts with key events in mineralization and differentiation, and (iii) effects of calvarial osteoblast ERPs on mineralization behavior and bone marker gene expression (Diekwisch, 2011). To address these questions, we assessed ERP expression levels in developing MC3T3 cells and calvarial osteoblasts and compared them with ERP expression levels in developing tooth enamel. To investigate whether ERPs were of functional relevance for bone development, we cultured osteoblasts from wild-type and Amel- and Ambn-deficient mice and examined them for mineralization potential and bone marker gene expression. In addition, skulls of Amel- and Ambn-deficient mice were analyzed.
Our study revealed that all ERPs studied here were detected in calvarial tissues and MC3T3 cells, and appeared to correlate with key time-points in osteoblast differentiation and mineralization. ERP expression patterns in MC3T3 osteoblast long-term culture peaked either at day 7 (Ambn, Enam, Amtn, Odam, and Klk4) or at day 28 (Amel and Mmp20). Based on parallel studies, these time-points correlated with either the early differentiation of osteoblasts, as indicated by a rise in Col1, Ibsp, and Ocn at day 7, or with the beginning of matrix mineralization at day 28. Analysis of these data indicates that Ambn, Enam, Amtn, and Odam expression peaks are functionally correlated with events in osteoblast differentiation, while Amel might play a role in bone matrix mineralization. According to our immunoreactions, ERPs were detected not only in the bone matrix but also in the surrounding periosteum, suggesting that they might affect calvarial growth at both locations.
In response to the question about functional relevance of ERP expression in calvarial osteoblasts, our study revealed that several bone-related parameters were significantly reduced in both Amel- and Ambn-deficient models when compared with those in wild-type controls, including: (i) Runx2, Sp7, and Ibsp expression levels; (ii) osteoblast mineral nodule formation; and (iii) size of developing calvariae and suture closure. Together, these findings illustrate that both Amel and Ambn ERPs exhibit highly visible effects on bone matrix mineralization and bone marker gene expression. The concept that ERPs are involved in osteoblast differentiation or bone matrix mineralization is supported by several previous studies related to the role of Amel in the modulation of mineralized tissue homeostasis, including activation of osteoclastogenesis (Hatakeyama et al., 2006), modulation of cementogenesis (Swanson et al., 2006), and bone marrow stem cell proliferation (Huang et al., 2010). Recent studies suggested that Ambn binds CD63 and may act through integrin β1 and C Src kinase inhibition to promote osteogenic differentiation (Iizuka et al., 2011; Zhang et al., 2011). Unique dysregulation profiles of major suture-patency-regulating genes such as Msx2, Twist1, Alx4, and Fgfr2 (Melville et al., 2010) in Amel- vs. Ambn-deficient mice suggest that individual ERPs affect individual transcription factor cascades to affect downstream effects on suture morphogenesis.
Five of the ERP genes studied here, Ambn, Enam, Amtn, Odam, and Amel, are members of the secretory calcium-binding phosphoprotein (SCPP) gene cluster of evolutionarily related skeletal mineralization molecules (Kawasaki and Weiss, 2008). According to our analysis, individual ERPs displayed 20% to 40% similarity with other SCPP family members, and bone proteins such as IBSP and OPN had more negatively charged amino acid residues than ERPs, suggesting that bone proteins may play a greater role in the regulation of crystal growth regulation (nucleation and/or inhibition) than ERPs. However, based on their sequence homologies, expression pattern overlap, and functional similarities, we suggest that all SCPP-derived proteins including ERPs contribute to the functional complexities of developing bone and other mineralized tissues. Evidence presented in this paper indicates that even after acquiring novel functions in enamel matrix organization, ERPs have retained their functional significance in bone development and homeostasis. Recent reports have shown a molecular decay of enamel matrix protein genes in turtles and other edentulous amniotes (Meredith et al., 2013). Analysis of these data suggests that the presence of enamel proteins in bone and other mineralized tissues may not be essential for the survival of organisms, but may simply fine-tune the biochemical make-up of the mineralization scaffold of bones in tooth-bearing vertebrates.
Supplementary Material
Footnotes
This study was supported by the NIDCR (grants DE18057 and DE19155 to XL; grant DE18900 to TGHD) and by the Thomas M. Graber AAOF fellowship (to PA).
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
A supplemental appendix to this article is published electronically only at http://jdr.sagepub.com/supplemental.
