Abstract
Both histone-acetylations and histone deacetylases have been shown to play a key role in cardiac remodeling. Recently, it has become abundantly clear that many non-histone proteins are modified by post-translational lysine acetylations and that these acetylations regulate protein activity, conformation, and binding. In the present study, non-histone acetylated proteins associated with heart failure were identified. Global screening for lysine acetylated proteins was performed using 2-dimensional gel electrophoresis coupled with immunoblotting with a primary monoclonal anti-acetyl-lysine antibody. Lysine acetylated proteins were compared in two rodent models of hypertensive heart failure, the Dahl salt-sensitive (SS) and spontaneously hypertensive heart failure prone (SHHF) rats with those in corresponding controls, i.e., the Dahl salt-resistant (SR) and W (W) rat strains, respectively. Forty-one and 66 acetylated proteins were detected in SS and SHHF failing hearts, respectively, but either not detected or detected with less abundance in corresponding control hearts. Twelve of these acetylated proteins were common to both models of heart failure. These were identified using matrix-assisted laser desorption/ionization time of flight (MALDI-TOF/TOF) mass spectrometry followed by Mascot Analysis and included mitochondrial enzymes: ATP synthase, long-chain acyl-CoA dehydrogenase, creatine kinase, malate dehydrogenase, and pyruvate dehydrogenase. The abundance of NAD-dependent deacetylase sirtuin-3 (Sirt3), a mitochondrial deacetylase was reduced in SS and SHHF failing hearts. This is the first description of non-histone protein acetylations associated with heart failure and raises the prospect that acetylations of mitochondrial proteins linked to reduced Sirt3 mediate, in part, metabolic changes in heart failure.1
Keywords: Heart failure, Lysine Acetylation, Sirtuin-3, Mitochondrial Proteins, Global Screening
1. INTRODUCTION
Histone protein acetylations, which stimulate gene expression by destabilizing the histone-histone and histone-DNA interactions that limit access of transcription factors to DNA, regulate cardiac remodeling. Both class 1 and class 2 histone deacetylases (HDACs) have been closely linked to cardiac hypertrophy [16]. Class 1 HDACs play a pro-hypertrophic role in the heart via the suppression of anti-hypertrophic pathways. Cardiac over-expression of HDAC2 induces hypertrophy by regulating the PI3K-Akt-Gsk3β growth control pathway [30]. Class 2 HDACs, on the other hand, prevent cardiac hypertrophy by repressing the activity of several pro-hypertrophic transcription factors such as serum response factor (SRF), GATA4, nuclear factor of activated T-cells (NFAT), and myocardin [1]. HDAC inhibitors (HDACis) are emerging as a therapeutic potential for cardiac hypertrophy and failure. Treatment with pan-HDACis can effectively halt, or even reverse, the disease process [5]. Histone acetyltransferases (HATs), specifically p300, have also been shown to mediate agonist-induced cardiac hypertrophy [7] and act as an adaptor for hypertrophy-responsive transcription factors, including GATA4, SRF, and myocyte enhancer factor 2 (MEF2) [1].
It has been increasingly recognized that post-translational lysine acetylation of nonhistone proteins may also play an important role in cellular signaling [27] and hundreds of nonhistone proteins modified by acetylation have been identified [6]. These lysine acetylated proteins participate in a range of processes including transcription, cytoskeleton dynamics, DNA repair and replication, metabolism, apoptosis, and nuclear transport. Furthermore, more than 20% of mitochondrial proteins controlling cellular metabolism have been reported to undergo lysine acetylation [18]. Non-histone protein acetylations regulate enzyme activity, e.g., in p300, ATM, PTEN, and ACS; protein-protein interactions, e.g., in STAT3, AR, EKLF, Importin A, STAT1, and actin; and protein stability, e.g., in p53, Smad7, c-Myc, Runx3, H2A.z, E2F1, GATA1, HIF-1α and SV40 T-Ag [27].
The present study was performed to determine if there are characteristic non-histone acetylated proteins associated with cardiac failure. In order to accomplish this, acetylated proteins in two different rodent models of pressure-overload cardiac remodeling were profiled and ones common to both models were identified.
2. METHODS
2.1 Animals
Dahl salt-resistant (SR) and Dahl salt-sensitive (SS) rats were obtained from Harlan Laboratories (Indianapolis, IN). W (W) and spontaneously hypertensive heart failure prone (SHHF) rats were obtained from Charles River (Wilmington, MA). SR and SS rats were placed on a high salt ad lib diet of 8% NaCl Harlan Teklad rat chow at 8 weeks of age. W and SHHF rats were maintained on a normal rat chow diet. Cardiac size and function was monitored by echocardiography and the SR and SS rats were euthanized under anesthesia at 29 weeks and the W and SHHF rats at 18 months.
2.2 Echocardiography
Rats were anesthetized with 3% isofluorane prior to echocardiography. Transthoracic 2D-targeted M-mode and pulsed Doppler echocardiography (ECHO) were performed with a 15-MHz linear array transducer (Acuson Sequoia C256 system). M-mode images of the left ventricle were obtained from the parasternal short axis view at the level of the papillary muscles. Left ventricular posterior wall thickness (LV PWT) and left ventricular internal dimensions at the end of diastole (EDD) and systole (ESD) were measured by the American Society of Echocardiography leading-edge method on the M-mode tracings. Left ventricular ejection fraction (LV EF) was calculated as follows: EF (%) = (EDD3-ESD3)/EDD3*100%.
2.3 Protein Preparation from Tissue
Flash frozen tissues (left ventricular free wall + septum) were rinsed with PBS washing buffer three times to remove contaminated blood. For 1D Western blots, 350 µL of Protein Lysis Buffer (50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 0.25% sodium deoxycholate, 1% Nonidet P-40) containing 1x protease inhibitor cocktail set III (CalBiochem) and phosphatase inhibitor cocktail A (Santa Cruz Biotechnology) was added to approximately 30 mg of sample tissue. For 2D Western blots, 200 µL of 2D Lysis Buffer (30 mM Tris-HCl, pH 8.8, 7 M urea, 2 M thiourea, and 4% CHAPS) containing 1x protease inhibitor cocktail set III (CalBiochem) and phosphatase inhibitor cocktail A (Santa Cruz Biotechnology) was added to a 2 mm × 2 mm × 2 mm tissue sample. The tissue samples were sonicated at 4° C, incubated on a shaker for 30 min at room temperature, and spun at 4° C for 30 min (12,000 rpm). The supernatant was collected and protein concentration determined using the Bio-Rad protein assay method. Protein samples were diluted with the appropriate buffer to equal concentrations between 5 and 8 mg/ml.
