Abstract
Polymeric materials have been used in a range of pharmaceutical and biotechnology products for more than 40 years. These materials have evolved from their earlier use as biodegradable products such as resorbable sutures, orthopaedic implants, macroscale and microscale drug delivery systems such as microparticles and wafers used as controlled drug release depots, to multifunctional nanoparticles (NPs) capable of targeting, and controlled release of therapeutic and diagnostic agents. These newer generations of targeted and controlled release polymeric NPs are now engineered to navigate the complex in vivo environment, and incorporate functionalities for achieving target specificity, control of drug concentration and exposure kinetics at the tissue, cell, and subcellular levels. Indeed this optimization of drug pharmacology as aided by careful design of multifunctional NPs can lead to improved drug safety and efficacy, and may be complimentary to drug enhancements that are traditionally achieved by medicinal chemistry. In this regard, polymeric NPs have the potential to result in a highly differentiated new class of therapeutics, distinct from the original active drugs used in their composition, and distinct from first generation NPs that largely facilitated drug formulation. A greater flexibility in the design of drug molecules themselves may also be facilitated following their incorporation into NPs, as drug properties (solubility, metabolism, plasma binding, biodistribution, target tissue accumulation) will no longer be constrained to the same extent by drug chemical composition, but also become in-part the function of the physicochemical properties of the NP. The combination of optimally designed drugs with optimally engineered polymeric NPs opens up the possibility of improved clinical outcomes that may not be achievable with the administration of drugs in their conventional form. In this critical review, we aim to provide insights into the design and development of targeted polymeric NPs and to highlight the challenges associated with the engineering of this novel class of therapeutics, including considerations of NP design optimization, development and biophysicochemical properties. Additionally, we highlight some recent examples from the literature, which demonstrate current trends and novel concepts in both the design and utility of targeted polymeric NPs (444 references).
1. Introduction
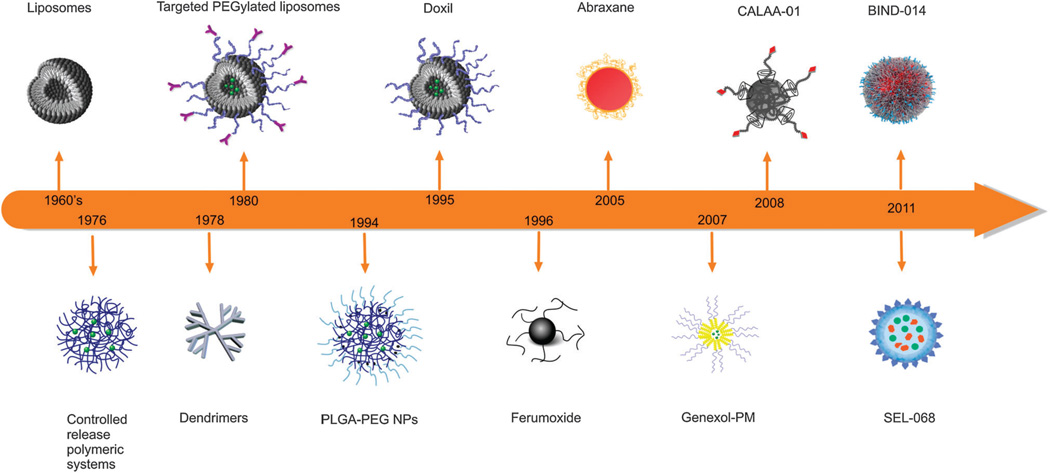
The application of nanotechnology to developing safer and more effective medicines (nanomedicine) is set to substantially influence the landscape of both pharmaceutical and biotechnology industries for decades to come.1–3 This increasing interest in nanomedicine is driven in large part by a fast pace of innovation and emerging successes of nanoparticle (NP) based drug delivery systems.4 Discoveries in the field of nanomedicine have so far proven to be both evolutionary and revolutionary in nature.5 The promotion of interdisciplinary research and the discovery of colloidal mechanisms of drug delivery in the 1960’s and 1970’s led to the development of the earlier nanomedicines; liposomes6 and polymer-drug conjugates.7, 8 The evolution of these NPs was followed by their successful “stealth” rendition by modifying the NP surface using polyethylene glycol (PEG) polymers in order to prevent non-specific binding of NP surfaces to blood components and reduce their rapid uptake and clearance in vivo by the cells of the mononuclear phagocytic system (MPS), leading to prolonged blood circulation times.21 Following the development of antibody technologies came the ability to potentially increase NP specificity through bioconjugation of affinity ligands, such as antibodies, antibody fragments, peptides, aptamers (Apts), sugars, and small molecules to their surface in order to create targeted NPs.12, 21–23 Fig. 1 presents a timeline for the development of several distinct NPs, which have been either approved for human use or are undergoing clinical trials including: liposome, albumin, and polymeric NPs. In addition to these, polymer coated iron oxide NPs have also been approved by the Food and Drug Administration (FDA) for use as magnetic resonance imaging (MRI) contrast agents.
Fig. 1.
Time line of clinical stage nanomedicine firsts. Liposomes,9 controlled release polymeric systems for macromolecules,10 dendrimers,11 targeted-PEGylated liposomes,12 first FDA approved liposome (DOXIL),13 long circulating poly(lactic-co-glycolic acid)-polyethyleneglycol (PLGA-PEG) NPs,14 iron oxide MRI contrast agent NP (Ferumoxide),15 protein based drug delivery system (Abraxane; nab technologyt),16 polymeric micelle NP (Genexol-PM),17 targeted cyclodextrin-polymer hybrid NP (CALAA-01),18 targeted polymeric NP (BIND-014; Accurint™ Technology),19 fully integrated polymeric nanoparticle vaccines (SEL-068, t SVPt™ Technology).20
Potential advantages of therapeutic NPs include: (1) the ability to improve the pharmaceutical and pharmacological properties of drugs, potentially without the need to alter drug molecules, (2) enhancement of therapeutic efficacy by targeted delivery of drugs in a tissue- or cell-specific manner, (3) delivery of drugs across a range of biological barriers including epithelial and endothelial, (4) delivery of drugs to intracellular sites of action, (5) the ability to deliver multiple types of therapeutics with potentially different physicochemical properties, (6) the ability to deliver a combination of imaging and therapeutic agents for real-time monitoring of therapeutic efficacy and, (7) possibilities to develop highly differentiated therapeutics protected by a unique set of intellectual properties.7, 24
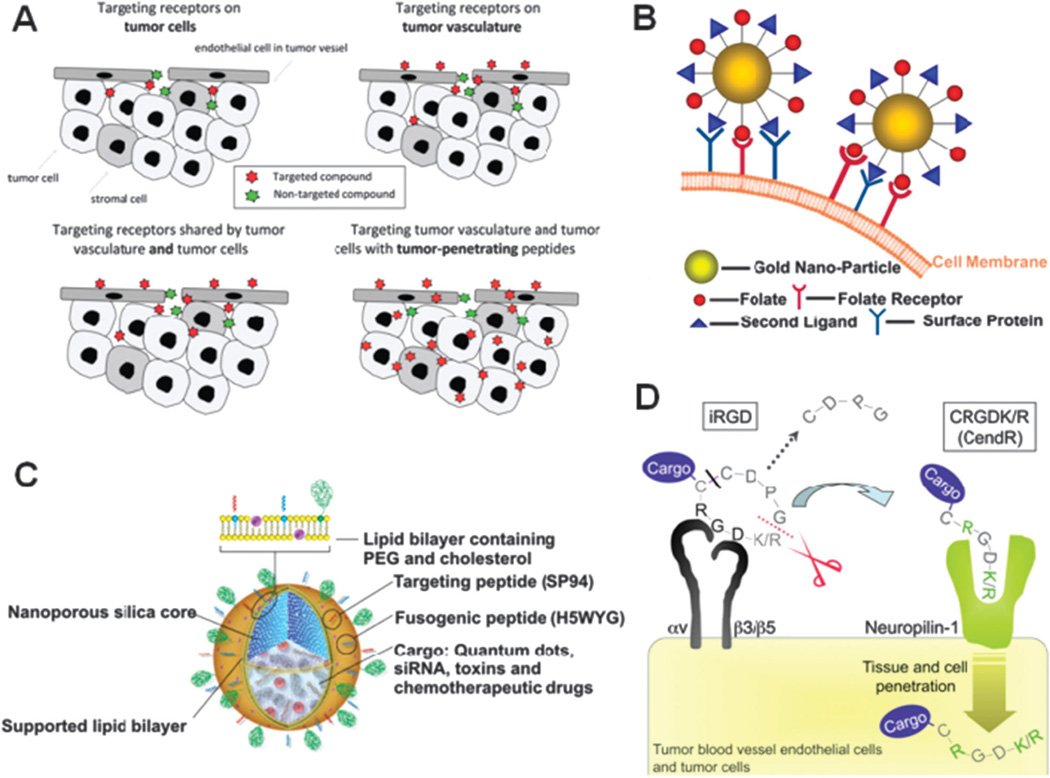
With respect to NP research, targeting refers to differential spatial localization and describes the intentional homing of NPs to active sites in disease conditions and is distinct from molecularly targeted drugs. While molecularly targeted drugs preferentially modulate the function of proteins abnormally expressed or activated in a disease state, they are not designed for spatial localization and indiscriminately distribute within the body, contributing to off-target adverse effects.25 This differential spatial localization of NPs encompasses two different approaches, which are “passive” or “active” targeting. Passive targeting refers to the preferential accumulation of NPs (bearing no affinity ligands) at active sites and is directly related to the inherent biophysicochemical properties of the NP (size, shape, charge and flexibility etc.). These biophysicochemical properties may also impede the effective concentration of NPs at active sites due to competitive events manifested by the MPS system leading to the sequestration of NPs, thereby limiting their systemic concentration and potential to extravasate into target tissues or bind to target cell populations.26 Active targeting is a term used to describe the mode of action of NPs with surface modification to incorporate affinity ligands with specificity to disease tissues and cells. These NPs differentially bind to target molecules as a result of the binding properties of the ligands on the NP surface (passive and active targeting are further described in section 3). Although more than 30 years have gone by since the implementation of the concept of targeted NPs, only a handful of these targeted NPs have reached clinical development and none have been clinically approved (Table 1).29–31 Limiting factors such as: (1) insufficient understanding of events at the nano-bio interface both in vitro and in vivo, (2) inadequate knowledge of the fate of NPs at the body, organ, and cellular levels, (3) difficulty in achieving reproducible and controlled synthesis of NPs at scales suitable for clinical development and commercialization, and, (4) lack of technologies enabling screening of a large number of NP candidates under biologically relevant conditions that could be reliably correlated to clinical performance are some possible explanations for the slow clinical translation of these nanomedicines. Though a vast array ofmaterials have been used to formulate NPs for drug delivery, including polymers, lipids, carbon, silica oxides, metal oxides and semiconductor nanocrystals,32 this review focuses specifically on targeted controlled release polymeric NPs for therapeutic applications. This focus stems from the potential impact of polymers on medicine as evidenced through previous clinical successes of polymers as biomedical materials,2, 10, 14, 33 as well as their potential as targeted therapeutic NPs.34
Table 1.
Targeted NPs in clinical development
| Identity | Ligand | Target | Nanoparticle | Active Pharmaceutical Ingredient (API) |
Indication | Status | Reference |
|---|---|---|---|---|---|---|---|
| BIND-014 | Small molecule | PSMAa | Polymeric | Docetaxel | Solid tumours | Phase I | 19 |
| SEL-068 | Small molecule | Antigen presenting cells | Polymeric | Nicotine antigen T-helper cell peptide, TLRb agonist |
Smoking cessation and relapse prevention vaccine |
Phase I | 20 |
| CALAA-01 | Transferrin | Transferrin receptor | Polymeric | siRNA | Solid tumours | Phase I | 28 |
| MBP-426 | Transferrin | Transferrin receptor | Liposome | Oxaliplatin | Gastric, esophageal, gastroesophageal adenocarcinoma |
Phase Ib/II | 29 |
| MCC-465 | Antibody fragment | Tumour antigen | Liposome | Doxorubicin | Metastatic stomach cancer |
Phase I | 30 |
| SGT53-01 | Antibody fragment | Transferrin receptor | Liposome | p53 gene | Solid tumours | Phase Ib | 31 |
PSMA: prostate specific membrane antigen.
TLR: Toll-Like Receptor agonist.
1.1 Polymeric therapeutic nanoparticles: drug delivery vehicles or a novel class of therapeutics?
The first generation clinically approved NP drug delivery technologies (liposomes, micelles, proteins etc.) lacked controlled release and active targeting properties, and were able to generally improve the safety and efficacy of the active drugs they carried. Among these first generation NPs, DOXIL, Abraxane and Genexol-PM were developed for cancer therapy. DOXIL was the first FDA approved liposome nanomedicine to reach clinical approval in 1995 for AIDS related Kaposi’s syndrome.35 By encapsulating doxorubicin (Dox) within liposomes, DOXIL changed the pharmacokinetics (PK) and biodistribution (BD) of Dox, facilitating a longer circulation half-life and therefore higher tumour dose accumulation of this drug. Although the maximum tolerated dose (MTD) of DOXIL (50 mg m−2 every 4 weeks) was lower than that of standard Dox (60 mg m−2 every 3 weeks) and DOXIL exhibited a new toxicity of hand-foot syndrome (palmar-plantar erythrodysesthesia), however, in this case the therapeutic index of Dox was enhanced. This enhancement was due to the fact that the cardiotoxicity associated with the free drug (Dox) was reduced and efficacy was demonstrated in taxane/platinum-resistant ovarian cancers.36 Despite the clinical validation of liposome technology, this class of NPs generally lack controlled release properties that can control the kinetics of drug exposure at the target tissue, and liposomes are also comparatively less stable as compared to polymeric NPs (discussed further in section 2).37, 38 The approval of Abraxane (nab-paclitaxel) in 2005 by the FDA, which is based on the NP albumin-bound (nab) platform, led to the second class of therapeutic NPs to be clinically validated.39 In comparison to paclitaxel (Ptxl) formulated with Cremophor EL (Taxol), Abraxane demonstrated significantly higher tumour response rates (33% vs. 19%) and longer times to tumour progression (23.0 vs. 16.9 weeks) among metastatic breast cancer patients who did not respond to combination therapy.40 Upon administration of Abraxane, the NP formulation rapidly dissociates into its constituents of albumin and Ptxl molecules and therefore does not materially impact the circulation half-life or BD profile of Ptxl.41 However, the nab platform significantly improved the MTD of Ptxl (260 mg m−2 vs. 175 mg m−2 for Taxol every 3 weeks) by removing the need for the use of the toxic excipient—Cremophor EL.16, 42 Therefore, nab-technology does not dramatically improve drug PK or BD, and the utility of this technology is largely limited to improving the therapeutic index of hydrophobic drugs that are currently formulated with poorly tolerated solvents. Furthermore, not all validated drugs can bind to albumin which further limits the utility of the nab-technology. Genexol-PM (a Ptxl loaded polymeric micelle) was approved in Korea in 2007. This polymeric micelle technology also removed the need for the use of Cremophor EL leading to an increase of Ptxl MTD up to 300 mg m−2 every 3 weeks for breast cancer treatment.39 Genexol-PM is currently in phase II clinical development in the USA.17, 43
Each clinically validated NP platform has made sufficient improvement to drug safety and efficacy for successful approval, yet each platform has unique limitations, and most clinicians would argue that none have made a marked improvement in clinical outcomes. Today there are nearly 250 nanomedicine products in various stages of preclinical and clinical development.44 For a NP platform to maximally improve drug pharmaceutical and pharmacological properties resulting in a highly differentiated new therapeutic with superior safety and efficacy, in most cases it will need to predictably change drug PK, BD, and tissue exposure kinetics-in a tunable and predictable manner. The successful development of such NP platforms is expected to create an entirely novel class of therapeutic NPs. The combination of one or more drugs with controlled release polymeric biomaterials for tuneable drug exposure, and molecular targeting for differential delivery has the potential to create novel therapeutic NPs for a range of medical applications.18, 45, 46 These efforts may yield NPs with highly differentiated drug pharmacology and efficacy, analogous to creating novel drugs through conventional medicinal chemistry. Polymeric NPs have the capability to: (1) release drugs at an experimentally predetermined rate over a prolonged period of time, (2) release drugs preferentially at target sites with the possibility of controlled release rates, (3) maintain drug concentrations within therapeutically appropriate ranges in circulation and within tissues and; (4) protect drugs (small molecules, proteins, nucleic acids or peptides) from hepatic inactivation, enzymatic degradation and rapid clearance in vivo. Polymeric NPs encapsulate various drugs and release them in a regulated manner via diffusion of the drug molecules through the polymer matrix or via differential surface and bulk erosion rates of the particles. The systematic design of these systems allows for the fine-tuning and optimization of the exact polymeric NP composition that can lead to increased efficacy in vivo. By careful selection of the composition of polymeric NPs resulting in optimal PK/BD, the total amount of drug and the duration of drug exposure in target tissue can be altered and improved substantially. Additionally, the incorporation of targeting ligands on NPs can lead to their increased uptake and their active agents, leading to enhanced therapeutic outcomes. In this regard, targeted polymeric NPs have the potential to be highly differentiated therapeutics, distinct from the original active agents used in their composition and distinct from first generation NPs that largely facilitated drug formulation. Additionally, the higher intracellular concentration of drugs delivered by targeted polymeric NPs can potentially maximize therapeutic efficacy by overcoming drug resistance mediated by multidrug resistance (MDR) proteins.47–49 The sub-cellular targeting of NPs can result in highly specific delivery of drug payloads to intracellular targets.50 Several MDR transporters exist, of which the p-glycoprotein, human multidrug resistance protein (MRP1) and the breast cancer resistance protein have been widely studied.51 Drug delivery using targeted polymeric NPs can result in continued release of drugs at a high concentration directly within the cell, potentially overcoming drug efflux.49 Several studies in drug-resistant mouse models have demonstrated an enhanced antitumour activity for targeted NPs (e.g. folate-receptor targeted polymeric micelles, transferrin-conjugated Ptxl NPs etc.),52–54 in comparison to their non-targeted equivalents which were shown to be not as effective. This review focuses on the development of targeted polymeric NPs and aims to highlight and discuss benefits and challenges of this novel class of therapeutics.
2. Controlled release polymeric NPs from discovery to the clinic
Controlled release systems generally refer to technologies or biomaterials that can be engineered to release drugs at predetermined and/or tuneable rates, or in response to external stimuli and triggers. Polymeric materials have emerged as a major class of controlled release systems since their unique physicochemical, synthetic, biocompatibility, and degradation properties can be readily manipulated using well-established techniques.55–57 Additionally, polymeric NP systems may be able to overcome the limitations of lipidic NPs such as liposomes. For example some key limitations of liposomes include: their propensity to burst release cargo in vivo, a lack of compatibility with various active agents, a limited drug loading volume, the oxidation of liposomal phospholipids, and poor shelf-life stability.3, 18, 58 In contrast, polymeric drug delivery systems are comparably stable in vivo, have high drug loading capacities, and can employ both controlled or triggered release of drugs.59 Due to these properties, polymeric nanomaterials are well positioned to continue to provide a diversity of solutions to a range of problems in medicine. In this section, we will provide an overview of selected developments that have led the way for the clinical translation of controlled release polymeric NPs.60
Polymers have been recognized for their potential in drug delivery applications since the 1960’s.61 However, at that time, many biomaterials were simply repurposed from industrial or household applications and therefore had inherent limitations. This began to change in the 1970’s, particularly after seminal work by Langer and Folkman in 1976 which demonstrated that controlled release of macromolecules from biodegradable polymers in a temporal manner was possible.10 A variety of macromolecules, which previously could not have been used as drugs due to PK or toxicity concerns could now be encapsulated into slow-releasing polymeric drug reservoirs and thereby developed into therapeutics suitable for use in humans. Over the past 4 decades controlled release polymer technology has impacted virtually every branch of medicine including ophthalmology, pulmonary, pain medicine, endocrinology, cardiology, orthopaedics, immunology, neurology and dentistry.62 The annual worldwide market of controlled release polymer systems which extends beyond drug delivery is now estimated at $60 billion and these systems are used by over 100 million people each year.62
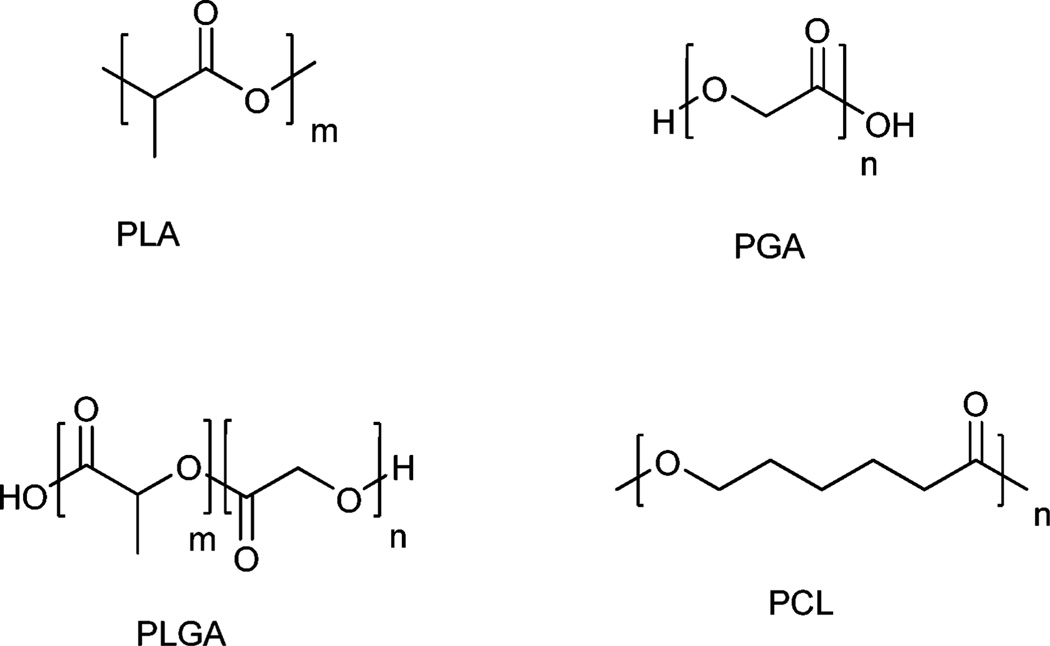
Today, the most commonly used polymers for controlled drug release applications include poly(d,l-lactide-co-glycolide) (PLGA), poly(lactic acid) (PLA), poly(glutamic acid) (PGA), poly(caprolactone) (PCL), N-(2-hydroxypropyl)-methacrylate copolymers (HPMA), and poly(amino acids).63 In particular, PLGA, PGA and PLA (Fig. 2) have been widely used in an impressive number of controlled release products, particularly due to their favourable biocompatibility and biodegradability properties. These stem, in part, from simple clearance of the polymer matrix by the body’s homeostatic metabolic pathways.64
Fig. 2.
Common biodegradable polymers utilized in controlled-release drug delivery applications. Poly(lactic acid) (PLA), poly(glutamic acid) (PGA), poly(d,l-lactic-co-glycolide) (PLGA), poly(caprolactone) (PCL).
Following the earlier work of Langer and Folkman, interest began to grow in developing slow releasing drug depots including surgical implants and injectable microparticles. The focus of these controlled release systems was principally to enable the potential use of macromolecules that had short half-lives as therapeutics, to enhance patient compliance, to improve drug efficacy and reduce side effects by delivering agents locally, and to simplify dosing in cases where prolonged drug exposure was necessary.65 Years of preclinical work done in parallel and in collaboration by several groups in the 1970s, 1980s, and 1990s ultimately led to a number of clinical successes. For example, Zoladex, a PLGA copolymer impregnated with Goserelin acetate for treating breast and prostate cancers was approved by the FDA in 1998. It was designed to be injected subcutaneously so that the active agent could be released slowly into systemic circulation and reach its target sites.66 The same year Lupron Depot, a PLGA microsphere formulation of leuprolide acetate, was approved by the FDA to treat advanced prostate cancers.67 Among other notable controlled release formulations that followed is Gliadel, a biodegradable Polifeprosan 20 carmustine-embedded wafer for the treatment of gliomas that became the first new treatment for gliomas in 20 years on its approval in 1996,68 and Atridox, a polylactide and N-methyl-2-pyrrolidone (NMP) polymer blend containing doxycycline hyclate for subgingival delivery, which was FDA approved in 1998 to treat periodontal disease.69 Other controlled release formulations of note include Sandostatin LAR, a PLGA slow release formulation of octreotide acetate for tumour control in neuroendocrine disorders approved in 1998.70 Trelstar Depot, a PLGA based microparticle formulation of triptorelin pamoatea used for prostate cancer and other indications, was FDA approved in 2000.71 The evolving ability to manufacture and control the assembly of polymers to nanoscale dimensions combined with growing interest in applying nanotechnology to medicine drove the downsizing of controlled release drug depots from macro or micro-scale products to the nano-scale.72 Indeed the clinical success of these initial formulations both validated the concept of controlled release from polymers and set the stage for the coming of the polymeric NP era.
3. NP differential spatial localization; by passive or active means
3.1 Passive targeting
Currently, all of the clinically validated therapeutic and imaging NPs are considered passively targeted first generation nanomedicines. 7 The majority of these NPs exhibit prolonged circulation times in vivo and accumulate at particular sites simply due to blood hemodynamic forces and diffusive mechanisms. Passive targeting is widely exploited in oncology applications since, in particular, tumours facilitate accumulation of NPs through the widely reported “enhanced permeation and retention” (EPR) effect. This was a milestone discovery made by Maeda et al., who in the 1980’s demonstrated the principle of passive targeting of colloidal particles to tumours.73 In their initial studies, significantly higher concentrations of the cytotoxic drug neocarzinostatin was discovered in tumour tissue post administration of the polymer-drug conjugate poly(styreneco-maleic acid)-neocarzinostatin (SMANCS), in comparison to control experiments where the drug was administered in its free form.73 This led Maeda et al. to postulate that the enhanced accumulation of the colloidal particles in the tumour was attributed to the structural features of the tumour vasculature, an observation, which was termed the EPR effect.74 The EPR effect has been observed with a wide range of macromolecular agents such as proteins; including immunoglobulin G (IgG), drug-polymer conjugates, micelles, liposomes, polymeric NPs and many other types of NPs.63, 75–77
Tumour tissue is highly heterogeneous and is perfused by an aberrant and leaky microvasculature. Indeed, tumour microvasculature has been shown to be characterized by excessive branching, chaotic structures, enlarged inter-endothelial gaps with associated break-down of tight junctions between endothelial cells, and a disrupted basement membrane.78 These large gaps between endothelial cells facilitate the extravasation of particulate material from the surrounding vessels into the tumour.79 In addition to large leaky endothelial gaps, an impaired lymphatic drainage system further entraps macromolecular particles and delays their clearance. EPR is most effective for colloidal material of >40 kDa and can occur even in the absence of targeting ligands on NPs.73 The size cut-off thresholds between endothelial cells varies between tumour type, though permeability and extravasation of NPs up to 400 nm through endothelial gaps has been observed (in mouse xenograft models of human cancers).80 In addition to abnormal architecture, tumour blood vessels also have impaired receptors for angiotensin II which controls vessel constriction.81 Solid tumours often produce large concentrations of vascular permeability factors as a result of rapidly growing tumour cells that require an increased supply of nutrients and oxygen. There are a number of vascular mediators which facilitate the EPR effect and these include; bradykinin, nitric oxide (NO), peroxynitrite (ONOO−), prostaglandins, angiotensin-converting enzyme (ACE) inhibitors, vascular endothelial permeability factor (VEGF) and numerous other cytokines.82 These factors are all indeed mediators of inflammatory processes and as such it is not surprising that the EPR effect may also manifest in other inflammatory scenarios such as arthritis, infection and advanced atherosclerotic plaques.73, 83
Currently the observations of EPR are the main premise for the design of tumour specific nanomedicines for drug delivery or imaging applications, however there are a number of caveats that need to be considered. For instance, the fact that large tumours show pathophysiological heterogeneity is a problem, as NPs cannot effectively accumulate throughout the tumour, in particular, the central regions of metastatic tumours do not exhibit the EPR effect which leads to lowered accumulation of colloidal NPs.84 Furthermore, the degree of vascular permeability which ultimately leads to heterogeneity between tumour models and variable tumour microenvironments can affect the cut-off size for NP accumulation in tumours, restricting their effective penetration range, and additionally, also accounts for the lack of observable EPR effects in certain tumour types.85, 86 Moreover, the negative pressure gradient present within the tumour interstitium can substantially limit the convection of NPs from the intravascular to the extravascular space within tumours, regardless of the presence of leaky vasculature.85, 87 Since interstitial pressure is higher at the tumour core and diminishes outwards towards the tumour periphery rim, this can cause NPs to flow outwards from the tumour leading to a loss of effective drug dose within tumours. To circumvent these problems targeted NPs can be used for more efficient tumour or target tissue retention and cellular uptake, resulting in improved efficacy. Additionally, methods of elevating blood pressure or introducing NO-secreting compounds have been investigated by means of administering adjuvants in addition to NP injections.82, 84 For example, VEGF can increase vascular permeability, and was shown to enhance the extravasation of NPs across tumour vasculature when co-administered with liposome NPs.88 In addition to bradykinin, NO and prostaglandins that are factors involved in the regulation of vascular permeability, the administration of a number of kinase inhibitors has also led to an enhanced EPR effect.89 The coadministration of a transforming growth factor beta (TGFβ) receptor inhibitor led to an enhancement of EPR mediated accumulation of both liposomal and micelle NPs, which was a direct result of reduction of pericyte coverage on tumour neovasculature.89 By enhancing vascular permeability and lowering the pressure difference by raising blood pressure, the overall “leakiness” of tumour vessels and therefore passive accumulation of NPs can be increased.
The majority of passively targeted NPs possess a surface coated with PEG polymer for biocompatibility; however, this highly hydrophilic surface does not result in optimal endocytic uptake by cancer cells within the tumour. This problem which has been referred to by some as the “PEG dilemma”90, 91 has been suggested to hamper efficient drug delivery in tumours as passively targeted NPs end up releasing their therapeutic payload into the tumour milieu rather than within cancer cells. However, in the case of cytotoxic drugs—many have been shown to have longer elimination half-lives in tumours vs. normal tissue. Therefore, the delivery of higher amounts of drugs to tumours can lead to longer durations of drug exposure at higher concentrations and enhanced efficacy.92–95 For example, docetaxel (Dtxl) has an elimination half-life of 2.2–4.5 h in normal tissue and 22 h in tumours, demonstrating long tumour site retention relative to non-tumoural tissues.96 For drugs that are not readily retained in tumours or macromolecular drugs that are not readily taken up by cancer cells, then extracellular drug release may be less effective at maintaining a differentially high tumour drug concentration over an extended period of time. This problem is further compounded with NP systems that lack controlled drug release properties. For example, micelle NPs can demonstrate a very rapid “burst” release post administration (releasing up to 50% of their encapsulated drug within 30 min) leading to premature drug release prior to effective EPR mediated tumour accumulation.82 Similar problems exist for liposome based NPs, which can lead to either very slow or fast release of their therapeutic content. Furthermore, the administration of PEGylated liposomes has led to the production of PEG-specific antibodies,97 causing the rapid clearance of a further administered dose—leading to an accelerated blood clearance (ABC) phenomena—which further diminishes effective drug concentrations at tumour sites, but can be rectified by careful tuning of dose (discussed in more detail in section 3.2).98
Extensive efforts in forming PEGylated block copolymers have ultimately resulted in the clinical translation of a number of passively targeted polymeric NPs including; SP1049C,99 NK911,100 Genexol-PM and others, which are now in early phase clinical trials for treating a variety of cancers.17 In general, these NPs are PEGylated polymeric micelle formulations. Polymer micelles are polymeric NPs that form from the self assembly of amphiphilic polymers at concentrations above the critical micelle concentration (CMC), yielding NPs which can encapsulate poorly water soluble drugs.101 SP1049C is a pluronic polymeric micelle NP that is composed of a Dox-entrapping hydrophobic core and a hydrophilic polymer, and is currently undergoing phase II studies in patients with metastatic cancer of the esophagus and esophageal junction that have been refractive to standard chemotherapy treatments.99 SP1049C was observed to be effective in bypassing p-glycoprotein-mediated drug resistance.102 In this study, patients were treated with a single dose of SP1049C, 75 mg m−2 (Dox) given as an intravenous infusion every 3 weeks.99 The results of this study and preclinical studies demonstrated superior anti-tumour efficacy for SP1049C when compared to free Dox administration.99
Two other passively targeted polymeric NPs are NK911, a micellar NP comprising PEG, Dox and poly(aspartic acid), and Genexol-PM, which is a Ptxl-encapsulated PEG-PLA micelle formulation currently in phase II development for various cancers.17, 43, 103 As mentioned previously, Genexol-PM does not require the use of Cremphor EL, and has therefore led to an increase in Ptxl MTD for breast cancer therapy.42, 104 Additionally, Genexol-PM administration demonstrated increased treatment response rates when given to patients who were not responsive to standard taxane therapy with Ptxl/carboplatin therapies, further suggesting improved outcomes for MDR cases. Xyotax (Ptxl-poliglumex), also a passively targeted polymeric NP in which Ptxl is conjugated to poly(l-glutamic acid), was shown to preferentially target ovarian tumours.105, 106 Another example of a passively targeted polymeric NP undergoing phase trials is IT-101, a camptothecin-cyclodextrin polymer conjugate that has shown prolonged circulation times and slow drug release kinetics in vivo, both in pre-clinical and clinical studies.107 These first generation polymeric NPs have so far demonstrated activity against tumours that have been resistant to standard therapies, and show promise in stabilizing disease in patients. The containment of drugs within these NPs leads to significantly reduced off target effects, which can lead to wider therapeutic windows and lower systemic toxicities. Passive targeting strategies are not without limitations and therefore considerable efforts are now underway to investigate actively targeted NPs that can further retain NPs at active sites. Currently there are three targeted polymeric NPs undergoing clinical trials which include: BIND-014, CALAA-01 and SEL-068; these targeted clinical stage NPs will be discussed further in section 3.3.