References
- Addison WN, McKee MD. (2010). ASARM mineralization hypothesis: a bridge to progress. J Bone Miner Res 25:1191-1192. [DOI] [PubMed] [Google Scholar]
- Deutsch D, Haze-Filderman A, Blumenfeld A, Dafni L, Leiser Y, Shay B, et al. (2006). Amelogenin, a major structural protein in mineralizing enamel, is also expressed in soft tissues: brain and cells of the hematopoietic system. Eur J Oral Sci 114(Suppl 1):183-189. [DOI] [PubMed] [Google Scholar]
- Diekwisch T. (2011). Evolution and ameloblastin. Eur J Oral Sci 119(Suppl 1):293-297. [DOI] [PubMed] [Google Scholar]
- Fong CD, Cerny R, Hammarström L, Slaby I. (1998). Sequential expression of an amelin gene in mesenchymal and epithelial cells during odontogenesis in rats. Eur J Oral Sci 106(Suppl 1):324-330. [DOI] [PubMed] [Google Scholar]
- Fukumoto S, Kiba T, Hall B, Iehara N, Nakamura T, Longenecker G, et al. (2004). Ameloblastin is a cell adhesion molecule required for maintaining the differentiation state of ameloblasts. J Cell Biol 167:973-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson CW, Yuan ZA, Hall B, Longenecker G, Chen E, Thyagarajan T, et al. (2001). Amelogenin-deficient mice display an amelogenesis imperfecta phenotype. J Biol Chem 276:31871-31875. [DOI] [PubMed] [Google Scholar]
- Hao J, He G, Narayanan K, Zou B, Lin L, Muni T, et al. (2005). Identification of differentially expressed cDNA transcripts from a rat odontoblast cell line. Bone 37:578-588. [DOI] [PubMed] [Google Scholar]
- Hatakeyama J, Philp D, Hatakeyama Y, Haruyama N, Shum L, Aragon MA, et al. (2006). Amelogenin-mediated regulation of osteoclastogenesis, and periodontal cell proliferation and migration. J Dent Res 85:144-149. [DOI] [PubMed] [Google Scholar]
- Haze A, Taylor AL, Blumenfeld A, Rosenfeld E, Leiser Y, Dafni L, et al. (2007). Amelogenin expression in long bone and cartilage cells and in bone marrow progenitor cells. Anat Rec (Hoboken) 290:455-460. [DOI] [PubMed] [Google Scholar]
- Huang YC, Tanimoto K, Tanne Y, Kamiya T, Kunimatsu R, Michida M, et al. (2010). Effects of human full-length amelogenin on the proliferation of human mesenchymal stem cells derived from bone marrow. Cell Tissue Res 342:205-212. [DOI] [PubMed] [Google Scholar]
- Huq NL, Cross KJ, Ung M, Reynolds EC. (2005). A review of protein structure and gene organisation for proteins associated with mineralised tissue and calcium phosphate stabilisation encoded on human chromosome 4. Arch Oral Biol 50:599-609. [DOI] [PubMed] [Google Scholar]
- Iizuka S, Kudo Y, Yoshida M, Tsunematsu T, Yoshiko Y, Uchida T, et al. (2011). Ameloblastin regulates osteogenic differentiation by inhibiting Src kinase via cross talk between integrin beta1 and CD63. Mol Cell Biol 31:783-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inai T, Inai Y, Kurisu K. (1993). Immunohistochemical detection of an enamel protein-related epitope in rat bone at an early stage of osteogenesis. Histochemistry 99:355-362. [DOI] [PubMed] [Google Scholar]
- Kawasaki K, Weiss KM. (2008). SCPP gene evolution and the dental mineralization continuum. J Dent Res 87:520-531. [DOI] [PubMed] [Google Scholar]
- Li Y, Yuan ZA, Aragon MA, Kulkarni AB, Gibson CW. (2006). Comparison of body weight and gene expression in amelogenin null and wild-type mice. Eur J Oral Sci 114(Suppl 1):190-193. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- Lu X, Ito Y, Atsawasuwan P, Dangaria S, Yan X, Wu T, et al. (2013). Ameloblastin modulates osteoclastogenesis through the integrin/ERK pathway. Bone 54:157-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melville H, Wang Y, Taub PJ, Jabs EW. (2010). Genetic basis of potential therapeutic strategies for craniosynostosis. Am J Med Genet A 152A:3007-3015. [DOI] [PubMed] [Google Scholar]
- Meredith RW, Gatesy J, Springer MS. (2013). Molecular decay of enamel matrix protein genes in turles and other edentulous amniotes. BMC Evol Biol 13:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez J, Sanz M, Hoz-Rodriguez L, Zeichner-David M, Arzate H. (2010). Human cementoblasts express enamel-associated molecules in vitro and in vivo. J Periodontal Res 45:809-814. [DOI] [PubMed] [Google Scholar]
- Quarles LD, Yohay DA, Lever LW, Caton R, Wenstrup RJ. (1992). Distinct proliferative and differentiated stages of murine MC3T3-E1 cells in culture: an in vitro model of osteoblast development. J Bone Miner Res 7:683-692. [DOI] [PubMed] [Google Scholar]
- Spahr A, Lyngstadaas SP, Slaby I, Haller B, Boeckh C, Tsoulfidou F, et al. (2002). Expression of amelin and trauma-induced dentin formation. Clin Oral Investig 6:51-57. [DOI] [PubMed] [Google Scholar]
- Swanson C, Lorentzon M, Conaway HH, Lerner UH. (2006). Glucocorticoid regulation of osteoclast differentiation and expression of receptor activator of nuclear factor-kappaB (NF-kappaB) ligand, osteoprotegerin, and receptor activator of NF-kappaB in mouse calvarial bones. Endocrinology 147:3613-3622. [DOI] [PubMed] [Google Scholar]
- Ye L, Le TQ, Zhu L, Butcher K, Schneider RA, Li W, et al. (2006). Amelogenins in human developing and mature dental pulp. J Dent Res 85:814-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeichner-David M, Chen LS, Hsu Z, Reyna J, Caton J, Bringas P. (2006). Amelogenin and ameloblastin show growth-factor like activity in periodontal ligament cells. Eur J Oral Sci 114(Suppl 1):244-253. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhang X, Lu X, Atsawasuwan P, Luan X. (2011). Ameloblastin regulates cell attachment and proliferation through RhoA and p27. Eur J Oral Sci 119(Suppl 1):280-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.