2.4 Acetyl-lysine Enrichment
Left ventricular protein extract (100 µg) from the SHHF failing heart was diluted in 750 µl of Protein Lysis Buffer. 20 µl of protein A/G plus agarose beads (Santa Cruz Biotechnology) were added to the sample followed by incubation on a rotator for 1 hour at 4° C to remove any non-specific binding of protein to the beads. The sample was then spun for 1 min at 1600 rpm and the supernatant was transferred to a new tube. 10 µl of rabbit monoclonal anti-acetyl-lysine antibody (Cell Signaling) was added and the mixture was incubated on a rotator at 4° C overnight. The next day, 50 µl of protein A/G plus agarose beads (Santa Cruz) was added to the sample followed by incubation on a rotator for 2 hours at 4° C. The sample was then spun at 1600 rpm for 1 min and the supernatant was removed. The remaining beads were washed three times with 1 ml of PBS containing decreasing amounts (1%, 0. 5%, and 0.05%) of Triton-X100. The protein was then eluted from the beads by adding 2D Sample Buffer (8 M urea, 4% CHAPS, 20 mg/ml DTT, 2% pharmalytes pH range 3 to 10 (GE Healthcare), and trace amounts of bromophenol blue). The samples were spun at 1600 rpm for 1 min and then the supernatant was harvested.
2.5 Detection of Lysine Acetylated Proteins by 2D Western Blot
1.0 µL of diluted CyDye (1 nmol/µL stock diluted 1:5 with dimethylformamide) (GE Healthcare) was added to 100 µg of protein. The samples were vortexed and then incubated for 30 min on ice, under dark conditions. 1.0 µL of 10 mM Lysine was added to each sample. The samples were vortexed and incubated for an additional 15 min on ice, under dark conditions. 2D Sample Buffer, 100 µL of destreak solution (GE Healthcare), and Rehydration buffer (7 M urea, 2 M thiourea, 4% CHAPS, 20 mg/ml DTT, 1% pharmalytes, and trace amounts of bromophenol blue) were added to reach a total volume of 250 µL. The samples were mixed, spun, and equal amounts of protein were loaded onto 13 cm IPG strips (pH 3–10 linear) (GE Healthcare) under 1 ml mineral oil. IEF was performed for 12 hrs at 20° C with 50 µA/strip. The focused IPG strips were incubated in freshly made Equilibration Buffer 1 (50 mM Tris-HCl, pH 8.8, containing 6 M urea, 30% glycerol, 2% SDS, 10 mg/ml DTT, and trace amounts of bromophenol blue) for 15 min with slow shaking. The strips were rinsed with Equilibration Buffer 2 (50 mM Tris-HCl, pH 8.8, containing 6 M urea, 30% glycerol, 2% SDS, 45 mg/ml iodoacetamide, and trace amounts of bromophenol blue) and then incubated with Equilibration Buffer 2 for 10 min with slow shaking. The IPG strips were rinsed once in SDS-gel running buffer, loaded into the 12% SDS-gels, and sealed with 0.5% w/v agarose solution (in SDS-gel running buffer). Paired samples were run simultaneously at 15° C until the dye front ran out of the gels. Gels were scanned immediately following SDS-PAGE using Typhoon TRIO (Amersham Biosciences) following the protocols provided. The scanned images were then analyzed by Image QuantTL software (GE Healthcare).
Proteins were transferred to PVDF membrane using a semidry transfer unit. The membranes were blocked with Blocking Buffer (5% w/v BSA in 1x TBST) for 3 hours and then incubated with primary rabbit monoclonal anti-acetyl-lysine antibody (1 µg/ml in Blocking Buffer) (Cell Signaling) for 3 hrs. The membranes were then washed 3 times with TBST, incubated with CF647-labeled goat anti-rabbit IgG secondary antibody (diluted 1:1000) (Biotium Inc.) for 2 hrs, and then washed 5 times with TBST. Membranes were scanned using Typhoon TRIO and image overlays were done using Image QuantTL software.
2.6 MALDI-TOF/TOF Mass Spectrometry
Protein spots chosen for analysis were excised by Ettan Spot Picker (GE Healthcare) and washed multiple times to remove staining dye and other inhibitory chemicals. Gel spots were dried and then rehydrated in digestion buffer containing sequencing grade modified trypsin. Proteins were digested in-gel at 37°C and digested peptides were extracted from the gel with TFA extraction buffer and shaking. The digested tryptic peptides were desalted using C-18 Zip-tips (Millipore) and then mixed with CHCA matrix (alpha-cyano-4-hydroxycinnamic acid) and spotted into the wells of a MALDI plate. Mass spectra (MS) of the peptides in each sample were obtained using an Applied Biosystems Proteomics Analyzer and five of the most abundant peptides in each sample were further subjected to fragmentation and tandem mass spectrometry (MS/MS) analysis. The combined MS and MS/MS spectra were submitted for database search using GPS Explorer software equipped with the MASCOT search engine to identify proteins from the NCBI non-redundant mammalian protein database. Proteins that were identified with a confidence interval (C.I.) greater than 99% were reported.
2.7 Sirt3 Protein Expression and 1D western blot to identify protein acetylations
Protein (50 µg) from left ventricular tissue extracts was boiled for 5 minutes in Laemmli buffer and subjected to SDS-PAGE using a 10% gel. The proteins were transferred to a PVDF membrane and washed 3 times with Tris-buffered saline containing Tween 20 (TBST) solution (20 mM Tris, 500 mM NaCl, and 0.1% Tween-20 [pH 7.5]). The membrane was then blocked in TBST containing 5% w/v non-fat dry milk for 1 hour on a shaker at room temperature. After 3 washes in TBST, the membrane was incubated overnight at 4° C on a shaker with either Sirt3 rabbit monoclonal antibody (Cell Signaling) or anti-acetylated lysine monoclonal antibody (Cell signaling) in TBST containing 5% w/v bovine serum albumin (BSA) at a dilution of 1:500. The membrane was then washed 3 times in (TBST) and incubated for 2 hours with peroxidase conjugated goat anti-rabbit IgG secondary antibody (Santa Cruz Biotechnology) at a dilution of 1:2000 in TBST containing 5% w/v non-fat dry milk. After 4 additional washes in TBST, the membrane was incubated for 1 minute with Amersham ECL Western Blotting detection reagent (GE Healthcare) and developed using the Kodak Image Station 4000R Pro for varying times to obtain desirable band intensity within a linear range and optimal saturation. The bands were quantified by ImageJ analysis and band densities were normalized to GAPDH. GAPDH levels were determined as described above using a rabbit monoclonal antibody (Cell Signaling).
2.8 Validation of proteins with increased acetylations by immunoprecipitation
The left ventricular protein extracts from W and SHHF rats are immunoprecipitated with anti-acetylated lysine monoclonal antibody (Cell Signaling) and subjected to western blotting with antibodies against ATP synthase β, long chain acyl-CoA dehydrogenase (LCAD), aspartate aminotransferase (AAT), and sarcomeric creatine kinase (sMtCK) (Santa Cruz Biotechnology).
2.9 Statistical Analysis
Student’s t-test for two samples assuming unequal variance was used to determine statistical significance between two compared groups. The two-tail P-value is reported for each analysis and P-values of less than 0.05 were considered statistically significant. Outliers (> 2 SD from mean) were dropped.