3.1.1 Long circulating polymeric NPs
Following the discovery of themany inherent advantages for the use of polymericmaterials in drug delivery applications, a landmark paper by Langer and colleagues in 1994 demonstrated that forming diblock copolymers of controlled release polymers with PEG could dramatically increase the circulation half-lives of polymeric NPs.14 Since then, there have been a myriad of PEGylated polymericNPs reported in the literature with the benefits of PEGylation demonstrated across a broad range of polymer molecular architectures and macromolecular assemblies.108–110 In addition to this extensive preclinical work, PEG has been validated clinically in many different applications, and is currently listed as “Generally Recognized as Safe” (GRAS) by the FDA, making it particularly attractive to translational researchers.109 The success of PEG in transforming polymeric NP drug delivery has not been without its challenges however, some of which remain ongoing areas of investigation. For example, the induction of the aforementioned ABC phenomena by PEGylated liposomes has been shown to be influenced by NP size, surface charge, constituents, and time period prior to second dose, and has been observed with other types of NPs, and even been shown to be dependent on NP therapeutic load and type.111–114 However, observations of ABC phenomena have been conflicting in the literature so far as the induction of PEG specific antibodies have been observed in some cases and not in others—therefore given the variable design and composition of NPs, these effects should be investigated on a case-by-case basis.112, 115, 116 A number of studies have now demonstrated that PEG appears to activate complement in a concentration and molecular weight dependent manner, through classical (C1q dependent), lectin, or alternative pathways.117 While PEG is capable of both activating complement and eliciting anti-PEG antibody responses, the manner and extent of these immune responses can be modulated. Further, it has been shown that modifying the density of PEG on a NP surface alters the complement activation pathway, perhaps by altering the conformation of the PEG chains on the NP surface.118 These data suggest that surface optimization of PEG density and molecular weight will be critical to avoid unwanted immune (non-IgE) hypersensitivity reactions. In addition to complement activation, as mentioned previously, PEGylated liposomes have been shown to elicit immunoglobulin M (IgM) antibodies in a number of reports, leading to ABC phenomena post-repeat dosing in a short time interval following initial dose administration.98 Production of short term anti-PEG IgM appears to be dependent on the species tested, dose, PEG density, NP charge, and type of drug encapsulated. It is noted, that to attenuate the anti-PEG immune response, it appears important to tune the dose, shorten the PEG molecular weight, increase the density of PEG on the NP surface, tune NP surface charge close to neutral, and/or encapsulate an agent that attenuates macrophage function, such as Dox.98 The clinical significance of these reports in terms of affecting PEGylated NP drug carriers has not been critically evaluated, though further preclinical and clinical data (Table 1) will at least provide an interim answer. At this point, with known methods to attenuate the anti-PEG immune response, and the well-established clinical success of the DOXIL (PEGylated liposome) and many other clinically validated PEGylated proteins, one can conclude that the anti-PEG immune response is not an intractable issue for polymeric NP carriers. Furthermore, alternatives to PEG are currently being developed, including new polymers or zwitterionic surfaces that are ultra-low fouling in nature.119 The preclinical data for these systems is encouraging, and their further development and study in the context of NP drug delivery is widely anticipated.109, 120, 121
In order to achieve effective EPR mediated targeting, NPs must have long-circulating half-lives that facilitate more opportunities for the passage of NPs from the systemic circulation into the disordered and permeable regions of tumour vasculature. As mentioned previously, passive targeting strategies are not without limitations and therefore considerable efforts are now underway to investigate actively targeted NPs that can further retain NPs at active sites. Targeted NPs facilitate receptor-mediated endocytosis (RME), releasing therapeutic agents in a more effective manner once inside target cell populations,122, 123 which can significantly increase drug efficacy.51, 124, 125
3.2 Active targeting
Active targeting involves the use of affinity ligands to direct the binding of NPs to antigens, differentially overexpressed on the plasma membrane of diseased cells or to the extra-cellular matrix proteins that are differentially overexpressed in the disease tissue. The first reports of targeted NPs date back to 1980’s and involved the surface modification of liposomes with monoclonal antibodies (mAbs) that recognized antigens on the target cells.22, 23, 126 There are 30 mAbs approved for clinical use to date.127 Muromonab-CD3 (OKT3, immunosuppressive agent) was the first antibody to be approved in 1986.128 Since then a myriad of antibody platforms have been developed including murine, chimeric, humanized and human mAbs.51 For example, the chimeric mAb rituximab (Rituxan), which binds to the CD20 antigen, was approved for the treatment of non-Hodgkin’s lymphoma in 1997.129 The humanized mAb trastuzumab (Herceptin) which binds to the HER2/neu antigen was approved for the treatment of breast cancer in 1998.130 Stemming from the success of mAbs, several other classes of binding ligands were developed against many target antigens, including antibody mimetics, peptides, nucleic acid ligands and small molecules (see section 5). Many of these ligands have been conjugated to radioisotopes or drug molecules to create more effective targeted imaging and therapeutic modalities.131–133 Subsequently, many of these ligands were also conjugated to the surface of NPs in order to achieve antigen-specific active targeting.134 In contrast to ligand-drug conjugates which typically carry 1–8 drug molecules, ligand targeted NPs may carry up to 103 to 104 drug molecules, allowing for potentially a higher amount of drug delivery per bio-recognition or binding event.
Actively targeted NPs can be utilized in applications where drug release is either extracellular or intracellular. Therapies that act on intracellular sites of action are most effectively delivered with targeted NPs.3, 135 Actively targeted NPs may be internalized via clathrin-dependent endocytosis pathways, caveolin-assisted, cell adhesion molecule directed, or lipid raft associated mechanisms, leading to endosome formation, which ultimately leads to lysosomes.136 For hydrophobic small molecule drugs that can readily permeate through the lipid bilayer of the endosomal membrane, drug release within the endosome will result in permeation within the intracellular compartments. For delivery of bioactive macromolecules such as nucleic acids (DNA, siRNA, miRNA) or charged hydrophilic small molecules that are relatively impermeable to the endosomal membrane, the NPs need to escape the endosome prior to fusion with lysosomes if NPs are to reach their desired subcellular compartments.137 Many efforts have led to the investigation of mechanisms that lead to endosomal escape based on pH buffering, osmotic swelling leading to endosome bursting or endosomal membrane destabilization.138, 139 Ligand mediated cell internalization can result in enhanced therapeutic benefits as compared to equivalent non-targeted NPs.124, 140 Experiments comparing targeted and non-targeted NPs have confirmed that the primary role of the targeting ligand is to enhance cellular uptake into target cells.141, 142 For example, accumulation of siRNA-loaded NPs at tumour sites is largely a function of effective EPR via passive targeting; however, cellular internalization and effective gene silencing are largely a function of targeting ligand where targetedNPs are significantly more efficacious as compared to equivalent non-targeted NPs.143, 144 This behaviour suggests that the colloidal properties of NPs determine their biodistribution, whereas the targeting ligand serves to facilitate and enhance cellular uptake at targeted sites.145
Ligand mediated targeting is also beneficial in the case of vascular endothelial targeting for both oncology and cardiovascular applications, and the identification of high affinity ligands for this purpose is an active area of research.146 Recent studies have shown that small peptide targeted polymeric NPs showed substantial accumulation to injured vasculature following angioplasty compared to non-targeted NPs.147 Interest in the use of short peptides as targeting ligands has increased. In comparison to larger mAb’s peptide ligands have the advantage of being (i) smaller in size, (ii) less immunogenic, (iii) more stable; and (iv) easier to manufacture.148 Peptides however, have relatively lower affinity for their target site, and this deficiency is in-part mitigated through ligand avidity which is achieved by incorporating multiple peptides on the NP surface.149 The establishment of a wide range of phage display libraries and screening technologies has resulted in isolation of peptide ligands against many important targets (targeting ligands are discussed further in section 5).150–152
While the potential benefit of ligand-mediated NP targeting is clear, this technology has not resulted in a clinically validated product so far. Within the 32 years since the first description of targeted NPs, only six targeted NPs have progressed to clinical trials (Table 1). From these six NPs, three are targeted polymeric NPs and three are targeted liposomes. MCC-465 was the first of these to be developed and consists of liposome encapsulated Dox, with a surface decorated with both PEG and dimers of antigen-binding fragments (F(ab′)2) for immune shielding and targeting respectively.213 The F(ab′)2 used in the development of this NP is a fragment of the human mAb, GAH which has shown affinity to >90% of human stomach cancer cells.213 Additionally, antibody fragments may be preferred for certain applications since they retain the high affinity and specificity of antibodies but are smaller in size and therefore potentially less immunogenic.51 MCC-456 was shown to exhibit significant antitumour response against GAH-positive xenografts resulting in up to 80% reduction in tumour mass in comparison to controls.214 Phase I trials with MCC-465 were carried out in order to determine the MTD and further dosing regimens for Phase II analysis. In this study patients with metastatic cancer or recurrent stomach cancer were administered 6.5 mg m−2 of MCC-465 as a 1 h infusion every 3 weeks for up to 6 treatment cycles. It was concluded that MCC-465 was well tolerated and similar pharmacokinetic outcomes were observed as compared to DOXIL. However, MCC-465 does not appear to have progressed through clinical development after phase I completion.
SGT53-01 is a transferrin receptor (TfR)-targeted liposome designed to carry the p53 tumour suppressor gene to cancer cells.215 SGT53-01 targets the TfR on the surface of cancer cells using single-chain antibody fragments (TfRscFv) and results in the expression of p53 gene in the targeted cancer cells. Pre-clinical studies have indicated that SGT53-01 could sensitize tumours to the effects of radiation and chemotherapy.215 SGT53-01 is currently undergoing phase I clinical trials in combination with Dox for treatment of solid tumours.
MBP-426 is also a TfR-targeted liposome that encapsulates oxaliplatin and is designed to preferentially target the delivery of oxaliplatin to cancer cells.216 Transferrin (Tf) is widely used as a targeting ligand since the TfR is significantly upregulated on most cancer cells.217 In a phase I study, patients with advanced or metastatic solid tumours refractory to conventional therapy received MBP-426 as 2–4 h infusions every 3 weeks in cohorts of 3 to 6 patients, and this targeted liposome was demonstrated to be well tolerated (with thrombocytopenia as the main dose limiting toxicity (DLT)).216
The last decade has seen a variety of strategies involving conjugation of targeting ligands to the surface of NPs in order to provide molecular interaction points between the NPs and antigens present on target cells and tissues. What has emerged from these studies is that a variety of different targeting ligands can trigger NP internalization into cells, and that internalization can significantly enhance treatment efficacy.51, 62 Table 2 highlights from the literature the wide range of targeted polymeric NPs along with their available physicochemical properties developed for numerous therapeutic applications.
Table 2.
Examples of preclinical targeted polymeric nanoparticles
| Material | Physicochemical Characteristics (Size, ζ-potential) |
Targeting Strategy | Drug/Disease or Indication |
Ref. |
|---|---|---|---|---|
| Poly(2-methacryloyloxyethyl | 220–240 nm, 02212;2 mV | Phosphorylcholine | Doxorubicin/Cancer | 153 |
| phosphorylcholine-co-butyl methacrylate) and poly(methacryloyloxyethylphos- phorylcholine-co-butylmeth- acrylate-co-methacryloyl hydrazide) |
||||
| Poly(lactic acid)-selectin conjugates | 170 nm, −20 mV | Small molecule; Selectin ligand | Inflammation | 154 |
| Galactosylated-chitosan polymer | 120 nm, +5 mV | Small molecule; Galactose | DNA/Various | 155 |
| Chitosan | 200 nm, + 40 mV | RGD; Charge | siRNA/Cancer | 156 |
| Chitosan-PEG | 150 nm, +16 mV | Antibody | Caspase inhibitor pep tide/Stroke |
157 |
| Poly(caprolactone) and poly(ethylene glycol) or poly(2-N,N-dimethylamino)ethyl methacrylate) |
25–200 nm | Passive | Various | 158 |
| (Allyloxy)12cucurbit[6]uril polymer | 70–90 nm | Triggered; Reducing environment sensitive | Cancer | 159 |
| Cyclodextrin polymer | 100–150 nm, +15mV | Transferrin | DNA/Cancer | 160 |
| Acetal modified dextran | 250–300 nm, −5 mV to + 12 mV | Pep tide | Various | 161 |
| DNA | 410 nm | Passive | Various | 162 |
| Elastin-like polypeptides | 20 nm | Triggered; pH-sensitive | Doxorubicin/Cancer | 163 |
| 60 nm | RGD; T-sensitive | Cancer | 164 | |
| Gelatin | 250–300 nm, −20 mV | Antibody | antiCD3 mAb/Cancer | 165 |
| poly(β-amino esters) | 200 nm, −5 mV | RGD | DNA/Gen therapy | 166 |
| Heparin | 60 nm, −16 mV | Small molecule; Folate | Paclitaxel/Cancer | 167 |
| Hyaluronic acid | 250–400 nm | Intrinsic | Cancer | 168 |
| Hyaluronic acid-ceramide/ pluronic 85 |
110–140 nm, −20 mV | Passive | Docetaxel/Cancer | 169 |
| Hydrophobically modified glycol chitosan |
360 nm, +22mV | Charge | Cancer | 170 |
| Oligoethylene glycol pyridine disulfide nanogels | 190 nm | Reducing environment sensitive |
Hydrophobic drugs | 171 |
| Poly(methyldiethene- aminesebacate)- co-[(cholesterylox- ocarbonylamidoethyl) methylbis(ethylene) ammonium bromide]sebacate |
80–180 nm, +70mV | Charge | Paclitaxel, DNA/Cancer | 172 |
| Poly(ethyleneoxide)-modified | 100–150 nm, +40mV | Triggered; pH-sensitive | Paclitaxel/Cancer | 173 |
| poly(beta-amino ester) | 60 nm | Triggered; pH-sensitive | Doxorubicin/Cancer | 174 |
| Modified poly(caprolactone)copolymer |
120 nm, −60 mV | Small molecule; Galactose | Various | 175 |
| Poly(carboxybetaine methacrylate) | 110 nm | Triggered; Reducing environment sensitive; RGD |
Reducing environments | 176 |
| PEG (PRINT) | 290 nm, −30 mV | Transferrin | Cancer | 177 |
| Poly(caprolactone)- | 25–60 nm, −5 mV | Large peptide; EGF | Cancer | 178 |
| poly(ethyleneglycol) | 70 nm, −3 mV | Pep tide | Brain | 179 |
| PEGylated Gelatin | 200 nm | Passive | DNA/Various | 180 |
| Poly(methacrylic acid) | 150–170 nm, −20 mV | Small molecule; Folate | Doxorubicin/Cancer | 181 |
| Poly(lactic acid) | 45 nm | Peptide; RGD | Doxorubicin/Cancer | 182 |
| 70–95 nm, −30 mV to + 45 mV | Charge | Various | 183 | |
| 80 nm, −25 mV | Triggered; pH-sensitive | Cisplatin/Cancer | 184 | |
| Poly(d,l-lactide-co-glycolide) | 110–190 nm | Antibody | Camptothecin/Cancer | 185 |
| 260 nm, −8 mV | Peptide | Inflammation | 186 | |
| 140–180 nm, −20 mV | Peptide | Loperamide/Analgesia | 187 | |
| poly(d,l-lactide-co-glycolide)- lipid hybrid |
80–120 nm | Passive | Doxorubicin, Combretastatin-4/ Cancer |
188 |
| 60 nm | Peptide | Injured vasculature | 147 | |
| poly(d,l-lactide-co-glycolide)- poly(ethyleneglycol) |
180 nm, −3 mV | Peptide; Tetanus toxin C fragment |
Neurons and Neuroblastoma |
189 |
| 40–60 nm | Small molecule; Alendronate | Estrogen/Bone hydroxyapatite |
190 | |
| 80–200 nm | Aptamer | Docetaxel/Cancer | 191 | |
| 140 nm | Aptamer | Cisplatin prodrug/Cancer | 192 | |
| 100 nm | Passive | MAPK signaling/ Cancer |
193 | |
| 100–120 nm, −20 mV | Peptide | Brain | 194 | |
| 80 nm | Small molecule | Epigallocatechin 3-Gallate/Cancer |
195 | |
| poly(d,l-lactide-co-glycolide)- poly(ethyleneglycol)-Aptamer |
160–240 nm, −25 mV | Aptamer | Docetaxel/Cancer | 196 |
| Poly(L-lysine) | 80 nm, + 1 mV | Triggered; pH-sensitive | Acidic tumours | 197 |
| Poly(lactic acid)-poly(ethyelene glycol) and poly(caprolactone)- poly(ethyeleneglycol) |
20–200 nm | Ultrasound triggered | Doxorubicin/Cancer | 198 |
| Pluronic | 40 nm, + 18 mV | Peptide | Cartilage | 199 |
| poly(N-isopropylacrylamide- b-methyl methacrylate) |
190 nm | Triggered; T-sensitive | Prednisone/Inflammation | 200 |
| poly((1-ethoxycarbonyl)-vinyl- phosphonic diacid and poly(n-butyl acrylate) |
80–120 nm | Protein; Annexin-A5 | Inflammation | 201 |
| Poly(ethylene glycol)- poly(aspartate hydrazone adriamycin) | 65 nm | Triggered; pH-sensitive | Doxorubicin/Cancer | 89 |
| Poly(γ-glutamic acid)-PL | 115–126 nm, −20 mV | Small molecule; Galactosamine | Paclitaxel/Cancer | 202 |
| Poly(L-glutamic acid) | 50 nm | Small molecule; Biotin | Doxorubicin/Cancer | 203 |
| poly(2-methyl-2-carboxy- trimethylene carbonate- co-d,l-lactide) |
130 nm | RGD | Corneal epithelial cells | 204 |
| Poly(β-malic acid) | 7–25 nm, −5 mV | Multiple; Antibody; Triggered | Antisense ON/Brain Tumour |
205 |
| 15–25 nm, −5 mV | Multiple; Antibody | Antisense oligonucleotides Hercep tin/Cancer |
206 | |
| Poly(γ-benzyl-L-glutamate)- Poly(vinylybenzyllactonamide) |
40–300 nm | Small molecule; Galactose | Various | 207 |
| Poly(acrylamide) | 20–30 nm | Peptide | Cisplatin/Cancer | 208 |
| Poly(hydroxyalkanoates) | 100–200 nm | Polypeptide | Cancer | 209 |
| Pullulan acetate/sulfadimethoxine conjugate |
70 nm | Triggered; pH-sensitive | Doxorubicin/Cancer | 210 |
| Ribonucleoprotein | 40–70 nm | Passive | Various | 211 |
| Styrene-maleic acid copolymers | 175 nm | Zinc protoporphyrin | Cancer | 212 |
3.3 Clinical stage targeted polymeric NPs
Conventional methods of preparing targeted NPs involve a series of chemical processes whereby the NP core is initially formed, followed by the bioconjugation of targeting ligands to the surface of the NP. This post-coupling of targeting ligands does not allow tuning of ligand density for optimal efficacy, requires excess amounts of reagent in order to achieve high coupling efficiencies, and is associated with purification techniques to remove unbound ligands. Due to this kind of complexity in the synthesis of NPs, difficulties may arise in the reproducibility of NP surface properties, resulting in batch-to-batch variability, which is not amenable to clinical translation and subsequent commercialization. Indeed, by reducing the number of components to the minimum, and employing a modular self-assembly approach using pre-functionalized polymeric materials,218 it is possible to create libraries of targeted NPs that vary narrowly from each other in their biophysicochemical properties. Using this strategy, BIND Biosciences recently developed and screened a library of targeted self-assembled polymeric NPs resulting in the development of BIND-014, the first targeted and controlled release polymeric NP for cancer chemotherapy to reach clinical development.219 BIND-014 is a prostate specific membrane antigen (PSMA)-targeted Dtxl-encapsulated polymeric NP, which entered phase I clinical trials in January 2011.219 PSMA is a transmembrane protein overexpressed on the surface of prostate cancer cells and tumour-associated neovasculature of virtually all solid tumours.220, 221 Dtxl is a semi-synthetic taxane approved for treatment of a number of major solid tumour cancers, including breast, prostate, lung, gastric, and head and neck.222 BIND-014 has been shown to deliver up to 10 times more Dtxl to tumours relative to an equivalent dose of Dtxl in multiple animal models.219 Initial clinical data in patients with advanced solid tumours indicate that BIND-014 displays a pharmacological profile differentiated from Dtxl, including pharmacokinetic properties consistent with long circulation half-life of BIND-014 and retention of Dtxl in the vascular compartments, and multiple cases of tumour shrinkage at doses up to 5 times below the Dtxl dose typically administered clinically.219
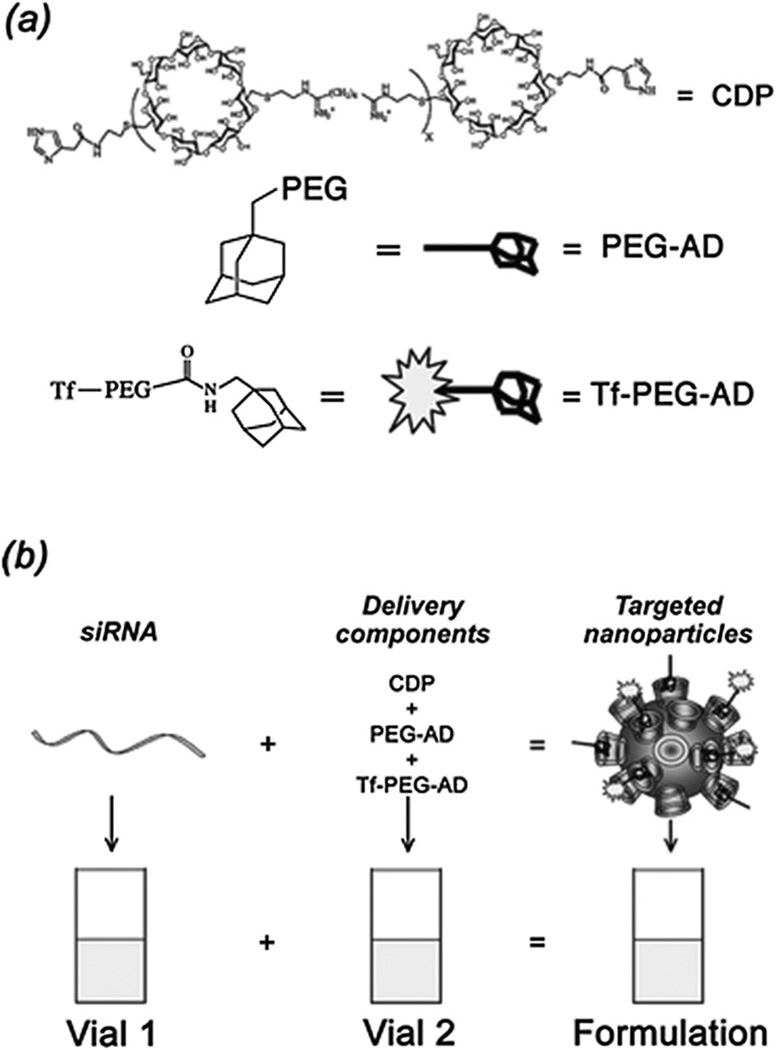
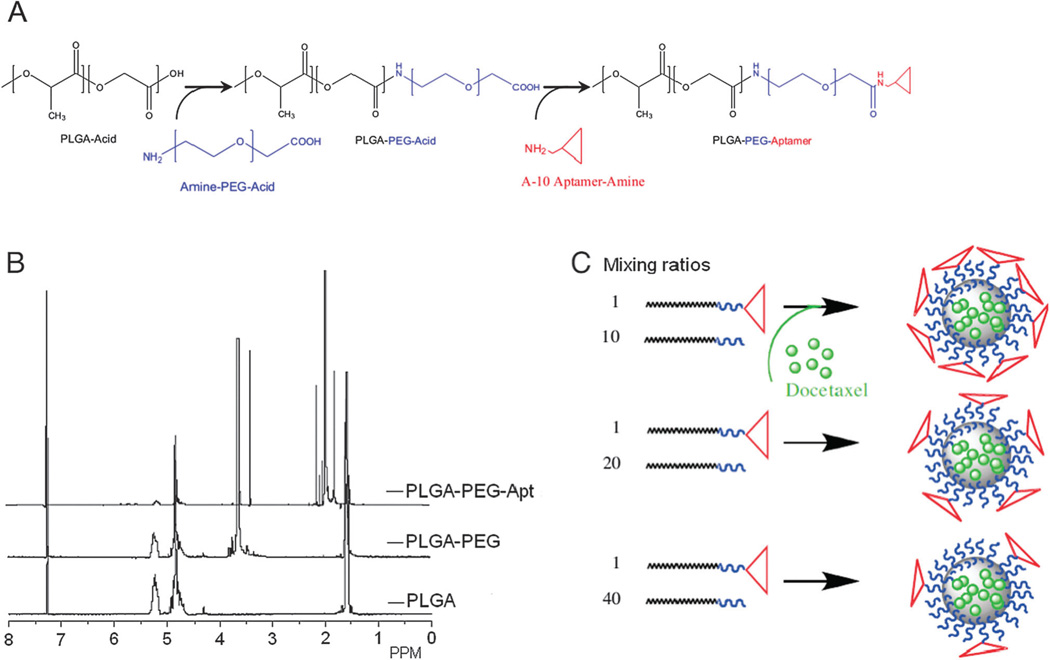
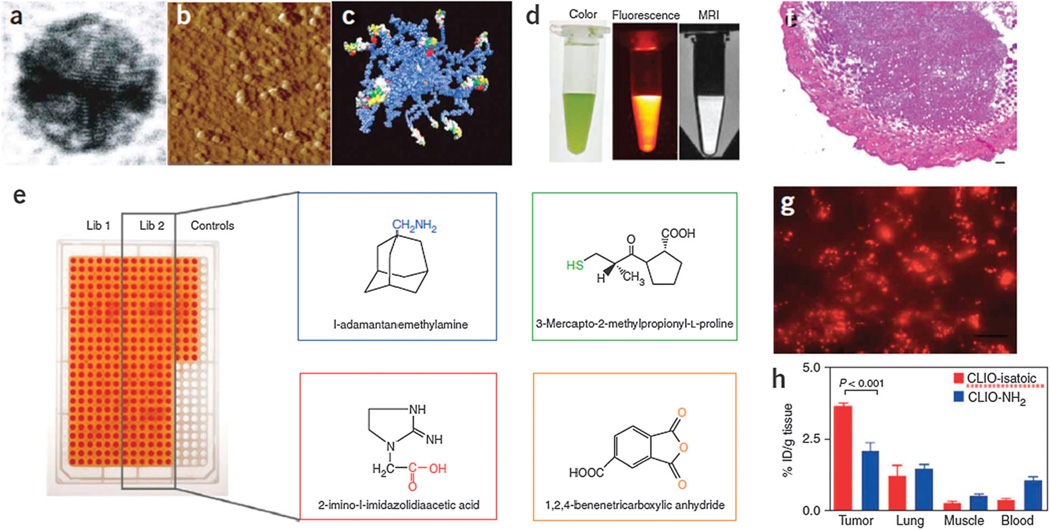
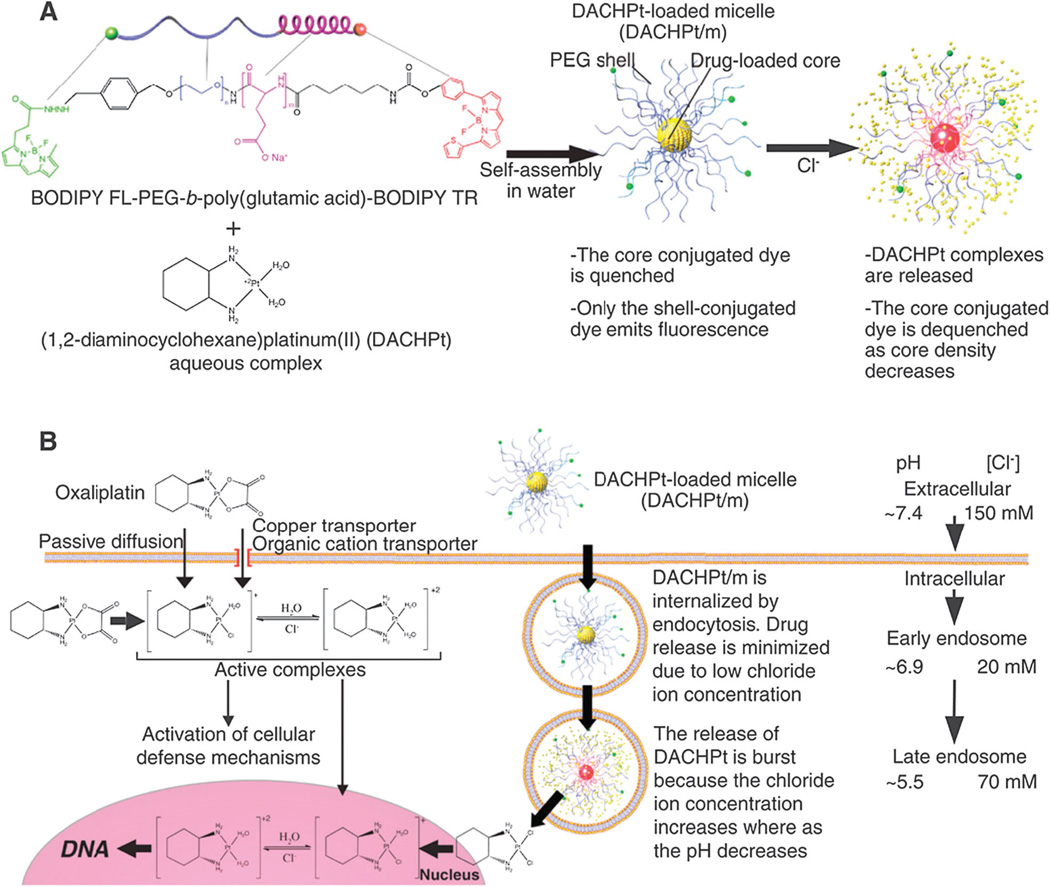
CALAA-01 is the first targeted NP to reach clinical development for siRNA delivery in 2008.18 The CALAA-01 NP consists of siRNA to reduce the expression of the M2 subunit of ribonucleotide reductase (R2), cyclodextrin containing polymer (CDP) for siRNA condensation, adamantine-PEG (AD-PEG) for steric stabilization, and adamantine-PEG conjugated to human Tf (AD-PEG-Tf) to target the TfR overexpressed on the surface of most cancer cells.223 CALAA-01 employs a unique two-vial formulation strategy, which allows for the rapid self-assembly of the NP (50–70 nm) delivery system components (CDP, AD-PEG, AD-PEG-Tf) with siRNA, at the point of care (Fig. 3).18 This formulation is also capable of high siRNA payload delivery and endosomal pH (<6.0) triggered release of siRNA once NPs are endocytosed.224
Fig. 3.
Components of CALAA-01 (Calando Pharmaceuticals-01) – a targeted NP for siRNA delivery: (a) CDP: water-soluble, linear cyclodextrin-containing polymer, AD: adamantane (AD)-PEG conjugate (PEG MW of 5000) (AD-PEG), and Tf-PEG-AD: an adamantane conjugate of PEG (PEGMWof 5000) conjugated with human transferrin (Tf) ligand. (b) CALAA-01 is formulated via a single self-assembly process of four individual components. Figure taken from Davis, M et al.18
SEL-068 is a first-in-class synthetic and integrative targeted polymeric NP vaccine to reach phase I clinical development in November 2011. SEL-068 contains nicotine as antigen, T-helper cell peptides, TLR agonists as adjuvant, and is currently under development for smoking cessation and relapse prevention.27 Post smoking, nicotine usually enters the lung and the systemic circulation and reaches the brain by crossing the blood-brain barrier (BBB), and binds to nicotine receptors resulting in release of stimulants such as dopamine leading to reinforcement of addiction.225 The administration of SEL-068, which is based on modular self-assembly NP technology,196 results in high anti-nicotine antibody concentrations and a high anti-nicotine antibody affinity; leading to the sequestration of nicotine molecules in circulation and largely blocking central nervous system exposure, thereby diminishing the addictive effects of nicotine.
The clinical translation of the above technologies has marked a new era in the development of multi-functional therapeutic NPs capable of targeting, controlled release, and co-delivery of multiple active agents.
4. Preparation of targeted polymeric NPs
4.1 Methods for preparing polymeric NPs
A number of top-downmethods are available for the preparation of polymeric NPs using biodegradable polymers. Most of these involve self-assembly of block copolymers that are composed of two distinct polymer chains with different solubilities. These methods include nanoprecipitation (also called solvent displacement method),226 various types of emulsification/solvent evaporation,227 and the salting out method.228 In addition to these conventional methods, new approaches used to create polymeric NPs, including supercritical technology, electro-spraying, premix membrane emulsification and aerosol flow reactor methods are also under investigation which are further discussed in a recent review.229 The choice of NP formulation method is usually dependent on the drug physicochemical properties along with the requirements for encapsulation and particle size. Nanoprecipitation, oil-in-water (O/W) emulsification-solvent evaporation (single emulsion), and water-in-oil-in-water (W/O/W) emulsification-solvent evaporation (double emulsion), are three of the most commonly utilized methods to prepare a variety of polymeric NPs and will be described in the next paragraphs.
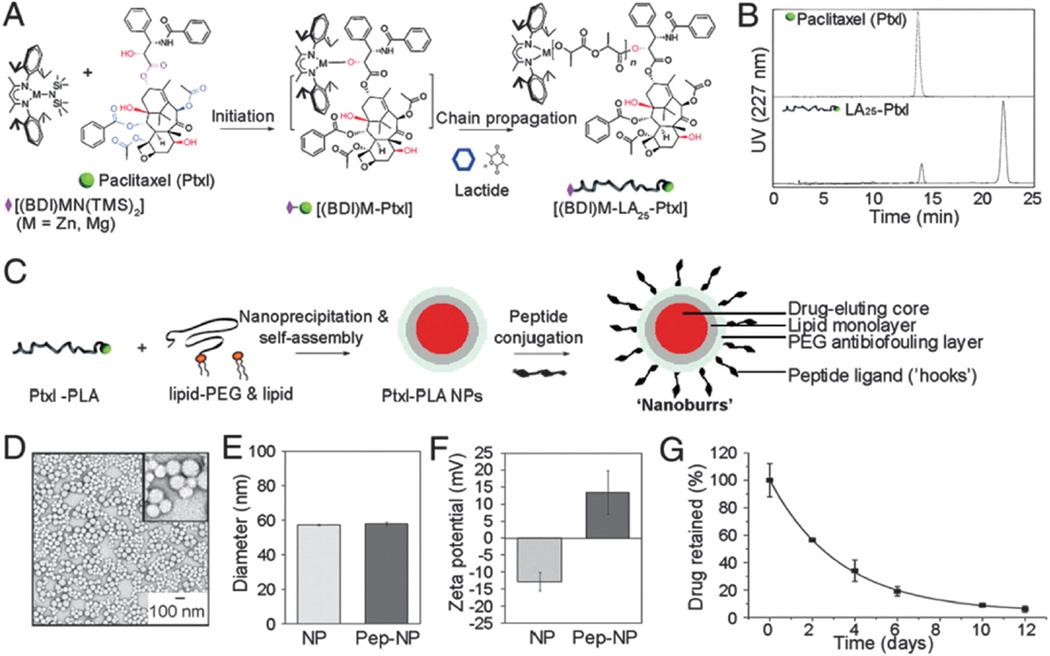
Nanoprecipitation is a method that involves the use of an organic solvent that is miscible with an aqueous phase.230 In this technique, the polymer and drug are dissolved in the organic solvent and this solution is then added dropwise to an aqueous (non-solvent) solution under stirring. Once in contact with water, the hydrophobic polymers and drug precipitate and self-assemble into core-shell like spherical structures in order to reduce the system’s free energy.45 After self-assembly, the organic solvent is evaporated either by reduced-pressure evaporation, or simply by continuous mixing at atmospheric pressure if the solvent is relatively volatile. The instantaneous formation of particles is governed by the principles of the Marangoni effect and has been attributed to interfacial interactions between liquid phases.231
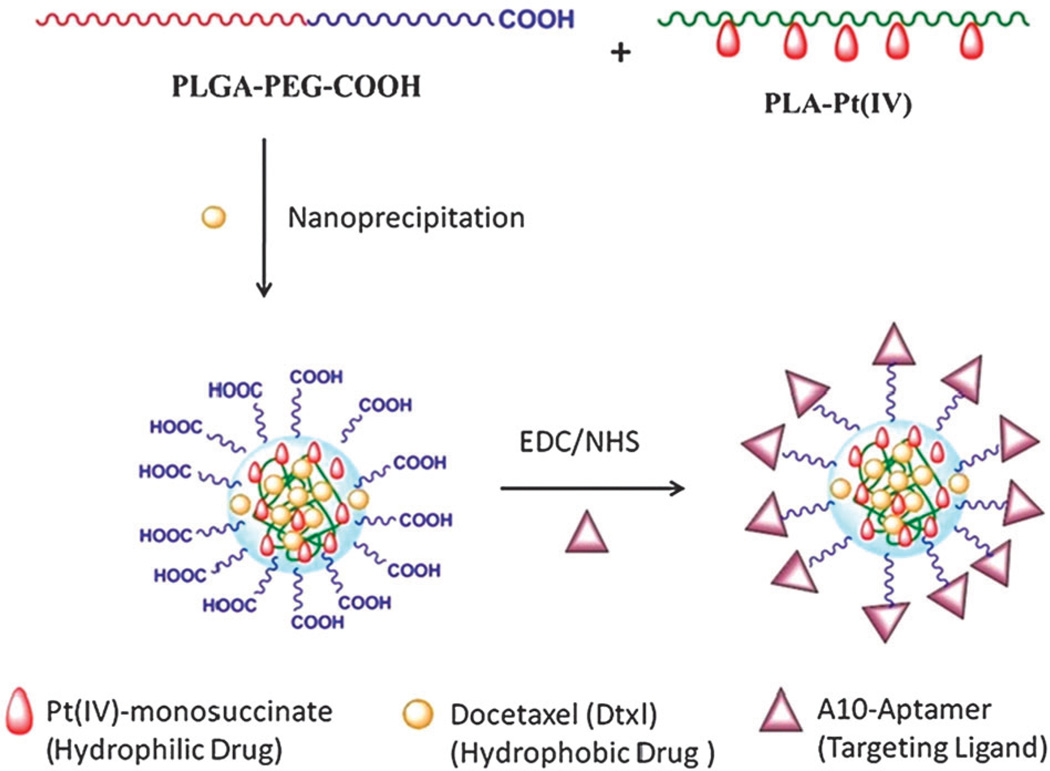
The use of diblock hydrophobic-PEGylated polymers in nanoprecipitation leads to NPs that consist of a hydrophobic core, with entrapped hydrophobic drugs, surrounded by a hydrophilic shell for steric stabilization.232 For example, Dtxl-loaded NPs were prepared using nanoprecipitation, whereby the hydrophobic drug Dtxl was mixed and co-precipitated with the diblock polymer poly(lactic-co-glycolic acid)-polyethylene glycol-carboxylic acid (PLGA-PEG-COOH).233 The resulting NPs had a hydrophobic core composed of PLGA, wherein Dtxl was encapsulated, and a hydrophilic shell composed of PEG. Nanoprecipitation is a simple method, amenable to scale-up at an industrial scale requiring only mild stirring under minimal shear stress. In general, smaller NPs are obtained through this method when compared to other methods at equivalent conditions. In contrast, some drawbacks include the poor entrapment of hydrophilic drugs (hydrophilic drugs can remain in the aqueous phase),229 lower entrapment efficiencies compared to other methods and difficulty in complete removal of the organic solvent after self-assembly.232 Although recent advances whereby the aqueous water phase (non-solvent) is replaced with other organic solvents (methanol, ethanol) has facilitated the use of this technique for the use of hydrophilic drugs also.229
Emulsification techniques (water-in oil: W/O, oil-in water: O/W and double emulsion: W/O/W) require the formation of emulsion, followed by the homogenization of this mixture, and although originally used to formulate microparticles, can now also be utilized to prepare nano-emulsions.234 The type of emulsification method utilized ultimately depends upon the properties of the polymer, drug and also the degree of miscibility of the organic (oil) solvent with the water phase.
The single emulsion technique (O/W) requires the drug to be soluble in a water-immiscible organic solvent. In this method, the polymer and the drug are dissolved in a volatile water-immiscible solvent such as dichloromethane or ethyl acetate, and the organic phase is emulsified under intense shear stress into an aqueous phase containing appropriate amounts of a surfactant, such as sodium cholate or polyvinyl alcohol (PVA). The organic solvent is allowed to evaporate, allowing the self-assembly of NPs.235 The O/W emulsification technique is suitable for entrapping hydrophobic drugs and generally results in higher drug loading and encapsulation efficiency compared to nanoprecipitation, as well as achieving complete solvent removal. However it requires an additional input of energy such as sonication or homogenization and the resulting NPs are often larger than those obtained through nanoprecipitation.235
Double emulsion (W/O/W) is generally used for encapsulation of hydrophilic drugs and proteins. In this method the drug is dissolved in a small volume of an aqueous phase together with a surfactant and this is emulsified in an organic phase containing the polymer. The W/O emulsion formed is then dispersed in a larger volume of an aqueous phase with or without surfactant to form the double W/O/W emulsion. Finally, the solution undergoes evaporation of the remaining organic solvent yielding NPs.235 This method normally yields NPs with larger size than nanoprecipitation or O/W methods, with moderate drug loading and encapsulation efficiency.232
In the aforementioned NP preparation methods, several factors affect the physicochemical properties of the NPs, such as the solvent of choice, the solubility of the drugs (e.g. log Po/w), the mixing time of the aqueous and organic solvents, the type of surfactant used, the concentration of polymer in the organic solution, the ratio of organic to aqueous solution in addition to others.45, 229, 232 For instance, Cheng et al.191 systematically studied the effect of some of these factors on the physicochemical properties of PLGA-PEG NPs. Their data suggested that an increase in the water miscibility of the solvent led to a decrease in mean NP size, and a linear relationship between polymer concentration and size was further shown. The solvent/water ratio, however, did not have a clear relationship with NP size. In addition, the effect of drug loading on resulting NP size distributions was also investigated.191 While plenty of examples exist that demonstrate the reproducible production of polymeric NPs using the range of aforementioned therapeutic NP preparation techniques—the ultimate challenge arises with the translation of these methods to industrial scale production levels. Despite the revolutionary developments in nanotechnology and the clinical translation of a range of NPs containing active pharmaceutical ingredients (APIs), greater strides are needed in NP synthesis, processing, scale-up and manufacturing.229
4.2 Microfluidic methods
Microfluidics, the science and technology of manipulating nanoliter volumes in microscale fluidic channels, has shown that several labour-intensive and time-consuming steps such as sample preparation, mixing, reactions, purification, separations, and detection could be performed on a single monolithic microfabricated device.238 In the last few years, applications of microfluidics have expanded from conventional chemical and biological analysis to other fields such as chemical reactions, biochemical assays, and cell handling.239 Two particularly important contributions have been the development of soft lithography in PDMS as a method for fabricating prototype devices, and the simple fabrication of pneumatically activated valves, rapid mixers and pumps on the basis of soft-lithographic procedures. This has resulted in the fabrication of prototype devices to test new ideas in approximately 2 days (from design to working device), whereas the same applications for silicon technology may take a month ormore for non-specialists to carry out.With respect to nanoparticles, the ability of microfluidic systems to mix reagents rapidly, provide homogenous reaction environments, continuously vary reaction conditions, enable rapid temperature control, and allow addition of reagents at precise time intervals—are some of the key features that have made microfluidic systems useful for the synthesis of NPs.240 Furthermore, synthesis carried out in microchannels allows for in-line characterization,241 feedback control,242 and high-throughput continuous synthesis,243 which potentially enables screening and optimization of libraries of nanoparticles with different properties.
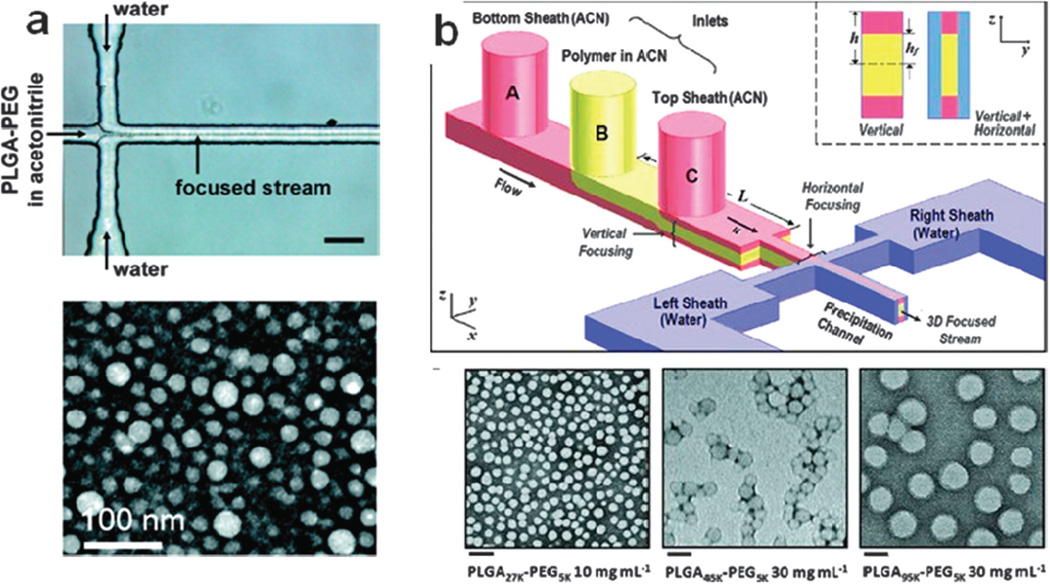
Recently, technologies developed for the synthesis of polymeric NPs have demonstrated the tremendous potential for microfluidics to dramatically improve on current bulk synthesis methods. Polymeric NPs prepared by bulk synthesis tend to have variable physicochemical properties (size, surface composition, and drug loading) due to the inability to control the mixing of precursors.236 Further post-processing by extrusion, freeze–thaw, sonication, and/or high-pressure homogenization is often required. Using rapid mixing techniques in micro-channels such as hydrodynamic flow focusing, polymeric NPs exhibiting narrow size distributions compared to bulk synthesis have been prepared in a reproducible manner (Fig. 4).236, 237 In these systems size can be tuned by either varying the mixing time of precursors, which is achieved by varying the flow ratio of the precursor streams, by varying the molecular weight of the polymer, or by simply varying the concentration of the polymer in the organic solution. Remarkably, for polymeric NPs prepared through microfluidics, higher drug encapsulation without increase in NP size has been observed,236 which is highly desired for therapeutic NPs. Another method to prepare NPs takes advantage of the rapid mixing microenvironment that occurs in micro-droplets formed inside microfluidic channels.244 For instance, cross-linked alginate NPs were synthesized in a micro-channel using aqueous alginate droplets as templates, followed by the shrinkage of the drops. This method exhibited remarkable control over the NP properties, specifically size and size distribution. These are just a few examples showing the advantages of microfluidics for nanoparticle synthesis, and recent reviews discuss these concepts further.240, 245 Given the volume of research currently involving the microfluidic synthesis of NPs, it is expected that as more therapeutic NPs reach a clinical stage, the need for improved synthesis methods would also increase, at which point microfluidic technologies could likely become an important tool in their development of NPs.
Fig. 4.
(a) Microfluidic synthesis of polymeric nanoparticles prepared under rapid mixing conditions in 2D flow focusing. (b) 3D flow focusing. Figure adapted from Karnik et al. and Rhee et al.236, 237
4.3 Drug loading methods
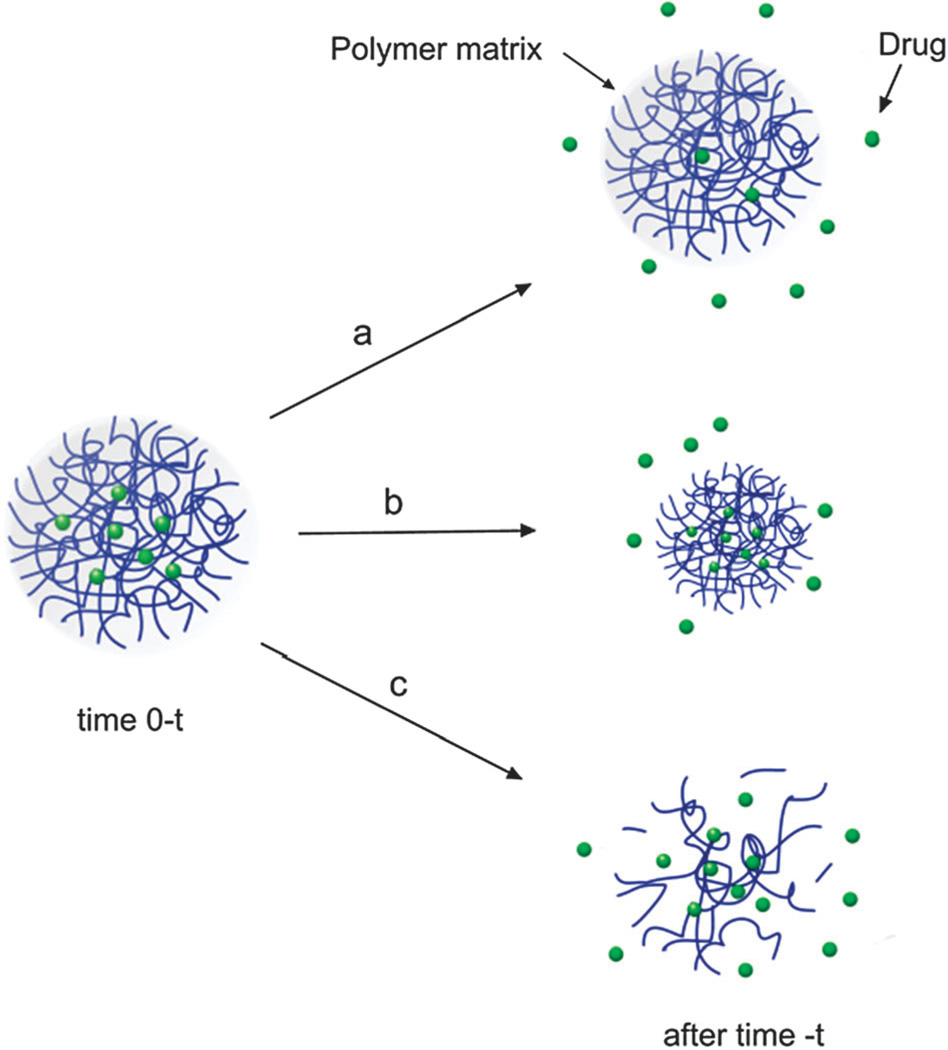
In general, drug loading into polymeric NPs can be achieved according to three techniques: (1) the drug is covalently attached to the polymer backbone, (2) the drug is adsorbed to the polymer surface, or, (3) the drug is entrapped in the polymer matrix during preparation of the NP.246 In turn, drug release rates from NPs depend upon a number of parameters including: (a) diffusion through the NP matrix, (b) erosion of the NP surface and, (c) polymer matrix degradation (see Fig. 5).51 In the following paragraphs we will discuss drug encapsulation by entrapping the drug during preparation and conjugation of the drug to the polymer backbone. Entrapping a drug during NP preparation is the most common technique used for incorporating drugs into NPs. For hydrophobic drugs the fact that they simply need to be mixed with the polymer in an organic solvent, makes it very attractive for formulation purposes. In fact, most NP formulations that are at the clinical stage rely on this method of encapsulation. However, this method suffers from some disadvantages mainly that of relatively low drug entrapment accompanied with low encapsulation efficiency at high loadings.247 In addition, maximum encapsulation efficiencies tend to vary with drug type, and are also affected by the type of polymer, solvents, temperature, and mixing time of NP precursors among other factors.248 Moreover, the encapsulation of two or more drugs with this method may be difficult to achieve, especially for drugs with different chemical properties. Conjugating drugs to the polymer backbone is an attractive alternative that minimizes some of the disadvantages encountered for drug entrapment.249–252 This method involves chemically modifying the polymer (or the drug) to allow for chemical conjugation of the drug to the polymer. Originally, this method was accomplished by conjugating drugs to a functionalized end of a polymer chain. For instance, Sengupta et al. conjugated Dox, to PLGA, and were able to reproducibly prepare Dox-loaded NPs.188 Similarly, Tong et al. developed a method to prepare Ptxl-conjugated PLA by carrying out a ring opening polymerization of LA units onto a Ptxl-metal complex.253 This drug-conjugated polymer resulted in the reproducible synthesis of NPs incorporating high loadings of Ptxl with encapsulation efficiencies close to 100%.253 While the last two examples involved conjugation of drug molecules at the distal end of a polymer chain, recently Kolishetti et al. demonstrated both the encapsulation of Dtxl and the conjugation of a cisplatin pro-drug to formulate polymeric NPs for combination therapy.254 This resulted in an increase in encapsulation together with a potential of tuning drug release kinetics by varying the number of drugs attached to the polymer.
Fig. 5.
Drug release mechanisms from polymeric NPs: (a) diffusion from polymer matrix with time varying diffusivity, (b) surface erosion/degradation of polymer matrix, and (c) biodegradation of polymer matrix due to hydrolytic degradation leading to drug release.
In order to achieve optimal polymer-drug conjugate NP systems, the following considerations should be noted: (i) the chemistry implemented in modifying the drug should not affect the chemical groups or moieties that endow the drug with therapeutic effects; (ii) the drug needs to be cleavable from the polymer backbone under biological conditions, and upon cleavage the drug should retain its functional and therapeutic properties; (iii) chemistries used for conjugation must be carefully chosen since some residues of, for instance, certain catalysts, might contaminate the resulting NPs leading to unexpected toxicity. Nevertheless, conjugation of drugs to polymer backbones is an attractive strategy to incorporate multiple drugs with varying physicochemical properties in the same NP, enabling integrated and controlled combination chemotherapy.
4.4 Incorporation of targeting ligands on NPs
The widespread interest in the surface attachment of targeting ligands to various drug delivery NPs ranging from liposomes, micelles, polymers and dendrimers has spurred chemists to actively research coupling methods that are safe, tuneable, biocompatible, and reproducible. However, unlike the limited range of chemistries available for coupling to protein surfaces, a larger number of chemical modifications are now possible in order to attach targeting ligands to NPs. This can be done through either covalent attachment of targeting ligands to the surface of the NP or through electrostatic, dative or coordinate bonds. It is important to identify the conjugation technique that will ultimately provide the most efficient coupling chemistry that does not lead to undesirable products or side reactions, and can be produced on large-scales in a reproducible manner. In the subsequent sections we will discuss common chemical strategies utilized in the development of targeted NPs, mainly conjugation of a targeting ligand after NP formation and pre-conjugation of targeting ligands to polymeric precursors followed by self-assembly of NPs.
4.4.1 Chemical strategies for incorporating targeting ligands on NPs
The most traditional approach for the development of targeted NPs involves the conjugation of targeting ligands to the surface of NPs using facile coupling chemistries such as carbodiimide-mediated amide and maleimide-thiol couplings. Amide bond formation is a popular cross-linking reaction and carboxylic acids are frequently activated using a variety of carbodiimides such as 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide (EDC). In this case, either the surface of the NP can bear carboxylic acids or the targeting ligand may have this functional group available for coupling. Although the formation of an amide bond using carbodiimide activation of carboxylic acids is a straightforward reaction, however, several side reactions can lead to undesired side reactions that produce N-acylureas, in addition to rapid hydrolysis of the reactive O-acylisourea intermediate. In order to avoid this hydrolysis an excess of the carbodiimide can be used, however considering that the surface carboxylic acid moieties are the solubility determining anchors of the NPs, then over-activation of these acids and formation of poorly soluble O-acylisourea intermediates can lead to a lack of solubility and colloidal instability and aggregation. These events can be minimized through the use of N-hydroxy-succinimide (NHS) carboxylic acid activated intermediates and can improve reaction efficiencies as the NHS intermediate is more resistant to hydrolysis. The formation of amide bonds between NP carboxylic acids and amino groups of targeting ligands is a popular bioconjugation method; however ligand conjugation numbers and surface densities cannot be effectively controlled. Maleimide coupling with thiols is highly specific, less prone to hydrolysis, and can occur at neutral pH; this method has been very useful for coupling to cysteine residues of proteins or other thiol containing ligands, and avoids the formation of undesirable side reactions.255, 256
The search for highly specific chemical reactions that create unique linkages and avoid susceptibility to competing reactions has led to the development of “bioorthogonal” reactions. This class of reactions aremainly concerned with [3+2] cycloadditions between azides and alkynes, and is more commonly known as click chemistry.257 In order to avoid the use of toxic copper catalysts in these reactions, various copper-free click chemistry reactions have also been developed and many studies are now underway that demonstrate the versatility of this type of bioconjugation technique.258–261 A further bioorthogonal [4+2] cycloaddition reaction was recently described by Devaraj et al. and was applied to the labelling of cancer cells, peptides and small molecules.262 This transition occurs between a 1,2,4,5-tetrazene (Tz) and trans-cyclooctene (TCO) and proceeds rapidly at room temperature and under physiological conditions without the need for a catalyst.263 This conjugation reaction was successfully used in a novel labelling methodology termed “bioorthogonal nanoparticle detection” or “BOND” to conjugate antibodies to nanoparticles.264 Antibody proteins were initially conjugated with TCO moieties and then reacted with Tz-functionalized NPs leading toNPs with multiple antibodies bound to their surface. This strategy can be used for efficient targeting and signal amplification and has been mainly applied to diagnostic applications for the highly sensitive detection of cancer cells.263 From a synthetic perspective, the development of targeted NPs poses many challenges which include the following: (1) the exact ligand conjugation stoichiometry is not easy to control and therefore only average ratios of coupled ligands to NP surfaces can be estimated, (2) ligand conjugated NPs will not always have a low polydispersity and some NPs may possess higher numbers of ligands than others, (3) targeting ligands may not always attach to the surface of NPs through predictable covalent bonds and may interact with the surface of NPs through aggregation phenomenon, (4) the bioactivity of targeting ligands may be compromised following attachment to the NP surface, which could also affect the NP therapeutic performance, (5) the coupling chemistries involved must be highly specific, occur rapidly and ideally be amenable to biological environments, (6) the conjugation linkages employed between ligands and NPs must be durable in the highly complex in vivo environment and not become prone to competing reactions that may be brought about by changes in pH or hydrolysis unless these are intended mechanisms used for ligand and/or NP protective layer shedding and finally, (7) the bioconjugation techniques employed in ligand conjugation to NPs should be scalable, reproducible and economical.265
4.4.2 Targeted NPs through polymer self-assembly
As we have seen in the previous section, the conventional approach to the development of targeted NPs involves the conjugation of bioactive molecules to the surface of NPs via coupling chemistries. Even though excess amounts of reactants are often used to drive these reactions to completion, and given the heterogeneous nature of NP samples, it is difficult to control the stoichiometry of functional biomolecules on the surface of NPs, thus leading to poor reproducibility in manufacturing.
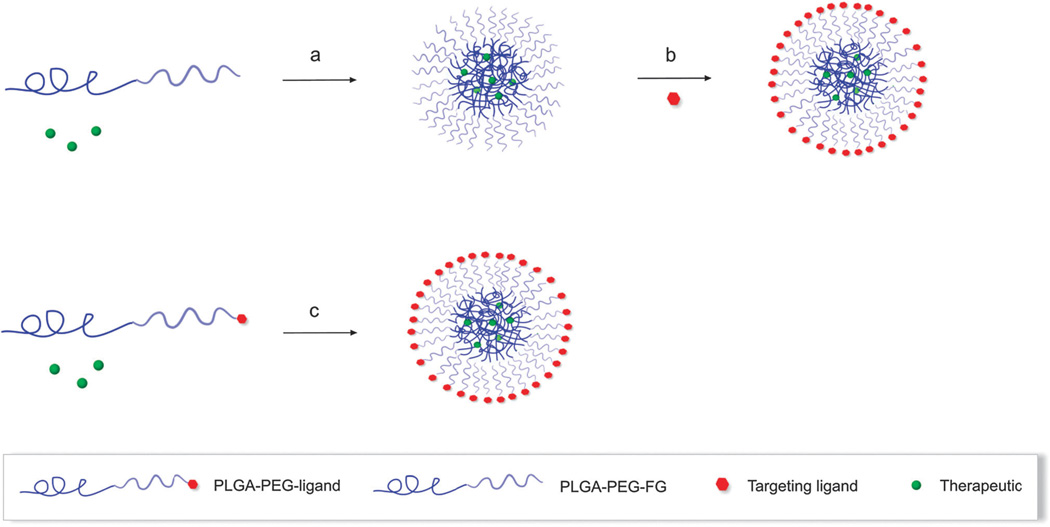
Recently, alternative approaches such as the self-assembly of targeted NPs using pre-functionalized components such as PEG polymers, and polymer-drug and polymer-targeting ligands have been developed to precisely engineer NP surfaces tending to uniformity (Fig. 6c).
Fig. 6.
Self-assembly of triblock PLGA-b-PEG copolymers in aqueous solution: (a) Polymeric NP formation via nanoprecipitation, FG = functional group (b) Conjugation of targeting ligand to the surface of pre-formed polymeric NPs (c) Pre-functionalized diblock polymer with hydrophilic targeting ligand.
The design of pre-functionalized triblock copolymers allows for the reproducible creation of optimal targeted NPs, whereby controlling the self-assembly and ratio of each constituent can lead to targeted polymeric NPs with precisely tuned biophysicochemical properties.196 Fig. 7 presents the development and characterization of PLGA-PEG-Apt triblock polymers, and the self-assembly of targeted NPs produced by the nanoprecipitation of a mixture of PLGA-PEG, PLGA-PEGApt and Dtxl drug. The concept of modular self-assembly of pre-functionalized material was demonstrated by Gu et al. who conjugated the A10 2′-fluorophyrimidine RNA Apt (Apt which binds to PSMA on prostate cancer cells), to PLGA-PEG block polymers in order to create triblock polymers that form targeted NPs by macromolecular self-assembly in a single step.218 In this study, targeted NPs with varying ligand surface densities were developed which led to the identification of the exact narrow range of Apt density yielding the highest tumour accumulation in vivo. Interestingly, there was a non-linear inverse relationship between ligand density and tumour targeting in vivo and discordance between in vitro and in vivo optimal ligand densities. While in vitro data demonstrated enhancement of cellular binding and uptake as ligand density increased, the in vivo data demonstrated that tumour accumulation increased with the addition of ligands only to a certain point. As ligand density continued to increase on the surface of NPs, there was a paradoxical reduction in tumour accumulation and in parallel an increase in liver accumulation. Therefore, while the NPs were becoming more targeted in nature with increased Apt ligands on their surfaces, their surface was also becoming less stealth-like, resulting in rapid liver accumulation and poor EPR. These findings, for the first time, demonstrated the importance of narrowly optimizing NP biophysicochemical properties for in vivo success and clinical translation, and formed the foundation of the discovery and development of BIND-014 and SEL-068 targeted NPs (Fig. 1 and section 3.3).
Fig. 7.
Self-assembling targeted polymeric NPs. A-B: Synthesis and characterization of PLGA-PEG-Apt triblock polymers. C: Nanoprecipitation leading to the self-assembly of PLGA-PEG-Apt NPs. Aptamer surface density is precisely controlled using distinct ratios of PLGA-PEG-Apt and PLGA- PEG. Figure taken from Gu et al.218
Recently, Valencia et al. investigated the self-assembly and properties of two distinct targeted NPs (Fig. 8) decorated with two widely used small molecule targeting ligands with varying solubilities (folate and RGD).266 While RGD is relatively hydrophilic, the folate molecule is relatively hydrophobic. Two different triblock copolymers of PLGA-PEG-RGD and PLGA-PEG-folate were synthesized using NHS/EDC amide coupling chemistry. NPs were developed using the nanoprecipitation method and this procedure demonstrated the ability to develop targeted polymeric NPs with different ligand densities by varying the ratios of the ligand bearing triblock polymers relative to PLGA-PEG copolymer lacking the targeting ligands. As predicted, further studies revealed the RGD targeting molecules to preferentially reside on the surface of the NPs, and the theoretically predicted RGD density on NP surfaces was shown to correlate to the experimentally derived RGD density. In contrast, the experimentally derived folate ligand density on NP surfaces was only 20% of the theoretically predicted folate density, presumably due to the fact that during the NP self-assembly process folate is in-part trapped in the hydrophobic NP core and cannot be presented on the NP surface. These results revealed the impact that ligand chemical properties can have on the targeting capabilities of self-assembled targeted PLGA NPs, and underscore the challenges associated with developing suitable targeted NPs and the need to optimize NP biophysicochemical properties on an individual basis.