3. RESULTS
3.1 Cardiac Measurements (Table 1)
Table 1. Cardiac parameters.
The cardiac parameters for the SS and SHHF rats are shown along with the parameters for their respective control groups, SR and W. n = 4 for each model. Left ventricular posterior wall thickness (LV PWT) and left ventricular ejection fraction (LV EF) are represented as the average for each group ± the standard deviation.
| Heart | n | Mean LV PWT ± Std. Dev. (mm) | Mean LV EF ± Std. Dev. (%) |
|---|---|---|---|
| Failure - SS | 4 | 1.718 ± 0.162 | 34.96 ± 2.57* |
| Control - SR | 4 | 1.606 ± 0.313 | 59.48 ± 2.90 |
| Failure – SHHF | 4 | 1.806 ± 0.133 | 42.78 ± 7.27* |
| Control – W | 4 | 1.822 ± 0.170 | 66.39 ± 5.74 |
P < 0.05 for SS vs. SR and SHHF vs. W.
Echocardiography was performed on a weekly interval. The SS and SHHF rats were sacrificed upon reaching heart failure at 29 weeks and 18 months, respectively, consistent with previous reports [9,14]. Upon sacrifice, the rats exhibited a significant decrease in ejection fraction compared to controls (P < 0.05) with an accompanying return in left ventricular posterior wall thickness to control levels (Table 1). Furthermore, the SHHF rats exhibited dyspnea and upon sacrifice had marked pleural effusion indicative of cardiac failure.
3.2 Lysine Acetylated Proteins in Heart Failure
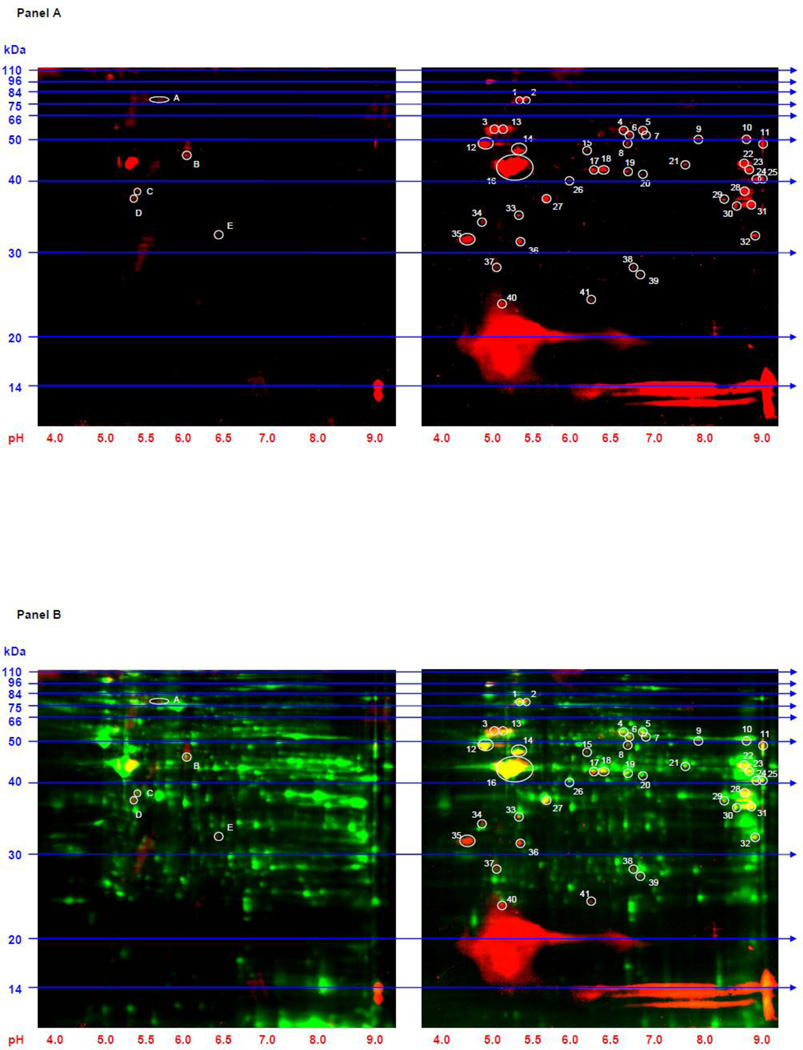
Whole protein extracts from the left ventricle and septum of the rats were subjected to 2D gel electrophoresis followed by immunoblotting with an anti-acetyl-lysine antibody (Figures 1 and 2). Forty-one and 66 acetylated protein spots were detected by immunoblotting in SS and SHHF failing hearts that were less abundant in the respective controls. Total protein from each spot was also detected by CyDye labeling. Five protein spots demonstrated increased lysine acetylation in the control models compared to their respective heart failure models. These spots, however, were not consistent across the two control models and thus were not investigated further. Twelve of the acetylated protein spots in heart failure were common to both models.
Figure 1. 2D Western blots to identify lysine acetylated proteins in whole tissue extracts from SS failing and SR control hearts.
Panel A: 2D gels (100 µg of protein extract was loaded on each gel) from SR control (left gel) and SS heart failure (right gel) immunoblotted with anti-acetyl-lysine antibody are shown. Forty-one spots (circled and numbered in white on the right gel) were more intense in the heart failure model while five spots (circled and lettered on the left gel) were more intense in the control model. Panel B: The 2D gels from Panel A are depicted along with a representation of all resolved proteins. SR control (left gel) and SS heart failure (right gel) are shown. The red indicates proteins that reacted with the anti-acetyl-lysine antibody. The green represents total protein from each extract obtained from CyDye labeling. The yellow indicates areas of overlap. Molecular weight (kDa) markers are displayed on the left side of the gels and pH markers are displayed underneath each gel.