Fig. 8.
PLGA-PEG-RGD and PLGA-PEG-folate triblock polymers used to prepare targeted NPs with different surface ligand densities. Figure taken from Valencia et al.266
Given the importance of the nature of the targeting ligands used in the development of targeted NPs, the various forms of targeting ligands, the isolation of high affinity ligands and ligand properties will be further discussed in the subsequent section.
5. Targeting ligands
Representative ligands for targeted NP development mainly include mAbs and their fragments, proteins or protein-like molecules, peptides, nucleic acid ligands (including Apts), small molecules and sugars.122 In the following section we firstly look at specific examples using various ligands to summarize recent efforts in the development of ligand-based targeted NPs, and secondly we highlight strategies to isolate these targeting ligands for use in a variety of medical applications. Additionally, various primary ligand properties that affect NP targeting will also be discussed.
5.1 Antibodies and their fragments
As discussed previously, mAbs have been most commonly used in the development of targeted NPs. For example, trastuzumab and rituximab which are mAbs currently in the clinic have been conjugated to PLA NPs resulting in conjugates that exhibit a 6-fold increase in the rate of particle uptake compared with similar particles lacking mAb targeting molecules.267, 268 Nevertheless, mAb-targeted NPs still encounter many challenges and limitations since these molecules are large (~150 kDa), unstable in organic solvents (not amenable to incorporation on NP surfaces by self-assembly), are potentially immunogenic resulting in rapid NP clearance, and create engineering challenges for NP scale-up and manufacturing.269, 270 Additionally, with an average hydrodynamic size of ~ 15 nm, when conjugated to NP surfaces they have the tendency to increase overall NP sizes by up to 30 nm.141, 142 Therefore, antibody fragments including antigen-binding fragments (Fab) and scFv’s have been developed to reduce these undesirable effects.51
Affibody molecules are small polypeptides derived from an antibody binding domain of staphylococcal protein A, that are used as scaffolds for the construction of combinatorial phage libraries.271 From these libraries, affibody variants that target a specific cell marker can be selected using phage display technology. A 6 kDa affibody molecule has been reported with selective binding to the HER2 receptor with subnanomolar affinity.140 Alexis et al. conjugated this affibody to the surface of polymeric NPs and showed increased uptake of these particles as compared to non-targeted NPs by breast cancer cells.140
Nanobodies are fully functional antigen-binding fragments evolved from the variable domain of heavy-chain antibodies. Similar to affibody molecules, nanobodies are also low molecular weight <15 kDa polypeptides that show similar antigen binding affinity which rivals that of the traditional mAbs.272 Nanobodies are typically evolved from single-domain antibodies (antibodies carrying only a functional heavy chain without the light chain). In comparison with mAbs, these smaller molecules have been shown to have lower immunogenicity. Recently, a 15 kDa nanobody was isolated that demonstrates specific uptake in vitro and in vivo using xenograft-tumour-bearing mice expressing a carcinoembryoic antigen.273
5.2 Proteins
Many endogenous proteins that selectively bind to specific membrane-bound receptors on cells can be used for targeting via receptor-mediated endocytosis.274 For example, the iron-transporting protein Tf, which binds specifically to the TfR, has been used to deliver NPs into different cell types. Choi et al. discovered that ligands targeting the TfR exert their influence by increasing uptake of targeted NPs by cancer cells and not by increasing particle accumulation in the tumour region.275 Alternatively, growth factors such as epidermal growth factor (EGF)276 and neural growth factor (NGF)277 attached to NPs are other examples of protein-targeted NPs which have been shown to enter cells and elicit specific molecular responses. Proteins however, are commonly immunogenic and susceptible to early clearance by different mechanisms in the body, which may limit their effectiveness. Additionally, the receptors of these protein ligands are endogenous and commonly expressed on many types of non-targeted cells, which can lead to off-target adverse effects.300
5.3 Peptides
Peptides have gained interest as targeting ligands due to their small size, relatively low immunogenicity as compared to larger proteins, high stability, and ease of conjugation to polymers, lipids and NP surfaces.301 Specifically, the development of highly specific peptide phage libraries, bacterial peptide display libraries, plasmid peptide libraries, and new screening technologies have made possible the synthesis of peptide ligands to a myriad of targets.302, 303 Combinatorial libraries have led to the discovery of short peptides (10–15 amino acids) that are able to bind to targeted proteins, cells, or tissues specifically. The most studied peptide sequence is that of RGD which binds to αvβ3 integrin receptors which are highly upregulated on both tumour cells and angiogenic endothelial cells.304 RGD is chemically synthesized, and is commercially available with different choices of linkers that allow for its conjugation to NP surfaces, biomaterials, or drugs. Despite the excitement of RGD-targeted therapies, some challenges still remain, including nonspecific adhesion, as αvβ3 integrin is also expressed on normal tissue and non-cancerous inflamed tissues.304
Development of phage display screening methods has successfully isolated peptide ligands with high specificity and affinity to various targets such as injured vasculature,147 cellsurface hormone receptors such as luteinizing hormone-releasing hormone gene receptors (LHRHR),305 and tumour vasculature antigens.306 For instance, Chan et al. developed a NP system that targets vascular antigens exposed in cardiovascular-related diseases such as angioplasty-injured vasculature (Fig. 9, discussed in more detail in section 7).147, 307 Similarly, Arap et al. have isolated peptides that bind specifically to tumour vasculature in patients and shown the potential of such in vivo phage library selections.308 Other examples of peptides used to develop targeted NPs include Cys-Arg-Glu-Lys-Ala (CREKA),221 Asn-Gly-Arg (NGR),309 and Ile-Thr-Asp-Gly-Glu-Ala-Thr-Asp-Ser-Gly (LABL).186 As discussed in section 3, the arrival of targeted NPs directly to tumour cells may be impeded by poor vascularisation and permeability within certain tumour regions. This problem is further confounded by the high interstitial pressure of tumours due to the leaky nature of their blood vessels and their dysfunctional lymphatic vessels which can diminish tumour tissue penetration and retention of NPs.310 To further retain NPs within tumour tissue and to enhance cellular uptake, cell-penetrating peptides have been used to facilitate translocation of cargoes across the plasma membrane and to specific organelles within the cell.311 Many of these sequences are derived from natural sequences, such as the protein-transduction domains of viruses.274 For example, the Tat peptide derived from the HIV-1 virus has been popularly used to deliver a variety of NPs into cells. In particular, Tat-conjugated fluorescent quantum dots were used asmodel systems to investigate the trajectory of Tat-functionalized NPs within cells.312
Fig. 9.
Polymer-lipid hybrid ‘Nanoburr’ particles. A: Polymer-drug conjugate synthesis (Ptxl-PLA), B: HPLC characterization of Ptxl and Ptxl-PLA polymer, C: Nanoburr synthesis via polymer lipid, and lipid-PEG self-assembly, D: TEM image of Nanoburrs (stained with 3% uranyl acetate), E: Dynamic light scattering measurements (DLS) pre and post peptide conjugation, F: Zeta potential measurements pre and post peptide onjugation, and G: in vitro drug release profile of Ptxl from Nanoburr NPs. Figure taken from Chan et al.147
While the Tat cell penetrating peptide does not display cellular specificity,313 recently, other classes of peptides that interact with proteins upregulated on tumour vessels have been discovered. These peptides may ultimately assist the extravasation of NPs into tumours. Three representatives of this new class of peptides are: (1) iRGD, which binds αnβ3 integrins upregulated in tumours on the endothelial cell surfaces,314 (2) LyP-1, which targets lymphatic vessels in some tumours,315 and, (3) F3, which is an N-terminal fragment of human high-mobility group protein 2 that can bind to tumour vasculature.316 All three peptides contain the cell penetration motif, R/KXXR/K or CendR sequence that is thought to interact with neurophilin-1 facilitating extravasation, tissue penetration, and cell entry of NPs.314 Although high levels of these peptides have to be administered (e.g. reaching >100 µM levels in blood), it has been reported that this approach can potentially increase tumour uptake of a wide spectrum of small to large agents including NPs by several fold.151 For example Sugahara et al. showed that systemic injection of iRGD peptide enhanced cancer drug delivery and activity when this peptide was administered as a combination therapy (without chemical conjugation) with either Dox, nab-Ptxl, Dox-liposomes or trastuzumab mAbs, suggesting this combination therapy to be a valuable method to enhance anti-cancer drug efficacy.281
5.4 Nucleic acid ligands
Nucleic acid ligands (i.e. Aptamers (Apts)) are oligonucleotides that fold by intramolecular interactions into unique conformations with ligand-binding characteristics.317 Apts can be DNA or RNA oligonucleotides, or modified oligonucleotides resistant to nuclease degradation. Apts can be selected from large pools of random oligonucleotide libraries by virtue of selective binding to antigens with high specificity and affinity. The in vitro Apt selection process is referred to as Systemic Evolution of Ligands by Exponential enrichment (SELEX) and was described independently by the groups of Larry Gold318 and Jack Szostak319 in 1990. Apts isolated from the SELEX process have approximate molecular weights of ~15 kDa, are chemically stable in a range of solvents, pH, and can be developed by chemical processes such as solid phase synthesis.320–323
NP-Apt conjugates targeting the PSMA were originally developed several years ago.282 In these studies the Apt used as the targeting ligand was A10 Apt, first isolated by Lupold et al.35 These targeted NPs were loaded with Dtxl and evaluated in vivo in a tumour model of LNCaP prostate cancer cells which overexpressed PSMA antigens. It was shown that the administration of these targeted NPs led to effective tumour size regression, following a single intra-tumour injection over a 109-day study.124 In another study, Bagalkot et al. demonstrated a novel strategy for the targeted delivery of anthracyclic agents including Dox, directly to cancer cells through the formation of an Apt-Dox physical conjugate.324 Recently Guo et al. presented studies that suggest PLA-PEG NPs targeted with the AS1411 Apt could cross the BBB and target nucleolin, a protein highly expressed in the plasma membrane of cancer cells, and specifically on glioma cells.325 One key characteristic that endows Apts with high specificity against targets is their secondary structure that arises from their specific nucleotide sequence. However, this secondary structure may be affected by heat, exonuclease or endonuclease degradation, and other environmental factors which could limit the stability of Apts and their binding properties.326
5.5 Small molecules
The use of small molecules to create targeted NPs remains an attractive targeting strategy given their small size and ease of handling (less prone to degradation than biomolecular ligands). Some further advantages of small molecule targeting ligands include: (1) the availability of a range of facile coupling chemistries for their conjugation, (2) achievement of higher ligand densities on NP surfaces due to the small size of ligands, (3) availability of a wide range of targeting ligands with variable solubilities and functional groups, as facilitated by advances in diversity-oriented synthesis, (4) less immunogenic effects in vivo (compared to macromolecular ligands) and, (5) reproducible and scalable manufacturing.327, 328
One of the most extensively studied small molecule targeting moieties for drug delivery is folic acid (or folate). This highaffinity vitamin is a commonly used ligand for cancer targeting as folate receptors (FRs) are frequently over-expressed in a range of tumour cells. Folate specifically binds to FRs with a high affinity (Kd = ~10−9 M), enabling a variety of folate derivatives and conjugates to deliver molecular complexes to cancer cells without causing harm to normal cells.329 For example, EC145 is conjugate of folate and a vinca alkaloid (desacetylvinblastine monohydrazide (DAVLBH)) currently in phase III development for cancer treatment.330, 331 In addition to drug conjugates, folate has been used as a targeting moiety combined with a wide array of drug delivery vehicles (including liposomes, protein toxins, polymeric NPs, linear polymers, and dendrimers), to deliver drugs selectively into cancer cells using FR-mediated endocytosis.332 Given that FRs are expressed not only in tumour tissue but also in normal epithelia in the choroids plexus, placenta, lung, intestine, and kidney, FR-targeted NP delivery systems need to be further refined to increase tumour selectivity.333 For example, as discussed previously (section 4.6) the optimal density of the targeting ligand should be investigated on a case-by-case basis given that this parameter affects the targeting efficacy of NPs to target cell populations. In these cases, greater ligand density does not necessarily translate to higher levels of tumour concentration—as NPs tend to lose their “stealth” surface characteristics and become increasingly prone to sequestration by cells of the MPS.196 Therefore, optimal folate density on NP surfaces needs to be investigated and determined on a case-by-case basis.334 Additionally, folate is relatively hydrophobic in nature and strategies to maximize its binding properties on the NP surface need to be carefully considered and investigated.266 Additionally, an increased surface density of folate ligands on NP surfaces has been shown to give rise to dimers, trimers or tubular quartet self-assembled folate structures which can hamper the binding efficiency of folate to its receptors.335
In one study, Pomper et al. were able to identify a small hydrophilic molecule that could target the PSMA receptor in prostate cancer cells.336 They showed that this molecule induced NP internalization by cells through endocytosis and preferential accumulation of NPs in tumours overexpressing PSMA receptors. Given that PSMA is not only overexpressed on the surface of prostate cancers but also on the neovasculature of all solid tumours, this targeting strategy is very attractive for therapy and diagnosis applications.337, 338
Carbohydrates, or sugar moieties, have also gained attention as targeting ligands due to their ease of production, low molecular weight and high abundance in nature. Some of these carbohydrate ligands target the membrane carbohydrate-binding proteins (membrane lectins) differentially expressed on the cellular and intracellular membranes of a number of cells. Their multiplicity, high affinity, and effective endocytosis after receptor binding as well as the biocompatibility of carbohydrate ligands make them potentially suitable ligands for carriers in cell-selective delivery of drugs and nucleic acids.339 Among the different targeting ligands mannose,340 glucose,341 galactose,342 and their derivatives have been successfully tested.
5.6 Ligand selection
5.6.1 Selection of high affinity ligands against complex targets
As a result of the need for highly efficient targeting ligands for use in a variety of biomedical applications as well as conjugation to NPs, a number of novel ligands with high affinities, internalization and transcytosis capabilities have been isolated recently, examples of which are presented in Table 3. The majority of the ligands currently available for targeting applications have been developed against well characterized purified proteins. However, selections against purified proteins are hampered by the limited number of purified receptors available, especially when the protein targets are insoluble or the targets are functionally part of multiprotein complexes.343 To overcome these limitations, selection protocols based on living cells have been developed as alternative methods.344, 345 In contrast to the selection processes against purified proteins, cell-based selections can be performed without prior knowledge of targets or multi-protein complexes expressed on the cell surface. Moreover, intact living cells with many native receptor proteins can be used as targets during the selection procedure. This allows for a variety of ligands targeting several proteins on the same cell to be isolated from such screenings.346 Since this strategy relies on the differences between the target cell population and the control cell population used for counter-selection (e.g. defined phenotype, protein expression levels, different protein conformations), multiple ligands recognizing only target cells and not the control cells can be identified. For example, de Kruif et al. extended phage display technology to living cells, whereby they isolated human scFv antibodies specific against subsets of blood leukocytes, and obtained two phage antibodies from such screening with specific binding to B-lineage cells.345 Similarly, Shangguan et al. successfully isolated a panel of Apts that can distinguish leukemia T-cells from B-Cells using cell-based SELEX.347
Table 3.
Novel ligands screened for targeted NP development
| Category | Ligand type | Receptor | Ref. | |
|---|---|---|---|---|
| High affinity ligands | Peptides | LHDH | αvβ3 integrin | 278 |
| SP5-2 (TDSILRSYDWTY) | Non-small cell lung cancer | 279 | ||
| NGR peptide | Angiogenic endothelial cell | 280 | ||
| iRGD (CRGDK/RGPD/EC) |
αvβ3 integrin and neuropilin-1, on tumour vessels |
281 | ||
| Aptamers | A 10 RNA Apt | Prostate-specific membrane antigen | 282 | |
| Sgc8c DNA Apt | Protein tyrosine kinase 7 (PTK7) receptor | 283 | ||
| 35-mer DNA Apt | Platelet-derived growth factor | 284 | ||
| 88-mer DNA Apt | CCRF-CEM (Human T cell lymphoblast-like cell line) | 285 | ||
| Antibodies | Monoclonal antibody A7 | Colorectal carcinoma | 286 | |
| Transferrin antibody | Transferrin receptor | 287 | ||
| DI17E6 | αvβ3 integrin receptor | 288 | ||
| 2C5 antibody | nuclesome (NS)-restricted activity | 289 | ||
| 5D4 antibody | Prostate cancer | 290 | ||
| Anti-HER2 scFv | ErbB2 receptor | 291 | ||
| Anti-VCAM-1 | Vascular cell adhesion molecule-1 (VCAM-1) on activated endothelial cells |
292 | ||
| Anti-CD22 scFv | CD22 antigen B-cell lymphomas | 293 | ||
| Other targeting molecules | Affibody (e.g. Anti-EGFR affibody) | EGFR | 294 | |
| Avimer | Human extracellular receptor | 295 | ||
| Nanobody | Human tumour-associated carcinoembryonic antigen | 273 | ||
| Internalizing ligands | Peptides | PC3 cells | 296 | |
| scFv antibodies | PC3 cells | 297 | ||
| Transcytosis ligands | Peptides | ACTTPHAWLCG | BBB | 298 |
| GLA and GYR | 299 | |||
| TGNYKALHPHNG | 194 |
Given that the binding of ligands is dependent on their target’s conformation, which in turn is affected by the target’s environment, selection strategies that can generate targeting ligands capable of specifically localizing to tissues or organs in vivo have been developed. In 1996, Pasqualini et al. introduced the technique of in vivo phage display and screened a group of peptides capable of specifically localizing to brain and kidney blood vessels.348 Similarly, this in vivo screening method was conducted to isolate prostate tissue-homing peptides.349 More recently in vivo phase display and screening was performed on 8 live patients with stage IV cancer resulting in the identification of several tumour-homing peptides and scFVs.303 In 2010, Mi et al. designed an “in vivo selection” approach to screen a library of nucleaseresistant RNA oligonucleotides in tumour-bearing mice in order to identify candidates with the ability to localize to hepatic colon cancer metastases. One of the selected molecules was an RNA Apt that binds to p68, an RNA helicase that has been shown to be upregulated in colorectal cancer.350 Further efforts are now underway to take these selection processes to the next level—that of identifying ligands capable of specific cellular internalization and transcytosis (transport across the cell interior).
5.6.2 Selection of cell internalizing and transcytosis ligands
Considering that intracellular delivery of drug loaded NPs could provide enhanced therapeutic effects over NPs that do not enter cells, selection techniques based on receptor-mediated endocytosis have been developed to identify internalizing ligands that are useful for NP delivery. For example Burg et al. identified eight distinct prostate cancer (PC3) cell internalizing peptides from a diverse peptide-display library.296 Similarly, a subset of internalizing scFv antibodies has been identified by phage antibody selections against PC3 cells.297 These antibodies have been further conjugated to NPs to direct their specific delivery into PC-3 cells.297
Recently, Xiao et al. developed a “cell-uptake selection” strategy to isolate a group of cancer-cell specific internalizing RNA Apts (Fig. 10).351, 352 In this strategy, selection was carried out against PC3 or LNCaP prostate epithelial cell lines to identify Apts that differentially bound to either PC3 or LNCaP and were subsequently internalized. In order to increase the specificity of the selection process, several cell lines representing normal prostate and non-prostate cells were used in the counter-selection process to deplete the Apt library from ligands that also interacted to these other cell types. Different from previously reported Cell-SELEX processes, this selection was performed at physiological temperature (37 °C), where cells and their membrane receptors are biologically active and continue their endocytosis functions. In addition, the isolated RNA Apts were introduced with 2′ O-methyl (OMe) modifications during the selection process, which facilitates the resistance of nuclease degradation inside intra-cellular environments. More importantly, only internalizing Apts were selectively collected, since the non-internalized membranebound Apts were removed during the selection process. After 12 rounds of selections, a group of PC3 or LNCaP specific internalizing Apts were identified and further conjugated with polymeric NPs. Results demonstrated that the internalizing Apt-targeted polymeric NPs were specifically and efficiently taken up by targeted cells, and could drastically improve the cellular cytotoxicity of Dtxl compared to non-targeted NPs.
Fig. 10.
Schematic protocol of cell-uptake selection for evolving cancer cell-specific internalizing Apts. Figure taken from Xiao et al.351
Towards the delivery of NPs capable of crossing physical barriers, such as the BBB, transcytosis peptide ligands have been isolated by utilizing phage display techniques. Wan et al. screened a C7C phage display library of peptides that were intranasally administered to rats, and subsequently recovered phage from the brain tissue.298 This screening revealed a peptide sequence (ACTTPHAWLCG) that can bypass the BBB through the nasal-to-brain passage.298 Additionally, Rooy et al. selected two 15-amino acid-peptides (GLA and GYR) that can bind to the murine brain in an in situ brain perfusion model.299 Recently Li et al. isolated TGNYKALHPHNG peptide (denoted as Pep TGN) through in vivo phage display screening, and when conjugated on the surface of PLGA NPs, Pep TGN was shown to facilitate targeted delivery of these NPs across the BBB, leading to significant higher cellular uptake and in vivo brain accumulation. 194 These studies demonstrate both the feasibility and efficiency of in situ ligand selection techniques and can be applied to a range of applications where there is a need for high affinity binding ligands that can direct NPs to disease cells.
5.6.3 Ligand properties affecting NP targeting
Efficient argeting of NPs to the cell surface lipid bilayer may be dependent on two important ligand properties, which include; ligand affinity, and ligand density. Upon binding of targeted NPs to the cellular lipid bilayer, complementary membrane receptors fluidly diffuse together to initiate NP membrane wrapping.353 If the binding affinity of the ligand to its receptor is strong, sufficient thermodynamic energy is generated to overcome the elastic recoil of the cellular membrane back to its original equilibrium state resulting in membrane wrapping around NPs.353 For large NPs, high binding affinities are particularly required to prolong the residence time of NPs onto cellular surfaces. In this situation, binding affinities are majorly influenced by equilibria between enthalpic advantages due to ligand-receptor binding and entropic losses for tether chain stretching or compressibility around the cell surface environment.354 Controlling ligand-to-NP ratios has been traditionally difficult, leading to heterogenous distributions of ligands on the surface of NPs. In order to address this issue the effective self-assembly of pre-functionalized and pre-targeted tri-block polymers was accomplished by Gu et al.196 This study demonstrated the facile tuneability of the density of ligands on NP surfaces, which was achieved by the blending of variable ratios of the targeted triblock polymers with other polymer blocks. In this way, the overall ligand density of NPs can be controlled leading to more homogenously targeted NPs. Tassa et al. recently quantitatively studied the affinity and binding kinetics of NP bearing small molecules using surface plasmon resonance (SPR).355 In this study the interactions between a single protein target and a range of targeting ligands with variable intrinsic affinities was measured. It was concluded that even small ligands with weak affinities could still significantly enhance target-specific avidity via multivalent interactions. Knowledge of NP affinity and association and dissociation rates may ultimately predict how deeply NPs can penetrate into tissues once target cell populations become saturated.355 Therefore, NP size, target ligand density and tissue permeability as governed by capillary permeability are all factors that affect the efficacy of NP targeting in vivo.
In another study, the effect of ligand density on the internalization of PLGA NPs was recently investigated using the cyclic peptide ligand, cLABL which binds to the intercellular cell adhesion molecule-1 (ICAM-1).356 In this case an optimum of between 2 to 4 pmol cm−2 of the peptide ligand was found to be effective for targeting to A459 cells. Further discussions on the optimization of ligand densities can be found in a review by Pirollo et al.145
Throughout this review so far we have seen that the nature of the targeting ligand (i.e. whether small molecule or macromolecular ligands etc.), the size of the NP system and the degree of ligand density on the NP surface, are all factors that affect the binding efficiency of targeted NPs to target cell receptors or other antigens, and can alter the path of NPs in vivo. Given the wide nature of ligands and targeted NPs developed, at this point, ideal generalizations of relationships between ligand density, NP size and optimal ligand binding affinities cannot be made and must be investigated for each unique system.
6. Optimal biophysicochemical characteristics of NPs
NP physicochemical properties, such as size, geometry/shape, surface charge, surface chemistry, hydrophobicity, roughness, rigidity, and degree of composition (Fig. 11), can result in differential uptake and/or targeting to certain organs, tissues or cells and may be optimized through the effective design of NPs.357 In this section, we will focus on discussions of these primary properties of NPs, the optimization of which is essential for the development of efficient therapeutic NPs.
Fig. 11.
NPs and their biophysicochemical characteristics which affect their performance both in vitro and in vivo.
6.1 Influence of NP size
NP size is a key parameter affecting the cellular uptake rate of NPs as it influences their internalization mechanism, and it is also a key property for in vivo circulation half-life as discussed further below.
There are two major endocytic mechanisms by which cells take up particles and macromolecules, and these are referred to as phagocytosis and pinocytosis (or fluid-phase uptake).358 Large particles (>1 µm) are generally internalized by phagocytosis mechanisms, which are present only on professional phagocytic cells, such as macrophages, neutrophils, or dendritic cells.359 Therefore pinocytosis is more relevant to NP cellular uptake and can occur either via adsorptive pinocytosis (non-specific adsorption of NPs or macromolecules to the cell membrane followed by internalization) or via receptor-mediated endocytosis (RME, which describes the interaction of NPs andmacromolecules with receptors, followed by their internalization).360 Pinocytic mechanisms of uptake can be further divided into caveolae-mediated endocytosis or clathrin-mediated endocytosis, as well as clathrin-independent or caveolin-independent endocytosis (smaller NPs can be internalized through a number of these pathways).359 Detailed discussions on these pathways and their implications for NP cellular uptake can be found in other excellent recent reviews.359–363
Cellular internalization of nanoparticles is majorly dependent on the size of the NPs, and in general, particles in the 40–50 nm range exhibit maximal uptake in vitro.349, 362 Jiang et al. studied the interactions of targeted antibody conjugated silver and gold NPs and found that 40–50 nm particles exhibited the highest amount of cellular internalization and concluded that this optimal size range for NP uptake is likely due to an intricate balance between multivalent cross-linking of the membrane receptors and membrane wrapping processes taking part in RME.364 In this study Herceptin (Her) gold NPs (2–100 nm) were synthesized and the size-dependent binding and uptake of these NPs was investigated with ErbB2 receptor expressing cells.364 It was also shown that the number of Her antibody binding sites on the NPs was dependent on the NP surface area and increased with particle radius. Antibody density on the surface of the NPs also increased linearly with NP radius—demonstrating that these multivalent antibodyconjugated NPs can allow for a high degree of ErbB2 cross-linking which can be tuned by NP size.
In general, 10–100 nm is a generally accepted size range for the development of NPs for in vivo applications which relates to their in vivo clearance and biodistribution patterns.360 These upper and lower bounds are largely determined by interactions with the immune system and kidney filtration cut-offs, respectively. Larger NPs possess low radii of curvature which can lead to increased interactions of opsonins onto their surface and faster clearance rates in vivo.365 The predominant proteins involved in the opsonization process are plasma complement and immunoglobulin proteins which can lead to development of hypersensitivity towards NPs.366 Large NPs are prone to filtration through the sinusoids in the spleen and clearance by the MPS cells, which include the Kupffer cells of the liver.367 Additionally, NPs smaller than approximately 5.5 nm have been shown to be rapidly cleared by glomerular filtration in the kidneys.368
For the purposes of tumour accumulation, the upper limit for extravasation into solid tumours has been suggested at ~400 nm and it is generally observed that NPs <200 nm in size can accumulate effectively within tumour tissue, with the 70–200 nm range considered optimal for tumour passive targeting.369 However, recent studies have revealed that there is an optimum size for maximum uptake by various tumour cell types, and even a 10 nm deviation from this optimum results in a significant decrease in NP uptake.364 Concurrently, some researchers have found that NPs of about ~10–20 nm are ideal for maximum tumour penetration.370
Recently, a study by Schadlich et al. investigated the size dependent accumulation of fluorescently labelled PLA-PEG polymeric NPs using two different tumour xenograft models, HT20 colon and A2780 ovarian carcinoma, which result in different tumour structures, growth rates, and microenvironments. 371 Using an in vivo fluorescence imaging technique, the biodistribution and accumulation of NIR-loaded PLA-PEG NPs was tracked. NPs 111 and 141 nm’s in diameter were shown to accumulate efficiently in tumours and the larger NP (166 nm) was observed to undergo rapid clearance in the liver. The pattern of accumulation was shown to be different in both tumours, however, fluorescence was mostly observed from the tumour core region in the case of HT29 tumours, which was not observed in the A2780 tumours. Using different NP size batches the authors concluded that NP accumulation to the necrotic HT29 tumour core is size independent, but size dependent in the more vascularised A2780 tumours. Additionally, larger NPs led to lower tumour accumulation. Interestingly, this study concluded that size variations of between 20–30 nm (z-averages) led to highly distinct in vivo outcomes for NP distribution. Although the NPs used in this study utilized NPs with low polydispersity indexes (PDI: 0.013–0.16), these results help to shed light on the importance of tumour vascularity and narrow NP size distributions, since polymeric NPs can have high PDIs due to inherent variations in both the molecular weight of the polymers and the mixing time of precursors during preparation.51
With regards to polymeric NPs, our previous work has demonstrated that the molecular weights (MW) of PLGA together with the concentration of polymer in organic solution are key parameters that allow for independent tuning of NP size.372 We have also successfully demonstrated that NPs made from a PLGA-PEG polymer with PLGA MW of 15 KDa at low concentrations in microfluidic devices can be formed in the range 20–25 nm.236 Similarly, NPs made from PLGA-PEG with PLGAMWof 95 KDa at high concentrations were achieved in sizes of 200–250 nm.372 These promising methods demonstrate the feasibility of obtaining small sizes in addition to highly narrow PDIs and demonstrate the use of microfluidics to effectively devise libraries of homogenous NPs with a range of sizes.
Given the range of nanomaterials and cell types used to study the effects of NP size on cellular uptake rates and mechanisms, and the often contradicting results and claims that has been demonstrated in the literature so far, then it is important for an optimal NP size to be determined experimentally for a given NP and specific cell type. Furthermore, so far not many studies have been conducted that investigate the relationship between incorporation/density of targeting ligands and NP size on cellular uptake efficacy and indeed these studies merit further investigation.
6.2 Influence of NP shape
Recent studies have shown that particle shape may be an important factor in the rate of NP cellular internalization.362 This is mainly due to the fact that NP shapes that can accommodate cellular membrane wrapping processes become most effective at cellular uptake. Studies have shown that amongst NPs of either rod or sphere design, the spherical shaped NPs were taken up by cells more readily.361 In another example, Desimone et al. have produced a variety of NP shapes using their top-down fabrication method termed particle replication in nonwetting templates (PRINT) (Fig. 12).360 They found that the internalization of rod-like NPs with high aspect ratios (depth: 150 nm, height: 450 nm and volume: 0.00795 µm3) occurs faster in HeLa cells than that of cylindrical NPs regardless of NP volume.
Fig. 12.
Development of an array of nano and microparticles with variable shapes and aspect ratios using the PRINT technique by Desimone et al. Figure taken from Wang et al.360
NP shape is also an important factor for the biodistribution and circulation of NPs in vivo. Geng and Decuzzi et al. have reported that non-spherical particles with longitudinal lengths reaching cellular diameters and discoidal shapes can exhibit longer circulation times than spherical particles.373, 374 Given the typical processes used to fabricate therapeutic NPs (bottomup fabrication, self-assembly), the majority of these particles are spherical. As such studies determining optimal NP shapes are still at an early stage and further investigations are required to determine the effects of NP shape on cellular uptake.
6.3 Influence of NP surface charge
NP surface charge is a major factor contributing to the nonspecific binding of NPs to cells and proteins in blood circulation. Positively charged NPs are rapidly cleared from circulation by cells of the MPS.360
NPs with cationic surfaces may promote cellular binding, resulting in either uptake through endocytosis or direct penetration of the cellular surface membrane. This is because cationic surfaces will interact with the negatively charged phospholipid head groups, proteins and glycans on the surface of cells.359 Similarly, negatively charged NPs can also show selective cellular uptake compared to NPs with neutral surfaces.361 Indeed, NP surface charge is a predominant factor for endocytotic uptake of NPs into cells. Recent studies have shown the uptake of positively charged NPs to be an energy dependent process involving the proteins dynamin and F-actin, whereas negatively charged NPs were internalized in a dynamin-independent manner.375 On the other hand, highly cationic NP surfaces could also by-pass endocytic modes of entry into cells as they can enter cells by creating holes in the cellular bilayer.359
Charged NPs however, will inevitably have short half-lives in vivo and high non-specific cellular uptakes due to interaction with blood proteins, resulting in complement activation. Indeed a recent systematic study of the immunocompatibility properties of lipid-polymeric NPs was conducted by measuring the effects of carboxy, amino and methoxy terminated PEGylated NPs on the degree of complement system activation, human plasma protein binding and coagulation system activation.376 Amongst the surface functional groups studied, amino terminated surfaces induced the highest levels of complement activation. NPs with the more neutral methoxy surface groups were most immunocompatible. Another study showed that after systemic administration, particles with surface charges <15 mV showed minimal macrophage uptake and led to longer circulation times and hence tumour retention.363
From our previous experiments we have concluded that the surface charge of NPs can be easily tuned by modifying the functional end group of the PEG polymer.377 Specifically, NPs composed of PEG-NH2 exhibited a zeta potential of 10 to 15 mV while NPs composed of PEG-COOH exhibited a zeta potential of −10 to −15 mV, and those composed of PEG-OCH3 remained neutral. More interestingly, we have shown that by blending PEG polymers with different end groups at various ratios the NP surface charge can be modulated to a desired value.376
6.4 Influence of NP hydrophobicity, roughness, and rigidity
Hydrophobicity plays an important role in NP targeting to cells; NPs that are more hydrophobic than the cellular surface membrane are more easily taken up by cells.359 Likewise, protein adsorption on the surface of NPs is highly dependent on the hydrophilicity of NP surfaces. Hydrophobic surfaces tend to lead to higher levels of protein adsorption, and IgG proteins which are also opsonins, have high affinities for hydrophobic surfaces.378
In addition, surface effects such as smooth versus rough surfaces also influence the degree of NP surface binding to cells.379 Nanoscale “roughness” manifests as local protrusions or depressions on the NP surface forming a harsh surface that has been shown to lead to a minimization of repulsive forces between cellular and NP surfaces.353 Nanoscale surface roughness influences the surface topography of NPs and was recently exploited to enhance the growth and osteogenic differentiation of human-bone-marrow-derived mesenchymal progenitor cells.380
Particle rigidity can highly influence in vivo biodistribution profiles. Red blood cell like hydroxyethyl acrylate hydrogels were synthetically engineered with both rigid and deformable structures and their biodistribution evaluated in vivo.381 The more deformable synthetic blood cells were shown to eliminate up to 30 times more slowly than their rigid counterparts, with more rigid microparticles accumulating in the lungs 2 h post-injection. Given the highly flexible and deformable discoid shape of naturally occurring red blood cells, it is hardly surprising that the elastic modulus properties of NPs can also play an important role in the pharmacokinetics and biodistribution of particles, with more flexible NPs traversing vessels and pores more easily in vivo. Studies exploring the relationship between targeted NPs with variable degrees of structural rigidity and flexibility should be of high interest.
6.5 Influence of NP PEGylation
Due to their large surface-area-to-volume ratios, NPs can attract a ‘corona’ type binding of blood proteins to their extremely curved surfaces.353 These numbers of proteins are by no means few and far between, but in the hundreds.382 Although plasma protein binding by abundant proteins such as albumin and 1-acid glycoprotein may lead to enhanced bioavailability for traditional small drug molecules (through the reduction of first pass hepatic extraction), however in the case of NPs, this can lead to their enhanced blood clearance by cells of the immune system.26 The binding of plasma proteins onto the surface of NPs, also known as opsonization, occurs instantaneously once the NPs make contact with the blood-stream. To address this issue, one classical design parameter for effective in vivo circulation and immune system shielding is the surface addition of PEG, termed PEGylation.365 PEG is a highly hydrophilic polymer that ensures prolonged in vivo halflives. Indeed, uncoated NPs have been observed to be rapidly cleared by the MPS.378 The density and thickness of this PEG masking layer has also been found to affect opsonization and biodistribution of injected NPs, and should be studied with more high-throughput and combinatorial approaches that can accelerate the discovery and development of NPs in a reproducible manner, together with a comprehensive study that combinatorially investigates the interrelation of NP PEG lengths and densities leading to reduced clearance.365, 384 Currently, it is common practice to decorate the surfaces of NPs with PEG polymers, and these types of NPs can benefit from prolonged circulation times.385 As such, surface-grafted PEG NPs have reduced uptake by liver cells as these NPs are not effectively bound by plasma proteins.386 In addition, the density and configuration of PEG on the surface of NPs is also an important parameter for in vivo biodistribution. PEG configurations on the surfaces of NPs can exist as either extended brush-like structures or coiled mushroom or mushroom/brush intermediates. From the two configurations, predominant brush-like PEG surfaces have been shown to sterically suppress the approach and binding of opsonins such as the C3 protein.378
Studies have shown that NPs covered with PEGMWof 10 kDa circulate longer in the blood but accumulate less in the tumour, while NPs with PEGMWof 5 kDa remain for shorter duration in blood circulation but accumulate more in the tumour.387 In addition, there are articles that intend to elucidate an “optimal PEG coverage” that falls in between a larger number of PEG chains on the NP surface with low mobility (brush configuration), and a lower number of PEG chains on the NP surface with high mobility (mushroom configuration).388
In summary, the diversity involved in NP biophysicochemical properties significantly affects their cell uptake, cell cytotoxicity, PK and BD in vivo. Additionally, there is a strong interplay between each of these properties and an optimal combination needs to be experimentally determined for everyNP type. Given the many parameters that must be optimally engineered and the variability and polydispersity in properties due tomultiple synthesis steps, it is not surprising that optimal biophysicochemical properties of NPs have been difficult to ascertain, as evidenced by the fact that many novel nanotechnologies have failed to make an impact on human health to date. Therefore, it is clear that there is a need for optimization of NP biophysicochemical properties in order to achieve optimal efficacy. Interest in the exact identification and characterization of NP surfaces is on the rise and computational techniques including electronic structure methods, all atom Monte Carlo, molecular dynamics methods and coarse-grained methods are now in use in order to better predict and understand the dynamic interactions of NP surfaces with biological environments and systems.389
7. Novel trends in the development of targeted polymeric nanoparticles
7.1 Multi-targeting
Multi targeting systems (MTS) generally describe NP surfaces decorated with two or more targeting ligands that recognize different receptors on the same or different cells. This type of targeting has gained a substantial amount of interest as a way to increase NP uptake by cancer cells using a “two-punch” (or “three-punch”) approach. An example of this approach was presented by Kluza et al.390 where they conjugated RGD (targeting integrin receptors) and anginex (targeting galectin-1) to paramagnetic and fluorescent liposomes and investigated NP cell uptake. Interestingly it was found that RGD-anginex dual targeting synergistically enhanced the uptake of these liposomes by HUVEC cells. Another example is demonstrated by Li et al. who used two commonly utilized small molecules, folic acid and glucose, to target KB cells with overexpressed FRs (Fig. 13b).341 In this work, the results suggested that enhanced cell recognition and internalization was due to increased multivalent interactions of the gold NPs with the cell surface, compared to single-ligand targeted NPs. Patil et al.391 also prepared dual-targeted PLA-PEG NPs using folic acid and biotin as targeting agents and investigated the tumour accumulation and efficacy of this construct versus folate-targeted and biotin-targeted mono-targeted NPs. The targeting strategy resulted in improved efficacy and higher tumour accumulation when compared to controls. More recently Ashley et al.383 developed porous silica NPs coated with a lipid bi-layer decorated with both a targeting peptide (SP94 which targets an unknown receptor in human hepatocellular carcinoma) and a fusogenic peptide (H5WYG, which enables endosome escape after being protonated) (Fig. 13c). This strategy resulted in a 10 000-fold greater affinity for hepatocellular carcinoma than for hepatocytes and 106-fold improvement over comparable liposomes. A further example of a novel targeting system was demonstrated by Sugahara et al. using iRGD.281, 314 This cell penetrating peptide has the added property of targeting two different receptors on the same cell (integrin and neuropilin-1 receptors) (Fig. 13d). In this case, targeting occurs sequentially, where integrins are targeted first with the iRGD moiety, followed by proteolytic cleavage of the peptide exposing a CendR motif that subsequently targets neuropilin-1, a key receptor for penetration of biological barriers.281, 314 Additionally, when iRGD was conjugated to the surface of Abraxane (Albumin-paclitaxel NPs), the efficacy of the therapy in mice was improved by several folds.281
Fig. 13.
Different strategies for multi-ligand targeting of NPs: A: Various modes of targeting using single or multi-ligands (Figure taken from Ruoslahti et al.260), B: Dual-targeting where one NP has two different ligands that target receptors on the same cell (Figure adapted from Li et al.341), C: example of cellular and sub-cellular targeting (Figure taken from Ashley et al.383), and D: dual targeting of one peptide to two different receptors on the same cell (Figure adapted from K. Sugahara et al.257).
While dual targeting presents an attractive strategy to potentiate NP uptake by cancer cells, it brings to the NP design another level of complexity. For a targeted NP to be highly effective, all the physicochemical properties such as size, charge, surface hydrophobicity, and targeting ligand density need to be optimized. Considering the interplay of these NP properties and the difficulty to alter one without affecting the other, introducing multiple types of targeting ligands to the NP design could potentially further complicate the optimization of NPs. Furthermore, the greater the number of components a NP has, the more challenging its large-scale manufacturing, regulatory approval, and translation to the clinic. Although studies have shown that the presence of two different targeting ligands on a single NP led to potent cytotoxicity, whereas no toxicity was observed when only one type of ligand was used,177 nevertheless, the ultimate benefit of dual or multi targeted NP systems should continue to be investigated at least at the pre-clinical stage.
7.2 Screening NP libraries
Small-molecule drug discovery is generally conducted by screening libraries of molecules to find drugs with optimal pharmacokinetics, biodistribution, tolerance, disposition, and elimination profiles. In recent years researchers have also taken this approach for the development of targeted NPs. However, targeted NP screening increases complexity by several orders of magnitude since there are several ‘inputs’ that could be screened for, such as chemical diversity from the precursors, or biophysicochemical diversity from the NPs. Careful design of combinatorial methods for measuring and understanding these diversities is one solution. For instance, Siegwart et al. recently developed a semi-automated screening of over 1000 polymeric and lipid-like materials with wide chemical diversities that could efficiently deliver siRNA.392 Each material was formulated into a NP and gene silencing was evaluated in vitro, followed by in vivo evaluation of a set of ‘hit’ NPs.392
The utility of NP screening libraries was demonstrated by Hrkach et al. who developed a library of ~100 distinct targeted polymeric NP formulations varying with respect to particle size, targeting ligand density, surface hydrophilicity, drug loading, and drug release properties, leading to a range of NPs with narrowly controlled biophysicochemical diversities.394 PK studies in mice, rat and monkey; and tissue distribution studies in rats showed that the lead targeted NP, BIND-014, had properties atypical of previously described polymeric NPs, including a blood circulation half-life of approximately 20 h and minimal liver accumulation.394 BIND-014 demonstrated superior anti-tumour efficacy in animals, as well as differentiated pharmacology, tolerability, and promising anti-tumour responses in human clinical trial.394
Weissleder et al. reported the screening of 146 different small-molecule ligands attached onto a model NP.393 Their model NP was a ‘magneto-fluorescent’ dextran coated iron oxide NP that could readily report uptake by different cell types, as well as tumour accumulation. Their choice of small molecule targeting was focused on compounds (MW < 500 Da) containing various chemical functional groups such as primary amines, alcohols, carboxylic acids, sulfhydryls and anhydrides, which can effect overall NP features such as water solubility, conjugation ability, biocompatibility and chemical diversity (Fig. 14).393
Fig. 14.
Screening targeted NPs from a derived NP library: (a–b) Laser light scattering and atomic force microscopy of NPs. (c) Model of the crosslinked dextran coating NPs modified with small molecules. (d) Water solubility, as well as fluorescent and magnetic properties of NPs. (e) Different classes of small molecules with amino, sulfhydryl, carboxyl or anhydride functionalities anchored onto the NPs. (f) Hemotoxylin eosin–stained sections of the tumours targeting with NPs. (g) Tumour cross-sections observed using the Cy5.5 fluorescence channel indicate marked fluorescence of one identified NP; CLIO-isatoic within tumour cells. (h) Biodistribution study with 111In-labelled NPs confirmed tumoural targeting of CLIO-isatoic NPs. Figure adapted from Weissleder et al.393
Wang et al. also screened various NPs to identify those with the best gene transfection efficiencies in vitro.243 This system was composed of six different building blocks including two different targeting ligands linked to PEG, cyclodextran, and dendrimers, with the mixing ratio of the compounds varied combinatorially. One important feature of this work was the development and implementation of a microfluidic system that facilitated the rapid mixing and preparation of different building block combinations in a high-throughput manner.243 These examples demonstrate a rising trend towards the combinatorial development of NPs to find NPs that exhibit the optimal interplay of properties impacting their efficacy and fate in vivo. The screening of NP libraries is one approach to comprehensively investigate the interrelation of NP biophysicochemical properties with their final therapeutic outcome.
7.3 Combination therapies
Combination therapy by co-delivering multiple drugs encapsulated in the same targeted polymeric NP was proposed to address the challenges of single-agent chemotherapy.46, 395–397 Some of the advantages of this type of approach include: (1) synergistic therapeutic effects, (2) control of drug-combination pharmacokinetics (especially for drugs with different chemical properties) and, (3) control of combination-drug exposure in a temporal and spatial manner. Combination chemotherapy can be carried out in non-targeted NPs or targeted NPs.46, 398 Recently, Kolishetti et al. developed a targeted therapeutic NP system for co-delivery of cisplatin and Dtxl (two drugs with different chemical properties) to prostate cancer cells.46 The hydrophilic Pt(IV) cisplatin prodrug was first conjugated to a poly(lactide) polymer derivative with pendant hydroxyl groups (denoted PLA-OH) to yield a PLA-Pt(IV) copolymer, and subsequently blended with PLGA-PEG and Dtxl by a nanoprecipitation process (Fig. 15).46 The dual-drug encapsulated NPs were then conjugated with the A10 Apt to develop a targeted co-delivery NP platform.46 In vitro studies demonstrated that the Apt-targeted, dual-drug encapsulated NPs were ~5.5–10 times more cytotoxic than respective single drug encapsulating NPs (PLA-Pt-NP-Apt and Dtxl-NP-Apt).46
Fig. 15.
Strategy for co-encapsulating hydrophobic Dtxl and more hydrophilic Pt(IV)-monosuccinate prodrug on a single nanoparticle. Figure taken from Kolishetti et al.46
In another study by Milane et al. the therapeutic efficacy and safety of the drug combination paclitaxel/lodinamine loaded in an EGFR NP was evaluated.396 The rationale for the targeting agent together with the drug combination was to overcomeMDRwith cells over-expressing the EGFR receptor.396 The drug combination was evaluated using a mouse orthotopic model of MDR human breast cancer, and it was observed that the NP performed significantly better than the free drug combination.396 Research on the development of combinatorial therapies is on the rise, however, this area will benefit from further investigations involving: (1) the discovery of further molecular targets in cancer cells and better understanding of drug activity in these cells, (2) chemistries that allow for the dual or multi-conjugation of drugs to polymer backbones, (3) improvements in loading methods of two or more drugs with variable properties within single NPs and, (4) reproducible and efficacious toxicity data following the use of combination drug therapies.
7.4 Polymer-lipid hybrid NPs
Polymer-lipid hybrid NPs aim to combine the advantages of both polymers and lipidic vesicles and have recently gained interest for drug delivery applications. The main rationale concerning the design of these systems is to achieve stable and controllable NPs for prolonged drug release properties in vivo. This involves the inclusion of an amphiphilic lipid in the formulation, which preferentially associates with the hydrophobic corona surface of the polymeric core, for example a PLGA based core (see Fig. 9). Recently Chan et al. developed PLGA-lipid NPs that combined three different types of biomaterials: (1) a PLGA hydrophobic core for drug entrapment, (2) soybean phosphatidylcholine (lecithin) to create a monolayer around the hydrophobic core, and, (3) DSPE-PEG-COOH which intersperses in the lecithin monolayer in order to form a stabilizing PEG shell (Fig. 9).399 These polymeric NPs were loaded with Ptxl and conjugated to peptides where they showed effective vascular wall targeting and preferential localization to arterial wall injury. Additionally a post-angioplasty reduction in arterial stenosis was observed in animals treated with these Ptxl-load vascular wall targeted NPs.147, 307 Another example of combining polymers and lipids in a single delivery system is the “nanocell” developed by Sengupta et al.188 In this work, the authors were able to achieve delivery of two anticancer drugs in a spatiotemporally controlled manner by incorporating a polymeric nanoparticle inside of a liposomal structure. The liposomal portion released its antiangiogenic payload more rapidly, thereby attacking the tumour-associated vasculature, followed by a more sustained release profile of a cytotoxic agent from the inner polymeric matrix, directly attacking the tumour cells.
Recently Shi et al. reported the development of a simple and robust hybrid lipid-polymer NP for siRNA delivery.400 This system differs from classical lipid-hybrid polymer systems in that these NPs were engineered to contain a differentially charged hollow core/shell lipid-polymer-lipid hybrid nanostructure (Fig. 16). These NPs were formulated using a modified double-emulsion solvent evaporation method. The inner positively charged hollow core which is composed of cationic lipids was designed to encapsulate siRNA more efficiently than a PLGA core alone. These hybrid lipid-polymeric NPs were shown to effectively load and efficiently deliver siRNA both in vitro and in vivo.
Fig. 16.
Biodegradable and biocompatible polymers and lipids forming hybrid core/shell nanoparticles for siRNA delivery. The unique lipid–polymer–lipid nanostructure is demonstrated by TEM (top right) and fluorescence microscopy (bottom right) with microsized particles. Figure taken from Shi et al.400
Hybrid NPs, especially those made with clinically validated materials hold great promise as the next generation of therapeutic NPs. The fact that hybrid NPs combine some of the unique physical and chemical characteristics of two or more classes of materials, such as polymers and liposomes to create a versatile and robust new class of nanoparticles, make them very attractive to potentiate the performance of NPs. However, it would be key to obtain a deeper scientific understanding on hybrid NPs, both physical (for instance understanding their self-assembly mechanism) and biological (for instance, determining their toxicity profiles at the cell-level and organ level), in order to accelerate their clinical translation.
7.5 Sub-cellular targeting
It is often assumed that mediating cell cytosolic internalization is adequate to ensure the interaction of drug molecules with their final subcellular targets via simple diffusion and random interactions. However, it has become increasingly evident that such an assumption does not likely hold true.401–407 In addition to the presence of the cytoskeletal network and various dispersed organelles, the cytoplasm contains a large amount of dissolved macromolecules, with a concentration between 50 and 400 g L−1.408, 409 Consequently, transport or diffusion events in such a crowded solution cannot be expected to be the same as those in buffer solutions. In the case of a drug molecule with no defined specificity for a particular organelle, the molecule would need to have sufficiently long metabolic stability to allow for random interactions with the organelle. In the case of molecules with a stronger affinity for a non-target subcellular compartment, there exists a greater need for the ability to control subcellular disposition. In the case of drug molecules that are recognized by the drug efflux pump (e.g. P-glycoprotein), the design of NPs that can be internalized by endocytosis and thus release their active drugs inside subcellular organelles can be used to overcome multidrug resistance in cancer cells.410, 411 In addition, increasing attention has been focused on the pathological disorders of subcellular organelles (e.g. endosomes and lysosomes), which could potentially benefit from therapies targeting these pathways.412 For example, endosomal abnormalities in neurons have been associated with the etiology of Alzheimer’s disease.413 Lysosomes and in particular lysosomal hydrolases have been associated with several aspects of malignant transformation, including the loss of cell growth control, altered regulation of cell death, and acquisition of chemo-resistance and of metastatic potential.414 Endosomes and lysosomes have thus been proposed as potential target organelles for the chemotherapy of Alzheimer’s disease and cancers. Subsequently, focus has shifted towards subcellular drug targeting, that is, directing therapeutic agents to an individual organelle and is now becoming a new frontier for the development of therapeutic NPs.415
Two major approaches have been applied in the design of NPs with the potential for subcellular targeting. The first is based on attaching subcellular targeting ligands on the surface of NPs to redirect their accumulation to the desired compartment. The second is based on the inherent predisposition of the NPs for a particular compartment, which can be influenced at the preparation stage of NPs by using components that have a strong affinity for a subcellular compartment, and tuning the NP aspect ratio and size.
The type of targeting ligands displayed by NPs will determine the deposition location of these agents inside the cells. Well-characterized endocytic targeting ligands potentially useful for NP-mediated drug delivery are folic acid, low-density lipoprotein, cholera toxin B, mannose-6-phosphate, Tf, riboflavin, the tripeptide RGD, ICAM-1 antibody and nicotinic acid, as reviewed elsewhere.412 The cellular internalization mechanisms utilized by these ligands involve clathrin-dependent receptor mediated endocytosis, caveolin-assisted endocytosis, lipid raftassociated endocytosis and cell adhesionmolecule (CAM)-directed cellular uptake.412, 416, 417
Tfs,418 a family of large non-heme iron-binding glycoproteins, are the most widely used endocytic targeting ligands for the functionalization of NP drug delivery systems. Iron-loaded transferrin binds to a specific cell-surface receptor (TfR1) and is taken up via clathrin-coated pits. The TfR complex is routed into the endosomal compartment, avoiding lysosomal digestion. This is an important feature of TfR1 for NP delivery, as normally glycoproteins taken up by means of receptor-mediated endocytosis are destined eventually to fuse with lysosomes. Encapsulation of Dox into liposomes bearing Tf on the distal end of liposomal PEG chains resulted in significantly increased Dox uptake into glioma cells, which are known to overexpress the TfRs.419 Tf modification of Dox-loaded palmitoylated glycol chitosan (GCP) vesicles resulted in higher uptake and increased cytotoxicity as compared with GCP doxorubicin alone.420
Intercellular adhesion molecule (ICAM)-1, a glycoprotein expressed on diverse cell types, is upregulated and functionally involved in inflammation, which is a hallmark of many lysosomal disorders.421 Recombinant human acid sphingomyelinase (ASM) enzyme, deficient in types A and B Niemann-Pick disease, was loaded into NPs coated with anti-ICAM antibody. Anti-ICAM/ASM NPs were found to enter cells by means of CAM-mediated endocytosis and traffic to lysosomes. The delivered enzyme displayed stable activity and alleviated lysosomal lipid accumulation, suggesting that NPs targeted to ICAM-1 bypassed defunct pathways and could improve the efficacy of enzyme replacement therapy for lysosomal disorders, such as Niemann-Pick disease.421 A mitochondrial leader peptide (MLP), derived from the nucleocytosol-expressed but mitochondria-localized ornithine transcarbamylase, was used to render polyethylenimine (PEI)NPs mitochondriotropic.422 Lee et al.422 conjugated the mitochondrial leader peptide to PEI NPs by means of a disulfide bond and confirmed the complex formation of PEI–MLP with DNA by a gel retardation assay. In vitro delivery tests of rhodamine-labelled DNA into living cells demonstrated that PEI–MLP/DNA complexes were localized at mitochondrial sites. The data suggested that PEI–MLP can deliver DNA to the mitochondrial sites and may be useful for the development of direct mitochondrial gene therapy.
In an elegant recent study, Murakami et al. investigated the real-time subcellular fate of polymeric micelles formed from (1,2-diaminocyclohexane) platinum(II) (DACHPt/m), the parent complex of oxaliplatin, in tumour tissues using fluorescence aided based assessment of their kinetic stability (Fig. 17).423 They were able to show potent antitumour activity for their system and to prove that their designed polymeric micelles were able to overcome Pt resistance both in vitro and in vivo. The extravasation of DACHP/m NP was observed from blood vessels into tumours in addition to polymer dissociation intracellularly. It was hypothesized that these polymeric NPs selectively dissociated in the late endosomes and facilitated Pt drug delivery to the nucleus relative to free oxaliplatin. The authors proposed that this outcome is most likely due to circumvention of the cytoplasmic detoxification systems of metallothionein and methionine synthase. By developing oxaliplatin resistance in various cell lines, they were able to test this hypothesis as in each case the drug loaded micelles exhibited higher toxicities than the free oxaliplatin. This study suggests that therapeutic NPs have enormous potential in both intracellular targeting via compartmentalization, and drug delivery in an efficacious manner.
Fig. 17.
Design of fluorescent-labelled DACHPt/m (F-DACHPt/m) for visualization of localization and drug release in cancer cells: (A) F-DACHPt/m self-assembled through polymer-metal complex formation between DACHPt and boron dipyrromethene (BODIPY) FL–poly(ethylene glycol)-b-poly(glutamic acid)–BODIPY TR in distilled water. (B) Schematic representation of hypothetical subcellular pathways and action of DACHPt/m. Figure taken from Murakami et al.423
7.6 Stimuli responsive and surface switching NPs
An emerging method for targeting drugs to disease areas is to exploit the local changes that occur due to disease pathology and use these changes as triggers to improve targeting. This is accomplished by developing stimuli-responsive materials that change their physicochemical or drug-release properties upon encountering specific environmental cues, potentially leading to increased drug delivery to diseased tissues.424–426 These methods of drug targeting have grown out of both a greater understanding of the local changes that occur in different disease conditions and through a greater ability to design materials that have a dynamic range that is physiologically relevant for the specific disease application. These stimuli can be classified as extracellular, intracellular, or both. Extracellular conditions that may be altered in disease typically include decreased pH, such as in certain solid tumours, inflammation, and infections, and the presence of certain enzymes, such as the clotting cascade.426 However, extracellular conditions can also be artificially manipulated such as by applying local heating, ultrasound, or near infrared light to trigger an effect.427, 428 Intracellular conditions that can be exploited include low pH, presence of certain unique enzymes, and the reducing environment of endolysosomes.429, 430
Stimuli-responsive materials used for drug delivery either trigger a burst of drug release or increase the rate of drug release in response to a given stimuli.431–433 In addition, it may be possible to improve NP targeting to sites of disease by causing changes in NP surface properties at sites of disease.434
These strategies can be applied broadly across different types of NPs and are not limited to polymeric NPs. For example, liposomes that have multiple targeting functions that are unveiled sequentially have been developed to respond to extracellular low pH to improve intracellular delivery.435, 436 In a recent study, Poon et al. developed polylysine-coated quantum dots via layer-by-layer (LbL) assembly.197 The neutral layers on the surface of these NPs were designed to shed in response to the acidic tumour environment to reveal a highly charged surface that facilitated uptake by tumour cells. The results of this study demonstrated the potential of LbL NPs for both tumour targeting and cancer cell targeting.
Stimuli-responsive polymeric micelle NPs based on block copolymers have been developed extensively by Bae et al.57 A few illustrative examples include poly(l-histidine)-b-polyethylene glycol/poly(L-lactide)-b-polyethylene glycol mixed micelles that dissolve at low pH, leading to low-pH-targeted drug release.437 The pH at which these micelles dissolved could be tuned by changing the ratio of poly(L-lactide)-b-polyethylene glycol to poly(l-histidine)-b-polyethylene glycol, demonstrating the potential to apply this drug deliverymethod to a variety of different tumours or specific areas within tumours based on the local acidity.437 Another example is a TAT-peptide-modified triblock copolymer of poly(l-lactic acid)-b-poly(ethylene glycol)-b-poly-(l-histidine)-TAT mixed with poly(l-histidine)-b-poly(ethylene glycol) to yield micelles that present TAT-peptide on encountering acidity for targeted internalization.438 In this case, pH-sensitive TAT-peptide exposure on the micelle surface is believed to be significant for tumour cell-specific internalization, as TAT-peptide is known to interact with various different cell membranes, such as the BBB.439
Other stimuli-responsive polymeric NPs have been developed, a targeted, environment-responsive polymeric nanocapsule based on cucurbit[6]uril, was also designed to collapse in reducing environments, such as inside cells, to trigger burst drug release.159 Additionally, temperature-sensitive NPs based on poly(N-isopropylacrylamide)- b-poly(butyl methacrylate) were designed to release drugs rapidly at temperatures above the lower critical solution temperature (LCST) of the poly(N-isopropylacrylamide) segment (32 °C).440
These and other methods of using stimuli to improve drug targeting are likely to see continued development, particularly in instances where a high level of differentiation between pathologic and physiologic conditions exists.
8. Opportunities and challenges for targeted NPs
Oncology is one area where nanomedicine products are set tomake the most impact, where cell and tissue targeting approaches can be used to efficiently deliver cytotoxic and molecularly targeted drugs to cancer cells. Most cancer drugs today are administered either intravenously or orally and become systemically distributed without preferential partitioning to cancer tissue. The widespread biodistribution of cancer drugs results in both therapeutic anti-cancer effects as well as off target adverse effects on healthy and proliferating non-cancer cells. The ability to target therapeutics in a more controlled and specific manner is the primary goal of developing targeted NPs. The molecularly targeted cancer drugs typically modulate signalling pathways that are aberrantly activated in cancer cells. These drugs are most commonly: (i) administered orally, (ii) dosed daily, (iii) interact with their targets reversibly, and (iv) are limited in their efficacy as single agents due to factors such as network robustness, redundancy, crosstalk and emergence of resistance. Polymeric NP technologies may enable the co-delivery of multiple molecularly targeted drugs, creating an integrative pharmacologic effect among distinct drugs, thereby modulating multiple pathways that may translate into more prolonged and efficacious anti-cancer therapies.
While the potential advantages of delivering molecularly targeted drugs via targeted polymericNPs is relatively clear, one potential challenge arises due to the fact that polymeric NPs are most commonly administered intravenously. This necessitates efficient NP capabilities for encapsulating sufficiently high drug concentrations in order to ensure a constant release of drugs—in particular, between infrequent intravenous (IV) dosing regimens.
Numerous works have demonstrated the effectiveness of targeted NPs in becoming internalized into cells and delivering therapeutic and imaging agents in a highly specific manner (see Table 2). For example, targeted polymeric NPs were shown to elicit marked increase in intracellular uptake by prostate cancer cells in comparison to non-targeted NPs in vitro.195, 282, 351, 441, 442 Furthermore, the careful tuning of targeting ligands density and chemical properties on the surface of NPs was shown to further enhance the intracellular uptake of polymeric NPs in targeted cells.218, 266 Ultimately a number of parameters need to be investigated for successful design of targeted NPs, which include optimization of NP biophysicochemical properties and the demonstration of the efficacy of targeted NPs in a clinical setting on their impact on patient outcomes. Aside from identification of optimal ligands and ligand targets suitable for highly selective NP targeting, a whole host of other practical challenges in the development of targeted therapeutic NPs should also be considered including: (1) the use of biocompatible, biodegradable/bioeliminable materials, (2) the use of simple, robust and reproducible bioconjugation chemistries for the attachment of precursors and targeting ligands, (3) facile NP assemblies that avoid multi-step NP preparation and purification steps, (4) optimization of NP biophysicochemical properties to achieve optimal drug load/release, long circulation half-life, suitable biodistribution, differential target tissue accumulation, efficacious target tissue drug concentration and drug exposure kinetics, (5) validation of NP stability and predictable shelf-life; and, (6) development or adaptation of scalable processes and units of operations amenable to the manufacturing of large quantities of targeted NPs for clinical development and commercialization.
Recently interest has developed on how the properties of size, shape, surface area, roughness, porosity, surface functional groups, ligands, surface defects, hydrophobicity and hydrophilicity (Fig. 12) collectively influence NP behaviour at the ‘nano-bio’ interface, as this understanding is key to improved NP design.353 Indeed, the value of tailoring these parameters with the purpose of minimizing toxicity, unfavourable interactions with the immune system, rapid renal clearance, and accumulation in organs such as the liver and spleen is beginning to be more systematically recognized and increasingly adopted.443
Targeted NPs can lead to nanomedicines being specifically retained in tissues and/or cells, resulting in higher dose and duration of drug exposure within the target tissue. However, ultimately the question remains as to whether targeted NPs demonstrate marked improvement in clinical outcomes, which need to be demonstrated through well executed larger clinical trials. Beyond the regulatory requirements of demonstrating safety, efficacy, quality and cost-effectiveness, further challenges of each targeted NP technology need to be investigated on a case-by-case basis and these challenges must be met in order to harness their tremendous potential as a new class of targeted therapeutics.
9. Conclusions and outlook
Polymeric NPs have shown tremendous therapeutic potential at both research and clinical levels. Coupled with the fast pace of development in nanotechnology and our deeper understanding of biophysicochemical parameters that govern NP behaviour in biological systems, then it is not over reaching to expect NP technologies to create revolutionary therapies for a myriad of medical problems. This optimism is justified given our improved knowledge of disease pathways, better manipulation, characterization and control of matter at the nanoscale, and proven clinical outcomes so far. We are now in a good position to no longer look at targeted polymeric NPs as merely nanosized drug delivery vehicles, but to view them in a new light—as a new class of therapeutics that impact treatment efficacies beyond that of the action of the drug itself. In particular, this concept is now being realized with respect to the modes of drug release at active sites both extra and intracellularly, which is governed by the therapeutic NP capabilities of undergoing endocytosis assisted uptake, bypassing of MDR pathways and targeting subcellular compartments.