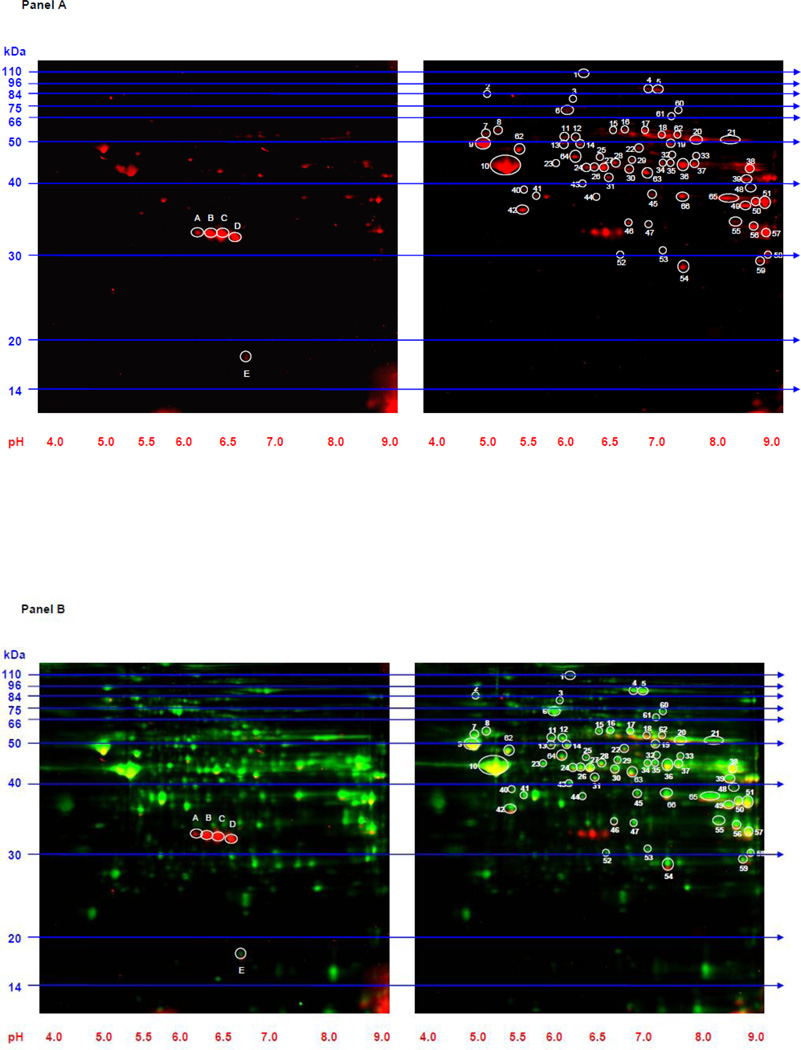
Figure 2. 2D Western blots to identify lysine acetylated proteins in whole tissue extracts from SHHF failing and W control hearts.
Panel A: 2D gels (100 µg of protein extract is loaded on each gel) from W control (left gel) and SHHF heart failure (right gel) immunoblotted with anti-acetyl-lysine antibody are shown. Sixty-six spots (circled and numbered in white on the right gel) were more intense in the heart failure model while five spots (circled and lettered on the left gel) were more intense in the control model. Panel B: The 2D gels from Panel A are depicted along with a representation of all resolved proteins. W control (left gel) and SHHF heart failure (right gel) are shown. The red indicates proteins that reacted with the anti-acetyl-lysine antibody. The green represents total protein from each extract obtained from CyDye labeling. The yellow indicates areas of overlap. Molecular weight (kDa) markers are displayed on the left side of the gels and pH markers are displayed underneath each gel.
These were identified from the SS gel using MALDI-TOF/TOF mass spectrometry coupled to Mascot Analysis (Table 2). The identity of each protein spot was determined with a confidence interval greater than 99%. To confirm that the common spots in the SHHF gel corresponded to the same proteins identified in the SS gel, two of these common spots were excised from the SHHF gel and correctly identified as mitochondrial malate dehydrogenase (peptide count: 16; protein score: 651; C.I.: 100%; sequence coverage: 59.17%) and pyruvate dehydrogenase (lipoamide) beta (peptide count: 16; protein score: 585; C.I.: 100%; sequence coverage: 67.13%).
Table 2. Lysine acetylated proteins in heart failure.
Proteins which demonstrated increased lysine acetylation in both the SS and SHHF model of heart failure are listed.
| Protein ID | Accession No. |
Pred. M.W. |
Pred .P.I. |
Peptide Count |
Protein Score |
C.I. % |
Seq. Cov. % |
Spot # in SS |
Spot # in SHHF |
|---|---|---|---|---|---|---|---|---|---|
| Dihydrolipoamide dehydrogenase | gi|38303871 | 54004.1 | 7.96 | 16 | 465 | 100 | 45 | 4* | 16 |
| 3-oxoacid CoA transferase 1 | gi|205829936 | 56168.1 | 8.7 | 20 | 748 | 100 | 55 | 5* | 17 |
| NADH dehydrogenase (ubiquinone) flavoprotein 1, 51kDa | gi|55741424 | 50698.6 | 8.37 | 23 | 398 | 100 | 57.11 | 9* | 20 |
| ATP synthase beta Subunit | gi|1374715 | 51170.6 | 4.92 | 23 | 658 | 100 | 66.53 | 12* | 9 |
| Ubiquinol-cytochrome c reductase core protein I | gi|51259340 | 52815.4 | 5.57 | 17 | 565 | 100 | 44.17 | 14* | 62 |
| Actin, alpha 1, skeletal Muscle | gi|149043182 | 51404.2 | 5.91 | 12 | 479 | 37.42 | 16* | 10 | |
| Long-chain acyl-CoA dehydrogenase | gi|205145 | 47842.4 | 7.63 | 23 | 463 | 100 | 51.16 | 17* | 26 |
| Long-chain acyl-CoA dehydrogenase | gi|205145 | 47842.4 | 7.63 | 22 | 736 | 100 | 53.02 | 18* | 27 |
| Muscle creatine kinase | gi|6978661 | 42991.8 | 6.58 | 23 | 656 | 100 | 62.21 | 19* | 30 |
| Aspartate aminotransferase, cytoplasmic | gi|122065118 | 46399.5 | 6.73 | 26 | 613 | 100 | 74.09 | 20* | 63 |
| Sarcomeric mitochondrial creatine kinase | gi|57537 | 47443.3 | 8.64 | 21 | 373 | 100 | 51.79 | 21* | 36 |
| Malate dehydrogenase, mitochondrial | gi|38648863 | 35660.8 | 8.93 | 16 | 517 | 100 | 57.4 | 29* | 65 |
| Malate dehydrogenase, mitochondrial | gi|38648863 | 35660.8 | 8.93 | 18 | 752 | 100 | 63.91 | 31* | 51* |
| Pyruvate dehydrogenase (lipoamide) beta | gi|50925725 | 38957 | 6.2 | 16 | 649 | 100 | 69.36 | 33* | 42* |
Spot numbers marked with an * indicate that the identity of the protein spot was determined by MALDI-TOF/TOF mass spectrometry and Mascot analysis (C.I. > 99%).
Pred. M.W. = Predicted Molecular Weight (Daltons); Pred. P.I. = Predicted P.I. (pH); C.I. = Confidence Interval (%); Seq. Cov. = Sequence Coverage (%).