We have confidence that with a well characterized system including: safe, effective, and specific targeting ligands, biocompatible, biodegradable and bioeliminable materials, and appropriate choice of therapeutics and disease models, targeted polymeric NPs could yield more effective treatments for a myriad of important human diseases. The exciting developments that are producing more “sensitive” NPs capable of triggered release of drugs at active sites under environmental cues further strengthen the therapeutic NP arsenal. Although targeted NPs have been slow to enter the clinic their potential as an entirely new class of therapeutics, remains tremendous.
A considerable amount of research and development is necessary from the proof-of-principle stage of developing novel targeted nanomedicines to their bench-to-bedside translation, and in particular, the multifunctionality and complexity of some targeted polymeric NPs should be investigated on a case-by-case basis. The processes for the engineering of targeted NPs must be carefully developed and controlled in order to facilitate reproducible and scalable NP production. The development of complementary technologies such as the identification of biochemically stable non-immunogenic ligands with higher affinities and specificity for clinically relevant targets is of potential importance. Selective adaptation of NP technologies early during drug discovery may also result in the development of improved drugs beyond what is achievable by medicinal chemistry alone. For example NP technologies can protect drugs from rapid metabolism and inactivation; improve drug solubility, PK, BD, and target tissue exposure. All of these properties are typically optimized by medicinal chemistry efforts and often occur at the expense of other desirable properties including for example, drug potency or specificity. The combination of medicinal chemistry and NP engineering early on during drug discovery holds enormous promise, and provides additional degrees of freedom to medicinal chemistry efforts for creating optimally developed targeted NP therapeutics. With continued infusion of capital from the public and private sectors toward the research and development in the area of nanomedicine we believe that targeted polymeric NP technologies will emerge as an important class of therapeutics in the next 20 years with broad therapeutic impact, analogous to the evolution and impact of mAb technologies over the last few decades. There is still “plenty of room at the bottom”.444
Acknowledgements
This work was supported by National Institutes of Health (NIH) grants CA151884, EB003647, and the David Koch-Prostate Cancer Foundation Award in Nanotherapeutics. This work was also supported by the National Heart, Lung, and Blood Institute, (NIH), as a Program of Excellence in Nanotechnology (PEN) Award, Contract #HHSN268201000045C. Dr Farokhzad declares financial interests in BIND Biosciences and Selecta Biosciences. We thank Liang Guo for photography.
Biographies

Nazila Kamaly
Dr Nazila Kamaly received an MSci in Medicinal Chemistry from University College London in 2002 and her PhD in Bioorganic Chemistry on the development of multimodal nanoparticles for tumour imaging and gene therapy applications from Imperial College London (ICL) Chemistry Department in 2007. She then carried out postdoctoral research on multifunctional theranostic nanomedicines at the Medical Research Council and ICL before joining the Laboratory for Nanomedicine and Biomaterials at Brigham Women’s Hospital, Harvard Medical School. Currently she is also a research affiliate at the David H. Koch Institute for Integrative Cancer Research at MIT. Her research interests involve the use of chemistry to develop “smart” nanomedicines for diagnostic and therapeutic purposes.

Zeyu Xiao
Dr Zeyu Xiao received her Bachelor in pharmacy from Peking University (China) in 2003. She received her PhD in chemistry in 2008 from the joint research program between Chinese Academy of Sciences (China) and University of Florida (USA). She is currently a postdoctoral fellow at the Laboratory of Nanomedicine and Biomaterials at Brigham and Women’s Hospital, Harvard Medical School, and a Research Affiliate at the David H. Koch Institute for Integrative Cancer Research at MIT. Her research is focused on the development and screening of targeted nanoparticles for therapeutic and imaging applications.

Pedro M. Valencia
Pedro Valencia completed his M.S. in Chemical engineering practice from MIT in 2009 and obtained a B.S. in Chemical Engineering from University of Wisconsin-Madison in 2007. Currently he is a PhD Candidate in Dr Robert Langer’s Laboratories at the MIT David H. Koch Institute for Integrative Cancer Research. His work has focused on development and understanding of targeted polymeric nanoparticles capable of trafficking and delivering thousands of drug molecules directly to diseased tissues. He has also developed platforms to synthesize and screen nanoparticles in a high-throughput fashion to accelerate their translation from bench side to bedside.

Aleksandar F. Radovic-Moreno
Aleks Radovic-Moreno completed his B.S. with Honors in Chemical Engineering at the Pennsylvania State University in 2005. He is currently a PhD Candidate and NSF fellow in the Harvard-MIT Division of Health Sciences & Technology and a member of the Langer Lab at MIT. His work has focused on developing targeted polymeric nanoparticles for treating cancer in addition to bacterial infections. More specifically, he has designed various new drug encapsulation and delivery strategies, including methods to improve drug targeting using aptamer-functionalized or surface charge-switching nanoparticles.

Omid C. Farokhzad
Dr Farokhzad is an associate professor at Harvard Medical School (HMS) and directs the Laboratory of Nanomedicine and Biomaterials at Brigham and Women’s Hospital (BWH); with a multi-disciplinary group of scientists focused on fundamental understanding of nanomaterials and development of novel nanotechnologies for medical applications. He received his M.D. and M.A. from Boston University School of Medicine and completed his post-doctoral clinical and research training at BWH/HMS and MIT in the laboratory of Dr Robert Langer. Dr Farokhzad was named among the Nano50 winners by the NASA Nanotech Briefs and also received the All Star Distinguished Achievement Award from Mass High Tech Journal. He is also a member of the College of the Fellows of the American Institute of Medical and Biological Engineering.
Footnotes
Part of the nanomedicine themed issue.
The rest of the authors declare no conflict of interest.
References
- 1.LaVan DA, McGuire T, Langer R. Nat. Biotechnol. 2003;21:1184–1191. doi: 10.1038/nbt876. [DOI] [PubMed] [Google Scholar]
- 2.Langer R. Science. 1990;249:1527–1533. doi: 10.1126/science.2218494. [DOI] [PubMed] [Google Scholar]
- 3.Shi J, Xiao Z, Kamaly N, Farokhzad OC. Acc. Chem. Res. 2011;44:1123–1134. doi: 10.1021/ar200054n. [DOI] [PubMed] [Google Scholar]
- 4.Davis ME, Chen ZG, Shin DM. Nat. Rev. Drug Discovery. 2008;7:771–782. doi: 10.1038/nrd2614. [DOI] [PubMed] [Google Scholar]
- 5.Farokhzad OC. Expert Opin. Drug Delivery. 2008;5:927–929. doi: 10.1517/17425247.5.9.927. [DOI] [PubMed] [Google Scholar]
- 6.Bangham AD, Standish MM, Watkins JC. J. Mol. Biol. 1965;13:238–252. doi: 10.1016/s0022-2836(65)80093-6. [DOI] [PubMed] [Google Scholar]
- 7.Farokhzad OC, Langer R. ACS Nano. 2009;3:16–20. doi: 10.1021/nn900002m. [DOI] [PubMed] [Google Scholar]
- 8.Wagner V, Dullaart A, Bock AK, Zweck A. Nat. Biotechnol. 2006;24:1211–1217. doi: 10.1038/nbt1006-1211. [DOI] [PubMed] [Google Scholar]
- 9.Bangham AD, Horne RW. J. Mol. Biol. 1964;8:660–668. doi: 10.1016/s0022-2836(64)80115-7. [DOI] [PubMed] [Google Scholar]
- 10.Langer R, Folkman J. Nature. 1976;263:797–800. doi: 10.1038/263797a0. [DOI] [PubMed] [Google Scholar]
- 11.Buhleier E, Wehner W, Vogtle F. Synthesis. 1978:155–158. [Google Scholar]
- 12.Torchilin VP. Immuno Methods. 1994;4:244–258. doi: 10.1006/immu.1994.1027. [DOI] [PubMed] [Google Scholar]
- 13.Gabizon A, Shmeeda H, Barenholz Y. Clin. Pharmacokinet. 2003;42:419–436. doi: 10.2165/00003088-200342050-00002. [DOI] [PubMed] [Google Scholar]
- 14.Gref R, Minamitake Y, Peracchia MT, Trubetskoy V, Torchilin V, Langer R. Science. 1994;263:1600–1603. doi: 10.1126/science.8128245. [DOI] [PubMed] [Google Scholar]
- 15.Ros PR, Freeny PC, Harms SE, Seltzer SE, Davis PL, Chan TW, Stillman AE, Muroff LR, Runge VM, Nissenbaum MA, et al. Radiology. 1995;196:481–488. doi: 10.1148/radiology.196.2.7617864. [DOI] [PubMed] [Google Scholar]
- 16.Gradishar WJ, Tjulandin S, Davidson N, Shaw H, Desai N, Bhar P, Hawkins M, O’Shaughnessy J. J. Clin. Oncol. 2005;23:7794–7803. doi: 10.1200/JCO.2005.04.937. [DOI] [PubMed] [Google Scholar]
- 17.Kim TY, Kim DW, Chung JY, Shin SG, Kim SC, Heo DS, Kim NK, Bang YJ. Clin. Cancer Res. 2004;10:3708–3716. doi: 10.1158/1078-0432.CCR-03-0655. [DOI] [PubMed] [Google Scholar]
- 18.Davis ME. Mol. Pharmaceutics. 2009;6:659–668. doi: 10.1021/mp900015y. [DOI] [PubMed] [Google Scholar]
- 19. http://clinicaltrials.gov/ct2/show/NCT01300533.
- 20. http://www.clinicaltrials.gov/ct2/show/NCT01478893.
- 21.Allen TM, Chonn A. FEBS Lett. 1987;223:42–46. doi: 10.1016/0014-5793(87)80506-9. [DOI] [PubMed] [Google Scholar]
- 22.Leserman LD, Barbet J, Kourilsky F, Weinstein JN. Nature. 1980;288:602–604. doi: 10.1038/288602a0. [DOI] [PubMed] [Google Scholar]
- 23.Heath TD, Fraley RT, Papahdjopoulos D. Science. 1980;210:539–541. doi: 10.1126/science.7423203. [DOI] [PubMed] [Google Scholar]
- 24.Burgess P, Hutt PB, Farokhzad OC, Langer R, Minick S, Zale S. Nat. Biotechnol. 2010;28:1267–1270. doi: 10.1038/nbt.1725. [DOI] [PubMed] [Google Scholar]
- 25.MacDonald ML, Lamerdin J, Owens S, Keon BH, Bilter GK, Shang Z, Huang Z, Yu H, Dias J, Minami T, Michnick SW, Westwick JK. Nat. Chem. Biol. 2006;2:329–337. doi: 10.1038/nchembio790. [DOI] [PubMed] [Google Scholar]
- 26.McNeil SE. J. Leukocyte Biol. 2005;78:585–594. doi: 10.1189/jlb.0205074. [DOI] [PubMed] [Google Scholar]
- 27. http://clinicaltrials.gov/ct2/show/NCT00689065.
- 28.Davis ME. Mol. Pharmaceutics. 2009;6:659–668. doi: 10.1021/mp900015y. [DOI] [PubMed] [Google Scholar]
- 29.Sankhala KK, Mita AC, Adinin R, Wood L, Beeram M, Bullock S, Yamagata N, Matsuno K, Fujisawa T, Phan A. J. Clin. Oncol. 2009;27:S2535. [Google Scholar]
- 30.Matsumura Y, Gotoh M, Muro K, Yamada Y, Shirao K, Shimada Y, Okuwa M, Matsumoto S, Miyata Y, Ohkura H, Chin K, Baba S, Yama T, Kannami A, Takamatsu Y, Ito K, Takahashi K. Ann. Oncol. 2004;15:517–525. doi: 10.1093/annonc/mdh092. [DOI] [PubMed] [Google Scholar]
- 31.Nemunaitis J, Senzer N, Bedell C, Nunan R, Sleer L, Chang E. Mol. Ther. 2009;17:S226. [Google Scholar]
- 32.Barreto JA, O’Malley W, Kubeil M, Graham B, Stephan H, Spiccia L. Adv. Mater. 2011;23:H18–40. doi: 10.1002/adma.201100140. [DOI] [PubMed] [Google Scholar]
- 33.Langer R, Tirrell DA. Nature. 2004;428:487–492. doi: 10.1038/nature02388. [DOI] [PubMed] [Google Scholar]
- 34.Langer R. Nature. 1998;392:5–10. [PubMed] [Google Scholar]
- 35.Udhrain A, Skubitz KM, Northfelt DW. Int. J. Nanomedicine. 2007;2:345–352. [PMC free article] [PubMed] [Google Scholar]
- 36.Campos SM, Penson RT, Mays AR, Berkowitz RS, Fuller AF, Goodman A, Matulonis UA, Muzikansky A, Seiden MV. Gynecol. Oncol. 2001;81:206–212. doi: 10.1006/gyno.2000.5980. [DOI] [PubMed] [Google Scholar]
- 37.Pinto-Alphandary H, Andremont A, Couvreur P. Int. J. Antimicrob. Agents. 2000;13:155–168. doi: 10.1016/s0924-8579(99)00121-1. [DOI] [PubMed] [Google Scholar]
- 38.Fattal E, Rojas J, Roblot-Treupel L, Andremont A, Couvreur P. J. Microencapsulation. 1991;8:29–36. doi: 10.3109/02652049109021855. [DOI] [PubMed] [Google Scholar]
- 39.Montana M, Ducros C, Verhaeghe P, Terme T, Vanelle P, Rathelot P. J. Chemother. 2011;23:59–66. doi: 10.1179/joc.2011.23.2.59. [DOI] [PubMed] [Google Scholar]
- 40.Gradishar WJ, Tjulandin S, Davidson N, Shaw H, Desai N, Bhar P, Hawkins M, O’Shaughnessy J. J. Clin. Oncol. 2005;23:7794–7803. doi: 10.1200/JCO.2005.04.937. [DOI] [PubMed] [Google Scholar]
- 41.Sparreboom A, Scripture CD, Trieu V, Williams PJ, De TP, Yang A, Beals B, Figg WD, Hawkins M, Desai N. Clin. Cancer Res. 2005;11:4136–4143. doi: 10.1158/1078-0432.CCR-04-2291. [DOI] [PubMed] [Google Scholar]
- 42.Lee KS, Chung HC, Im SA, Park YH, Kim CS, Kim SB, Rha SY, Lee MY, Ro J. Breast Cancer Res. Treat. 2008;108:241–250. doi: 10.1007/s10549-007-9591-y. [DOI] [PubMed] [Google Scholar]
- 43.Park SR, Oh DY, Kim DW, Kim TY, Heo DS, Bang YJ, Kim NK, Kang WK, Kim HT, Im SA, Suh JH, Kim HK. Oncol. Rep. 2004;12:1059–1064. [PubMed] [Google Scholar]
- 44.Marshall J. Nature News. 2011 [Google Scholar]
- 45.Chan JM, Valencia PM, Zhang L, Langer R, Farokhzad OC. Methods Mol. Biol. 2010;624:163–175. doi: 10.1007/978-1-60761-609-2_11. [DOI] [PubMed] [Google Scholar]
- 46.Kolishetti N, Dhar S, Valencia PM, Lin LQ, Karnik R, Lippard SJ, Langer R, Farokhzad OC. Proc. Natl. Acad. Sci. U. S. A. 2010;107:17939–17944. doi: 10.1073/pnas.1011368107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hearn EM, Patel DR, Lepore BW, Indic M, van den Berg B. Nature. 2009;458:367–U134. doi: 10.1038/nature07678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ferrari M. Nat. Rev. Cancer. 2005;5:161–171. doi: 10.1038/nrc1566. [DOI] [PubMed] [Google Scholar]
- 49.Gottesman MM, Fojo T, Bates SE. Nat. Rev. Cancer. 2002;2:48–58. doi: 10.1038/nrc706. [DOI] [PubMed] [Google Scholar]
- 50.Seow WY, Xue JM, Yang YY. Biomaterials. 2007;28:1730–1740. doi: 10.1016/j.biomaterials.2006.11.039. [DOI] [PubMed] [Google Scholar]
- 51.Peer D, Karp JM, Hong S, Farokhzad OC, Margalit R, Langer R. Nat. Nanotechnol. 2007;2:751–760. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- 52.Lee ES, Na K, Bae YH. J. Controlled Release. 2005;103:405–418. doi: 10.1016/j.jconrel.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 53.Sahoo SK, Ma W, Labhasetwar V. Int. J. Cancer. 2004;112:335–340. doi: 10.1002/ijc.20405. [DOI] [PubMed] [Google Scholar]
- 54.Wang X, Li J, Wang Y, Koenig L, Gjyrezi A, Giannakakou P, Shin EH, Tighiouart M, Chen ZG, Nie S, Shin DM. ACS Nano. 2011;5:6184–6194. doi: 10.1021/nn200739q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.York AW, Kirkland SE, McCormick CL. Adv. Drug Delivery Rev. 2008;60:1018–1036. doi: 10.1016/j.addr.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 56.Dhal PK, Polomoscanik SC, Avila LZ, Holmes-Farley SR, Miller RJ. Adv. Drug Delivery Rev. 2009;61:1121–1130. doi: 10.1016/j.addr.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 57.Bae Y, Kataoka K. Adv. Drug Delivery Rev. 2009;61:768–784. doi: 10.1016/j.addr.2009.04.016. [DOI] [PubMed] [Google Scholar]
- 58.Drummond DC, Meyer O, Hong K, Kirpotin DB, Papahadjopoulos D. Pharmacol. Rev. 1999;51:691–743. [PubMed] [Google Scholar]
- 59.Caldorera-Moore ME, Liechty WB, Peppas NA. Acc. Chem. Res. 2011;44:1061–1070. doi: 10.1021/ar2001777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hoffman AS. J. Controlled Release. 2008;132:153–163. doi: 10.1016/j.jconrel.2008.08.012. [DOI] [PubMed] [Google Scholar]
- 61.Kleber JW, Nash JF, Lee CC. J. Pharm. Sci. 1964;53:1519–1521. doi: 10.1002/jps.2600531219. [DOI] [PubMed] [Google Scholar]
- 62.Farokhzad OC, Langer R. Adv. Drug Delivery Rev. 2006;58:1456–1459. doi: 10.1016/j.addr.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 63.Acharya S, Sahoo SK. Adv. Drug Delivery Rev. 2011;63:170–183. doi: 10.1016/j.addr.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 64.Shive MS, Anderson JM. Adv. Drug Delivery Rev. 1997;28:5–24. doi: 10.1016/s0169-409x(97)00048-3. [DOI] [PubMed] [Google Scholar]
- 65.Langer R. Pharmacol. Ther. 1983;21:35–51. doi: 10.1016/0163-7258(83)90066-9. [DOI] [PubMed] [Google Scholar]
- 66.West CP, Lumsden MA, Lawson S, Williamson J, Baird DT. Fertil. Steril. 1987;48:45–51. doi: 10.1016/s0015-0282(16)59288-7. [DOI] [PubMed] [Google Scholar]
- 67.Sethi R, Sanfilippo N. Clin. Interv. Aging. 2009;4:259–267. doi: 10.2147/cia.s4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lin SH, Kleinberg LR. Expert Rev. Anti-Infect. Ther. 2008;8:343–359. doi: 10.1586/14737140.8.3.343. [DOI] [PubMed] [Google Scholar]
- 69.Johnson LR, Stoller NH. Compend. Contin. Educ. Dent. 1999;20:19–25. quiz 35. [PubMed] [Google Scholar]
- 70.Anthony L, Freda PU. Curr. Med. Res. Opin. 2009;25:2989–2999. doi: 10.1185/03007990903328959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Crawford ED, Phillips JM. Cancer Manage. Res. 2011;3:201–209. doi: 10.2147/CMR.S12700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kreuter J. Pharm. Acta Helv. 1978;53:33–39. [PubMed] [Google Scholar]
- 73.Maeda H. Bioconjugate Chem. 2010;21:797–802. doi: 10.1021/bc100070g. [DOI] [PubMed] [Google Scholar]
- 74.Matsumura Y, Maeda H. Cancer Res. 1986;46:6387–6392. [PubMed] [Google Scholar]
- 75.Maeda H, Bharate GY, Daruwalla J. Eur. J. Pharm. Biopharm. 2009;71:409–419. doi: 10.1016/j.ejpb.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 76.Saha RN, Vasanthakumar S, Bende G, Snehalatha M. Mol. Membr. Biol. 2010;27:215–231. doi: 10.3109/09687688.2010.510804. [DOI] [PubMed] [Google Scholar]
- 77.Torchilin VP. Handb. Exp. Pharmacol. 2010:3–53. doi: 10.1007/978-3-642-00477-3_1. [DOI] [PubMed] [Google Scholar]
- 78.Carmeliet P, Jain RK. Nature. 2000;407:249–257. doi: 10.1038/35025220. [DOI] [PubMed] [Google Scholar]
- 79.Danquah MK, Zhang XA, Mahato RI. Adv. Drug Delivery Rev. 2011;63:623–639. doi: 10.1016/j.addr.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 80.Yuan F, Dellian M, Fukumura D, Leunig M, Berk DA, Torchilin VP, Jain RK. Cancer Res. 1995;55:3752–3756. [PubMed] [Google Scholar]
- 81.Suzuki M, Hori K, Abe I, Saito S, Sato H. J. Natl. Cancer Inst. 1981;67:663–669. [PubMed] [Google Scholar]
- 82.Fang J, Nakamura H, Maeda H. Adv. Drug Delivery Rev. 2011;63:136–151. doi: 10.1016/j.addr.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 83.Mulder WJ, Douma K, Koning GA, van Zandvoort MA, Lutgens E, Daemen MJ, Nicolay K, Strijkers GJ. Magn. Reson. Med. 2006;55:1170–1174. doi: 10.1002/mrm.20883. [DOI] [PubMed] [Google Scholar]
- 84.Nagamitsu A, Greish K, Maeda H. Jpn. J. Clin. Oncol. 2009;39:756–766. doi: 10.1093/jjco/hyp074. [DOI] [PubMed] [Google Scholar]
- 85.Chrastina A, Massey KA, Schnitzer JE. Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol. 2011;3:421–437. doi: 10.1002/wnan.143. [DOI] [PubMed] [Google Scholar]
- 86.Hobbs SK, Monsky WL, Yuan F, Roberts WG, Griffith L, Torchilin VP, Jain RK. Proc. Natl. Acad. Sci. U. S. A. 1998;95:4607–4612. doi: 10.1073/pnas.95.8.4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jain RK. Adv. Drug Delivery Rev. 2001;46:149–168. doi: 10.1016/s0169-409x(00)00131-9. [DOI] [PubMed] [Google Scholar]
- 88.Monsky WL, Fukumura D, Gohongi T, Ancukiewcz M, Weich HA, Torchilin VP, Yuan F, Jain RK. Cancer Res. 1999;59:4129–4135. [PubMed] [Google Scholar]
- 89.Kano MR, Bae Y, Iwata C, Morishita Y, Yashiro M, Oka M, Fujii T, Komuro A, Kiyono K, Kaminishi M, Hirakawa K, Ouchi Y, Nishiyama N, Kataoka K, Miyazono K. Proc. Natl. Acad. Sci. U. S. A. 2007;104:3460–3465. doi: 10.1073/pnas.0611660104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hatakeyama H, Akita H, Harashima H. Adv. Drug Delivery Rev. 2011;63:152–160. doi: 10.1016/j.addr.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 91.Romberg B, Hennink WE, Storm G. Pharm. Res. 2008;25:55–71. doi: 10.1007/s11095-007-9348-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Horton JK, Houghton PJ, Houghton JA. Biochem. Pharmacol. 1988;37:3995–4000. doi: 10.1016/0006-2952(88)90085-8. [DOI] [PubMed] [Google Scholar]
- 93.Jordan MA, Wendell K, Gardiner S, Derry WB, Copp H, Wilson L. Cancer Res. 1996;56:816–825. [PubMed] [Google Scholar]
- 94.Kuh HJ, Jang SH, Wientjes MG, Weaver JR, Au JL. J. Pharmacol. Exp. Ther. 1999;290:871–880. [PubMed] [Google Scholar]
- 95.Zamboni WC, Strychor S, Joseph E, Parise RA, Egorin MJ, Eiseman JL. Cancer Chemother. Pharmacol. 2008;62:417–426. doi: 10.1007/s00280-007-0620-7. [DOI] [PubMed] [Google Scholar]
- 96.Bissery MC. Eur. J. Cancer. 1995;31(Suppl 4):S1–S6. doi: 10.1016/0959-8049(95)00357-o. [DOI] [PubMed] [Google Scholar]
- 97.Ishida T, Ichihara M, Wang X, Yamamoto K, Kimura J, Majima E, Kiwada H. J. Controlled Release. 2006;112:15–25. doi: 10.1016/j.jconrel.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 98.Ishida T, Kiwada H. Yakugaku Zasshi. 2008;128:233–243. doi: 10.1248/yakushi.128.233. [DOI] [PubMed] [Google Scholar]
- 99.Valle JW, Armstrong A, Newman C, Alakhov V, Pietrzynski G, Brewer J, Campbell S, Corrie P, Rowinsky EK, Ranson M. Invest. New Drugs. 2011;29:1029–1037. doi: 10.1007/s10637-010-9399-1. [DOI] [PubMed] [Google Scholar]
- 100.Matsumura Y, Hamaguchi T, Ura T, Muro K, Yamada Y, Shimada Y, Shirao K, Okusaka T, Ueno H, Ikeda M, Watanabe N. Br. J. Cancer. 2004;91:1775–1781. doi: 10.1038/sj.bjc.6602204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Adams ML, Lavasanifar A, Kwon GS. J. Pharm. Sci. 2003;92:1343–1355. doi: 10.1002/jps.10397. [DOI] [PubMed] [Google Scholar]
- 102.Danson S, Ferry D, Alakhov V, Margison J, Kerr D, Jowle D, Brampton M, Halbert G, Ranson M. Br. J. Cancer. 2004;90:2085–2091. doi: 10.1038/sj.bjc.6601856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Oerlemans C, Bult W, Bos M, Storm G, Nijsen JF, Hennink WE. Pharm. Res. 2010;27:2569–2589. doi: 10.1007/s11095-010-0233-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sutton D, Nasongkla N, Blanco E, Gao J. Pharm. Res. 2007;24:1029–1046. doi: 10.1007/s11095-006-9223-y. [DOI] [PubMed] [Google Scholar]
- 105.Boddy AV, Plummer ER, Todd R, Sludden J, Griffin M, Robson L, Cassidy J, Bissett D, Bernareggi A, Verrill MW, Calvert AH. Clin. Cancer Res. 2005;11:7834–7840. doi: 10.1158/1078-0432.CCR-05-0803. [DOI] [PubMed] [Google Scholar]
- 106.Galic VL, Herzog TJ, Wright JD, Lewin SN. Expert Opin. Invest. Drugs. 2011;20:813–821. doi: 10.1517/13543784.2011.576666. [DOI] [PubMed] [Google Scholar]
- 107.Schluep T, Hwang J, Cheng J, Heidel JD, Bartlett DW, Hollister B, Davis ME. Clin. Cancer Res. 2006;12:1606–1614. doi: 10.1158/1078-0432.CCR-05-1566. [DOI] [PubMed] [Google Scholar]
- 108.Jokerst JV, Lobovkina T, Zare RN, Gambhir SS. Nanomedicine. 2011;6:715–728. doi: 10.2217/nnm.11.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Knop K, Hoogenboom R, Fischer D, Schubert US. Angew. Chem., Int. Ed. 2010;49:6288–6308. doi: 10.1002/anie.200902672. [DOI] [PubMed] [Google Scholar]
- 110.Harris JM, Chess RB. Nat. Rev. Drug Discovery. 2003;2:214–221. doi: 10.1038/nrd1033. [DOI] [PubMed] [Google Scholar]
- 111.Kaminskas LM, McLeod VM, Porter CJ, Boyd BJ. J. Pharm. Sci. 2011;100:5069–5077. doi: 10.1002/jps.22682. [DOI] [PubMed] [Google Scholar]
- 112.Tagami T, Uehara Y, Moriyoshi N, Ishida T, Kiwada H. J. Controlled Release. 2011;151:149–154. doi: 10.1016/j.jconrel.2010.12.013. [DOI] [PubMed] [Google Scholar]
- 113.Koide H, Asai T, Hatanaka K, Urakami T, Ishii T, Kenjo E, Nishihara M, Yokoyama M, Ishida T, Kiwada H, Oku N. Int. J. Pharm. 2008;362:197–200. doi: 10.1016/j.ijpharm.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 114.Lu W, Wan J, She Z, Jiang X. J. Controlled Release. 2007;118:38–53. doi: 10.1016/j.jconrel.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 115.Ma H, Shiraishi K, Minowa T, Kawano K, Yokoyama M, Hattori Y, Maitani Y. Pharm. Res. 2010;27:296–302. doi: 10.1007/s11095-009-0018-9. [DOI] [PubMed] [Google Scholar]
- 116.Xu H, Wang KQ, Deng YH, Chen da W. Biomaterials. 2010;31:4757–4763. doi: 10.1016/j.biomaterials.2010.02.049. [DOI] [PubMed] [Google Scholar]
- 117.Hamad I, Hunter AC, Szebeni J, Moghimi SM. Mol. Immunol. 2008;46:225–232. doi: 10.1016/j.molimm.2008.08.276. [DOI] [PubMed] [Google Scholar]
- 118.Hamad I, Al-Hanbali O, Hunter AC, Rutt KJ, Andresen TL, Moghimi SM. ACS Nano. 2010;4:6629–6638. doi: 10.1021/nn101990a. [DOI] [PubMed] [Google Scholar]
- 119.Jiang S, Cao Z. Advanced materials (Deerfield Beach, Fla) 2010;22:920–932. doi: 10.1002/adma.200901407. [DOI] [PubMed] [Google Scholar]
- 120.Vaisocherova H, Yang W, Zhang Z, Cao Z, Cheng G, Piliarik M, Homola J, Jiang S. Anal. Chem. 2008;80:7894–7901. doi: 10.1021/ac8015888. [DOI] [PubMed] [Google Scholar]
- 121.Banerjee I, Pangule RC, Kane RS. Adv. Mater. 2010;23:690–718. doi: 10.1002/adma.201001215. [DOI] [PubMed] [Google Scholar]
- 122.Yu B, Tai HC, Xue W, Lee LJ, Lee RJ. Mol. Membr. Biol. 2010;27:286–298. doi: 10.3109/09687688.2010.521200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kolhatkar R, Lote A, Khambati H. Curr. Drug Discovery Technol. 2011;8:197–206. doi: 10.2174/157016311796799044. [DOI] [PubMed] [Google Scholar]
- 124.Farokhzad OC, Cheng J, Teply BA, Sherifi I, Jon S, Kantoff PW, Richie JP, Langer R. Proc. Natl. Acad. Sci. U. S. A. 2006;103:6315–6320. doi: 10.1073/pnas.0601755103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lammers T, Hennink WE, Storm G. Br. J. Cancer. 2008;99:392–397. doi: 10.1038/sj.bjc.6604483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Warenius HM, Galfre G, Bleehen NM, Milstein C. Eur. J. Cancer Clin. Oncol. 1981;17:1009–1015. doi: 10.1016/s0277-5379(81)80006-5. [DOI] [PubMed] [Google Scholar]
- 127. www.antibodysociety.org/news/approved_mabs.php.
- 128.Hooks MA, Wade andW. J.Millikan, Jr. CS. Pharmacotherapy. 1991;11:26–37. [PubMed] [Google Scholar]
- 129.James JS, Dubs G. AIDS Treat News. 1997:2–3. [PubMed] [Google Scholar]
- 130.Albanell J, Baselga J. Drugs Today (Barc) 1999;35:931–946. [PubMed] [Google Scholar]
- 131.Graham MM, Menda Y. J. Nucl. Med. 2011;52(Suppl 2):56S–63S. doi: 10.2967/jnumed.110.085746. [DOI] [PubMed] [Google Scholar]
- 132.Sudimack J, Lee RJ. Adv. Drug Delivery Rev. 2000;41:147–162. doi: 10.1016/s0169-409x(99)00062-9. [DOI] [PubMed] [Google Scholar]
- 133.Qian ZM, Li H, Sun H, Ho K. Pharmacol. Rev. 2002;54:561–587. doi: 10.1124/pr.54.4.561. [DOI] [PubMed] [Google Scholar]
- 134.Allen TM. Nat. Rev. Cancer. 2002;2:750–763. doi: 10.1038/nrc903. [DOI] [PubMed] [Google Scholar]
- 135.Sugano M, Egilmez NK, Yokota SJ, Chen FA, Harding J, Huang SK, Bankert RB. Cancer Res. 2000;60:6942–6949. [PubMed] [Google Scholar]
- 136.Bareford LM, Swaan PW. Adv. Drug Delivery Rev. 2007;59:748–758. doi: 10.1016/j.addr.2007.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Whitehead KA, Langer R, Anderson DG. Nat. Rev. Drug Discovery. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Schroeder A, Levins CG, Cortez C, Langer R, Anderson DG. J. Intern. Med. 2010;267:9–21. doi: 10.1111/j.1365-2796.2009.02189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Varkouhi AK, Scholte M, Storm G, Haisma HJ. J. Controlled Release. 2011;151:220–228. doi: 10.1016/j.jconrel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 140.Alexis F, Basto P, Levy-Nissenbaum E, Radovic-Moreno AF, Zhang L, Pridgen E, Wang AZ, Marein SL, Westerhof K, Molnar LK, Farokhzad OC. ChemMedChem. 2008;3:1839–1843. doi: 10.1002/cmdc.200800122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kirpotin DB, Drummond DC, Shao Y, Shalaby MR, Hong K, Nielsen UB, Marks JD, Benz CC, Park JW. Cancer Res. 2006;66:6732–6740. doi: 10.1158/0008-5472.CAN-05-4199. [DOI] [PubMed] [Google Scholar]
- 142.Bartlett DW, Su H, Hildebrandt IJ, Weber WA, Davis ME. Proc. Natl. Acad. Sci. U. S. A. 2007;104:15549–15554. doi: 10.1073/pnas.0707461104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Pun SH, Tack F, Bellocq NC, Cheng J, Grubbs BH, Jensen GS, Davis ME, Brewster M, Janicot M, Janssens B, Floren W, Bakker A. Cancer Biol. Ther. 2004;3:641–650. doi: 10.4161/cbt.3.7.918. [DOI] [PubMed] [Google Scholar]
- 144.Hu-Lieskovan S, Heidel JD, Bartlett DW, Davis ME, Triche TJ. Cancer Res. 2005;65:8984–8992. doi: 10.1158/0008-5472.CAN-05-0565. [DOI] [PubMed] [Google Scholar]
- 145.Pirollo KF, Chang EH. Trends Biotechnol. 2008;26:552–558. doi: 10.1016/j.tibtech.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 146.Samanta S, Sistla R, Chaudhuri A. Biomaterials. 2010;31:1787–1797. doi: 10.1016/j.biomaterials.2009.10.027. [DOI] [PubMed] [Google Scholar]
- 147.Chan JM, Zhang L, Tong R, Ghosh D, Gao W, Liao G, Yuet KP, Gray D, Rhee JW, Cheng J, Golomb G, Libby P, Langer R, Farokhzad OC. Proc. Natl. Acad. Sci. U. S. A. 2010;107:2213–2218. doi: 10.1073/pnas.0914585107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhang XX, Eden HS, Chen X. J. Control. Release. 2011 doi: 10.1016/j.jconrel.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Tkachenko AG, Xie H, Coleman D, Glomm W, Ryan J, Anderson MF, Franzen S, Feldheim DL. J. Am. Chem. Soc. 2003;125:4700–4701. doi: 10.1021/ja0296935. [DOI] [PubMed] [Google Scholar]
- 150.Smith GP, Petrenko VA. Chem. Rev. 1997;97:391–410. doi: 10.1021/cr960065d. [DOI] [PubMed] [Google Scholar]
- 151.Ruoslahti E, Bhatia SN, Sailor MJ. J. Cell Biol. 2010;188:759–768. doi: 10.1083/jcb.200910104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ruoslahti E. Drug Discovery Today. 2002;7:1138–1143. doi: 10.1016/s1359-6446(02)02501-1. [DOI] [PubMed] [Google Scholar]
- 153.Iwasaki Y, Maie H, Akiyoshi K. Biomacromolecules. 2007;8:3162–3168. doi: 10.1021/bm700606z. [DOI] [PubMed] [Google Scholar]
- 154.Banquy X, Leclair G, Rabanel JM, Argaw A, Bouchard JF, Hildgen P, Giasson S. Bioconjugate Chem. 2008;19:2030–2039. doi: 10.1021/bc800257m. [DOI] [PubMed] [Google Scholar]
- 155.Kim TH, Park IK, Nah JW, Choi YJ, Cho CS. Biomaterials. 2004;25:3783–3792. doi: 10.1016/j.biomaterials.2003.10.063. [DOI] [PubMed] [Google Scholar]
- 156.Han HD, Mangala LS, Lee JW, Shahzad MM, Kim HS, Shen D, Nam EJ, Mora EM, Stone RL, Lu C, Lee SJ, Roh JW, Nick AM, Lopez-Berestein G, Sood AK. Clin. Cancer Res. 2010;16:3910–3922. doi: 10.1158/1078-0432.CCR-10-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Aktas Y, Yemisci M, Andrieux K, Gursoy RN, Alonso MJ, Fernandez-Megia E, Novoa-Carballal R, Quinoa E, Riguera R, Sargon MF, Celik HH, Demir AS, Hincal AA, Dalkara T, Capan Y, Couvreur P. Bioconjugate Chem. 2005;16:1503–1511. doi: 10.1021/bc050217o. [DOI] [PubMed] [Google Scholar]
- 158.Xu P, Tang H, Li S, Ren J, Van Kirk E, Murdoch WJ, Radosz M, Shen Y. Biomacromolecules. 2004;5:1736–1744. doi: 10.1021/bm049874u. [DOI] [PubMed] [Google Scholar]
- 159.Kim E, Kim D, Jung H, Lee J, Paul S, Selvapalam N, Yang Y, Lim N, Park CG, Kim K. Angew. Chem., Int. Ed. 2010;49:4405–4408. doi: 10.1002/anie.201000818. [DOI] [PubMed] [Google Scholar]
- 160.Bellocq NC, Pun SH, Jensen GS, Davis ME. Bioconjugate Chem. 2003;14:1122–1132. doi: 10.1021/bc034125f. [DOI] [PubMed] [Google Scholar]
- 161.Beaudette TT, Cohen JA, Bachelder EM, Broaders KE, Cohen JL, Engleman EG, Frechet JM. J. Am. Chem. Soc. 2009;131:10360–10361. doi: 10.1021/ja903984s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lee JB, Roh YH, Um SH, Funabashi H, Cheng W, Cha JJ, Kiatwuthinon P, Muller DA, Luo D. Nat. Nanotechnol. 2009;4:430–436. doi: 10.1038/nnano.2009.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.MacKay JA, Chen M, McDaniel JR, Liu W, Simnick AJ, Chilkoti A. Nat. Mater. 2009;8:993–999. doi: 10.1038/nmat2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Simnick AJ, Valencia CA, Liu R, Chilkoti A. ACS Nano. 2010;4:2217–2227. doi: 10.1021/nn901732h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Balthasar S, Michaelis K, Dinauer N, von Briesen H, Kreuter J, Langer K. Biomaterials. 2005;26:2723–2732. doi: 10.1016/j.biomaterials.2004.07.047. [DOI] [PubMed] [Google Scholar]
- 166.Green JJ, Chiu E, Leshchiner ES, Shi J, Langer R, Anderson DG. Nano Lett. 2007;7:874–879. doi: 10.1021/nl062395b. [DOI] [PubMed] [Google Scholar]
- 167.Wang X, Li J, Wang Y, Cho KJ, Kim G, Gjyrezi A, Koenig L, Giannakakou P, Shin HJ, Tighiouart M, Nie S, Chen ZG, Shin DM. ACS Nano. 2009;3:3165–3174. doi: 10.1021/nn900649v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Choi KY, Chung H, Min KH, Yoon HY, Kim K, Park JH, Kwon IC, Jeong SY. Biomaterials. 2010;31:106–114. doi: 10.1016/j.biomaterials.2009.09.030. [DOI] [PubMed] [Google Scholar]
- 169.Cho HJ, Yoon HY, Koo H, Ko SH, Shim JS, Lee JH, Kim K, Kwon IC, Kim DD. Biomaterials. 2011;32:7181–7190. doi: 10.1016/j.biomaterials.2011.06.028. [DOI] [PubMed] [Google Scholar]
- 170.Nam HY, Kwon SM, Chung H, Lee SY, Kwon SH, Jeon H, Kim Y, Park JH, Kim J, Her S, Oh YK, Kwon IC, Kim K, Jeong SY. J. Controlled Release. 2009;135:259–267. doi: 10.1016/j.jconrel.2009.01.018. [DOI] [PubMed] [Google Scholar]
- 171.Ryu JH, Jiwpanich S, Chacko R, Bickerton S, Thayumanavan S. J. Am. Chem. Soc. 2010;132:8246–8247. doi: 10.1021/ja102316a. [DOI] [PubMed] [Google Scholar]
- 172.Wang Y, Gao S, Ye WH, Yoon HS, Yang YY. Nat. Mater. 2006;5:791–796. doi: 10.1038/nmat1737. [DOI] [PubMed] [Google Scholar]
- 173.Shenoy D, Little S, Langer R, Amiji M. Pharm. Res. 2005;22:2107–2114. doi: 10.1007/s11095-005-8343-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Ko J, Park K, Kim YS, Kim MS, Han JK, Kim K, Park RW, Kim IS, Song HK, Lee DS, Kwon IC. J. Controlled Release. 2007;123:109–115. doi: 10.1016/j.jconrel.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 175.Cade D, Ramus E, Rinaudo M, Auzely-Velty R, Delair T, Hamaide T. Biomacromolecules. 2004;5:922–927. doi: 10.1021/bm034504b. [DOI] [PubMed] [Google Scholar]
- 176.Zhang L, Xue H, Cao Z, Keefe A, Wang J, Jiang S. Biomaterials. 2011;32:4604–4608. doi: 10.1016/j.biomaterials.2011.02.064. [DOI] [PubMed] [Google Scholar]
- 177.Wang J, Tian S, Petros RA, Napier ME, Desimone JM. J. Am. Chem. Soc. 2010;132:11306–11313. doi: 10.1021/ja1043177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Lee H, Fonge H, Hoang B, Reilly RM, Allen C. Mol. Pharmaceutics. 2010;7:1195–1208. doi: 10.1021/mp100038h. [DOI] [PubMed] [Google Scholar]
- 179.Xin H, Jiang X, Gu J, Sha X, Chen L, Law K, Chen Y, Wang X, Jiang Y, Fang X. Biomaterials. 2011;32:4293–4305. doi: 10.1016/j.biomaterials.2011.02.044. [DOI] [PubMed] [Google Scholar]
- 180.Kaul G, Amiji M. Pharm. Res. 2005;22:951–961. doi: 10.1007/s11095-005-4590-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Nukolova NV, Oberoi HS, Cohen SM, Kabanov AV, Bronich TK. Biomaterials. 2011;32:5417–5426. doi: 10.1016/j.biomaterials.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Nasongkla N, Bey E, Ren J, Ai H, Khemtong C, Guthi JS, Chin SF, Sherry AD, Boothman DA, Gao J. Nano Lett. 2006;6:2427–2430. doi: 10.1021/nl061412u. [DOI] [PubMed] [Google Scholar]
- 183.Harush-Frenkel O, Rozentur E, Benita S, Altschuler Y. Biomacromolecules. 2008;9:435–443. doi: 10.1021/bm700535p. [DOI] [PubMed] [Google Scholar]
- 184.Aryal S, Hu CM, Zhang L. ACS Nano. 2009;4:251–258. doi: 10.1021/nn9014032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.McCarron PA, Marouf WM, Quinn DJ, Fay F, Burden RE, Olwill SA, Scott CJ. Bioconjugate Chem. 2008;19:1561–1569. doi: 10.1021/bc800057g. [DOI] [PubMed] [Google Scholar]
- 186.Zhang N, Chittasupho C, Duangrat C, Siahaan TJ, Berkland C. Bioconjugate Chem. 2008;19:145–152. doi: 10.1021/bc700227z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Tosi G, Fano RA, Bondioli L, Badiali L, Benassi R, Rivasi F, Ruozi B, Forni F, Vandelli MA. Nanomedicine. 2011;6:423–436. doi: 10.2217/nnm.11.11. [DOI] [PubMed] [Google Scholar]
- 188.Sengupta S, Eavarone D, Capila I, Zhao G, Watson N, Kiziltepe T, Sasisekharan R. Nature. 2005;436:568–572. doi: 10.1038/nature03794. [DOI] [PubMed] [Google Scholar]
- 189.Townsend SA, Evrony GD, Gu FX, Schulz MP, Brown, Jr. RH, Langer R. Biomaterials. 2007;28:5176–5184. doi: 10.1016/j.biomaterials.2007.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Choi SW, Kim JH. J. Controlled Release. 2007;122:24–30. doi: 10.1016/j.jconrel.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 191.Cheng J, Teply BA, Sherifi I, Sung J, Luther G, Gu FX, Levy-Nissenbaum E, Radovic-Moreno AF, Langer R, Farokhzad OC. Biomaterials. 2007;28:869–876. doi: 10.1016/j.biomaterials.2006.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Dhar S, Gu FX, Langer R, Farokhzad OC, Lippard SJ. Proc. Natl. Acad. Sci. U. S. A. 2008;105:17356–17361. doi: 10.1073/pnas.0809154105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Basu S, Harfouche R, Soni S, Chimote G, Mashelkar RA, Sengupta S. Proc. Natl. Acad. Sci. U. S. A. 2009;106:7957–7961. doi: 10.1073/pnas.0902857106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Li J, Feng L, Fan L, Zha Y, Guo L, Zhang Q, Chen J, Pang Z, Wang Y, Jiang X, Yang VC, Wen L. Biomaterials. 2011;32:4943–4950. doi: 10.1016/j.biomaterials.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Sanna V, Pintus G, Roggio AM, Punzoni S, Posadino AM, Arca A, Marceddu S, Bandiera P, Uzzau S, Sechi M. J. Med. Chem. 2011;54:1321–1332. doi: 10.1021/jm1013715. [DOI] [PubMed] [Google Scholar]
- 196.Gu F, Zhang L, Teply BA, Mann N, Wang A, Radovic- Moreno AF, Langer R, Farokhzad OC. Proc. Natl. Acad. Sci. U. S. A. 2008;105:2586–2591. doi: 10.1073/pnas.0711714105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Poon Z, Chang D, Zhao X, Hammond PT. ACS Nano. 2011;5:4284–4292. doi: 10.1021/nn200876f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Rapoport N, Gao Z, Kennedy A. J. Natl. Cancer Inst. 2007;99:1095–1106. doi: 10.1093/jnci/djm043. [DOI] [PubMed] [Google Scholar]
- 199.Rothenfluh DA, Bermudez H, O’Neil CP, Hubbell JA. Nat. Mater. 2008;7:248–254. doi: 10.1038/nmat2116. [DOI] [PubMed] [Google Scholar]
- 200.Wei H, Zhang XZ, Zhou Y, Cheng SX, Zhuo RX. Biomaterials. 2006;27:2028–2034. doi: 10.1016/j.biomaterials.2005.09.028. [DOI] [PubMed] [Google Scholar]
- 201.Schmidt V, Giacomelli C, Lecolley F, Lai-Kee-Him J, Brisson AR, Borsali R. J. Am. Chem. Soc. 2006;128:9010–9011. doi: 10.1021/ja062408n. [DOI] [PubMed] [Google Scholar]
- 202.Liang HF, Chen SC, Chen MC, Lee PW, Chen CT, Sung HW. Bioconjugate Chem. 2006;17:291–299. doi: 10.1021/bc0502107. [DOI] [PubMed] [Google Scholar]
- 203.Yuan H, Luo K, Lai Y, Pu Y, He B, Wang G, Wu Y, Gu Z. Mol. Pharmaceutics. 2010;7:953–962. doi: 10.1021/mp1000923. [DOI] [PubMed] [Google Scholar]
- 204.Lu J, Shi M, Shoichet MS. Bioconjugate Chem. 2009;20:87–94. doi: 10.1021/bc8003167. [DOI] [PubMed] [Google Scholar]
- 205.Ding H, Inoue S, Ljubimov AV, Patil R, Portilla-Arias J, Hu J, Konda B, Wawrowsky KA, Fujita M, Karabalin N, Sasaki T, Black KL, Holler E, Ljubimova JY. Proc. Natl. Acad. Sci. U. S. A. 2010;107:18143–18148. doi: 10.1073/pnas.1003919107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Inoue S, Ding H, Portilla-Arias J, Hu J, Konda B, Fujita M, Espinoza A, Suhane S, Riley M, Gates M, Patil R, Penichet ML, Ljubimov AV, Black KL, Holler E, Ljubimova JY. Cancer Res. 2011;71:1454–1464. doi: 10.1158/0008-5472.CAN-10-3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Cho CS, Kobayashi A, Takei R, Ishihara T, Maruyama A, Akaike T. Biomaterials. 2001;22:45–51. doi: 10.1016/s0142-9612(00)00143-5. [DOI] [PubMed] [Google Scholar]
- 208.Winer I, Wang S, Lee YE, Fan W, Gong Y, Burgos- Ojeda D, Spahlinger G, Kopelman R, Buckanovich RJ. Cancer Res. 2010;70:8674–8683. doi: 10.1158/0008-5472.CAN-10-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Yao YC, Zhan XY, Zhang J, Zou XH, Wang ZH, Xiong YC, Chen J, Chen GQ. Biomaterials. 2008;29:4823–4830. doi: 10.1016/j.biomaterials.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 210.Na K, Bae YH. Pharm. Res. 2002;19:681–688. doi: 10.1023/a:1015370532543. [DOI] [PubMed] [Google Scholar]
- 211.Kickhoefer VA, Garcia Y, Mikyas Y, Johansson E, Zhou JC, Raval-Fernandes S, Minoofar P, Zink JI, Dunn B, Stewart PL, Rome LH. Proc. Natl. Acad. Sci. U. S. A. 2005;102:4348–4352. doi: 10.1073/pnas.0500929102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Iyer AK, Greish K, Fang J, Murakami R, Maeda H. Biomaterials. 2007;28:1871–1881. doi: 10.1016/j.biomaterials.2006.11.051. [DOI] [PubMed] [Google Scholar]
- 213.Matsumura Y, Gotoh M, Muro K, Yamada Y, Shirao K, Shimada Y, Okuwa M, Matsumoto S, Miyata Y, Ohkura H, Chin K, Baba S, Yamao T, Kannami A, Takamatsu Y, Ito K, Takahashi K. Ann. Oncol. 2004;15:517–525. doi: 10.1093/annonc/mdh092. [DOI] [PubMed] [Google Scholar]
- 214.Hamaguchi T, Matsumura Y, Nakanishi Y, Muro K, Yamada Y, Shimada Y, Shirao K, Niki H, Hosokawa S, Tagawa T, Kakizoe T. Cancer Sci. 2004;95:608–613. doi: 10.1111/j.1349-7006.2004.tb02495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. http://clinicaltrials.gov/ct2/show/NCT00470613.
- 216.Sankhala KK, Mita AC, Adinin R, Wood L, Beeram M, Bullock S, Yamagata N, Matsuno K, Fujisawa T, Phan A. J. Clin. Oncol. 2009;27:15S. [Google Scholar]
- 217.Daniels TR, Delgado T, Helguera G, Penichet ML. Clin. Immunol. 2006;121:159–176. doi: 10.1016/j.clim.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 218.Gu F, Zhang L, Teply BA, Mann N, Wang A, Radovic- Moreno AF, Langer R, Farokhzad OC. Proc. Natl. Acad. Sci. U. S. A. 2008;105:2586–2591. doi: 10.1073/pnas.0711714105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. www.bindbio.com.
- 220.Elsasser-Beile U, Buhler P, Wolf P. Curr. Drug Targets. 2009;10:118–125. doi: 10.2174/138945009787354601. [DOI] [PubMed] [Google Scholar]
- 221.Slovin SF. Expert Opin. Ther. Targets. 2005;9:561–570. doi: 10.1517/14728222.9.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Taxoteres® B. prescribing information, sanofi-aventis. U.S. LLC, NJ: 2010. [Google Scholar]
- 223.Heidel JD, Liu JY, Yen Y, Zhou B, Heale BS, Rossi JJ, Bartlett DW, Davis ME. Clin. Cancer Res. 2007;13:2207–2215. doi: 10.1158/1078-0432.CCR-06-2218. [DOI] [PubMed] [Google Scholar]
- 224.Bartlett DW, Davis ME. Bioconjugate Chem. 2007;18:456–468. doi: 10.1021/bc0603539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Moreno AY, Janda KD. Pharmacol., Biochem. Behav. 2009;92:199–205. doi: 10.1016/j.pbb.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Jung T, Breitenbach A, Kissel T. J. Controlled Release. 2000;67:157–169. doi: 10.1016/s0168-3659(00)00201-7. [DOI] [PubMed] [Google Scholar]
- 227.Couvreur P, Kante B, Roland M, Guiot P, Bauduin P, Speiser P. J. Pharm. Pharmacol. 1979;31:331–332. doi: 10.1111/j.2042-7158.1979.tb13510.x. [DOI] [PubMed] [Google Scholar]
- 228.Allemann E, Leroux JC, Gurny R, Doelker E. Pharm. Res. 1993;10:1732–1737. doi: 10.1023/a:1018970030327. [DOI] [PubMed] [Google Scholar]
- 229.Grabnar PA, Kristl J. J. Microencapsulation. 2011;28:323–335. doi: 10.3109/02652048.2011.569763. [DOI] [PubMed] [Google Scholar]
- 230.Fessi H, Puisieux F, Devissaguet JP, Ammoury N, Benita S. Int. J. Pharm. 1989;55:R1–4. doi: 10.1002/jps.2600790902. [DOI] [PubMed] [Google Scholar]
- 231.Quintanar-Guerrero D, Allemann E, Fessi H, Doelker E. Drug Dev. Ind. Pharm. 1998;24:1113–1128. doi: 10.3109/03639049809108571. [DOI] [PubMed] [Google Scholar]
- 232.Mora-Huertas CE, Fessi H, Elaissari A. Int. J. Pharm. 2010;385:113–142. doi: 10.1016/j.ijpharm.2009.10.018. [DOI] [PubMed] [Google Scholar]
- 233.Farokhzad OC, Cheng JJ, Teply BA, Sherifi I, Jon S, Kantoff PW, Richie JP, Langer R. Proc. Natl. Acad. Sci. U. S. A. 2006;103:6315–6320. doi: 10.1073/pnas.0601755103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Anton N, Benoit JP, Saulnier P. J. Controlled Release. 2008;128:185–199. doi: 10.1016/j.jconrel.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 235.Avgoustakis K. Curr. Drug Delivery. 2004;1:321–333. doi: 10.2174/1567201043334605. [DOI] [PubMed] [Google Scholar]
- 236.Karnik R, Gu F, Basto P, Cannizzaro C, Dean L, Kyei- Manu W, Langer R, Farokhzad OC. Nano Lett. 2008;8:2906–2912. doi: 10.1021/nl801736q. [DOI] [PubMed] [Google Scholar]
- 237.Rhee M, Valencia PM, Rodriguez MI, Langer R, Farokhzad OC, Karnik R. Adv. Mater. 2011;23:H79–83. doi: 10.1002/adma.201004333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Whitesides GM. Nature. 2006;442:368–373. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 239.DeMello AJ. Nature. 2006;442:394–402. doi: 10.1038/nature05062. [DOI] [PubMed] [Google Scholar]
- 240.Marre S, Jensen KF. Chem. Soc. Rev. 2010;39:1183–1202. doi: 10.1039/b821324k. [DOI] [PubMed] [Google Scholar]
- 241.Fraikin JL, Teesalu T, McKenney CM, Ruoslahti E, Cleland AN. Nat. Nanotechnol. 2011;6:308–313. doi: 10.1038/nnano.2011.24. [DOI] [PubMed] [Google Scholar]
- 242.Gong J, Kim CJ. Lab Chip. 2008;8:898–906. doi: 10.1039/b717417a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Wang H, Liu K, Chen KJ, Lu Y, Wang S, Lin WY, Guo F, Kamei K, Chen YC, Ohashi M, Wang M, Garcia MA, Zhao XZ, Shen CK, Tseng HR. ACS Nano. 2010;4:6235–6243. doi: 10.1021/nn101908e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Song H, Tice JD, Ismagilov RF. Angew. Chem., Int. Ed. 2003;42:768–772. doi: 10.1002/anie.200390203. [DOI] [PubMed] [Google Scholar]
- 245.Zhao CX, He LZ, Qiao SZ, Middelberg APJ. Chem. Eng. Sci. 2011;66:1463–1479. [Google Scholar]
- 246.Dreis S, Rothweiler F, Michaelis M, Cinatl, Jr. J, Kreuter J, Langer K. Int. J. Pharm. 2007;341:207–214. doi: 10.1016/j.ijpharm.2007.03.036. [DOI] [PubMed] [Google Scholar]
- 247.Kumar V, Prud’homme RK. J. Pharm. Sci. 2008;97:4904–4914. doi: 10.1002/jps.21342. [DOI] [PubMed] [Google Scholar]
- 248.Gindy ME, Panagiotopoulos AZ, Prud’homme RK. Langmuir. 2008;24:83–90. doi: 10.1021/la702902b. [DOI] [PubMed] [Google Scholar]
- 249.Sanchis J, Canal F, Lucas R, Vicent MJ. Nanomedicine. 2010;5:915–935. doi: 10.2217/nnm.10.71. [DOI] [PubMed] [Google Scholar]
- 250.Greco F, Vicent MJ. Adv. Drug Delivery Rev. 2009;61:1203–1213. doi: 10.1016/j.addr.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 251.Duncan R. Adv. Drug Delivery Rev. 2009;61:1131–1148. doi: 10.1016/j.addr.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 252.Alexis F, Pridgen EM, Langer R, Farokhzad OC. Handb. Exp. Pharmacol. 2010:55–86. doi: 10.1007/978-3-642-00477-3_2. [DOI] [PubMed] [Google Scholar]
- 253.Tong R, Cheng J. Angew. Chem., Int. Ed. 2008;47:4830–4834. doi: 10.1002/anie.200800491. [DOI] [PubMed] [Google Scholar]
- 254.Kolishetti N, Dhar S, Valencia PM, Lin LQ, Karnik R, Lippard SJ, Langer R, Farokhzad OC. Proc. Natl. Acad. Sci. U. S. A. 2010;107:17939–17944. doi: 10.1073/pnas.1011368107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Di Marco M, Shamsuddin S, Razak KA, Aziz AA, Devaux C, Borghi E, Levy L, Sadun C. Int. J. Nanomedicine. 2010;5:37–49. [PMC free article] [PubMed] [Google Scholar]
- 256.Wang CH, Huang YF, Yeh CK. Langmuir. 2011;27:6971–6976. doi: 10.1021/la2011259. [DOI] [PubMed] [Google Scholar]
- 257.Prescher JA, Bertozzi CR. Nat. Chem. Biol. 2005;1:13–21. doi: 10.1038/nchembio0605-13. [DOI] [PubMed] [Google Scholar]
- 258.Canalle LA, van der Knaap M, Overhand M, van Hest JC. Macromol. Rapid Commun. 2011;32:203–208. doi: 10.1002/marc.201000507. [DOI] [PubMed] [Google Scholar]
- 259.Guo J, Chen G, Ning X, Wolfert MA, Li X, Xu B, Boons GJ. Chem. Eur. J. 2010;16:13360–13366. doi: 10.1002/chem.201002532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Debets MF, van der Doelen CW, Rutjes FP, van Delft FL. ChemBioChem. 2010;11:1168–1184. doi: 10.1002/cbic.201000064. [DOI] [PubMed] [Google Scholar]
- 261.Jewett JC, Bertozzi CR. Chem. Soc. Rev. 2010;39:1272–1279. doi: 10.1039/b901970g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Devaraj NK, Upadhyay R, Haun JB, Hilderbrand SA, Weissleder R. Angew. Chem., Int. Ed. 2009;48:7013–7016. doi: 10.1002/anie.200903233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Tassa C, Shaw SY, Weissleder R. Acc. Chem. Res. 2011;44:842–852. doi: 10.1021/ar200084x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Haun JB, Devaraj NK, Marinelli BS, Lee H, Weissleder R. ACS Nano. 2011;5:3204–3213. doi: 10.1021/nn200333m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Algar WR, Prasuhn DE, Stewart MH, Jennings TL, Blanco-Canosa JB, Dawson PE, Medintz IL. Bioconjugate Chem. 2011;22:825–858. doi: 10.1021/bc200065z. [DOI] [PubMed] [Google Scholar]
- 266.Valencia PM, Hanewich-Hollatz MH, Gao WW, Karim F, Langer R, Karnik R, Farokhzad OC. Biomaterials. 2011;32:6226–6233. doi: 10.1016/j.biomaterials.2011.04.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Nobs L, Buchegger F, Gurny R, Allemann E. Bioconjugate Chem. 2006;17:139–145. doi: 10.1021/bc050137k. [DOI] [PubMed] [Google Scholar]
- 268.Nobs L, Buchegger F, Gurny R, Allemann E. Eur. J. Pharm. Biopharm. 2004;58:483–490. doi: 10.1016/j.ejpb.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 269.Brennan FR, Shaw L, Wing MG, Robinson C. Mol. Biotechnol. 2004;27:59–74. doi: 10.1385/MB:27:1:59. [DOI] [PubMed] [Google Scholar]
- 270.Weinberg WC, Frazier-Jessen MR, Wu WJ, Weir A, Hartsough M, Keegan P, Fuchs C. Cancer Metastasis Rev. 2005;24:569–584. doi: 10.1007/s10555-005-6196-y. [DOI] [PubMed] [Google Scholar]
- 271.Orlova A, Tolmachev V, Pehrson R, Lindborg M, Tran T, Sandstrom M, Nilsson FY, Wennborg A, Abrahmsen L, Feldwisch J. Cancer Res. 2007;67:2178–2186. doi: 10.1158/0008-5472.CAN-06-2887. [DOI] [PubMed] [Google Scholar]
- 272.Willuda J, Honegger A, Waibel R, Schubiger PA, Stahel R, Zangemeister-Wittke U, Pluckthun A. Cancer Res. 1999;59:5758–5767. [PubMed] [Google Scholar]
- 273.Cortez-Retamozo V, Backmann N, Senter PD, Wernery U, De Baetselier P, Muyldermans S, Revets H. Cancer Res. 2004;64:2853–2857. doi: 10.1158/0008-5472.can-03-3935. [DOI] [PubMed] [Google Scholar]
- 274.Chou LY, Ming K, Chan WC. Chem. Soc. Rev. 2011;40:233–245. doi: 10.1039/c0cs00003e. [DOI] [PubMed] [Google Scholar]
- 275.Choi CH, Alabi CA, Webster P, Davis ME. Proc. Natl. Acad. Sci. U. S. A. 2010;107:1235–1240. doi: 10.1073/pnas.0914140107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Thomas TP, Shukla R, Kotlyar A, Liang B, Ye JY, Norris TB, Baker, Jr. JR. Biomacromolecules. 2008;9:603–609. doi: 10.1021/bm701185p. [DOI] [PubMed] [Google Scholar]
- 277.Cui B, Wu C, Chen L, Ramirez A, Bearer EL, Li WP, Mobley WC, Chu S. Proc. Natl. Acad. Sci. U. S. A. 2007;104:13666–13671. doi: 10.1073/pnas.0706192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Norberto SE, Kakar SS. Exp. Mol. Pathol. 2009 doi: 10.1016/j.yexmp.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 279.Chang DK, Lin CT, Wu CH, Wu HC. PLoS One. 2009;4:e4171. doi: 10.1371/journal.pone.0004171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Negussie AH, Miller JL, Reddy G, Drake SK, Wood BJ, Dreher MR. J. Controlled Release. 2010;143:265–273. doi: 10.1016/j.jconrel.2009.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Sugahara KN, Teesalu T, Karmali PP, Kotamraju VR, Agemy L, Greenwald DR, Ruoslahti E. Science. 2010;328:1031–1035. doi: 10.1126/science.1183057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Farokhzad OC, Jon S, Khademhosseini A, Tran TN, Lavan DA, Langer R. Cancer Res. 2004;64:7668–7672. doi: 10.1158/0008-5472.CAN-04-2550. [DOI] [PubMed] [Google Scholar]
- 283.Huang YF, Shangguan D, Liu H, Phillips JA, Zhang X, Chen Y, Tan W. ChemBioChem. 2009;10:862–868. doi: 10.1002/cbic.200800805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Huang CC, Huang YF, Cao Z, Tan W, Chang HT. Anal. Chem. 2005;77:5735–5741. doi: 10.1021/ac050957q. [DOI] [PubMed] [Google Scholar]
- 285.Herr JK, Smith JE, Medley CD, Shangguan D, Tan W. Anal. Chem. 2006;78:2918–2924. doi: 10.1021/ac052015r. [DOI] [PubMed] [Google Scholar]