3.3 Confirmation of increased acetylation of proteins in failure models by 1D gel analysis (Figs 3 and 4)
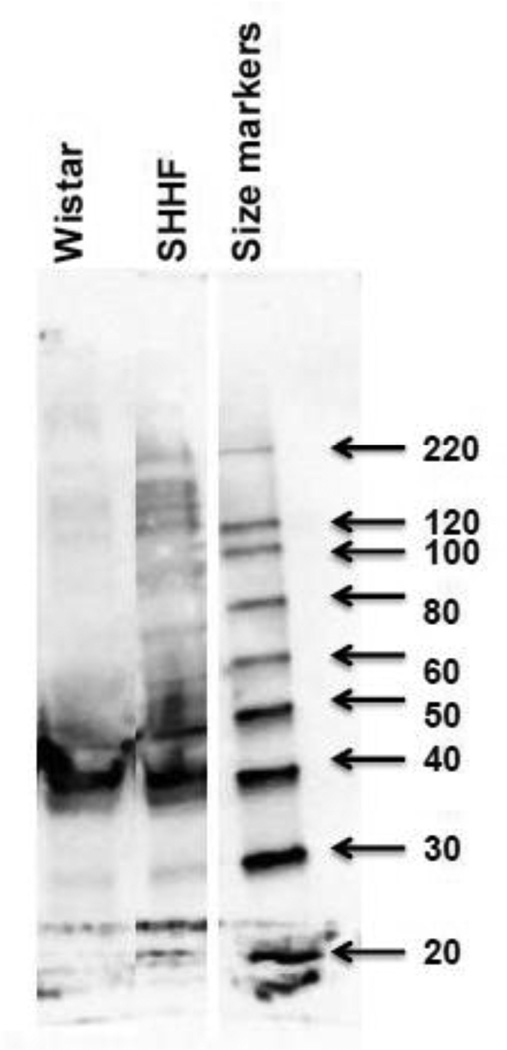
Figure 3. 1D western blot to evaluate increased acetylated proteins in whole cell extracts form W and SHHF rats.
Fifty µg of left ventricular extract is loaded on gel and subjected to western blot using monoclonal anti-acetylated lysine antibody (cell signaling Inc). Molecular weight (kDa) markers are displayed on the right side of the gel.
Figure 4. Acetyl-lysine enrichment in SHHF failing hear.
Left ventricular protein extract from the SHHF failing heart was immunoprecipitated with an anti-acetyl-lysine antibody, labeled with CyDye, and then subjected to 2D gel electrophoresis. A portion of the 2D gel is shown. Five of the proteins which demonstrated increased lysine acetylation in both models of heart failure were identified on this gel (circled and numbered in white). Green indicates resolved proteins obtained from CyDye labeling. Molecular weight (kDa) markers are displayed on the left side of the gel and pH markers are displayed underneath.
To confirm and complement the results of 2D western blot, 1D western blot on heart extracts of W and SHHF rats was performed with anti-acetyl-lysine antibody. Increased acetylation of proteins was detected in SHHF compared to its control counterpart W rats (Fig 3).
To confirm that our technique was reliably identifying proteins with lysine acetylations in heart failure, we subjected left ventricular tissue extracts from the SHHF failing heart to an acetyl-lysine enrichment followed by 2D gel electrophoresis (Figure 4). Five protein spots which we believed corresponded to the lysine acetylated proteins present in both models of heart failure were excised from the gel and subjected to MALDI-TOF/TOF mass spectrometry. They were correctly identified (C.I. > 99%) as ATP synthase, long chain acyl-CoA dehydrogenase, aspartate aminotransferase, muscle creatine kinase, and sarcomeric creatine kinase (Table 3), thus confirming the specificity of our global screening strategy. Corresponding spots to 11 of the 12 (PDH, spot 42, was detected in the non-enriched sample only) identified ones (section 3.2 and Fig 2)) were also present in the acetyl-lysine enrichment gels (Fig 4), indicating that these were specific.
Table 3. Acetyl-lysine enriched protein spots in the SHHF failing heart.
Five of the 12 proteins which demonstrated increased acetylation in heart failure were identified in an acetyl-lysine enrichment by MALDI-TOF/TOF mass spectrometry and Mascot analysis (C.I. > 99%).
| Protein ID | Access ion No. |
Pred. M.W. |
Pred .P.I. |
Peptide Count |
Protein Score |
C.I. % |
Seq. Cov. % |
Spot # |
|---|---|---|---|---|---|---|---|---|
| ATP synthase beta Subunit | gi|1374715 | 51170.6 | 4.92 | 21 | 1270 | 100 | 67.16 | 1 |
| Long-chain specific acyl-CoA dehydrogenase | gi|205145 | 47842.4 | 7.63 | 15 | 276 | 100 | 34.42 | 2 |
| Muscle creatine kinase | gi|6978661 | 42991.8 | 6.58 | 14 | 570 | 100 | 40.16 | 3 |
| Aspartate aminotransferase, cytoplasmic | gi|122065118 | 46399.5 | 6.73 | 12 | 300 | 100 | 31.72 | 4 |
| Sarcomeric mitochondrial creatine kinase | gi|57537 | 47443.3 | 8.64 | 16 | 565 | 100 | 38.66 | 5 |
Pred. M.W. = Predicted Molecular Weight (Daltons); Pred. P.I. = Predicted P.I. (pH); C.I. = Confidence Interval (%); Seq. Cov. = Sequence Coverage (%).
3.4 Validation of hyper-acetylated proteins identified by MALDI-TOF/TOF (Fig 5)
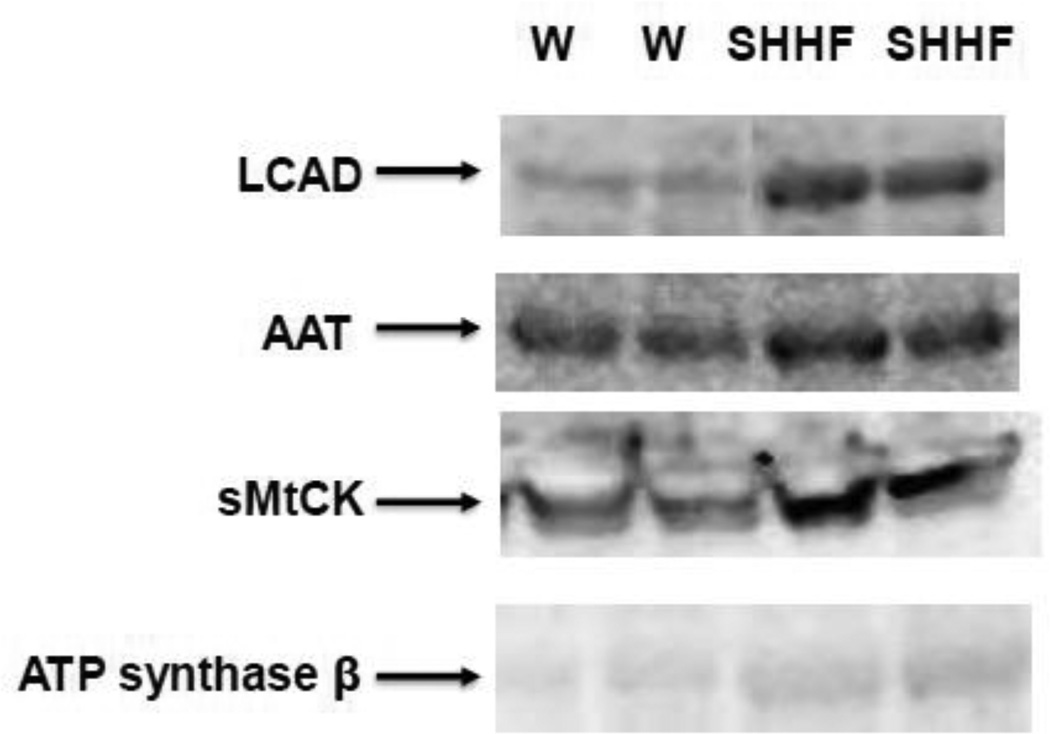
Figure 5. Validation of hyper-acetylated proteins identified with MADI-TOF/TOF by western blot analysis.