- 286.Toma A, Otsuji E, Kuriu Y, Okamoto K, Ichikawa D, Hagiwara A, Ito H, Nishimura T, Yamagishi H. Br. J. Cancer. 2005;93:131–136. doi: 10.1038/sj.bjc.6602668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Gosk S, Vermehren C, Storm G, Moos T. J. Cereb. Blood Flow Metab. 2004;24:1193–1204. doi: 10.1097/01.WCB.0000135592.28823.47. [DOI] [PubMed] [Google Scholar]
- 288.Wagner S, Rothweiler F, Anhorn MG, Sauer D, Riemann I, Weiss EC, Katsen-Globa A, Michaelis M, Cinatl, Jr. J, Schwartz D, Kreuter J, von Briesen H, Langer K. Biomaterials. 2010;31:2388–2398. doi: 10.1016/j.biomaterials.2009.11.093. [DOI] [PubMed] [Google Scholar]
- 289.Torchilin VP, Lukyanov AN, Gao Z, Papahadjopoulos-Sternberg B. Proc. Natl. Acad. Sci. U. S. A. 2003;100:6039–6044. doi: 10.1073/pnas.0931428100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Sawant RM, Cohen MB, Torchilin VP, Rokhlin OW. J. Drug Targeting. 2008;16:601–604. doi: 10.1080/10611860802228954. [DOI] [PubMed] [Google Scholar]
- 291.Laginha KM, Moase EH, Yu N, Huang A, Allen TM. J. Drug Targeting. 2008;16:605–610. doi: 10.1080/10611860802229978. [DOI] [PubMed] [Google Scholar]
- 292.Voinea M, Manduteanu I, Dragomir E, Capraru M, Simionescu M. Pharm. Res. 2005;22:1906–1917. doi: 10.1007/s11095-005-7247-3. [DOI] [PubMed] [Google Scholar]
- 293.Loomis K, Smith B, Feng Y, Garg H, Yavlovich A, Campbell-Massa R, Dimitrov DS, Blumenthal R, Xiao X, Puri A. Exp. Mol. Pathol. 2010;88:238–249. doi: 10.1016/j.yexmp.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Beuttler J, Rothdiener M, Muller D, Frejd FY, Kontermann RE. Bioconjugate Chem. 2009;20:1201–1208. doi: 10.1021/bc900061v. [DOI] [PubMed] [Google Scholar]
- 295.Silverman J, Liu Q, Bakker A, To W, Duguay A, Alba BM, Smith R, Rivas A, Li P, Le H, Whitehorn E, Moore KW, Swimmer C, Perlroth V, Vogt M, Kolkman J, Stemmer WP. Nat. Biotechnol. 2005;23:1556–1561. doi: 10.1038/nbt1166. [DOI] [PubMed] [Google Scholar]
- 296.Burg M, Ravey EP, Gonzales M, Amburn E, Faix PH, Baird A, Larocca D. DNA Cell Biol. 2004;23:457–462. doi: 10.1089/1044549041474760. [DOI] [PubMed] [Google Scholar]
- 297.Liu B, Conrad F, Cooperberg MR, Kirpotin DB, Marks JD. Cancer Res. 2004;64:704–710. doi: 10.1158/0008-5472.can-03-2732. [DOI] [PubMed] [Google Scholar]
- 298.Wan XM, Chen YP, Xu WR, Yang WJ, Wen LP. Peptides. 2009;30:343–350. doi: 10.1016/j.peptides.2008.09.026. [DOI] [PubMed] [Google Scholar]
- 299.van Rooy I, Cakir-Tascioglu S, Couraud PO, Romero IA, Weksler B, Storm G, Hennink WE, Schiffelers RM, Mastrobattista E. Pharm. Res. 2010;27:673–682. doi: 10.1007/s11095-010-0053-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Stern ST, McNeil SE. Toxicol. Sci. 2008;101:4–21. doi: 10.1093/toxsci/kfm169. [DOI] [PubMed] [Google Scholar]
- 301.Gu FX, Karnik R, Wang AZ, Alexis F, Levy-Nissenbaum E, Hong S, Langer RS, Farokhzad OC. Nano Today. 2007;2:14–21. [Google Scholar]
- 302.Brissette R, Prendergast JK, Goldstein NI. Curr. Opin. Drug Discovery Dev. 2006;9:363–369. [PubMed] [Google Scholar]
- 303.Krag DN, Shukla GS, Shen GP, Pero S, Ashikaga T, Fuller S, Weaver DL, Burdette-Radoux S, Thomas C. Cancer Res. 2006;66:7724–7733. doi: 10.1158/0008-5472.CAN-05-4441. [DOI] [PubMed] [Google Scholar]
- 304.Desgrosellier JS, Cheresh DA. Nat. Rev. Cancer. 2010;10:9–22. doi: 10.1038/nrc2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Randolph JT, Waid P, Nichols C, Sauer D, Haviv F, Diaz G, Bammert G, Besecke LM, Segreti JA, Mohning KM, Bush EN, Wegner CD, Greer J. J. Med. Chem. 2004;47:1085–1097. doi: 10.1021/jm030418i. [DOI] [PubMed] [Google Scholar]
- 306.Cortijo J, Sanz MJ, Iranzo A, Montesinos JL, Nabah YN, Alfon J, Gomez LA, Merlos M, Morcillo EJ. Br. J. Pharmacol. 2006;147:661–670. doi: 10.1038/sj.bjp.0706658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Chan JM, Rhee JW, Drum CL, Bronson RT, Golomb G, Langer R, Farokhzad OC. Proc. Natl. Acad. Sci. U. S. A. 2011;108:19347–19352. doi: 10.1073/pnas.1115945108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Arap W, Kolonin MG, Trepel M, Lahdenranta J, Cardo- Vila M, Giordano RJ, Mintz PJ, Ardelt PU, Yao VJ, Vidal CI, Chen L, Flamm A, Valtanen H, Weavind LM, Hicks ME, Pollock RE, Botz GH, Bucana CD, Koivunen E, Cahill D, Troncoso P, Baggerly KA, Pentz RD, Do KA, Logothetis CJ, Pasqualini R. Nat. Med. 2002;8:121–127. doi: 10.1038/nm0202-121. [DOI] [PubMed] [Google Scholar]
- 309.Pasqualini R, Koivunen E, Kain R, Lahdenranta J, Sakamoto M, Stryhn A, Ashmun RA, Shapiro LH, Arap W, Ruoslahti E. Cancer Res. 2000;60:722–727. [PMC free article] [PubMed] [Google Scholar]
- 310.Jain RK, Stylianopoulos T. Nat. Rev. Clin. Oncol. 2010;7:653–664. doi: 10.1038/nrclinonc.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Heitz F, Morris MC, Divita G. Br. J. Pharmacol. 2009;157:195–206. doi: 10.1111/j.1476-5381.2009.00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Ruan G, Agrawal A, Marcus AI, Nie S. J. Am. Chem. Soc. 2007;129:14759–14766. doi: 10.1021/ja074936k. [DOI] [PubMed] [Google Scholar]
- 313.Gump JM, Dowdy SF. Trends Mol. Med. 2007;13:443–448. doi: 10.1016/j.molmed.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 314.Sugahara KN, Teesalu T, Karmali PP, Kotamraju VR, Agemy L, Girard OM, Hanahan D, Mattrey RF, Ruoslahti E. Cancer Cell. 2009;16:510–520. doi: 10.1016/j.ccr.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Laakkonen P, Porkka K, Hoffman JA, Ruoslahti E. Nat. Med. 2002;8:751–755. doi: 10.1038/nm720. [DOI] [PubMed] [Google Scholar]
- 316.Porkka K, Laakkonen P, Hoffman JA, Bernasconi M, Ruoslahti E. Proc. Natl. Acad. Sci. U. S. A. 2002;99:7444–7449. doi: 10.1073/pnas.062189599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Wilson DS, Szostak JW. Annu. Rev. Biochem. 1999;68:611–647. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- 318.Tuerk C, Gold L. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 319.Ellington AD, Szostak JW. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 320.Keefe AD, Pai S, Ellington A. Nat. Rev. Drug Discovery. 2010;9:537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Mayer G. Angew. Chem., Int. Ed. 2009;48:2672–2689. doi: 10.1002/anie.200804643. [DOI] [PubMed] [Google Scholar]
- 322.Jayasena SD. Clin. Chem. 1999;45:1628–1650. [PubMed] [Google Scholar]
- 323.Cohen C, Forzan M, Sproat B, Pantophlet R, McGowan I, Burton D, James W. Virology. 2008;381:46–54. doi: 10.1016/j.virol.2008.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Bagalkot V, Farokhzad OC, Langer R, Jon S. Angew. Chem., Int. Ed. 2006;45:8149–8152. doi: 10.1002/anie.200602251. [DOI] [PubMed] [Google Scholar]
- 325.Guo J, Gao X, Su L, Xia H, Gu G, Pang Z, Jiang X, Yao L, Chen J, Chen H. Biomaterials. 2011;32:8010–8020. doi: 10.1016/j.biomaterials.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 326.Wu Z, Tang LJ, Zhang XB, Jiang JH, Tan W. ACS Nano. 2011;5:7696–7699. doi: 10.1021/nn2037384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Drabovich AP, Berezovski MV, Musheev MU, Krylov SN. Anal. Chem. 2009;81:490–494. doi: 10.1021/ac8023813. [DOI] [PubMed] [Google Scholar]
- 328.Talekar M, Kendall J, Denny W, Garg S. Anti-Cancer Drugs. 2011;22:949–962. doi: 10.1097/CAD.0b013e32834a4554. [DOI] [PubMed] [Google Scholar]
- 329.Xia W, Low PS. J. Med. Chem. 2010;53:6811–6824. doi: 10.1021/jm100509v. [DOI] [PubMed] [Google Scholar]
- 330.Leamon CP, Reddy JA. Adv. Drug Delivery Rev. 2004;56:1127–1141. doi: 10.1016/j.addr.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 331.Reddy JA, Dorton R, Westrick E, Dawson A, Smith T, Xu LC, Vetzel M, Kleindl P, Vlahov IR, Leamon CP. Cancer Res. 2007;67:4434–4442. doi: 10.1158/0008-5472.CAN-07-0033. [DOI] [PubMed] [Google Scholar]
- 332.Liang C, Yang Y, Ling Y, Huang Y, Li T, Li X. Bioorg. Med. Chem. 2011;19:4057–4066. doi: 10.1016/j.bmc.2011.05.016. [DOI] [PubMed] [Google Scholar]
- 333.Hilgenbrink AR, Low PS. J. Pharm. Sci. 2005;94:2135–2146. doi: 10.1002/jps.20457. [DOI] [PubMed] [Google Scholar]
- 334.Kamaly N, Kalber T, Thanou M, Bell JD, Miller AD. Bioconjugate Chem. 2009;20:648–655. doi: 10.1021/bc8002259. [DOI] [PubMed] [Google Scholar]
- 335.Ohguchi Y, Kawano K, Hattori Y, Maitani Y. J. Drug Targeting. 2008;16:660–667. doi: 10.1080/10611860802201464. [DOI] [PubMed] [Google Scholar]
- 336.Chandran SS, Banerjee SR, Mease RC, Pomper MG, Denmeade SR. Cancer Biol. Ther. 2008;7:974–982. doi: 10.4161/cbt.7.6.5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Chen Y, Dhara S, Banerjee SR, Byun Y, Pullambhatla M, Mease andM. G. Pomper RC. Biochem. Biophys. Res. Commun. 2009;390:624–629. doi: 10.1016/j.bbrc.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Farokhzad OC, Jon SY, Khadelmhosseini A, Tran TNT, LaVan DA, Langer R. Cancer Res. 2004;64:7668–7672. doi: 10.1158/0008-5472.CAN-04-2550. [DOI] [PubMed] [Google Scholar]
- 339.Wijagkanalan W, Kawakami S, Hashida M. Front. Biosci. 2011;17:2970–2987. doi: 10.2741/3892. [DOI] [PubMed] [Google Scholar]
- 340.Hashida M, Nishikawa M, Yamashita F, Takakura Y. Adv. Drug Delivery Rev. 2001;52:187–196. doi: 10.1016/s0169-409x(01)00209-5. [DOI] [PubMed] [Google Scholar]
- 341.Li X, Zhou H, Yang L, Du G, Pai-Panandiker AS, Huang X, Yan B. Biomaterials. 2011;32:2540–2545. doi: 10.1016/j.biomaterials.2010.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Bergen JM, von Recum HA, Goodman TT, Massey AP, Pun SH. Macromol. Biosci. 2006;6:506–516. doi: 10.1002/mabi.200600075. [DOI] [PubMed] [Google Scholar]
- 343.Sawyers CL. Nature. 2008;452:548–552. doi: 10.1038/nature06913. [DOI] [PubMed] [Google Scholar]
- 344.Guo KT, Ziemer G, Paul A, Wendel HP. Int. J. Mol. Sci. 2008;9:668–678. doi: 10.3390/ijms9040668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.de Kruif J, Terstappen L, Boel E, Logtenberg T. Proc. Natl. Acad. Sci. U. S. A. 1995;92:3938–3942. doi: 10.1073/pnas.92.9.3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Fang X, Tan W. Acc. Chem. Res. 2009;43:48–57. doi: 10.1021/ar900101s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Shangguan D, Cao ZC, Li Y, Tan W. Clin. Chem. 2007;53:1153–1155. doi: 10.1373/clinchem.2006.083246. [DOI] [PubMed] [Google Scholar]
- 348.Pasqualini R, Ruoslahti E. Nature. 1996;380:364–366. doi: 10.1038/380364a0. [DOI] [PubMed] [Google Scholar]
- 349.Arap W, Haedicke W, Bernasconi M, Kain R, Rajotte D, Krajewski S, Ellerby HM, Bredesen DE, Pasqualini R, Ruoslahti E. Proc. Natl. Acad. Sci. U. S. A. 2002;99:1527–1531. doi: 10.1073/pnas.241655998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Mi J, Liu Y, Rabbani ZN, Yang Z, Urban JH, Sullenger BA, Clary BM. Nat. Chem. Biol. 2010;6:22–24. doi: 10.1038/nchembio.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Xiao Z, Levy-Nissenbaum E, Alexis F, Luptak A, Teply BA, Chan JM, Shi J, Digga E, Cheng J, Langer R, Farokhzad OC. ACS Nano. 2012;6:696–704. doi: 10.1021/nn204165v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352. http://cen.acs.org/articles/90/web/2012/01/Aptamer-Guided-Nanoparticles-Slip-Cells.html.
- 353.Nel AE, Madler L, Velegol D, Xia T, Hoek EM, Somasundaran P, Klaessig F, Castranova V, Thompson M. Nat. Mater. 2009;8:543–557. doi: 10.1038/nmat2442. [DOI] [PubMed] [Google Scholar]
- 354.Wang S, Dormidontova EE. Biomacromolecules. 2010;11:1785–1795. doi: 10.1021/bm100248e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Tassa C, Duffner JL, Lewis TA, Weissleder R, Schreiber SL, Koehler AN, Shaw SY. Bioconjugate Chem. 2010;21:14–19. doi: 10.1021/bc900438a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Fakhari A, Baoum A, Siahaan TJ, Le KB, Berkland C. J. Pharm. Sci. 2011;100:1045–1056. doi: 10.1002/jps.22342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Adiseshaiah PP, Hall JB, McNeil SE. Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol. 2010;2:99–112. doi: 10.1002/wnan.66. [DOI] [PubMed] [Google Scholar]
- 358.Conner SD, Schmid SL. Nature. 2003;422:37–44. doi: 10.1038/nature01451. [DOI] [PubMed] [Google Scholar]
- 359.Zhao F, Zhao Y, Liu Y, Chang X, Chen C. Small. 2011;7:1322–1337. doi: 10.1002/smll.201100001. [DOI] [PubMed] [Google Scholar]
- 360.Wang J, Byrne JD, Napier ME, Desimone JM. Small. 2011;7:1919–1931. doi: 10.1002/smll.201100442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Verma A, Stellacci F. Small. 2010;6:12–21. doi: 10.1002/smll.200901158. [DOI] [PubMed] [Google Scholar]
- 362.Gratton SE, Ropp PA, Pohlhaus PD, Luft JC, Madden VJ, Napier ME, DeSimone JM. Proc. Natl. Acad. Sci. U. S. A. 2008;105:11613–11618. doi: 10.1073/pnas.0801763105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.He C, Hu Y, Yin L, Tang C, Yin C. Biomaterials. 2010;31:3657–3666. doi: 10.1016/j.biomaterials.2010.01.065. [DOI] [PubMed] [Google Scholar]
- 364.Jiang W, Kim BY, Rutka JT, Chan WC. Nat. Nanotechnol. 2008;3:145–150. doi: 10.1038/nnano.2008.30. [DOI] [PubMed] [Google Scholar]
- 365.Vonarbourg A, Passirani C, Saulnier P, Simard P, Leroux JC, Benoit JP. J. Biomed. Mater. Res. A. 2006;78:620–628. doi: 10.1002/jbm.a.30711. [DOI] [PubMed] [Google Scholar]
- 366.Petros RA, DeSimone JM. Nat. Rev. Drug Discovery. 2010;9:615–627. doi: 10.1038/nrd2591. [DOI] [PubMed] [Google Scholar]
- 367.Moghimi SM, Hunter AC, Murray JC. Pharmacol. Rev. 2001;53:283–318. [PubMed] [Google Scholar]
- 368.Choi HS, Liu W, Misra P, Tanaka E, Zimmer JP, Itty Ipe B, Bawendi MG, Frangioni JV. Nat. Biotechnol. 2007;25:1165–1170. doi: 10.1038/nbt1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Torchilin VP. AAPS J. 2007;9:E128–E147. doi: 10.1208/aapsj0902015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Wong C, Stylianopoulos T, Cui J, Martin J, Chauhan VP, Jiang W, Popovic Z, Jain RK, Bawendi MG, Fukumura D. Proc. Natl. Acad. Sci. U. S. A. 2011;108:2426–2431. doi: 10.1073/pnas.1018382108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Schadlich A, Caysa H, Mueller T, Tenambergen F, Rose C, Gopferich A, Kuntsche J, Mader K. ACS Nano. 2011;5:8710–8720. doi: 10.1021/nn2026353. [DOI] [PubMed] [Google Scholar]
- 372.Rhee M, Valencia PM, Rodriguez MI, Langer R, Farokhzad OC, Karnik R. Adv. Mater. 2011;23:H79–H83. doi: 10.1002/adma.201004333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Geng Y, Dalhaimer P, Cai S, Tsai R, Tewari M, Minko T, Discher DE. Nat. Nanotechnol. 2007;2:249–255. doi: 10.1038/nnano.2007.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Decuzzi P, Godin B, Tanaka T, Lee SY, Chiappini C, Liu X, Ferrari M. J. Controlled Release. 2010;141:320–327. doi: 10.1016/j.jconrel.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 375.Dausend J, Musyanovych A, Dass M, Walther P, Schrezenmeier H, Landfester K, Mailander V. Macromol. Biosci. 2008;8:1135–1143. doi: 10.1002/mabi.200800123. [DOI] [PubMed] [Google Scholar]
- 376.Salvador-Morales C, Zhang L, Langer R, Farokhzad OC. Biomaterials. 2009;30:2231–2240. doi: 10.1016/j.biomaterials.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Valencia PM, Basto PA, Zhang L, Rhee M, Langer R, Farokhzad OC, Karnik R. ACS Nano. 2010;4:1671–1679. doi: 10.1021/nn901433u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Vonarbourg A, Passirani C, Saulnier P, Benoit JP. Biomaterials. 2006;27:4356–4373. doi: 10.1016/j.biomaterials.2006.03.039. [DOI] [PubMed] [Google Scholar]
- 379.Hoek EM, Agarwal GK. J. Colloid Interface Sci. 2006;298:50–58. doi: 10.1016/j.jcis.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 380.Shastri VP, Lipski AM, Jaquiery C, Choi H, Eberli D, Stevens M, Martin I, Chen IW. Adv. Mater. 2007;19:553. [Google Scholar]
- 381.Merkel TJ, Jones SW, Herlihy KP, Kersey FR, Shields AR, Napier M, Luft JC, Wu H, Zamboni WC, Wang AZ, Bear JE, DeSimone JM. Proc. Natl. Acad. Sci. U. S. A. 2011;108:586–591. doi: 10.1073/pnas.1010013108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Anderson NL, Polanski M, Pieper R, Gatlin T, Tirumalai RS, Conrads TP, Veenstra TD, Adkins JN, Pounds JG, Fagan R, Lobley A. Mol. Cell. Proteomics. 2004;3:311–326. doi: 10.1074/mcp.M300127-MCP200. [DOI] [PubMed] [Google Scholar]
- 383.Ashley CE, Carnes EC, Phillips GK, Padilla D, Durfee PN, Brown PA, Hanna TN, Liu J, Phillips B, Carter MB, Carroll NJ, Jiang X, Dunphy DR, Willman CL, Petsev DN, Evans DG, Parikh AN, Chackerian B, Wharton W, Peabody DS, Brinker CJ. Nat. Mater. 2011;10:389–397. doi: 10.1038/nmat2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Aggarwal P, Hall JB, McLeland CB, Dobrovolskaia MA, McNeil SE. Adv. Drug Delivery Rev. 2009;61:428–437. doi: 10.1016/j.addr.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Moghimi SM, Szebeni J. Prog. Lipid Res. 2003;42:463–478. doi: 10.1016/s0163-7827(03)00033-x. [DOI] [PubMed] [Google Scholar]
- 386.Scherphof GL, Velinova M, Kamps J, Donga J. Liposome research days: Towards new products for human health, Shizuoka, Japan. 1996 [Google Scholar]
- 387.Perrault SD, Walkey C, Jennings T, Fischer HC, Chan WC. Nano Lett. 2009;9:1909–1915. doi: 10.1021/nl900031y. [DOI] [PubMed] [Google Scholar]
- 388.Owens DE, 3rd, Peppas NA. Int. J. Pharm. 2006;307:93–102. doi: 10.1016/j.ijpharm.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 389.Makarucha AJ, Todorova N, Yarovsky I. Eur. Biophys. J. 2011;40:103–115. doi: 10.1007/s00249-010-0651-6. [DOI] [PubMed] [Google Scholar]
- 390.Kluza E, van der Schaft DW, Hautvast PA, Mulder WJ, Mayo KH, Griffioen AW, Strijkers GJ, Nicolay K. Nano Lett. 2010;10:52–58. doi: 10.1021/nl902659g. [DOI] [PubMed] [Google Scholar]
- 391.Patil YB, Toti US, Khdair A, Ma L, Panyam J. Biomaterials. 2009;30:859–866. doi: 10.1016/j.biomaterials.2008.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Siegwart DJ, Whitehead KA, Nuhn L, Sahay G, Cheng H, Jiang S, Ma M, Lytton-Jean A, Vegas A, Fenton P, Levins CG, Love KT, Lee H, Cortez C, Collins SP, Li YF, Jang J, Querbes W, Zurenko C, Novobrantseva T, Langer R, Anderson DG. Proc. Natl. Acad. Sci. U. S. A. 2011;108:12996–13001. doi: 10.1073/pnas.1106379108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Weissleder R, Kelly K, Sun EY, Shtatland T, Josephson L. Nat. Biotechnol. 2005;23:1418–1423. doi: 10.1038/nbt1159. [DOI] [PubMed] [Google Scholar]
- 394.Hrkach J, Von Hoff D, Ali M, Andrianova E, Auer J, Campbell T, de Witt D, Figa M, Figueiredo M, Horhota A, Low S, McDonell K, Peeke E, Retnarajan B, Sabnis A, Schnipper E, Song J, Song YH, Summa J, Tompsett D, Troiano G, van Geen T, Wright J, LoRusso P, Kantoff PW, Bander NH, Sweeney CJ, Farokhzad OC, Langer R, Zale S. Preclinical Development and Clinical Translation of a PSMA-Targeted Docetaxel Nanoparticle with a Differentiated Pharmacological Profile. Sci. Transl. Med. 2012 doi: 10.1126/scitranslmed.3003651. (in press) [DOI] [PubMed] [Google Scholar]
- 395.Zhang L, Radovic-Moreno AF, Alexis F, Gu FX, Basto PA, Bagalkot V, Jon S, Langer RS, Farokhzad OC. ChemMedChem. 2007;2:1268–1271. doi: 10.1002/cmdc.200700121. [DOI] [PubMed] [Google Scholar]
- 396.Milane L, Duan Z, Amiji M. PLoS One. 2011;6:e24075. doi: 10.1371/journal.pone.0024075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Hu CM, Zhang L. Biochem. Pharmacol. 2012 Jan 18; [Google Scholar]
- 398.Hu C, Aryal S, Zhang L. Ther. Delivery. 2010;1:323–334. doi: 10.4155/tde.10.13. [DOI] [PubMed] [Google Scholar]
- 399.Chan JM, Zhang L, Yuet KP, Liao G, Rhee JW, Langer R, Farokhzad OC. Biomaterials. 2009;30:1627–1634. doi: 10.1016/j.biomaterials.2008.12.013. [DOI] [PubMed] [Google Scholar]
- 400.Shi J, Xiao Z, Votruba AR, Vilos C, Farokhzad OC. Angew. Chem., Int. Ed. 2011;50:7027–7031. doi: 10.1002/anie.201101554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Torchilin VP. Annu. Rev. Biomed. Eng. 2006;8:343–375. doi: 10.1146/annurev.bioeng.8.061505.095735. [DOI] [PubMed] [Google Scholar]
- 402.Li S-D, Huang L. Mol. Pharmaceutics. 2008;5:496–504. doi: 10.1021/mp800049w. [DOI] [PubMed] [Google Scholar]
- 403.Kaufmann AM, Krise JP. J. Pharm. Sci. 2007;96:729–746. doi: 10.1002/jps.20792. [DOI] [PubMed] [Google Scholar]
- 404.Panyam J, Labhasetwar V. Adv. Drug Delivery Rev. 2003;55:329–347. doi: 10.1016/s0169-409x(02)00228-4. [DOI] [PubMed] [Google Scholar]
- 405.Weissig V. Crit. Rev. Ther. Drug Carrier Syst. 2003;20:1–62. doi: 10.1615/critrevtherdrugcarriersyst.v20.i1.10. [DOI] [PubMed] [Google Scholar]
- 406.Weissig V. Expert Opin. Drug Delivery. 2005;2:89–102. doi: 10.1517/17425247.2.1.89. [DOI] [PubMed] [Google Scholar]
- 407.Weissig V, Boddapati SV, Jabr L, D’Souza GG. Nanomedicine. 2007;2:275–285. doi: 10.2217/17435889.2.3.275. [DOI] [PubMed] [Google Scholar]
- 408.Minton AP. J. Cell Sci. 2006;119:2863–2869. doi: 10.1242/jcs.03063. [DOI] [PubMed] [Google Scholar]
- 409.Ellis RJ, Minton AP. Nature. 2003;425:27–28. doi: 10.1038/425027a. [DOI] [PubMed] [Google Scholar]
- 410.Kim D, Lee ES, Oh KT, Gao ZG, Bae YH. Small. 2008;4:2043–2050. doi: 10.1002/smll.200701275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Cho K, Wang X, Nie S, Chen ZG, Shin DM. Clin. Cancer Res. 2008;14:1310–1316. doi: 10.1158/1078-0432.CCR-07-1441. [DOI] [PubMed] [Google Scholar]
- 412.Bareford LM, Swaan PW. Adv. Drug Delivery Rev. 2007;59:748–758. doi: 10.1016/j.addr.2007.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Tate BA, Mathews PM. Sci. Aging Knowl. Environ. 2006:re2. doi: 10.1126/sageke.2006.10.re2. [DOI] [PubMed] [Google Scholar]
- 414.Castino R, Demoz M, Isidoro C. J. Mol. Recognit. 2003;16:337–348. doi: 10.1002/jmr.643. [DOI] [PubMed] [Google Scholar]
- 415.Lim CS. Adv. Drug Delivery Rev. 2007;59:697. [Google Scholar]
- 416.Seibel P, Trappe J, Villani G, Klopstock T, Papa S, Reichmann H. Nucleic Acids Res. 1995;23:10–17. doi: 10.1093/nar/23.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Weissig V, Torchilin VP. Adv. Drug Delivery Rev. 2001;49:127–149. doi: 10.1016/s0169-409x(01)00131-4. [DOI] [PubMed] [Google Scholar]
- 418.Qian ZM, Li H, Sun H, Ho K. Pharmacol. Rev. 2002;54:561–587. doi: 10.1124/pr.54.4.561. [DOI] [PubMed] [Google Scholar]
- 419.Eavarone DA, Yu X, Bellamkonda RV. J. Biomed. Mater. Res. 2000;51:10–14. doi: 10.1002/(sici)1097-4636(200007)51:1<10::aid-jbm2>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 420.Dufes J-M, Muller C, Couet J-C, Olivier W, Uchegbu IF, Schaetzlein AG. Pharm. Res. 2004;21:101–107. doi: 10.1023/b:pham.0000012156.65125.01. [DOI] [PubMed] [Google Scholar]
- 421.Muro S, Schuchman EH, Muzykantov VR. Mol. Ther. 2006;13:135–141. doi: 10.1016/j.ymthe.2005.07.687. [DOI] [PubMed] [Google Scholar]
- 422.Lee M, Choi JS, Choi MJ, Pak YK, Rhee BD, Ko KS. J. Drug Targeting. 2007;15:115–122. doi: 10.1080/10611860600953555. [DOI] [PubMed] [Google Scholar]
- 423.Murakami M, Cabral H, Matsumoto Y, Wu S, Kano MR, Yamori T, Nishiyama N, Kataoka K. Sci. Transl. Med. 2011;3:64ra62. doi: 10.1126/scitranslmed.3001385. [DOI] [PubMed] [Google Scholar]
- 424.Preiss MR, Bothun GD. Expert Opin. Drug Delivery. 2011;8:1025–1040. doi: 10.1517/17425247.2011.584868. [DOI] [PubMed] [Google Scholar]
- 425.Delcea M, Mohwald H, Skirtach AG. Adv. Drug Delivery Rev. 2011;63:730–747. doi: 10.1016/j.addr.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 426.Gao WW, Chan JM, Farokhzad OC. Mol. Pharmaceutics. 2010;7:1913–1920. doi: 10.1021/mp100253e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Hribar KC, Lee MH, Lee D, Burdick JA. ACS Nano. 2011;5:2948–2956. doi: 10.1021/nn103575a. [DOI] [PubMed] [Google Scholar]
- 428.Peng K, Tomatsu I, van den Broek B, Cui C, Korobko AV, van Noort J, Meijer AH, Spaink HP, Kros A. Soft Matter. 2011;7:4881–4887. [Google Scholar]
- 429.la Rica RD, Aili D, Stevens MM. Adv. Drug Delivery Rev. 2011 doi: 10.1016/j.addr.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 430.Thornton PD, Heise A. J. Am. Chem. Soc. 2010;132:2024–2028. doi: 10.1021/ja9094439. [DOI] [PubMed] [Google Scholar]
- 431.Zhu LJ, Tu CL, Zhu BS, Su Y, Pang Y, Yan DY, Wu JL, Zhu XY. Polym. Chem. 2011;2:1761–1768. [Google Scholar]
- 432.Esser-Kahn AP, Odom SA, Sottos NR, White SR, Moore JS. Macromolecules. 2011;44:5539–5553. [Google Scholar]
- 433.Jin Y, Song L, Su Y, Zhu LJ, Pang Y, Qiu F, Tong GS, Yan DY, Zhu BS, Zhu XY. Biomacromolecules. 2011;12:3460–3468. doi: 10.1021/bm200956u. [DOI] [PubMed] [Google Scholar]
- 434.O’Neill BE, Rapoport N. Ther. Delivery. 2011;2:1165–1187. doi: 10.4155/tde.11.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Mamasheva E, O’Donnell C, Bandekar A, Sofou S. Mol. Pharmaceutics. 2011;8:2224–2232. doi: 10.1021/mp200079y. [DOI] [PubMed] [Google Scholar]
- 436.Chen D, Liu W, Shen Y, Mu H, Zhang Y, Liang R, Wang A, Sun K, Fu F. Int. J. Nanomed. 2011;6:2053–2061. doi: 10.2147/IJN.S24344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Yin H, Lee ES, Kim D, Lee KH, Oh KT, Bae YH. J. Controlled Release. 2008;126:130–138. doi: 10.1016/j.jconrel.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Lee ES, Gao Z, Kim D, Park K, Kwon IC, Bae YH. J. Controlled Release. 2008;129:228–236. doi: 10.1016/j.jconrel.2008.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Schwarze SR, Ho A, Vocero-Akbani A, Dowdy SF. Science. 1999;285:1569–1572. doi: 10.1126/science.285.5433.1569. [DOI] [PubMed] [Google Scholar]
- 440.Chung JE, Yokoyama M, Yamato M, Aoyagi T, Sakurai Y, Okano T. J. Controlled Release. 1999;62:115–127. doi: 10.1016/s0168-3659(99)00029-2. [DOI] [PubMed] [Google Scholar]
- 441.Zhang L, Chan JM, Gu FX, Rhee JW, Wang AZ, Radovic-Moreno AF, Alexis F, Langer R, Farokhzad OC. ACS Nano. 2008;2:1696–1702. doi: 10.1021/nn800275r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Katsogiannou M, Peng L, Catapano CV, Rocchi P. Curr. Cancer Drug Targets. 2011;11:954–965. doi: 10.2174/156800911797264770. [DOI] [PubMed] [Google Scholar]
- 443.Alexis F, Pridgen E, Molnar LK, Farokhzad OC. Mol. Pharmaceutics. 2008;5:505–515. doi: 10.1021/mp800051m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Feynman RP. Caltec Engineering and Science. 1960;23:22–36. [Google Scholar]