Hundred µg of left ventricular protein extracts from W and SHHF rats are immunoprecipitated with monoclonal anti-acetylated lysine antibody followed by western blot analysis with antibodies against proteins indicated on left.
The hyper-acetylated proteins that were identified by MALDI-TOF/TOF were validated by immunoprecipitation with anti-acetyl-lysine antibody followed by western blotting with antibodies against respective proteins (Fig 5). Increased acetylations of ATP synthase β, long chain acyl-CoA dehydrogenase (LCAD), aspartate aminotransferase (AAT), and sarcomeric creatine kinase (sMtCK) were observed in heart failure (SHHF) rats compared to those of control groups (Fig 5).
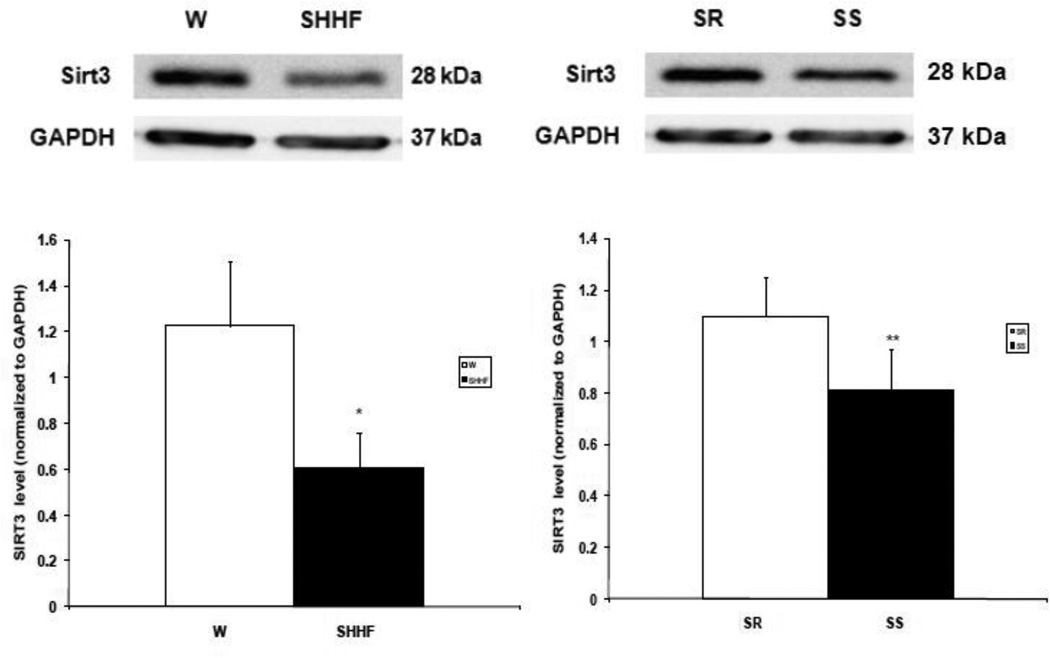
3.5 Sirt3 Abundance (Figure 6)
Figure 6. Sirt3 abundance in heart failure.
Fifty µg of left ventricular protein extracts from hearts were subjected to 1D SDS-PAGE and then immunoblotted with monoclonal anti-Sirt3 antibody. Upper Panel: representative Western blot for short form of Sirt3 (28 kDa) and GAPDH. Lower The abundance of short form of Sirt3, normalized to GAPDH for failing SHHF and control W rat hearts (left) and failing SS and control SR rats (right). n=3 for each rat model. * P < 0.05 **P = 0.07.
Since the majority of acetylated proteins identified were mitochondrial, the abundance of the mitochondrial deacetylase Sirt3 was measured by Western blot analysis in whole tissue extracts from failing SS and SHHF hearts as well as control SR and W hearts. The Sirt3 levels, normalized to GAPDH, were lower in SS and SHHF failure rats compared to their counterparts SR and W rats (Fig 6; P < 0.05).
3.6 Acetylation of LCAD, a Sirt3 target, is increased in failure models
To establish whether down-regulation of Sirt3 might be a potential mechanism for increased acetylation of proteins in SHHF rats compared to W rats we evaluated the acetylation status of a known Sirt3 target LCAD [2,12]. LCAD is more acetylated in heart failure models compared to controls (Fig 5 LCAD).
4. DISCUSSION
Using a global screening strategy, twelve non-histone lysine acetylated proteins were identified that are more abundant in failing than normal hearts. To our knowledge, this is the first instance where increased lysine acetylation of non-histone proteins has been shown in two models of pressure-overload induced heart failure. The acetylated proteins include multiple mitochondrial enzymes involved in all aspects of cardiac energy metabolism, such as pyruvate dehydrogenase, 3-oxoacid CoA transferase, NADH dehydrogenase, ATP synthase, ubiquinol-cytochrome c reductase, long-chain acyl-CoA dehydrogenase (LCAD), mitochondrial creatine kinase (mCK), and mitochondrial malate dehydrogenase (mMDH). These appear to be linked to a decrease in mitochondrial Sirt3 levels.
The abundance of lysine acetylated proteins was compared by immunoblotting of 2D gels. Possible mechanisms for changes in abundance include either parallel changes in total protein expression and/or degree of acetylation. Since lysine acetylations shift isoelectric points to the more acidic side, the non-acetylated forms are not expected at the same spot as the acetylated protein. Multiple spots on the 2D gel may exist for a given protein with various degrees of acetylation. The possibility that corresponding spots represent the same protein with different, but the same total amount of lysines acetylated, cannot be excluded.
MDH has lysine acetylations on four residues, Lys 185, 301, 307 and 314 [32]. Furthermore, inhibition of deacetylase activity, via treatment with trichostatin A and nicotinamide, increased activity of both endogenously and ectopically expressed MDH, indicating that acetylations regulate its enzymatic activity [32]. MDH activity is reduced in patients with cardiac insufficiency due to valvular heart disease [23], while MDH levels were unaltered in an animal model of pressure-overload failure [4]. However, the role of MDH acetylation and its altered activity in heart failure remains to be determined.
LCAD purified from liver mitochondria of Sirt3 knock-out mice has been reported to be acetylated at eight lysine residues, Lys 42, 156, 189, 240, 254, 318, 322, and 358. Sirt3 deacetylates LCAD at Lys 42, resulting in a two fold increase in the enzymatic activity of LCAD [10]. Furthermore, increased LCAD activity has been linked to increased state 3 respiration and improved contractility in heart failure [25].
Bugger et al. [4] showed a strong reduction in fatty acid oxidation without a concomitant decrease in LCAD expression levels in pressure-overload induced failure. Acetylation of LCAD has been reported to reduce its enzymatic activity. [11, 26]. These reports coupled to our finding of increased acetylation of LCAD raise the prospect that reduced LCAD activity, due to acetylation, may be an important aspect of perturbed fatty acid oxidation in cardiac failure. This suggests a model in which fatty acids increase acetyl-coA acid, which in turn increases LCAD acetylation, leading to reduced LCAD activity and mitochondrial function.
Acetylation of the amino terminus and epsilon amino group of lysine of a variety of mitochondrial outer membrane proteins has been reported [17]. Over 1300 mitochondrial acetylated proteins were reported by Zhao [31]. While the effect of only a few of these acetylations has been determined, there is significant evidence that they affect protein function. For example, Trichostatin A, an inhibitor of class 1 and class 2 histone deacetylases and nicotinamide (NAM), an inhibitor of Sirt family deacetylases, increases the activity of enoyl-CoA hydratase/3-hydroxyacyl-CoA dehydrogenase and malate dehydrogenase [31]. On the other hand, argininosuccinate catalytic activity is inhibited by acetylation. It has been hypothesized that acetylation regulates the activity of AMP-forming enzymes [28]. Acetylation of both mitochondrial and cytosolic acetyl-CoA synthase isoforms results in loss of function [8,28]. Acetylation of hepatic carnitine palmitoyltransferase-1 (CPT1) has been identified in fed, but not starved mice, and postulated in increased sensitivity to malonyl-CoA inhibition [19]. Acetylation of the three forms of the voltage-dependent anion channel (VDAC1-3) and Long-chain acyl-CoA synthase 1 (ACS1) has been reported, however, the effect on activity is not known.
Acetylations of several of the proteins found in our global screen of failing hearts have been reported in other contexts (Table 4). Acetylation of creatine kinase has been demonstrated in mouse organs [15]. In heart failure, creatine kinase activity is significantly decreased [13]. Previous work has focused upon post-translational phosphorylation(s) of creatine kinase and its regulation of activity in the heart [20]. In our study, acetylation of both the mitochondrial and cytosolic forms of creatine kinase were associated with heart failure, however the role that this modification plays in the activity of the enzyme has not been established. Acetylation of several components of the electron transport chain, which have decreased activity in heart failure, i.e., complex I, III, and V [3,21], were found to be associated with heart failure in our study, raising the possibility that acetylation of these proteins may regulate mitochondrial respiration and ATP production.
Table 4. Previous reports of lysine acetylations for the acetylated proteins associated with heart failure.
The lysine acetylated proteins common to both models of heart failure are listed with any known acetylation sites previously reported in the literature and the model in which the acetylations were documented.
| Protein | Reported Acetylation Site in Literature |
Reference | Model |
|---|---|---|---|
| Dihydrolipoamide dehydrogenase | Lysine 127 | [8] | mouse liver |
| NADH dehydrogenase (ubiquinone) flavoprotein 1, 51kDa | Lysine 81, 104, 375 | [8] | mouse liver |
| ATP synthase beta subunit | Lysine 133, 259, 522 | [8] | mouse liver |
| Ubiquinol-cytochrome c reductase core protein I | Lysine acetylation detected but no specific site was determined | [25] | mouse liver |
| Actin, alpha 1, skeletal muscle | Lysine acetylation detected but no specific site was determined | [7] | human |
| Long-chain acyl-CoA dehydrogenase | Lysine 42 (functionally significant), 156, 189, 240, 254, 318, 322, 358 | [14] | SIRT3 KO mouse liver |
| Muscle creatine kinase | Lysine acetylation detected but no specific site was determined | [16] | mouse skeletal tissue |
| Aspartate aminotransferase, cytoplasmic | Lysine 154 | [7] | human |
| Malate dehydrogenase | Lysine 165, 185, 301, 307, 314, 329, 335 | [7],[8],[11] | mouse liver, human |
| Pyruvate dehydrogenase (lipoamide) beta | Lysine 354 | [7] | human |
In the present study, Sirt3 abundance was decreased in both heart failure models. Reduced expression of mitochondrial Sirt3 has been shown in two models of pressure-overload induced cardiac hypertrophy and linked to the hypertrophic response to pressure-overload by Gupta et al. in a series of experiments in which it was shown that Sirt3 plays an anti-hypertrophic role through LKB1-AMPK and forkhead box O3a-dependent signaling pathways [24,29]. Sirt3 has also been shown to regulate mitochondrial proteins through deacetylation. The activity of manganese superoxide dismutase, the primary mitochondrial reactive oxygen species scavenger, is increased by Sirt3-mediated deacetylation [22]. Acetyl-CoA synthetase 2 is also activated by Sirt3 mediated deacetylation at Lysine 642 [26]. The present results raise the prospect that a potential mechanism for increased mitochondrial protein acetylations in heart failure is a reduction in the mitochondrial deacetylase Sirt3.
Highlights.
Non-histone acetylated proteins associated with heart failure were identified.
Dahl salt-sensitive and spontaneously hypertensive heart failure rats were used.
Global screening was done using 2D gel electrophoresis coupled to immunoblotting.
Twelve lysine acetylated proteins were common to both models of heart failure.
Mitochondrial Sirt3 was reduced in failing hearts.
Acknowledgements
We appreciate the helpful discussions with Drs. Subhash Pandey (University of Illinois at Chicago) and Pieter deTombe (Loyola University, Chicago).
Role of the Funding Source: Work supported by Veterans Administration Merit Award (RSD), NIH R21HL096031 (RSD), NIH KO1 DK071641 (KK), NIH T32 HL 07692 (KRJ).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
C.I.: confidence interval; GAPDH: glyceraldehyde 3-phosphate dehydrogenase; HATs: histone acetyltransferases; HDACs: histone deacetylases; HDACis: histone deacetylase inhibitors; LCAD: long-chain acyl-CoA dehydrogenase; LV EF: left ventricular ejection fraction; LV PWT: left ventricular posterior wall thickness; MDH: malate dehydrogenase; SHHF: spontaneously hypertensive heart failure prone; Sirt3: Sirtuin-3; SR: Dahl salt-resistant; SS: Dahl salt-sensitive; W: W.
Disclosures: none declared
Reference
- 1.Backs J, Olson EN. Control of cardiac growth by histone acetylation/deacetylation. Circ. Res. 2006;98:15. doi: 10.1161/01.RES.0000197782.21444.8f. [DOI] [PubMed] [Google Scholar]
- 2.Bell EL, Guarente L. The SirT3 divining rod points to oxidative stress. Mol. Cell. 2011;42:561. doi: 10.1016/j.molcel.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchwald A, Till H, Unterberg C, Oberschmidt R, Figulla HR, Wiegand V. Alterations of the mitochondrial respiratory chain in human dilated cardiomyopathy. Eur. Heart J. 1990;11:509. doi: 10.1093/oxfordjournals.eurheartj.a059743. [DOI] [PubMed] [Google Scholar]
- 4.Bugger H, Schwarzer M, Chen D, Schrepper A, Amorim PA, Schoepe M, Nguyen TD, Mohr FW, Khalimonchuk O, Weimer BC, Doenst T. Proteomic remodelling of mitochondrial oxidative pathways in pressure overload-induced heart failure. Cardiovasc. Res. 2010;85:376. doi: 10.1093/cvr/cvp344. [DOI] [PubMed] [Google Scholar]
- 5.Bush EW, McKinsey TA. Protein acetylation in the cardiorenal axis: the promise of histone deacetylase inhibitors. Circ. Res. 2010;106:272. doi: 10.1161/CIRCRESAHA.109.209338. [DOI] [PubMed] [Google Scholar]
- 6.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 7.Gusterson RJ, Jazrawi E, Adcock IM, Latchman DS. The transcriptional co-activators CREB-binding protein (CBP) and p300 play a critical role in cardiac hypertrophy that is dependent on their histone acetyltransferase activity. J. Biol. Chem. 2003;278:6838. doi: 10.1074/jbc.M211762200. [DOI] [PubMed] [Google Scholar]
- 8.Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc. Natl. Acad. Sci. U. S. A. 2006;103:10230. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heyen JR, Blasi ER, Nikula K, Rocha R, Daust HA, Frierdich G, Van Vleet JF, De CP, McMahon EG, Rudolph AE. Structural, functional, and molecular characterization of the SHHF model of heart failure. Am. J. Physiol Heart Circ. Physiol. 2002;283:H1775–H1784. doi: 10.1152/ajpheart.00305.2002. [DOI] [PubMed] [Google Scholar]
- 10.Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S, Ilkayeva OR, Stevens RD, Li Y, Saha AK, Ruderman NB, Bain JR, Newgard CB, Farese RV, Jr, Alt FW, Kahn CR, Verdin E. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464:121. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S, Ilkayeva OR, Stevens RD, Li Y, Saha AK, Ruderman NB, Bain JR, Newgard CB, Farese RV, Jr, Alt FW, Kahn CR, Verdin E. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464:121. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S, Ilkayeva OR, Stevens RD, Li Y, Saha AK, Ruderman NB, Bain JR, Newgard CB, Farese RV, Jr, Alt FW, Kahn CR, Verdin E. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464:121. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ingwall JS. Energy metabolism in heart failure and remodelling. Cardiovasc. Res. 2009;81:412. doi: 10.1093/cvr/cvn301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inoko M, Kihara Y, Morii I, Fujiwara H, Sasayama S. Transition from compensatory hypertrophy to dilated failing left ventricles in Dahl salt-sensitive rats. Am. J. Physiol. 1994;267:H2471–H2482. doi: 10.1152/ajpheart.1994.267.6.H2471. [DOI] [PubMed] [Google Scholar]
- 15.Iwabata H, Yoshida M, Komatsu Y. Proteomic analysis of organ-specific post-translational lysine-acetylation and -methylation in mice by use of anti-acetyllysine and -methyllysine mouse monoclonal antibodies. Proteomics. 2005;5:4653. doi: 10.1002/pmic.200500042. [DOI] [PubMed] [Google Scholar]
- 16.Kee HJ, Kook H. Roles and targets of class I and IIa histone deacetylases in cardiac hypertrophy. J. Biomed. Biotechnol. 2011;2011:928326. doi: 10.1155/2011/928326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kerner J, Lee K, Hoppel CL. Post-translational modifications of mitochondrial outer membrane proteins. Free Radic. Res. 2011;45:16. doi: 10.3109/10715762.2010.515218. [DOI] [PubMed] [Google Scholar]
- 18.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang XJ, Zhao Y. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell. 2006;23:607. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 19.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang XJ, Zhao Y. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell. 2006;23:607. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 20.Lin G, Liu Y, MacLeod KM. Regulation of muscle creatine kinase by phosphorylation in normal and diabetic hearts. Cell Mol. Life Sci. 2009;66:135. doi: 10.1007/s00018-008-8575-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marin-Garcia J, Goldenthal MJ, Moe GW. Abnormal cardiac and skeletal muscle mitochondrial function in pacing-induced cardiac failure. Cardiovasc. Res. 2001;52:103. doi: 10.1016/s0008-6363(01)00368-6. [DOI] [PubMed] [Google Scholar]
- 22.Ozden O, Park SH, Kim HS, Jiang H, Coleman MC, Spitz DR, Gius D. Acetylation of MnSOD directs enzymatic activity responding to cellular nutrient status or oxidative stress. Aging (Albany. NY) 2011;3:102. doi: 10.18632/aging.100291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peters TJ, Brooksby IA, Webb-Peploe MM, Wells G, Jenkins BS, Coltart DJ. Enzymic analysis of cardiac biopsy material from patients with valvular heart-disease. Lancet. 1976;1:269. doi: 10.1016/s0140-6736(76)91401-x. [DOI] [PubMed] [Google Scholar]
- 24.Pillai VB, Sundaresan NR, Jeevanandam V, Gupta MP. Mitochondrial SIRT3 and heart disease. Cardiovasc. Res. 2010;88:250. doi: 10.1093/cvr/cvq250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rennison JH, McElfresh TA, Okere IC, Patel HV, Foster AB, Patel KK, Stoll MS, Minkler PE, Fujioka H, Hoit BD, Young ME, Hoppel CL, Chandler MP. Enhanced acyl-CoA dehydrogenase activity is associated with improved mitochondrial and contractile function in heart failure. Cardiovasc. Res. 2008;79:331. doi: 10.1093/cvr/cvn066. [DOI] [PubMed] [Google Scholar]
- 26.Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc. Natl. Acad. Sci. U. S. A. 2006;103:10224. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spange S, Wagner T, Heinzel T, Kramer OH. Acetylation of non-histone proteins modulates cellular signalling at multiple levels. Int. J. Biochem. Cell Biol. 2009;41:185. doi: 10.1016/j.biocel.2008.08.027. [DOI] [PubMed] [Google Scholar]
- 28.Starai VJ, Celic I, Cole RN, Boeke JD, Escalante-Semerena JC. Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science. 2002;298:2390. doi: 10.1126/science.1077650. [DOI] [PubMed] [Google Scholar]
- 29.Sundaresan NR, Gupta M, Kim G, Rajamohan SB, Isbatan A, Gupta MP. Sirt3 blocks the cardiac hypertrophic response by augmenting Foxo3a-dependent antioxidant defense mechanisms in mice. J. Clin. Invest. 2009;119:2758. doi: 10.1172/JCI39162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trivedi CM, Luo Y, Yin Z, Zhang M, Zhu W, Wang T, Floss T, Goettlicher M, Noppinger PR, Wurst W, Ferrari VA, Abrams CS, Gruber PJ, Epstein JA. Hdac2 regulates the cardiac hypertrophic response by modulating Gsk3 beta activity. Nat. Med. 2007;13:324. doi: 10.1038/nm1552. [DOI] [PubMed] [Google Scholar]
- 31.Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, Li Y, Shi J, An W, Hancock SM, He F, Qin L, Chin J, Yang P, Chen X, Lei Q, Xiong Y, Guan KL. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]